Abstract
The M2-1 protein of respiratory syncytial virus (RSV) is a transcription processivity factor that is essential for virus replication. The function of RSV M2-1 protein can be examined by using an RSVlacZ minigenome assay in vitro since the expression of the lacZ gene is dependent on M2-1. The M2-1 protein of pneumonia virus of mice (PVM), also a member of the Pneumovirus genus, functions poorly in the RSVlacZ minigenome assay despite conservation of the Cys3-His1 motif at its N terminus and an overall 40% amino acid identity with RSV M2-1. To identify the amino acids responsible for the differences between these two proteins, two chimeric proteins were constructed. The RSV/PVM (RP) M2-1 chimera that contains the N-terminal 30 amino acids from RSV and the remaining C-terminal 148 amino acids from PVM maintained a level of activity at an ca. 36% of RSV M2-1. However, the PVM/RSV (PR) M2-1 chimera with the N-terminal 29 amino acids from PVM and 164 amino acids from RSV had an activity of <5% of RSV M2-1, indicating that the functional determinants are mainly located in the N terminus of M2-1. Mutagenesis of the N terminus of PR M2-1 and RSV M2-1 identified that Leu-16 and Asn-17 of RSV M2-1 are critical to the M2-1 function. In addition, several charged residues in the N terminus of RSV M2-1 also contributed to the functional integrity of M2-1.
Human respiratory syncytial virus (RSV), a member of the Pneumovirus genus of the Paramyxoviridae family, is the most important viral agent of serious pediatric respiratory tract disease worldwide (7). The genome of the RSV A2 strain is 15,222 nucleotides (nt) in length and contains 10 transcriptional units encoding 11 proteins (NS1, NS2, N, P, M, SH, G, F, M2-1, M2-2, and L). The genome is tightly bound by the N protein to form the nucleocapsids and serves as the template for the viral RNA polymerase, a complex of the N, P and L proteins (16, 30). Each transcription unit is flanked by a highly conserved 10-nt gene-start signal, where mRNA synthesis begins, and ends with a semiconserved 12- to 13-nt gene-end signal that directs polyadenylation and the release of mRNAs (20, 21, 23). Transcription of RSV genes is sequential that produces a gradient of decreasing mRNA synthesis due to transcription attenuation (4, 12). The viral RNA polymerase must terminate synthesis of the upstream mRNA before initiating synthesis of the downstream mRNA.
Unlike other members in Paramyxoviridae family, efficient transcription of RSV mRNA requires an additional protein, M2-1 (8). M2-1 is encoded by the first of two overlapping open reading frames on the single M2 mRNA (2, 10). M2-1 functions as a transcriptional processivity factor to prevent premature termination during transcription (8, 13, 14) and enhances transcriptional readthrough at gene junctions (17-19), which permits access of the RSV polymerase to the downstream transcriptional units. Functional M2-1 is essential for RSV replication; alteration of its sequence destroys virus infectivity (26).
The M2-1 protein of RSV A2 is 194 amino acids in length with a molecular weight of ca. 22,150 (9, 10). It contains a Cys3-His1 motif in the N terminus from residues 7 to 25 that is highly conserved among human, bovine, ovine, and murine strains of pneumoviruses (2, 3, 27, 30). The M2-1 function requires its interaction with the N and P proteins. Recent studies have demonstrated a direct interaction between the M2-1 and N proteins that is mediated through RNA (5, 11). Substitutions of the three cysteines and one histidine in this motif significantly reduced the ability of M2-1 to enhance transcription readthrough and disrupted the interaction between the M2-1 and N proteins (18), which is lethal to virus replication (26). M2-1 is phosphorylated at Ser-58 and Ser-61, and this phosphorylation is also important for M2-1 function (5, 11).
Other distantly related pneumoviruses have homologies to the RSV M2-1 protein. In order to determine whether these divergent proteins can substitute for each other functionally, we evaluated whether the M2-1 protein of pneumonia virus of mice (PVM) would substitute for RSV M2-1 in minigenome assays. PVM M2-1 functioned poorly in the RSV minigenome assay. However, its relatedness enabled the identification of function residues in RSV M2-1.
MATERIALS AND METHODS
Cells and viruses.
Monolayers of HEp-2 cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum. Modified vaccinia virus Ankara (MVA) expressing T7 RNA polymerase, MVA-T7, was obtained from Bernard Moss (25, 28) and propagated in CEK cells (SPAFAS).
