Abstract
The activating protein 1 (AP-1) family of regulatory proteins is characterized as immediate-early inducible transcription factors which were shown to be activated by a variety of stress-related stimuli and to be involved in numerous biological processes, including cellular and viral gene expression, cell proliferation, differentiation, and tumorigenesis. We have recently demonstrated the involvement of the AP-1 family members c-Jun and c-Fos in transcriptional regulation of the human polyomavirus, JC virus (JCV), genome. Here, we further examined their role in JCV gene regulation and replication through their physical and functional interaction with JCV early regulatory protein large T antigen (T-Ag). Transfection and replication studies indicated that c-Jun and c-Fos can significantly diminish T-Ag-mediated JCV gene transcription and replication. Affinity chromatography and coimmunoprecipitation assays demonstrated that c-Jun and T-Ag physically interact with each other. Results from band shift assays showed that the binding efficiency of c-Jun to the AP-1 site was reduced in the presence of T-Ag. In addition, we have mapped, through the use of a series of deletion mutants, the regions of these proteins which are important for their interaction. While the c-Jun interaction domain of T-Ag is localized to the middle portion of the protein, the T-Ag interacting domain of c-Jun maps to its basic-DNA binding region. Results of transient-transfection assays with various c-Jun mutants and T-Ag expression constructs further confirm the specificity of the functional interaction between c-Jun and T-Ag. Taken together, these data demonstrate that immediate-early inducible transcription factors c-Jun and c-Fos physically and functionally interact with JCV major early regulatory protein large T-Ag and that this interaction modulates JCV transcription and replication in glial cells.
The activating protein 1 (AP-1) family of transcription factors was shown to be involved in a wide variety of cellular processes, including cell proliferation, cellular and viral gene expression, cell death, survival, and differentiation, and tumorigenesis (52). In particular, c-jun knockout studies resulted in an embryonically lethal phenotype. Furthermore, mouse embryonic fibroblasts established from the c-Jun knockout mouse showed severe proliferative defects and can be propagated only once or twice before entering a premature senescence (23). Biochemical purification showed that AP-1 is not a single transcription factor but instead is a series of related dimeric complexes of the Jun (c-Jun, JunB, and JunD) and Fos (c-Fos, FosB, Fra-1, and Fra-2) families (4, 62). Each family member is a phospho-nuclear protein and composed of three distinct functional domains, including a carboxy-terminal leucine-zipper domain followed by an adjacent basic DNA binding domain and an amino-terminal transactivation domain. The family members form homo- and heterodimers within the family and outside the family with those factors that contain basic-leucine zipper (bZIP) motifs, such as the CREB and ATF2 families (50). Dimerization occurs through leucine repeats that are clustered proximally to the carboxy-terminal region. It is interesting that unlike c-Jun family members, the Fos family members form only heterodimers. DNA binding activity of AP-1 is mediated by the basic DNA binding domain and occurs in a hierarchical manner. Following dimerization, the specific residues in the basic region (62) make base contacts with target sequences on DNA, which are known as the 12-O-tetradecanoyl-13-phorbol acetate-response element (TGACTCA, TRE). These sequences are present within the promoter regions of many inducible genes (3, 58).
Transcriptional activity of this family of factors is regulated by the N-terminal transactivation domain, largely in a phosphorylation-dependent manner. For example, phosphorylation of serine63 and serine73 residues of c-Jun by the Jun N-terminal kinase (JNK) family of kinases results in a large increase in its ability to interact with the CBP/p300 family of cofactors and, to a similar extent, in the transcriptional activation potential of the protein (4, 62). AP-1 family members are induced by a wide variety of signals, including, but not limited to, UV light, ionizing radiation, oxidative stress, neuronal depolarization, cytokines (tumor necrosis factor α, gamma interferon, and interleukin-1), and viral infection (9, 13, 17, 18, 21, 50, 51, 63). They are collectively known as proto-oncogenes because of their high sequence homology to some retroviral encoded oncogenic proteins and their involvement in many cellular processes, including cell proliferation, survival, and apoptosis (8, 30, 50, 56, 62).
JC virus (JCV) is a human polyomavirus with a double-stranded covalently linked circular genome and is the etiological agent of a fatal demyelinating disease, progressive multifocal leukoencephalopathy (PML), in immunocompromised individuals (6). JCV lytically infects oligodendrocytes, the myelin-producing cells of the central nervous system, and utilizes a clathrin-mediated pathway to enter the cells (31, 36, 61). The viral genome is composed of regulatory and coding regions. The regulatory region contains DNA target sequences for both viral and cellular transcription factors, including NF-κB (39, 45), Tst-1 (41, 60), NF-1 (1, 2, 35), Sp-1 (20), GBP-i (38), YB-1 (24, 44, 46, 48), and Purα (11, 46). The viral coding regions encode early regulatory proteins (small t, large T, and isoforms of early proteins, T′) and late structural capsid proteins (VP-1, VP-2, and VP-3). In addition to structural proteins, the leader sequences of late transcripts also encode a basic regulatory protein, Agnoprotein, which has been recently shown to play a role in viral DNA replication and transcription (43, 48). Although little is known about the function of small t antigen, the large T antigen (T-Ag) was shown to be a multifunctional phosphoprotein involved in both viral DNA replication (32-34, 54) and viral gene transcription (25, 29). Additionally, T-Ag is oncogenic; its expression can lead to the induction of tumors of neuronal origin in experimental animals (27, 53, 57), and its genome has been detected in several human tumors (26, 28, 40). In addition, the several spliced variants of early proteins were shown to differentially interact with the retinoblastoma family of tumor suppressor proteins (14, 55).
JCV exhibits significant sequence homology (70%) to its counterpart simian virus 40 (SV40) in coding regions (15), and the early gene product, large T-Ag, for each virus displays similar functions in viral DNA replication and transcription. Previous studies indicated that SV40 large T-Ag cooperates with c-Jun in down-regulation of myelin Po gene expression in secondary Schwann cells (7). It was also demonstrated that cells undergo apoptosis when SV40 T-Ag and c-Jun coexpressed in HaCat cells (12). In addition, we and other investigators demonstrated that c-Jun interacts with its target DNA sequences present within the control region of JCV and positively regulates viral gene transcription (2, 42). Altogether, these studies suggested the possibility of a functional interaction between T-Ag and AP-1 family members such as c-Jun and perhaps with other family members as well. In this report, we used JCV as a model system and investigated the effect of AP-1 family members c-Jun and c-Fos on JCV T-Ag-mediated functions and provide experimental evidence that AP-1 family members physically and functionally interact with JCV large T-Ag and negatively affect both T-Ag-dependent viral gene transcription and replication.
MATERIALS AND METHODS
Cell lines.
U-87MG (ATCC HTB14), a human glioblastoma cell line, was grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum and antibiotics (penicillin-streptomycin, 100 μg/ml) and was maintained at 37°C in a humidified atmosphere with 7% CO2. HJC-15b cells (37) derived from hamster brain tumors (59) which were induced by JCV were grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum and antibiotics (penicillin-streptomycin, 100 μg/ml).
Plasmid constructs.
The pBLCAT3-Mad-1L reporter construct containing the regulatory region of JCV Mad-1 strain in the late orientation has been previously described (11). Expression plasmids for RSV-c-Jun and RSV-c-Fos were kindly provided by B. E. Sawaya (Temple University). pGEX2T-T-Ag (1-688) and its deletion mutants, pGEX2T-T-Ag (1-411), pGEX2T-T-Ag (1-265), pGEX2T-T-Ag (1-81), pGEX2T-T-Ag (266-688), pGEX2T-T-Ag (412-688), and pGEX2T-T-Ag (629-688), were previously described (44). pGEX2T-c-Jun (1-333) and its deletion mutants, pGEX2T-c-Jun (1-257), pGEX2T-c-Jun (1-150), pGEX2T-c-Jun (1-151-333), pGEX2T-c-Jun (258-333), and pGEX2T-c-Jun (281-333), were created by PCR amplification utilizing the following specific primers.