Plasmids construction.
pRSVLacZ minigenome encodes the negative sense lacZ gene flanked by the RSV leader and trailer sequences under the control of the T7 promoter (26). The protein expression plasmids encoding the N, P, L, or M2-1 genes under the control of the T7 promoter in pCITE2a vector (Novagen) were described previously (22, 26). The PVM M2-1 gene was amplified from PVM M2-1 cDNA (1) by using primers of 5′BsmBI (GCATCGTCTCCCATGAGTGTGAGACCTTGC) and 3′BamHI (CTCGAGCTGCAGGGATCCG) and then cloned into the NcoI site of pCITE2a. To construct chimeric M2-1 plasmids, an MscI restriction enzyme site that is present in RSV M2-1 at nt 7693 of the RSV genome was introduced into the corresponding position of pPVM-M2-1 by using the QuikChange site-directed mutagenesis kit (Stratagene). RSV/PVM (RP) M2-1 was constructed by fusing the RSV N-terminal 30 amino acids with the C-terminal 148 amino acids of PVM M2-1 through the MscI site. Likewise, PVM/RSV (PR) M2-1 was constructed by replacing the PVM M2-1 MscI-to-BamHI restriction fragment with that of RSV. Introduction of mutations into RSV M2-1 or PVM M2-1 was performed by using the QuikChange site-directed mutagenesis kit (Strategene).
Processivity function of M2-1 mutants.
HEp-2 cells were infected with MVA-T7 at a multiplicity of infection of 1.0 and transfected with 0.4 μg of pP, 0.4 μg of pN, 0.2 μg of pL, and 0.4 μg of pRSVLacZ, together with 0.1 μg (or the amount indicated) of M2-1 expression plasmid. Transfection was performed in duplicates by using LipofecTACE or Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. The transfected cells were incubated at 35°C for 2 days, and cell extracts prepared by incubating them in cell permeabilization buffer that contained 0.5% NP-40 and 20 mM β-mercaptoethanol. Cell lysates were clarified by centrifugation at 2,500 rpm for 5 min at 4°C and analyzed for β-galactosidase activity by using 5 mM chlorophenol red-β-d-galactopyranoside (CPRG; Roche Molecular Biochemicals) as described previously (6, 26). The change in optical density at 550 nm was measured with a SPECTRAmax 340PC microplate spectrophotometer by using SOFTmax software (Molecular Devices). The assay was shown to be linearly responsive up to an optical density at 550 nm of 3.0 (26). The relative activity of each mutant was calculated compared to RSV M2-1, and the data obtained were an average of a minimum of three experiments.
The synthesis of lacZ RNA in transfected cells was analyzed by Northern blotting. At 48 h posttransfection, total intracellular RNA was extracted by RNeasy extraction kit (Qiagen) and electrophoresed on 1% agarose-urea gel. The RNA blot was hybridized with digoxigenin-labeled negative sense lacZ or M2-1 probe. The hybridized RNA was detected by using digoxigenin-RNA detection kit (Roche Molecular Biochemicals), followed by exposure to the X-ray film (Kodak).
Protein labeling and immunoprecipitation.
Phosphorylation of M2-1 and M2-1-N protein complex in transfected cells was examined by radiolabeling, followed by immunoprecipitation. MVA-T7-infected HEp-2 cells were transfected with pN, pP, pL, pM2-1, and pRSVLacZ minigenome and incubated at 37°C for 18 h. The transfected cells were radiolabeled either with 35S-Promix (100 μCi/ml) in DMEM lacking methionine and cysteine or with [33P]phosphate (100 μCi/ml) in DMEM lacking phosphate for 4 h. The cells were lysed in radioimmunoprecipitation assay buffer containing 0.15 M NaCl and immunoprecipitated with anti-M2-1 monoclonal antibodies (a gift of R. P. Yeo) or anti-RSV polyclonal antibody (Biogenesis). After incubation with protein G-agarose beads (Invitrogen) for 30 min, the immunoprecipitated complex was washed three times with radioimmunoprecipitation assay buffer containing 0.3 M NaCl and electrophoresed on 4 to 15% gradient polyacrylamide gel (Novagen). The immunoprecipitated proteins were visualized by radiography (Kodak).
RESULTS
Comparison of RSV and PVM M2-1 function.
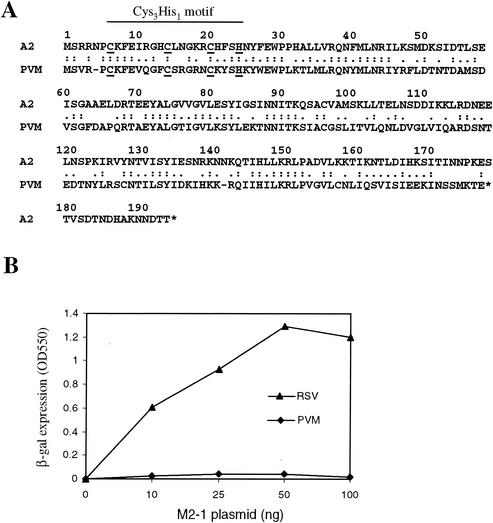
PVM M2-1 is 40% identical to RSV M2-1 and contains the Cys3-His1 motif at its N terminus (Fig. 1A); it is expected that this protein functions as a transcriptional processivity factor for PVM transcription. The RSV M2-1 processivity function can be measured by using the RSVlacZ minigenome assay (26). To examine whether PVM M2-1 protein could function in the RSV minigenome assay, HEp-2 cells were transfected with plasmids encoding the RSV N, P, and L proteins; pRSVLacZ; and either PVM M2-1 or RSV M2-1. As shown in Fig. 1B, the processivity function of RSV M2-1 was required for lacZ reporter expression. β-Galactosidase was not detected in the absence of M2-1 but was produced in a dose-dependent manner with increasing amounts of RSV M2-1 plasmid (between 0 and 50 ng). In contrast, PVM M2-1 showed a very low processivity in this assay. A level of 2 to 5% of that of RSV M2-1 was reproducibly detected at a concentration of 10 to 50 ng of PVM M2-1 plasmid. Therefore, in contrast to RSV M2-1, PVM M2-1 exhibited a very low level of processivity in the RSVlacZ minigenome assay.
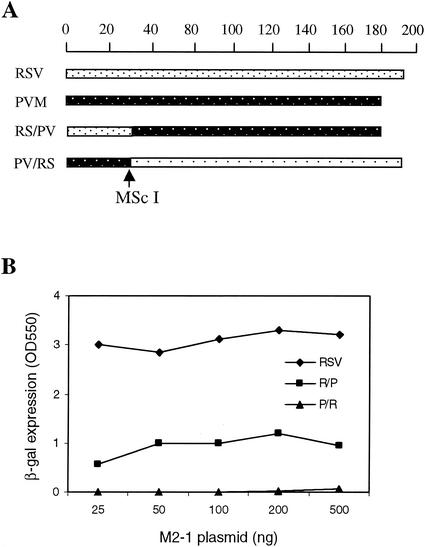
FIG. 1.
Amino acid sequence and functional comparison of A2 and PVM M2-1. (A) Protein sequence alignment of A2 and PVM M2-1. The conserved Cys3-His1 motif is indicated. (B) The indicated amount of RSV or PVM M2-1 plasmid was transfected into MVA-T7-infected HEp-2 cells, together with 0.2 μg of pN, 0.2 μg of pP, 0.1 μg of pL, and 0.2 μg of pRSVLacZ. At 2 days after transfection, the level of β-galactosidase activity was determined.
Processivity of RSV and PVM M2-1 chimeric proteins.
To dissect the essential functional domain of RSV M2-1 required for its processivity function, two chimeric protein expression plasmids derived from portions of the PVM and RSV M2-1 open reading frames were constructed (Fig. 2A). The N-terminal 30 amino acids of the RP M2-1 chimera were derived from RSV M2-1, and the remaining C-terminal sequence was derived from PVM. PR M2-1 represented the converse chimera in that its N-terminal 29 amino acids were derived from PVM and the C terminus from RSV. The one-amino-acid difference was accounted for by a lack of Asn-5 in the PVM sequence. These two chimeric proteins exhibited strikingly different activities in the RSVlacZ minigenome assay (Fig. 2B). PR M2-1 had an activity similar to PVM M2-1, at a level <5% of RSV M2-1. However, RP M2-1 maintained ca. 36% of M2-1 activity at concentrations between 50 and 500 ng, and a dose response was less apparent. Therefore, the N-terminal region of M2-1 played an important role in determining the protein's function.
FIG. 2.
(A) Schematic diagram of RP and PR M2-1 chimeric proteins in comparison with RSV and PVM M2-1. (B) The processivity function of RP and PR M2-1 was examined by the RSVLacZ minigenome assay, and their activities were indicated by the level of β-galactosidase protein produced in the transfected cells.
Identification of residues in the N-terminal region that are critical to M2-1 function.