The forward primers were c-Jun 5′ (5′-ATGACTGCAAAGATGGAAACGACCTTC-3′), FP aa 258 (5′-AAGCGCATGAGGAACCGCATC-3′), FF aa 151 (5′-ACTGCAAAGATGGAAACGACCTTC-3′), and FF aa 281 (5′-GAGGAAAAAGTGAAAACCTTG-3′). The reverse primers were c-Jun-3′ (5′-TCAAAATGTTTGCAACTGCTG-3′), RP aa 280 (5′TCACAGCCGGGCGATTCTCTCCAG-3′), RP aa 257 (5′-TCACCATGTCAGTGGGGGACAGGG-3′), and RP aa 150 (5′-TCACACCGAGGGTACCGCGGGAGC-3′). RSV-c-Jun plasmid was used as a template in PCR amplification. pGEX2T-c-Fos was created by PCR amplification utilizing 5′ primer (5′-ATGTTCTGCGGCTTCAACGCAGACTAC-3′) and 3′ primer (5′-TCACAGGGCCAGCAGCGTGGGTGAGCT-3′) by subcloning the PCR product into BamHI/EcoRI sites of pGEX2T vector. RT-PCR-amplified human c-Fos cDNA was used as a template in PCR. pcDNA3-HA-c-Jun (1-257) and pcDNA3-HA-c-Jun (258-333) expression plasmids were also created by PCR amplification. Corresponding regions were PCR amplified and subcloned into BamHI/EcoRI sites of pcDNA3 and tagged with hemagglutinin tag in-frame at the 5′-end. CMV-T-Ag expression plasmid has been previously described (10).
Reporter gene assays.
A reporter construct containing the JCV regulatory region in late (pBLCAT3-Mad-1L) orientation was transiently transfected into U-87MG cells by the calcium-phosphate precipitation method (19) either alone or in combination with c-Jun (RSV-c-Jun) and c-Fos (RSV-c-Fos) and JCV T-Ag (CMV-T-Ag) expression plasmids. Plasmid concentrations used in each transfection experiment are indicated in the text and/or in the respective figure legends. The total amount of DNA transfected into the cells was normalized by using respective empty vectors. A glycerol shock was applied at 3 h posttransfection, and the medium was replenished. At 48 h posttransfection, cells were lysed by freeze-thaw cycles. After clearance of cell debris, the protein concentration of the supernatants was normalized, and CAT (chloramphenicol acetyltransferase) activity of samples was determined with 100 μg of protein for each sample. Transfections were repeated more than three times with different plasmid preparations. Standard deviations are indicated by error bars.
Replication assay.
Replication assays were carried out as previously described (47). Briefly, a replication-competent plasmid, pBLCAT3-Mad-1L, containing the regulatory region of Mad-1 strain of JCV was transfected alone or in combination with expression vectors CMV-T-Ag, RSV-c-Jun, RSV-c-Fos, CMV-c-Jun (1-257), and CMV-HA-c-Jun (258-333) into U-87MG cells (0.4 × 106 cells per 60-mm-diameter plate) with the calcium phosphate precipitation method. Plasmid concentrations used in transfections are indicated in respective figure legends, and the total amount of DNA transfected into the cells was normalized with appropriate empty vectors. A glycerol shock was applied at 4 h posttransfection, and the medium was replenished. At 72 h posttransfection, low-molecular-weight DNA containing both input and replicated plasmids was isolated by the Hirt method (22), digested with BamHI and DpnI enzymes, resolved on 1% agarose gel, and analyzed by Southern blotting. The bands corresponding to the replicated DNA were quantitated by utilizing a densitometer (Bio-Rad Fx PhosphorImager) with Quantity One Software. The degree of inhibition of T-Ag-mediated JCV DNA replication by AP-1 was expressed as percent inhibition with respect to the degree of viral DNA replication in the presence of T-Ag alone.
Nuclear extract preparation.
Nuclear extracts from U-87MG (treated or untreated with UV) was prepared by a modification of the miniextract protocol, as described by Schreiber et al. (49). UV-treated and untreated cells were harvested by trypsinization, washed once with complete Dulbecco's modified Eagle medium and twice with phosphate-buffered saline (PBS), and transferred to an Eppendorf tube. The cells were then resuspended in cold hypotonic buffer [10 mM N-2-hydroxyethylpipezine-N-2-ethanesulfonic acid (HEPES) (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.1 mM ethylene glycol bis(β-aminoethyl ether)-N,N,N′,N,-tetraacetic acid (EGTA), 1 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF)] and allowed to swell on ice. Cellular membranes were cleared by the addition of Nonidet P-40 (0.5% final concentration) and vortexing. The nuclei were pelleted by centrifugation at 10,000 × g, resuspended in cold extraction buffer (containing 20 mM HEPES [pH 7.9], 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, and 1 mM DTT and supplemented with a cocktail of protease inhibitors), and extracted at 4°C for 15 min on a racking platform. The nuclear extract was centrifuged, and the supernatant was frozen at −70°C.
Mobility band shift assay.
Band shift assays were carried out as described previously (46). Briefly, a double-stranded synthetic oligonucleotide containing JCV AP-1 binding site (5′-CAAGCATGAGCTCATACCTA-3′) was end labeled with [γ-32P]ATP with T4 polynucleotide kinase and gel purified. Nuclear extracts (10 μg/lane) prepared from U-87MG cells, untransfected or transfected with a JCV large T-Ag expression plasmid (CMV-T-Ag), were incubated with labeled probe (40,000 cpm/lane) in a binding buffer containing 1.0 μg of poly(dI-dC), 12 mM HEPES (pH 7.9), 4 mM Tris (pH 7.5), 60 mM KCl, 5 mM MgCl2, and 1.0 mM DTT. The reaction mixture was incubated at 4°C for 30 min to allow assembly of DNA-protein complexes. Experimental conditions for competitive band shift and antibody supershift assays are described under respective figure legends. The complexes were resolved on a 6% polyacrylamide gel in 0.5× TBE (1× TBE is 89 mM Tris-HCl [pH 8.0], 89 mM boric acid, and 2 mM EDTA [pH 8.0]). Gels were dried, and complexes were detected by autoradiography.
UV treatment.
U-87MG cells were plated on 100-mm-diameter tissue culture dishes and grown to subconfluence. Cells were then washed twice with PBS and kept under a thin layer of PBS until treated with UV (254 nm, 40 J/m2). Cells were subsequently incubated in fresh media and harvested 16 h posttreatment for nuclear extract preparation.
In vitro transcription-translation assay.
Full-length c-Jun was radiolabeled with [35S]methionine by using a TNT coupled in vitro transcription-translation system (Promega, Madison, Wis.) in accordance with the recommendations of the manufacturer.
Coimmunoprecipitation and Western blot analysis.
Two micrograms of anti-c-Jun antibody (KM-1; Santa Cruz) or preimmune serum was incubated with 0.5 mg of nuclear extract prepared from U-87MG cells overnight at 4°C with continuous rocking. Immunocomplexes were precipitated with the addition of protein A-Sepharose beads (20 μl of 50% slurry) (Pharmacia, Piscataway, N.J.) for an additional 2 h and washed extensively with lysis buffer containing 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% NP-40, and a cocktail of protease inhibitors. Immunocomplexes were then resolved by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) and transferred onto an immunoblotting membrane. The blots were then probed with an anti-SV40 T-Ag antibody (Ab-2 416) which cross-reacts with JCV T-Ag, developed with an ECL detection kit (Amersham-Pharmacia, Piscataway, N.J.) in accordance with the manufacturer's recommendations, and analyzed for the presence of T-Ag.
GST affinity chromatography assays (GST pull-down).
All glutathione S-transferase (GST) and GST fusion proteins were expressed and purified as described previously (48). For GST pull-down assays, 2 μg of either GST alone or GST-c-Jun or the deletion mutants of c-Jun immobilized on glutathione-Sepharose beads were incubated with 0.2 mg of nuclear extract prepared from U-87MG cells treated with UV overnight at 4°C in lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.5% Nonidet P-40 supplemented with a cocktail of protease inhibitors (Sigma). Formed complexes were washed extensively with lysis buffer and resolved by SDS-10% PAGE followed by Western blot analysis with anti-Jun antibody (KM-1; Santa Cruz). Alternatively, 4 μl of 35S-labeled in vitro-translated full-length c-Jun was incubated with GST alone or full-length GST-c-Jun fusion protein immobilized on glutathione-Sepharose beads. All reactions were performed in 400 μl of total reaction volume in lysis buffer overnight at 4°C with continuous rocking. After incubation, the beads were washed extensively with lysis buffer, and complexes were resolved by SDS-10% PAGE. Gels were dried and analyzed for c-Jun by autoradiography. Similarly, GST pull-down assays were also performed for GST-c-Fos fusion protein with whole-cell extracts (0.2 mg) prepared from HJC cells constitutively expressing JCV T-Ag utilizing anti-SV40 T-Ag antibody (Ab-2 416).
For mapping studies, 0.3 mg of whole-cell extract from HJC cells constitutively expressing JCV T-Ag were incubated with GST, GST-c-Jun, or GST-c-Jun amino- and carboxy-terminal deletion mutants immobilized on glutathione-Sepharose beads. Bound complexes were washed with lysis buffer and analyzed by Western blotting with anti-SV40 T-Ag antibody (Ab-2 416) for the detection of T-Ag. In reciprocal mapping studies, nuclear extracts prepared from U-87MG cells treated with UV were incubated with 2 μg of GST, GST-T-Ag, or GST-T-Ag amino- and carboxy-terminal deletion mutants immobilized on glutathione-Sepharose beads. Bound complexes were analyzed by Western blotting with anti-c-Jun antibody (KM-1) for the detection of c-Jun. All reactions were performed in 400 μl of total reaction volume in lysis buffer overnight at 4°C with continuous rocking.
RESULTS
AP-1 functionally interacts with large T-Ag.
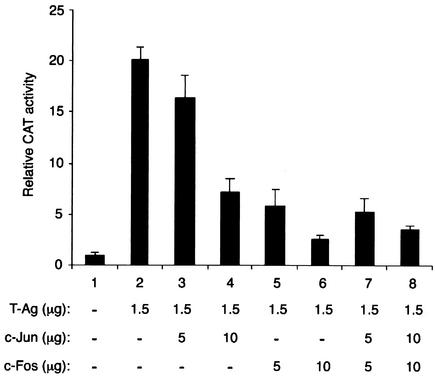
The members of the AP-1 family of transcription factors are involved in regulation of a wide range of cellular processes, including gene activation and repression (52). It has been previously shown that both c-Jun and SV40 large T-Ag are required for inhibition of Po gene promoter expression in secondary Schwann cells (7). Additionally, the induction of apoptosis by SV40 T-Ag correlates with c-Jun overexpression (12), suggesting that both proteins functionally interact with each other. Moreover, our recent findings indicated that an AP-1 family member, c-Jun, binds to its target sequences present within the regulatory region of JCV and regulates transcription from JCV early and late promoters (42). Altogether, these observations suggested the possibility that there would be a functional interaction between these two proteins. To that end, U-87MG cells, which support JCV transcription, were transiently transfected with a reporter construct containing the JCV late promoter alone or together with expression plasmids expressing either c-Jun, c-Fos, or T-Ag transgenes. As shown in Fig. 1A, a CAT reporter construct containing the JCV late promoter was poorly expressed in U-87MG cells in the absence of transactivators (lane 1). Cotransfection of the reporter construct with a T-Ag expression plasmid, as expected, resulted in a substantial (19.4-fold) increase in the transcriptional activity of the JCV late promoter (29) (compare lanes 1 and 2). However, when a constant amount of T-Ag was cotransfected with increasing amounts of either c-Jun (lanes 3 and 4) or c-Fos (lanes 5 and 6) or a combination of c-Jun and c-Fos (lanes 7 and 8), we observed a suppressive effect of c-Jun and c-Fos on the transcriptional activity of the late promoter. Of note, expression levels of T-Ag in the transfectants were analyzed by Western blotting, and results indicate that there appear to be equal levels of T-Ag expression. Therefore, the observed reduction in T-Ag-mediated activation of late promoter is not due to the transfection efficiency of transfectants or apoptotic effect of transactivators or the influence of c-Jun and c-Fos on CMV promoter driving T-Ag expression in U-87MG cells (data not shown). Altogether, these observations indicated that c-Jun and c-Fos negatively affect the T-Ag-dependent activation of JCV late promoter.
FIG. 1.
Effect of c-Jun and c-Fos on T-Ag-mediated transcription from JCV late promoter. A reporter plasmid (7 μg) containing the JCV late gene promoter was transfected into U-87MG cells alone or together with c-Jun (RSV-c-Jun), c-Fos (RSV-c-Fos), and T-Ag (CMV-JCV T-Ag) expression plasmids, as described in Materials and Methods. At 48 h posttransfection, cells were harvested and CAT enzymatic activity of each transfectant was determined by using equal 100-μg amounts of protein for each sample. Plasmid DNA concentrations used in the transfection are indicated at the bottom of the panel (in micrograms per 60-mm-diameter plate). Transfections were repeated several times with different plasmid preparations. Error bars indicate the standard deviation for each sample. The data are represented as CAT activity relative to basal level expression of the promoter.
c-Jun and c-Fos suppress T-Ag-mediated viral DNA replication.
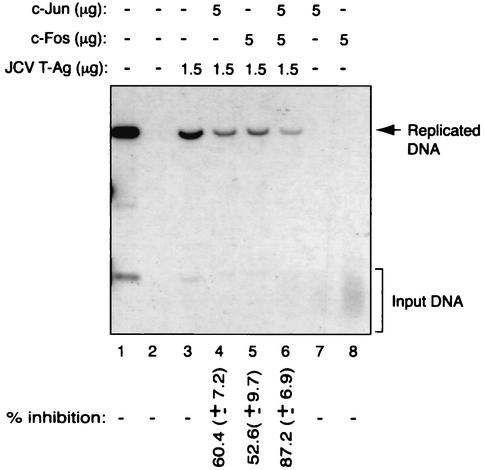
T-Ag is a multifunctional phosphoprotein and is required for initiation of JCV DNA replication (15, 32, 33). Transcriptional analysis of the JCV late promoter in the presence of c-Jun and c-Fos and T-Ag by cotransfection experiments showed a functional interaction between these proteins. We further investigated this functional interaction by employing a DpnI-DNA replication assay (10). A replication-competent plasmid containing the JCV origin of DNA replication was transfected alone or in combination with expression plasmids for c-Jun, c-Fos, and T-Ag into U-87MG cells. At 72 h posttransfection, low-molecular-weight DNA was isolated by the Hirt procedure (22) and newly replicated DNA was analyzed by Southern blotting. As shown in Fig. 2, in the presence of T-Ag, the plasmid containing the viral regulatory region was efficiently replicated (lane 3). However, overexpression of either c-Jun (lane 4) or c-Fos (lane 5) or c-Jun and c-Fos together (lane 6) significantly inhibited T-Ag-mediated viral DNA replication. c-Jun or c-Fos proteins alone showed no ability to induce viral DNA replication (lanes 7 and 8, respectively). These findings indicate that the AP-1 family members c-Jun and c-Fos can negatively affect T-Ag-induced replication of JCV DNA, perhaps through their physical interaction with T-Ag.