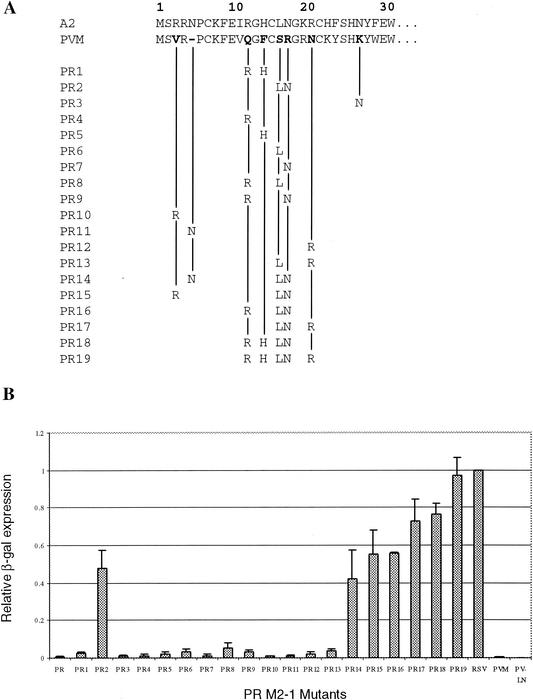
In order to identify critical residues in the N-terminal region, a systematic analysis was performed to introduce RSV sequences into PR M2-1 that restored processivity function. The 29 amino acids of the N-terminal PVM M2-1 differ from RSV by 13 amino acids and lack the Asn residue corresponding to the fifth amino acid of RSV M2-1 (Fig. 3A). Among the 13 amino acids different between RSV and PVM, five residues (I11V, K19R, H22K, F23Y, and F29W) have similar biochemical properties and were not selected for substitution mutagenesis. The remaining eight amino acids in the N-terminal 30 residues of PR M2-1 were mutagenized individually or in combination to change the PVM residues to those of RSV. A total of 19 mutants were constructed (Fig. 3A), and their functions were analyzed by the RSVlacZ minigenome assay (Fig. 3B). Expression of the M2-1 protein was monitored by immunoprecipitation to ensure that an equivalent level of M2-1 protein was expressed. Of the 13 single and double mutations initially constructed (PR1 to PR13), only mutants that contained substitutions of both S15L and R16N had a significant increase in their processivity. Individual substitution of either S15L (PR6) or R16N (PR7) had very little positive effect on the PR M2-1 function. However, PR2 M2-1 containing only these two changes together had an increase in protein processivity to levels ca. 48% of RSV M2-1. Therefore, it was considered that other residues differed between RSV M2-1 and PVM M2-1 might also influence the protein function.
FIG. 3.
Mutagenesis of the N-terminal 29 amino acids of PR M2-1. (A) The residues that were changed from PVM to RSV for each PR M2-1 mutant are indicated. (B) The processivity of each mutant was analyzed by the RSVLacZ minigenome assay, and the level of β-galactosidase expressed by each mutant was normalized to RSV M2-1.
Substitutions of the PVM residues at positions 3, 11, 13, 19, and 25 by those of RSV M2-1 or insertion of Asn-5 did not increase PR M2-1 protein function. Thus, more mutagenesis was performed in PR2 M2-1 (containing S15L and R16N) to determine whether other amino acid changes could further increase its activity approaching the level of RSV M2-1 (Fig. 3A). An increase in protein function was observed for mutations that involved charged residues. Introduction of V3R (PR15) and Q11R (PR16) increased PR2 M2-1 processivity by ca. 7%, and the N19R (PR17) mutation increased PR2 M2-1 activity to ≥25%. Double Q11R and F13H mutations (PR18) increased PR2 M2-1 function by 27%, and the triple mutations (Q11R, F13H, and N19R) introduced into PR2 M2-1 resulted in a protein that had an activity almost identical to that of RSV M2-1. Thus, in addition to S15L and R16N residues that are critical to PR M2-1 function, several charged residues in addition are also required to produce a fully functional protein.
In order to determine whether PVM M2-1 protein processivity could be increased by introduction of Leu and Asn, mutagenesis was performed in PVM M2-1 to substitute Ser-15 and Arg-16 by Leu and Asn. Unexpectedly, PVM M2-1 bearing S15L and R16N changes (Fig. 3, PV-LN) did not have increased processivity function in the RSV minigenome assay. Thus, the C-terminal region of PVM M2-1 may have a greater influence on its function.
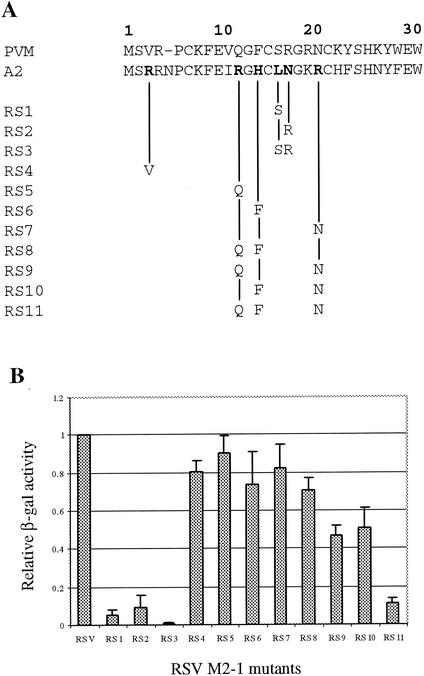
To confirm the role of Leu-16, Asn-17, and the charged residues at the N terminus of RSV M2-1 to its function, mutagenesis was further performed in the RSV M2-1 molecule, and a total of 11 mutants were generated (Fig. 4A). The single-substitution mutations L16S (RS1) or N17R (RS2) greatly reduced RSV M2-1 protein function by 97 and 94%, respectively (Fig. 4B). The double-substitution mutations L16S and N17R (RS3) further reduced the protein function, and only 1% of normal lacZ activity could be detected for RS3 in the RSVlacZ minigenome assay (Fig. 4B). These data demonstrated that Leu-16 and Asn-17 are the two residues critical to the RSV M2-1 function. Substitution of single charged residues at positions of 3 (RS4), 12 (RS5), 14 (RS6), or 20 (RS7) each resulted in reduced M2-1 protein activity by 10 to 25%; however, none were as critical as Leu-16 and Asn-17. Substitutions of multiple charged residues had a greater effect on the RSV M2-1 function. Double substitution mutations reduced protein function by 30% for RS8, 53% for RS9, and 50% for RS10. The triple mutations bearing R12Q, H14F, and R20N reduced the M2-1 function by ca. 90%. Thus, consistent with the mutagenesis analysis of PR M2-1, the charged residues in the N terminus of RSV M2-1 protein were important to the protein processivity function in addition to the Leu-16 and Asn-17 residues.