FIG. 2.
Both c-Jun and c-Fos inhibit T-Ag-mediated JCV DNA replication. A replication-competent plasmid, pBLCAT3-Mad-1L (5 μg), containing the regulatory region of the Mad-1 strain of JCV was transfected alone or in combination with c-Jun (RSV-c-Jun), c-Fos (RSV-c-Fos), and T-Ag (CMV JCV T-Ag) expression plasmids into U-87MG cells, as described in Materials and Methods. Plasmid DNA concentrations used in transfections are indicated on top (in micrograms per 60-mm-diameter plate). The total amount of DNA transfected into the cells was normalized with appropriate empty vectors. At 72 h posttransfection, low-molecular-weight DNA was isolated by the Hirt method (22), digested with both BamHI and DpnI enzymes, and analyzed by Southern blotting. The bands corresponding to the replicated DNA bands were quantitated by a densitometric method (see Materials and Methods) and expressed as percent inhibition with respect to the viral DNA replication in the presence of T-Ag alone. Representative data for the DpnI assay are shown, and variability among different experimental data is indicated by standard deviation. In lane 1, pBLCAT3-Mad-1L plasmid linearized with BamHI enzyme digestion was loaded as a control. The results are represented as percent inhibition and are shown at the bottom. The variability between different replication assays is indicated by standard deviation.
T-Ag inhibits c-Jun binding to its target sequences.
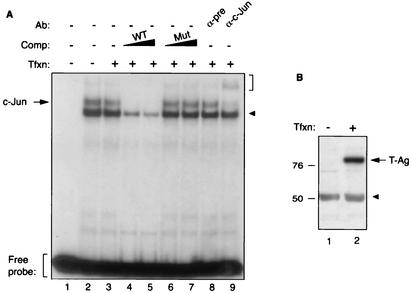
Our observations from functional assays suggested that c-Jun and T-Ag may associate with each other and influence each other's activity. To investigate the mechanism(s) involved in these observations, we performed electrophoretic mobility band shift assays using both double-stranded 32P end-labeled probe (JCV AP-1 WT, whose sequence is shown in the legend to Fig. 3) and nuclear extracts prepared from U-87MG cells transfected with a T-Ag expression plasmid. As demonstrated in Fig. 3A, when JCV AP-1 probe was incubated with nuclear extracts prepared from either untransfected cells (lane 2) or transfected cells (lane 3), we observed the formation of two distinct protein-DNA complexes, namely, a major complex and a minor complex with a slower migration property. It was surprising that the intensity of the band for the minor complex formed with nuclear extracts prepared from untransfected cells was significantly higher (69.3%) (lane 2) by densitometric analysis than that formed with extracts from transfected cells (lane 3), suggesting that T-Ag may interfere with the ability of c-Jun to efficiently interact with its target sequences on DNA. We have repeated these experiments with different nuclear extract preparations and observed similar results (data not shown). The specificity of observed complexes was further investigated by competitive band shift and antibody supershift experiments. Addition of the unlabeled JCV AP-1 oligonucleotide to the reaction mixture abolished the formation of minor bands and significantly decreased the efficiency of the protein-DNA complex formation for the major band (lanes 4 and 5). In contrast, unlabeled mutant JCV AP-1 oligonucleotide had no effect on both complexes. The nature of these complexes was further examined by antibody supershift experiments. Addition of preimmune antisera (lane 8) to the reaction mixture failed to show any effect on the formation of either band. However, anti-c-Jun antibody completely supershifted the minor band complex yet had no effect on the formation of the major protein-DNA complex, indicating that the observed minor complex is specific for c-Jun binding. Figure 3B demonstrates the analysis of T-Ag expression in transfected cells by Western blotting. Taken together, the results from band shift assays indicate that T-Ag interferes with c-Jun binding to its target sequences.
FIG. 3.
T-Ag inhibits c-Jun binding to its target sequences in an electrophoretic mobility shift assay. (A) Band shift assay. A double-stranded synthetic oligonucleotide containing JCV AP-1 binding site (5′-CAAGCATGAGCTCATACCTA-3′) spanning nucleotides 155 to 162 of JCV Mad-1 regulatory region was end labeled with [γ-32P]ATP with T4 polynucleotide kinase and gel purified. Nuclear extracts (10 μg/lane) prepared from U-87MG cells and untransfected (lane 2) or transfected with a T-Ag expression plasmid (lane 3) were incubated with labeled probe (40,000 cpm/lane) in a binding buffer, as described in Materials and Methods. In addition, probe plus nuclear extract from T-Ag-transfected cells was also incubated with either unlabeled wild-type oligonucleotide (WT) (lanes 4 and 5) or 25- or 150-fold molar excesses of its mutant variant competitor oligonucleotides (Mut) (5′-CAAGCATTAGCTTGTACCTA-3′; bold indicates base substitutions relative to the wild type) (lanes 6 and 7, respectively). Probe plus nuclear extract mixture was also incubated either with a preimmune (α-pre; 2 μg) (lane 8) or an anti-c-Jun (α-c-Jun, KM-1; 2 μg) (lane 9) antibody. Formed DNA-protein complexes were then resolved on a 6% polyacrylamide gel under native conditions and visualized by autoradiography. The specific DNA-protein complexes are indicated by an arrow, and nonspecific complexes are indicated by a solid arrowhead. A bracket indicates antibody supershifted complexes. (B) Western blot analysis. Nuclear extracts prepared from either untransfected U-87MG cells (lane 1) or U-87MG cells transfected with CMV-T-Ag expression plasmid (lane 2) were analyzed by Western blotting using an anti-SV40 T-Ag antibody (Ab-2 416) which is cross-reactive with JCV T-Ag. Comp, competitor; Ab, antibody; Tfxn, transfection.
c-Jun and c-Fos physically associate with T-Ag.
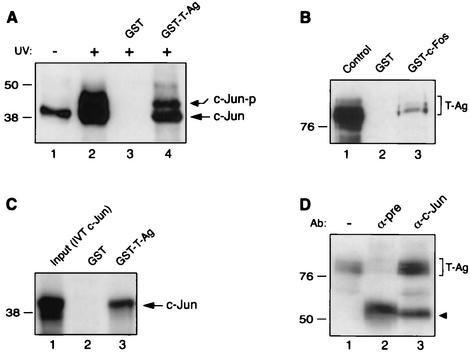
Results from both functional and band shift assays indicated that c-Jun and c-Fos may in fact physically interact with JCV large T-Ag. To test this possibility, we performed affinity chromatography (GST pull-down) experiments in which one of the two proteins was bacterially expressed as a GST fusion protein and bound to a glutathione resin, while the other protein was passed over the resin and analyzed for its ability to be specifically retained by the GST fusion protein. GST or GST-T-Ag fusion protein was immobilized on glutathione-Sepharose beads and incubated with nuclear extracts prepared from U-87MG cells either nontreated or treated with UV, which is known to potently induce c-Jun upregulation and phosphorylation (4, 62). Protein complexes bound to GST or GST-T-Ag were washed extensively and analyzed by Western blotting with an anti-c-Jun antibody, which detects both phosphorylated and nonphosphorylated forms of c-Jun on Western blots. As shown in Fig. 4A, surprisingly, both phosphorylated and nonphosphorylated forms of c-Jun protein are retained on the Sepharose column containing GST-T-Ag fusion protein (lane 4), indicating that both forms of c-Jun are capable of interacting with T-Ag in vitro. An interaction of this type between GST alone and c-Jun was not observed (lane 3), which indicates the specificity of association of c-Jun with T-Ag.
FIG. 4.
Both c-Jun and c-Fos physically interact with T-Ag. (A) c-Jun associates with T-Ag in GST pull-down assays. Nuclear extracts (0.5 mg) prepared from U-87MG cells treated with UV and incubated with either GST alone (lane 3) or GST-T-Ag (lane 4), both of which are already immobilized on GST beads. After being washed extensively with incubation buffer, proteins interacting with GST or GST-T-Ag were analyzed by Western blotting with an anti-c-Jun antibody, KM-1, which detects nonphosphorylated and phosphorylated forms of c-Jun. Nuclear extracts prepared from nontreated U-87MG cells (lane 1) or U-87MG cells treated with UV (lane 2) were loaded as negative and positive controls, respectively (20 μg/lane). Phosphorylated (c-Jun-P) and nonphosphorylated (c-Jun) forms of c-Jun are indicated by arrows. (B) Similarly, whole-cell extracts (0.2 mg) prepared from HJC cells constitutively expressing JCV T-Ag were incubated with either GST alone (lane 2) or GST-c-Fos (lane 3), and after being washed, proteins interacting with GST or GST-c-Fos were analyzed by Western blotting with an anti-SV40 T-Ag antibody, Ab-2 416, which is also cross-reactive with JCV T-Ag. Whole-cell extract (10 μg/lane) from HJC cells was loaded as a positive control (lane 1). T-Ag is indicated by a bracket. (C) In vitro-translated [35S]methionine-labeled c-Jun (IVT-c-Jun) interacts with GST-T-Ag in a cell-free system. In vitro-labeled full-length c-Jun was incubated with either GST (lane 2) or GST-T-Ag fusion protein (lane 3). After extensive washing of the column with binding buffer, bound proteins were resolved by SDS-10% PAGE and analyzed by autoradiography. A small portion of IVT-c-Jun was loaded as a positive control (lane 1). (D) T-Ag coimmunoprecipitates with c-Jun. Coimmunoprecipitation experiments were performed as described in Materials and Methods. Antibodies used for respective lanes are shown at the top. An arrowhead points to the large subunit of α-c-Jun immunoglobulin G antibody used in immunoprecipitation assays. The positions of the molecular mass markers (in kilodaltons) are shown on the left side of each panel.
A similar GST pull-down assay was performed to demonstrate the association of c-Fos with T-Ag. Whole-cell extracts from hamster glial HJC-15b cells, constitutively expressing T-Ag (37, 59), were incubated with either GST or GST-c-Fos, and after washing, protein complexes retained in the columns were analyzed by Western blotting with an anti-SV40 T-Ag antibody (Ab-2 416). As demonstrated in Fig. 4B, T-Ag protein from whole-cell extracts was retained in the GST-c-Fos glutathione column (lane 3) but not in the GST column (lane 2), indicating the specificity of interaction between c-Fos and T-Ag.
We also investigated the protein-protein interaction between c-Jun and T-Ag by using a cell-free system. As shown in Fig. 4C, in vitro-transcribed and translated full-length c-Jun specifically and strongly interacts with GST-T-Ag fusion protein (lane 2) but not GST alone (lane 2) in a GST-pull-down assay, further confirming the observed interaction between c-Jun and T-Ag.
Next, to further examine the association of c-Jun with T-Ag, we performed coimmunoprecipitation experiments. Nuclear extracts prepared from HJC-15b cells were immunoprecipitated either with normal serum (control) or an anti-c-Jun antibody, and immunocomplexes were analyzed by Western blotting for the presence of T-Ag with an anti-T-Ag antibody. Of note, in addition to expressing large T-Ag, HJC-15b cells also express high levels of c-Jun constitutively. As demonstrated in Fig. 4D, T-Ag was coimmunoprecipitated with c-Jun and was detected by the anti-T-Ag antibody (lane 3). The specificity of this coimmunoprecipitation was verified when protein extracts from HJC-15b cells were incubated with normal mouse serum where no apparent coimmunoprecipitation of T-Ag was detected (lane 2). Expression of c-Jun in HJC-15b cells was verified by Western blotting (data not shown). Altogether, our findings from in vitro protein-protein interaction experiments demonstrated that the AP-1 family members c-Jun and c-Fos physically associate with T-Ag.
Mapping of T-Ag interaction domain of c-Jun.
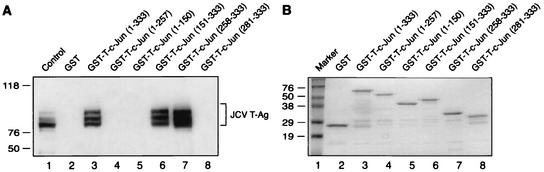
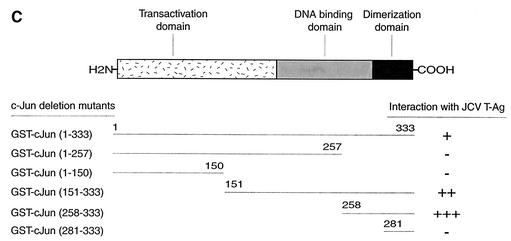
In the next series of experiments, we attempted to map the region(s) of c-Jun that are involved in the interaction with T-Ag. A series of deletion mutants of the c-Jun gene was created, and mutant c-Jun proteins fused to GST were incubated with whole-cell lysate from HJC-15b cells. Bound complexes were resolved by SDS-PAGE and analyzed by Western blotting with anti-T-Ag antibody. As shown in Fig. 5A, as expected, GST-c-Jun full-length protein (lane 3), but not GST alone (lane 2), efficiently binds to T-Ag. However, removal of the carboxy-terminal region of c-Jun spanning amino acid residues 258 to 333 (lane 4), 151 to 333 (lane 5), and 281 to 333 (lane 8) completely abolished the interaction between c-Jun and T-Ag. However, deletion of the amino-terminal region spanning residues 1 to 150 (lane 6) and 1 to 257 (lane 7) enhanced the binding ability of T-Ag to c-Jun in comparison to that observed with full-length c-Jun (compare lanes 6 and 7 to lane 3), suggesting that amino acid residues between positions 1 and 257 have a negative effect on the interaction between c-Jun and T-Ag. It is of note that T-Ag expressed in HJC-15b cells exists as several isoforms, depending on the phosphorylation state of the protein (37). Interestingly, c-Jun interacts with all isoforms of T-Ag expressed in HJC-15b cells. This is similar to the previously reported interaction of T-Ag with Purα and YB-1 (16, 44). Taken together, these mapping studies demonstrate that the T-Ag interaction domain of c-Jun is localized to amino acid residues between positions 258 and 280, which overlap with a portion of the basic-DNA binding domain of c-Jun protein, suggesting that T-Ag, by binding to the DNA binding domain of c-Jun, may interfere with its binding to DNA and therefore with its transcriptional activity. These findings correlate with our band shift assays (Fig. 3A) in which we observed that T-Ag negatively affects the DNA binding activity of c-Jun to its target sequences (Fig. 3A). Figure 5B illustrates the Coomassie blue-stained SDS-PAGE of the full-length and mutant proteins which were used in this study and verifies the integrity of the protein preparations. Figure 5C summarizes the results of the GST pull-down assay and depicts the regions of c-Jun that bind to T-Ag.
FIG. 5.