FIG. 4.
Mutagenesis of the N-terminal 30 amino acids of RSV M2-1. (A) The residues that were changed from RSV to PVM for each RSV M2-1 mutant are indicated. (B) The processivity of each mutant was analyzed by the RSVLacZ minigenome assay, and the level of β-galactosidase expressed by each mutant was normalized to wild-type RSV M2-1.
Phosphorylation of M2-1 and its processivity.
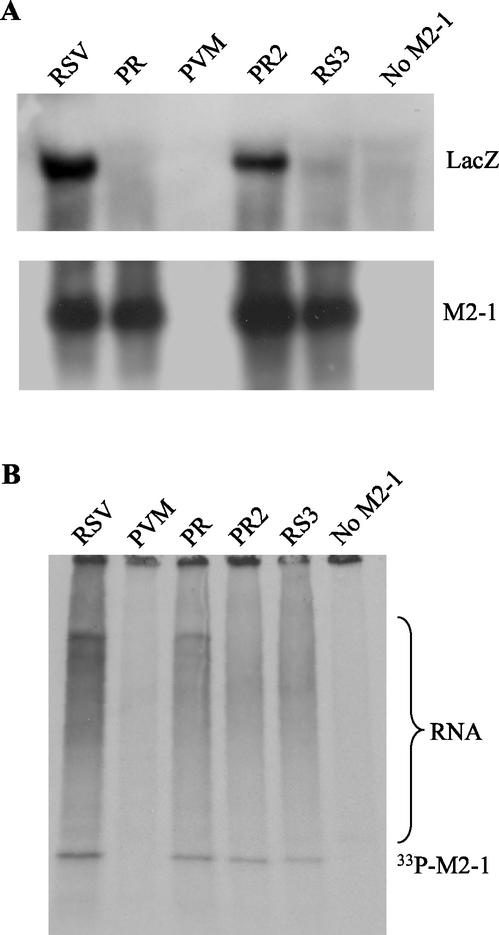
To examine whether M2-1 mutations affected M2-1 protein phosphorylation status, HEp-2 cells were transfected with plasmids encoding the N, P, and L proteins, pRSVLacZ, together with RSV, PVM, PR, PR2, or RS3 M2-1 expression plasmids in duplicate. At 24 h posttransfection, RNA was extracted from one set of cells, and a Northern blot was probed with a riboprobe specific for lacZ or M2-1 (Fig. 5A). Another set of cells was radiolabeled with [33P]phosphate and immunoprecipitated with anti-M2-1 monoclonal antibodies (Fig. 5B). Consistent with the β-galactosidase assay, lacZ mRNA was not detected in cells expressing PVM, PR, or RS3 M2-1 or in cells that had no M2-1 protein expressed (Fig. 5A). lacZ mRNA was detected in cells expressing PR2 M2-1 at a level ca. 50% of that of the RSV M2-1, which was also consistent with the level of β-galactosidase detected (Fig. 3B). Except for PVM M2-1 that was not detected by RSV M2-1 probe due to low sequence homology, a comparable level of M2-1 mRNA was produced in the cells transfected with all of the mutants. Again, except for PVM M2-1 that was not detected by anti-RSV M2-1 antibodies, PR, PR2, and RS3 M2-1 proteins were phosphorylated regardless of their processivity activity. Each of the proteins was also able to bind to RNA, as shown by the presence of 33P-labeled coimmunoprecipitated materials that were sensitive to RNase A treatment (data not shown and reference 5) and were at low levels or absent in the cells that expressed PVM M2-1 or had no M2-1 protein expressed. These data suggested that chimeric PR M2-1 or M2-1 mutations did not result in significant changes in mRNA synthesis, protein phosphorylation, or RNA-binding ability of M2-1 mutants.
FIG. 5.
(A) Northern blot analysis of RNA from MVA-T7-infected cells transfected with RSV N, P, and L; pRSVLacZ; and M2-1 plasmid as indicated. The blots were hybridized to a riboprobe specific to the lacZ or M2-1 gene. The lacZ gene was not detected in cells expressing PR, PVM, and RS3 M2-1. However, a reduced level of lacZ mRNA was detected in cells expressing PR2 M2-1. Except for PVM M2-1, M2-1 mRNA was detected in PV, PR2, and RS3 M2-1 transfected cells by the RSV M2-1-specific probe. (B) The transfected cells were labeled with [33P]phosphate and immunoprecipitated with anti-M2-1 monoclonal antibodies. The phosphorylated M2-1 (33P-M2-1) and 33P-labeled RNA are indicated.
Effect of M2-1 mutations on M2-1 and N interaction.
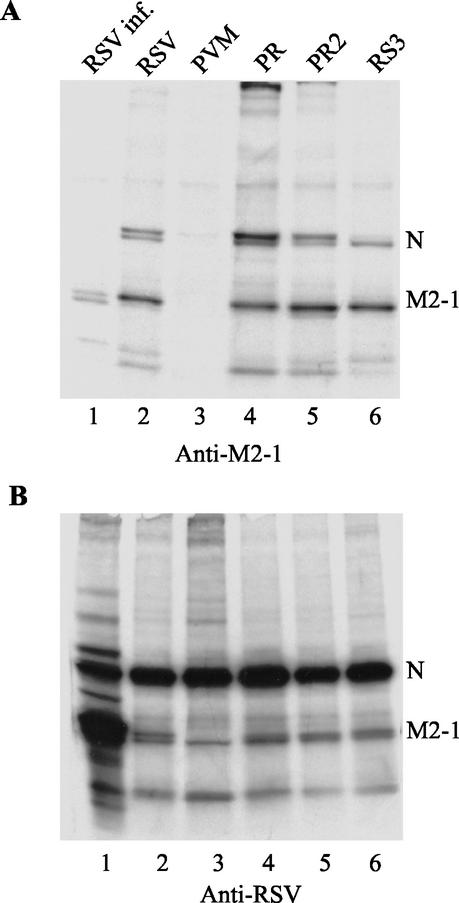
To determine whether the difference in M2-1 processivity was due to any changes in M2-1 and N protein interaction, HEp-2 cells were transfected with N, P, and L expression plasmids, pRSVLacZ, and M2-1 expression plasmids, radiolabeled with [35S]Met-[35S]Cys and immunoprecipitated with monoclonal antibodies to RSV M2-1 or a polyclonal antibody to RSV. RSV-infected cells produced less N protein than the transfected cells in this experiment (Fig. 6B, lane 1), and the N protein immunoprecipitated by anti-M2-1 antibodies was detected in a longer exposure. A comparable level of N and M2-1 proteins was detected in each transfected cell, as shown by immunoprecipitation with anti-RSV antibody (Fig. 6B). Except for PVM M2-1 that was not recognized by anti-M2-1 antibodies, the N proteins were coimmunoprecipitated with the M2-1 protein of RSV, PR, PR2, or RS3 (Fig. 6A). The slower-migrating M2-1 represented the phosphorylated form, which is more abundant in the transfected cells (lanes 2 to 6) than in the RSV-infected cells (lane 1). Although the level of the N proteins coprecipitated by each M2-1 protein varied between experiments, it did not appear to have direct correlation with the M2-1 protein function.
FIG. 6.
Coimmunoprecipitaton of M2-1 and N proteins. The transfected cells as described in Fig. 5 were labeled with [35S]Met-[35S]Cys at 18 h posttranfection and immunoprecipiated with monoclonal antibodies to RSV M2-1 (A) or anti-RSV polyclonal antibody (B). The RSV-infected cells were included as a control. The N proteins were coprecipitated with M2-1 of PR, PR2, and RS3 from the cotransfected cells.
DISCUSSION
We examined here the transcriptional processivity of M2-1 of RSV and M2-1 from the distally related PVM in an RSV minigenome assay. We demonstrated that the functional difference between RSV and PVM mainly resides at their N termini. The residues that are critical for RSV M2-1 function are Leu-16 and Asn-17. In addition, several charged residues in the N terminus of RSV M2-1 also play an important role.
Both RSV and PVM M2-1 proteins contain the conserved Cys3-His1 motif at a similar location. It has been hypothesized that the Cys3-His1 motif of M2-1 binds zinc to present the protein in a conformation that is fully functional for RSV RNA transcription (5). The striking difference in the processivity of PVM and RSV M2-1 indicated that the Cys3-His1 motif alone is not sufficient for its function. Since RP M2-1 but not PR M2-1 maintained a good level of activity despite conservation of the Cys3-His1 motif, it was ascertained that other residues at the N terminus must have an important role in contributing to the protein function. Leu-15 and Asn-16 together were required for increasing PR M2-1 function. In addition, three charged residues (Q11R, F13H, and N19R) were required for optimal M2-1 activity. The contribution of these five identified residues to M2-1 processivity function was also confirmed by mutagenesis of the RSV M2-1 protein. Therefore, Leu-16, Asn-17 and the charged residues in the N terminus of RSV M2-1 are important for maintaining the structural and functional integrity of RSV M2-1.