Localization of c-Jun domains important for interaction with T-Ag. (A) Whole-cell extracts prepared from HJC-15b cells (0.25 mg) were incubated with either GST alone (lane 2), GST-c-Jun (lane 3), or deletion mutants of c-Jun fused to GST (lanes 4 to 8) (2 μg each) which were immobilized on glutathione-Sepharose beads. The Sepharose beads were washed extensively, and bound proteins were resolved by SDS-10% PAGE, transferred onto nitrocellulose membrane, and detected with an anti-SV40 T-Ag antibody (Ab-2 416). (B) SDS-10% PAGE analysis of GST, GST-T-Ag, and T-Ag deletion mutants. In lane 1, 10 μg of whole-cell extract from HJC-15b cells was loaded as positive control. (C) Summary of the results obtained from mapping assays. A schematic representation of c-Jun is shown at the top (not to scale). The abilities of c-Jun and its deletion mutants to interact with T-Ag are shown on the right. +++, very strong interaction; ++, strong interaction; +, reduced or weak interaction; −, no interaction.
Localization of c-Jun interaction domain of T-Ag.
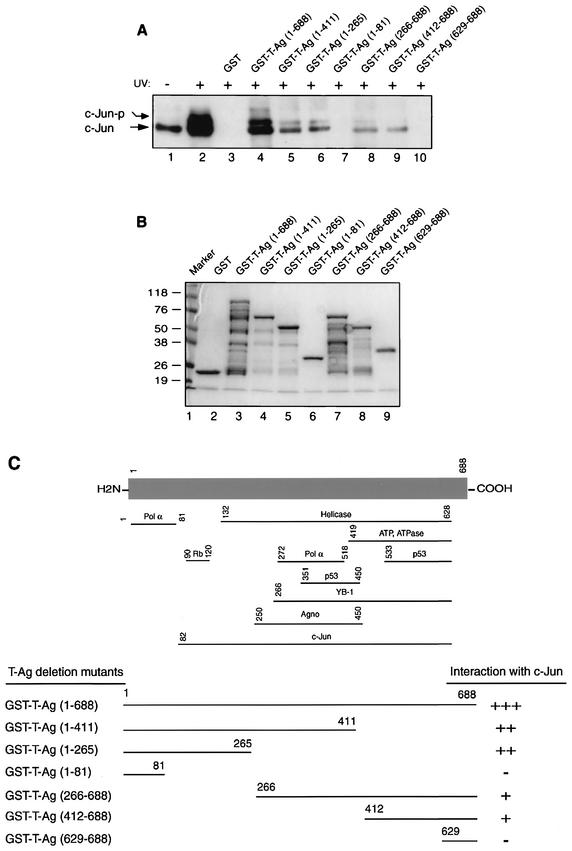
To identify the protein domain(s) of T-Ag which confers the interaction with c-Jun, a series of T-Ag carboxy-terminal and amino-terminal deletion mutants fused with GST were prepared and incubated with nuclear extracts prepared from U-87MG cells treated with UV. In agreement with the results presented in Fig. 4A, GST-T-Ag, but not GST, bound to both nonphosphorylated and phosphorylated forms of c-Jun (Fig. 6A, lanes 4 and 3, respectively). The binding activities of two carboxy-terminal deletion mutants were slightly reduced, compared to that of the full-length protein, when the residues between positions 412 and 688 (lane 5) and 266 and 688 (lane 6) were deleted. Similarly, the removal of the amino acids 1 to 265 (lane 8) and 1 to 411 (lane 9) further decreased the binding efficiency of amino-terminal deletion mutants of T-Ag. The remaining two deletion mutants retaining residues 1 to 81 (lane 7) and 629 to 688 (lane 10) showed no c-Jun binding activity. Taken together, these mapping experiments demonstrate that the c-Jun interaction domain of T-Ag lies within the middle portion of the protein spanning amino acid residues 82 to 628. Figure 6B illustrates the Coomassie blue-stained SDS-PAGE gel of the full-length T-Ag and T-Ag mutants which were used in this study. A scheme for these observations is shown in Fig. 6C.
FIG.6.
Mapping of interaction domain of T-Ag with c-Jun. (A) Nuclear extracts (0.2 mg) prepared from U-87MG cells treated with UV were incubated with either GST alone (lane 3), GST-T-Ag (lane 4), or deletion mutants of GST-T-Ag fusion proteins (lanes 5 to 10) immobilized on glutathione-Sepharose beads. Bound proteins were washed extensively, resolved by SDS-10% PAGE, and analyzed by Western blotting with an anti-T-Ag antibody (Ab-2 416). C-Jun-P indicates the phosphorylated form of c-Jun. Nuclear extracts from nontreated U-87MG cells (lane 1) and U-87MG cells treated with UV (lane 2) were loaded as positive controls. (B) Analysis of the GST and GST-T-Ag proteins and GST-T-Ag deletion mutants by SDS-10% PAGE. (C) Summary of the binding assays. Upper portion, schematic representation of T-Ag with various functional domains and binding regions for different proteins. Lower portion, the ability of T-Ag and its deletion mutants to interact with c-Jun is depicted. +++, very strong interaction; ++, strong interaction; +, reduced interaction; −, no interaction.
Effect of two c-Jun mutant proteins on T-Ag-mediated gene transcription and replication.
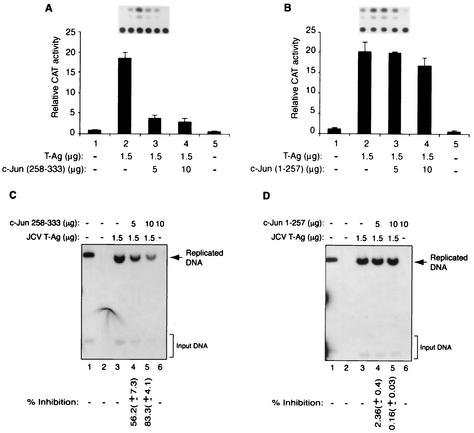
To further assess the functional interaction between c-Jun and T-Ag, we examined the ability of mutant c-Jun proteins, which have retained or lost their ability to interact with T-Ag in GST pull-down assays. We chose mutant c-Jun (258-333), which had the ability to strongly interact with T-Ag in in vitro GST pull-down assays, and mutant c-Jun (1-257), which showed no T-Ag binding activity. Transient-transfection assays were performed by utilizing a reporter JCV late CAT construct and expression plasmids for mutants c-Jun (258-333) and c-Jun (1-257). As shown in Fig. 7A, mutant c-Jun protein (258-333), which binds to T-Ag more efficiently than full-length protein (Fig. 5A, lane 7), showed a drastic negative effect on the T-Ag-mediated transcriptional activation of the JCV late promoter (Fig. 7A) compared to full-length protein (Fig. 1A, lanes 3 and 4). Under similar conditions, however, mutant c-Jun protein (1-257), which does not interact with T-Ag, had virtually no significant effect on the level of transcriptional activation of the JCV late promoter by T-Ag (Fig. 7B).
FIG. 7.
Functional interaction of T-Ag with two c-Jun deletion mutants. (A and B) Effects of c-Jun deletion mutants on T-Ag-mediated activation of JCV late promoter. A CAT reporter plasmid (7 μg) containing the JCV late promoter was transfected into U-87MG cells alone or in combination with c-Jun deletion mutants c-Jun (258-333) (A) or c-Jun (1-257) (B) and T-Ag expression plasmids, as described in legend to Fig. 1. The concentrations of expression plasmids are indicated at the bottom of the panels in micrograms per 60-mm-diameter plate. (C and D) Effects of c-Jun deletion mutants on T-Ag-mediated viral DNA replication. A replication-competent plasmid, pBLCAT3-Mad-1L (5 μg) containing the regulatory region of the Mad-1 strain of JCV was transfected alone or in combination with c-Jun deletion mutants CMV-HA-c-Jun (258-333) (C) and CMV-HA-c-Jun (1-257) (D) and CMV-T-Ag expression plasmids into U-87MG cells. Plasmid concentrations used in transfections are indicated at the top of the panels in micrograms. In lane 1, pBLCAT3 plasmid digested with BamHI enzyme was loaded as a positive control. Both the replication assays and the quantitation of the bands corresponding to the replicated viral DNA were carried out as described in Materials and Methods. Representative data for DpnI assays are shown, with variability indicated by standard deviation. The total amount of DNA transfected into the cells in all four panels was normalized with appropriate empty vectors.
We also examined the effects of these two mutants [c-Jun (1-257) and c-Jun (258-333)] on T-Ag-induced JCV DNA replication. In agreement with the above observations from transfection assays, c-Jun (258-333) strongly inhibited T-Ag-mediated JCV DNA replication (Fig. 7C). The mutant protein c-Jun (1-258) showed no appreciable inhibitory or stimulatory activity on the replication of viral DNA in glial cells (Fig. 7D). Interestingly, comparison of the results from the studies regarding the effect of full-length c-Jun on T-Ag-mediated transcription (Fig. 1A) with that obtained with mutant c-Jun (258-333) (Fig. 7A) suggests that the mutant c-Jun protein (258-333) is more potent in modulating T-Ag function than the full-length protein. These findings corroborate the results obtained from protein-protein interaction studies (Fig. 5A), where we observed that c-Jun (258-333) displayed stronger binding activity for T-Ag than the full-length protein. It should also be noted that the expression of c-Jun mutants in these transfection assays was demonstrated by Western blot analysis (data not shown), and therefore the absence of a functional interaction between c-Jun and T-Ag may not be attributed to its lack of expression in transfected cells. Taken together, the data from functional assays utilizing c-Jun mutants, one of which, c-Jun (258-333), physically and functionally interacts with T-Ag, and the other, c-Jun (1-257), which lacks such characteristics, further confirm the significance of interaction between c-Jun and T-Ag.
DISCUSSION
In this report, we examined the molecular mechanism(s) involved in the regulation of JCV gene transcription and replication by the AP-1 family members c-Jun and c-Fos and by the viral protein T-Ag; we showed that both c-Jun and c-Fos interact with T-Ag and negatively regulate T-Ag-mediated viral gene transcription and replication.
The members of the AP-1 family of transcription factors are known to regulate transcription from many cellular and viral promoters and are implicated in many important cellular and viral processes, including gene transcription. It has been reported recently that a member of the AP-1 family, c-Jun, specifically interacts with the target DNA binding sequences present within the control region of JCV and regulates transcription from JCV promoters (42). However, it was interesting to observe in this study that c-Jun and c-Fos, individually or in combination, negatively regulate T-Ag-dependent viral gene transcription and replication (Fig. 1 and 2). Consistent with this observation, the previous reports showed that Jun cooperates with SV40 virus large T-Ag in down-regulation of myelin Po gene expression in secondary Schwann cells in vitro (7). These observations suggested the possibility that c-Jun and c-Fos may form a complex with T-Ag which is functionally inactive in nature. With respect to this notion, we also observed that in the presence of T-Ag, DNA binding activity of c-Jun is diminished compared to normal levels in band shift assays (Fig. 3). We also carried out in vitro protein-protein interaction studies, including coimmunoprecipitation and GST pull-down assays, to support the findings from band shift assays and provided evidence that c-Jun and T-Ag in fact physically interact with each other (Fig. 4A, B and C). In addition, it was also evident from GST pull-down assays that the phosphorylated form of c-Jun is as capable of interacting with T-Ag as the nonphosphorylated form. It was previously shown that AP-1 family members are phosphorylated upon induction by a variety of extracellular stimuli, including UV and viral infection, and that phosphorylated forms are more active in terms of their transcriptional activity (5, 62).
In addition to performing protein-protein interaction studies to demonstrate physical interaction between c-Jun and T-Ag, we also carried out experiments to map the domain(s) of the interaction of these proteins with each other. The interaction domain of c-Jun with T-Ag maps to the basic DNA binding domain of the protein, which is juxtaposed with the leucine-rich dimerization domain of the protein. This is an important finding in light of our observations from functional assays, and this interaction appears to interfere with the DNA binding and therefore with transcriptional activity of c-Jun. It is known that the DNA binding activity of the AP-1 family of transcription factors is a prerequisite for their transcriptional activity. Results from DNA binding studies (Fig. 3) corroborate our findings from mapping studies in that T-Ag interacts with DNA binding domain of c-Jun and interferes with its association with DNA. It is also important to note that T-Ag, which is constitutively expressed in HJC-b15 cells, exists as several isoforms, depending on the phosphorylation state of the protein, and all isoforms are capable of interacting with c-Jun. Interestingly, the phosphorylation state of T-Ag does not appear to dictate the association between c-Jun and T-Ag, since bacterially expressed T-Ag, which does not contain phosphorylated residues, is able to interact with c-Jun (Fig. 6A). Nonetheless, further experiments will be required to demonstrate the contribution, if any, of the phosphorylation state of T-Ag to the efficiency of formation of the intermolecular complex between c-Jun and T-Ag. It was also interesting to observe that as the amino-terminal sequences of c-Jun were deleted, the binding efficiency of c-Jun deletion mutants (258-333) and (281-333) (Fig. 5A, lanes 6 and 7) significantly improved, implying that the sequences located within the amino-terminal region of c-Jun may negatively regulate the protein-protein interaction observed between c-Jun and T-Ag. Consistent with these observations, the use of c-Jun mutant (258-333) in transfection studies (Fig. 7A) showed that this mutant is capable of suppressing the T-Ag-mediated viral late gene transcription more drastically than full-length c-Jun (compare lanes 3 and 4 in Fig. 7A with those in Fig. 1). Moreover, the results from in vitro mapping experiments demonstrated that the c-Jun interaction domain of T-Ag is localized to its central region (Fig. 6A), which encompasses functionally important domains, including the polymerase α, ATP binding, ATPase, and helicase domains. This suggests that c-Jun may interfere with those functions by sequestering T-Ag in cells. Consistent with this hypothesis, our findings from viral DNA replication studies indicate that c-Jun, by binding to the central region of T-Ag, may interfere with its function (Fig. 2 and 6C) in viral DNA replication.
Transcriptional and replicational regulation of JCV involve a highly organized cascade of events that requires participation of both viral and cellular factors. The combination of cooperative and antagonistic regulatory activities of both viral and cellular transcription factors determines a successful outcome of the viral productive cycle. At the initial stages of viral infection, only host cellular factors are responsible for expression of viral early genes in the absence of the viral large T-Ag. When expressed, JCV large T-Ag in cooperation with cellular factors initiates viral DNA replication and transactivates viral late genes. In the early stages of infection cycle, the immediate-early inducible genes, including the c-Jun and c-Fos genes, which were shown to be induced by viral infection (17), are likely to participate in regulation of JCV early promoter. Consistent with this hypothesis, we have recently demonstrated that the AP-1 family members c-Jun and c-Fos activate transcription from JCV early promoter more strongly than from late promoter (42). In this report, we further investigated the regulatory function of AP-1 family members, in particular c-Jun and c-Fos, through their physical and functional interaction with JCV regulatory protein large T-Ag and presented evidence that both proteins display negative effects on T-Ag-mediated viral gene transcription and replication. The physiological consequence(s) of this negative regulation by AP-1 is currently unknown. However, one can hypothesize that the immediate-early inducible factors, including c-Jun and c-Fos, although displaying positive regulatory roles on the expression of JCV promoters in the absence of T-Ag and at the early phases of infection, may exhibit antagonistic effects on T-Ag-mediated activities during the late phases of infection. This negative regulatory activity by AP-1 may result in a positive effect on viral growth. AP-1 may prolong the survival of infected cells by slowing down both JCV transcription and replication and therefore may positively influence the process of maturation of the infectious viral particles. The study of JCV regulatory processes at the molecular level will shed more light on the molecular mechanisms governing the JCV-host interactions and thereby pave the way to understand the progression of the diseases associated with JCV infections.