The chimeric RP M2-1 protein manifested an activity at a level of 36% of RSV M2-1, indicating that the C-terminal region also played a role in protein function. Previously, we described an M2-1 mutant with 17 amino acids deleted from its C-terminal end that retained the M2-1 function at a level ca. 40% that of wild-type RSV M2-1; infectious virus bearing this deletion in the M2-1 protein replicated less efficiently in vitro and in vivo (26). Since PVM M2-1 is 16 amino acids shorter than RSV M2-1 from its C-terminal end, its shortened length may be responsible for the reduced activity of RP M2-1. Introduction of Leu-15 and Asn-16 into PR M2-1 but not PVM M2-1 greatly increased its activity, suggesting that the C-terminal region of RSV M2-1 is also important for the processivity function.
The M2-1 protein is phosphorylated at residues 58 and 64 (5), and the integrity of the Cys3-His1 motif is important for phosphorylation of the RSV M2-1 protein (18). Consistent with the previous report (18), we found that the major species of M2-1 expressed in the transfected cells was phosphorylated, whereas both phosphorylated and nonphosphorylated forms exist in the infected cells. Several lines of evidence have indicated that M2-1 interacts with the N and P proteins in RSV-infected cells. Garcia et al. (15) discovered that the M2-1, N, and P proteins are components of the cytoplasmic inclusion bodies observed in RSV-infected HEp-2 cells (15). In addition, coexpression of P with M2-1 in plasmids transfected cells changes the relative dominance of the phosphorylated M2-1 form (18). Although it is known that the M2-1 protein interacts with N through its binding to viral RNA (5), it remains to be determined whether M2-1 and P protein interaction is also mediated through RNA. In addition to Leu-16 and Asn-17 residues in RSV M2-1, at least three charged residues are also involved in M2-1 processivity function. Charged residues are usually involved in protein-protein interaction. Although we were unable to identify the defect of several M2-1 mutants in their interaction with the N protein or RNA in the cotransfected cells, it is possible that interaction of the M2-1 mutants with P could be affected. It is also likely that the poor processivity function of M2-1 mutants resides in their ability to function once bound to RNA or protein instead of their weekened interaction with RNA or proteins.
The minigenome used in the present study contains the cis elements derived from RSV instead of PVM. Although the cis elements of avian pneumovirus can be recognized by RSV replicative proteins in vitro (24), it remains to be determined whether PVM M2-1 would function better with the homologous PVM minignome together with the N, P, and L proteins derived from PVM. It is possible that replication and transcription of PVM genome is less dependent on the M2-1 processivity. Interestingly, the M2-1 proteins of human, bovine, and ovine pneumoviruses all contain Leu-16 and Asn-17, with the exception of PVM M2-1, which has Ser and Arg in place (1, 3, 9, 31). PVM appears to be more closely related to the metapneumoviruses in its M2-1 structure. Metapneumoviruses, also classified in the Pneumovirinae subfamily, have Asn-16 and Arg-17 (27, 29). Metapneumoviruses lack the NS1 and NS2 genes and have a gene order (N-P-M-F-M2-SH-G-L) different from that of RSV (NS1-NS2-N-P-M-SH-G-F-M2-L). Further work is needed to study the structural and functional relation between RSV M2-1 and the M2-1s of other pneumoviruses.
Acknowledgments
We are grateful to A. Easton for providing PVM M2-1 plasmid and to R. P. Yeo for providing anti-M2-1 monoclonal antibodies. We thank the tissue culture facility of MedImmune Vaccines for providing HEp-2 cells; HynJung Park for technical assistance; and G. Kemble, R. Spaete, and B. Lu for suggestions and critical review of the manuscript.
This work was supported in part by an NIH SBIR grant (2R44A145267-02/03).
REFERENCES
- 1.Ahmadian, G., P. Chambers, and A. J. Easton. 1999. Detection and characterization of proteins encoded by the second ORF of the M2 gene of pneumoviruses. J. Gen. Virol. 80:2011-2016. [DOI] [PubMed] [Google Scholar]
- 2.Ahmadian, G., J. S. Randhawa, and A. J. Easton. 2000. Expression of the ORF-2 protein of the human respiratory syncytial M2 gene is initiated by a ribosomal termination-dependent reinitiation mechanism. EMBO J. 19:2681-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alansari, H., and L. N. D. Potgieter. 1994. Molecular cloning and sequence analysis of the phosphoprotein, nucleocapsid protein, matrix protein and 22K (M2) protein of the ovine respiratory syncytial virus. J. Gen. Virol. 75:3597-3601. [DOI] [PubMed] [Google Scholar]
- 4.Barik, S. 1992. Transcription of human respiratory syncytial virus genome RNA in vitro: requirement of cellular factor(s). J. Virol. 66:6813-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cartee, T. L., and G. W. Wertz. 2001. Respiratory syncytial virus M2-1 protein requires phosphorylation for efficient function and binds viral RNA during infection. J. Virol. 75:12188-12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, X., M. Munoz, H. Zhou, and H. Jin. 2002. Expression of β-galactosidase by recombinant respiratory syncytial viruses for microneutralization assay. J. Virol. Methods 105:287-295. [DOI] [PubMed] [Google Scholar]
- 7.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Repiratory syncytial virus, p. 1443-1485. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, vol. 1. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 8.Collins, P. L., M. G. Hill, J. Cristina, and H. Grosfeld. 1996. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc. Natl. Acad. Sci. USA 93:81-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, P. L., M. G. Hill, and P. R. Johnson. 1990. The two open reading frames of the 22K mRNA of human respiratory syncytial virus: sequence comparison of antigenic subgroups A and B and expression in vitro. J. Gen. Virol. 71:3015-3020. [DOI] [PubMed] [Google Scholar]
- 10.Collins, P. L., and G. W. Wertz. 1985. The envelope-associated 22K protein of human respiratory syncytial virus: nucleotide sequence of the mRNA and a related polytranscript. J. Virol. 54:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuesta, I., X. Geng, A. Asenjo, and N. Villanueva. 2000. Structural phosphoprotein M2-1 of the human respiratory syncytial virus is an RNA binding protein. J. Virol. 74:9858-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickens, L. E., P. L. Collins, and G. W. Wertz. 1984. Transcriptional mapping of human respiratory syncytial virus. J. Virol. 52:364-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fearns, R., and P. L. Collins. 1999. Model for polymerase access to the overlapped L gene of respiratory syncytial virus. J. Virol. 73:388-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fearns, R., and P. L. Collins. 1999. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 73:5852-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia, J., B. Garcia-Barreno, A. Vivo, and J. A. Melero. 1993. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195:243-247. [DOI] [PubMed] [Google Scholar]
- 16.Grosfeld, H., M. G. Hill, and P. L. Collins. 1995. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L protein; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol. 69:5677-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy, R. W., S. B. Harmon, and G. W. Wertz. 1999. Diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J. Virol. 73:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, R. W., and G. W. Wertz. 2000. The Cys3-His1 motif of the respiratory syncytial virus M2-1 protein is essential for protein function. J. Virol. 74:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardy, R. W., and G. W. Wertz. 1998. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J. Virol. 72:520-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmon, S. B., A. G. Megaw, and G. W. Wertz. 2001. RNA sequences involved in transcriptional termination of respiratory syncytial virus. J. Virol. 75:36-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, Y. T., P. L. Collins, and G. W. Wertz. 1985. Characterization of the 10 proteins of human respiratory syncytial virus: identification of a fourth envelope-associated protein. Virus Res. 2:157-173. [DOI] [PubMed] [Google Scholar]
- 22.Jin, H., D. Clarke, Z.-Y. H. Zhou, X. Cheng, K. Coelingh, M. Bryant, and S. Li. 1998. Recombinant human respiratory syncytial viru (RSV) from cDNA and construction of subgroup A and B chimeric RSV. Virology 251:206-214. [DOI] [PubMed] [Google Scholar]
- 23.Kuo, L., H. Grosfeld, J. Cristina, M. G. Hill, and P. L. Collins. 1996. Effects of mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J. Virol. 70:6892-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marriott, A. C., J. M. Smith, and A. J. Easton. 2001. Fidelity of leader and trailer sequence usage by the respiratory syncytial virus and avian pneumovirus replication complexes. J. Virol. 75:6265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutter, G., M. Ohlmann, and V. Erfle. 1995. Nonreplicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 371:9-12. [DOI] [PubMed] [Google Scholar]
- 26.Tang, R. S., N. Nguyen, X. Cheng, and H. Jin. 2001. Requirement of cysteines and length of the human respiratory syncytial virus M2-1 protein for protein function and virus viability. J. Virol. 75:11328-11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van den Hoogen, B. G., T. M. Bestebroer, A. D. Osterhaus, and R. A. Fouchier. 2002. Analysis of the genomic sequence of a human metapneumovirus. Virology 295:119-132. [DOI] [PubMed] [Google Scholar]
- 28.Wyatt, L. S., B. Moss, and S. Rozenblatt. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202-205. [DOI] [PubMed] [Google Scholar]
- 29.Yu, Q., P. J. Davis, T. D. Brown, and D. Cavanagh. 1992. Sequence and in vitro expression of the M2 gene of turkey rhinotracheitis pneumovirus. J. Gen. Virol. 73:1355-1363. [DOI] [PubMed] [Google Scholar]
- 30.Yu, Q., R. W. Hardy, and G. W. Wertz. 1995. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J. Virol. 69:2412-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zamora, M., and S. K. Samal. 1992. Sequence analysis of M2 mRNA of bovine respiratory syncytial virus obtained from an F-M2 dicistronic mRNA suggests structural homology with that of human respiratory syncytial virus. J. Gen. Virol. 73:737-741. [DOI] [PubMed] [Google Scholar]