Acknowledgments
The first two authors contributed equally to this work.
We thank past and present members of the Center for Neurovirology and Cancer Biology for their insightful discussion and sharing of ideas and reagents. We also thank Cynthia Schriver for editorial assistance.
This work was made possible by grants awarded by NIH to K.K., S.A., and M.S.
REFERENCES
- 1.Amemiya, K., R. Traub, L. Durham, and E. O. Major. 1992. Adjacent nuclear factor-1 and activator protein binding sites in the enhancer of the neurotropic JC virus. A common characteristic of many brain-specific genes. J. Biol. Chem. 267:14204-14211. [PubMed] [Google Scholar]
- 2.Amemiya, K., R. Traub, L. Durham, and E. O. Major. 1989. Interaction of a nuclear factor-1-like protein with the regulatory region of the human polyomavirus JC virus. J. Biol. Chem. 264:7025-7032. [PubMed] [Google Scholar]
- 3.Angel, P., M. Imagawa, R. Chiu, B. Stein, R. J. Imbra, H. J. Rahmsdorf, C. Jonat, P. Herrlich, and M. Karin. 1987. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49:729-739. [DOI] [PubMed] [Google Scholar]
- 4.Angel, P., and M. Karin. 1991. The role of Jun, Fos and the AP-1 complex in cell proliferation and transformation. Biochim. Biophys. Acta 1072:129-157. [DOI] [PubMed] [Google Scholar]
- 5.Angel, P., and M. Karin. 1992. Specific members of the Jun protein family regulate collagenase expression in response to various extracellular stimuli. Matrix Suppl. 1:156-164. [PubMed] [Google Scholar]
- 6.Berger, J. R., and M. Concha. 1995. Progressive multifocal leukoencephalopathy: the evolution of a disease once considered rare. J. Neurovirol. 1:5-18. [DOI] [PubMed] [Google Scholar]
- 7.Bharucha, V. D., K. W. C. Peden, and G. I. Tennekoon. 1994. SV40 large T antigen with c-Jun down regulates myelin Po gene expression: a mechanism for papovaviral T antigen-mediated demyelination. Neuron 12:627-637. [DOI] [PubMed] [Google Scholar]
- 8.Bossy-Wetzel, E., L. Bakiri, and M. Yaniv. 1997. Induction of apoptosis by the transcription factor c-Jun. EMBO J. 16:1695-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner, D. A., M. O'Hara, P. Angel, M. Chojkier, and M. Karin. 1989. Prolonged activation of jun and collagenase genes by tumour necrosis factor-alpha. Nature 337:661-663. [DOI] [PubMed] [Google Scholar]
- 10.Chang, C. F., G. L. Gallia, V. Muralidharan, N. N. Chen, P. Zoltick, E. Johnson, and K. Khalili. 1996. Evidence that replication of human neurotropic JC virus DNA in glial cells is regulated by the sequence-specific single-stranded DNA-binding protein Pur alpha. J. Virol. 70:4150-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, N. N., and K. Khalili. 1995. Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Pur alpha in glial cells. J. Virol. 69:5843-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, S. L., Y. P. Tsoa, Y. L. Chen, S. J. Huang, J. L. Chang, and S. F. Wu. 1998. The induction of apoptosis by SV40 T antigen correlates with c-Jun overexpression. Virology 244:521-529. [DOI] [PubMed] [Google Scholar]
- 13.Devary, Y., R. A. Gottlieb, L. F. Lau, and M. Karin. 1991. Rapid and preferential activation of the c-jun gene during the mammalian UV response. Mol. Cell. Biol. 11:2804-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frisque, R. J. 2001. Structure and function of JC virus T' proteins. J. Neurovirol. 7:293-297. [DOI] [PubMed] [Google Scholar]
- 15.Frisque, R. J., and F. A. White. 1992. The molecular biology of JC virus, causative agent of progressive multifocal leukoencephalopathy. Humana Press Inc., Totowa, N.J.
- 16.Gallia, G. L., M. Safak, and K. Khalili. 1998. Interaction of the single-stranded DNA-binding protein Puralpha with the human polyomavirus JC virus early protein T-antigen. J. Biol. Chem. 273:32662-32669. [DOI] [PubMed] [Google Scholar]
- 17.Glenn, G. M., and W. Echart. 1990. Transcriptional regulation of early-response genes during polyomavirus infection. J. Virol. 64:2193-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldgaber, D., H. W. Harris, T. Hla, T. Maciag, R. J. Donnelly, J. S. Jacobsen, M. P. Vitek, and D. C. Gajdusek. 1989. Interleukin 1 regulates synthesis of amyloid beta-protein precursor mRNA in human endothelial cells. Proc. Natl. Acad. Sci. USA 86:7606-7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 20.Henson, J., J. Saffer, and H. Furneaux. 1992. The transcription factor Sp1 binds to the JC virus promoter and is selectively expressed in glial cells in human brain. Ann. Neurol. 32:72-77. [DOI] [PubMed] [Google Scholar]
- 21.Herrlich, P., H. Ponta, and H. J. Rahmsdorf. 1992. DNA damage-induced gene expression: signal transduction and relation to growth factor signaling. Rev. Physiol. Biochem. Pharmacol. 119:187-223. [DOI] [PubMed] [Google Scholar]
- 22.Hirt, B. J. 1976. Selective extraction of polyoma DNA from infected mouse cell culture. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, R. S., B. van Lingen, V. E. Papaioannou, and B. M. Spiegelman. 1993. A null mutation at the c-jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev. 7:1309-1317. [DOI] [PubMed] [Google Scholar]
- 24.Kerr, D., C. F. Chang, N. Chen, G. Gallia, G. Raj, B. Schwartz, and K. Khalili. 1994. Transcription of a human neurotropic virus promoter in glial cells: effect of YB-1 on expression of the JC virus late gene. J. Virol. 68:7637-7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khalili, K., L. Feigenbaum, and G. Khoury. 1987. Evidence for a shift in 5′-termini of early viral RNA during the lytic cycle of JC virus. Virology 158:469-472. [DOI] [PubMed] [Google Scholar]
- 26.Krynska, B., L. Del Valle, S. Croul, J. Gordon, C. D. Katsetos, M. Carbone, A. Giordano, and K. Khalili. 1999. Detection of human neurotropic JC virus DNA sequence and expression of the viral oncogenic protein in pediatric medulloblastomas. Proc. Natl. Acad. Sci. USA 96:11519-11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krynska, B., J. Otte, R. Franks, K. Khalili, and S. Croul. 1999. Human ubiquitous JCV(CY) T-antigen gene induces brain tumors in experimental animals. Oncogene 18:39-46. [DOI] [PubMed] [Google Scholar]
- 28.Laghi, L., A. E. Randolph, D. P. Chauhan, G. Marra, E. O. Major, J. V. Neel, and C. R. Boland. 1999. JC virus DNA is present in the mucosa of the human colon and in colorectal cancers. Proc. Natl. Acad. Sci. USA 96:7484-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lashgari, M. S., H. Tada, S. Amini, and K. Khalili. 1989. Regulation of JCVL promoter function: transactivation of JCVL promoter by JCV and SV40 early proteins. Virology 170:292-295. [DOI] [PubMed] [Google Scholar]
- 30.Le-Niculescu, H., E. Bonfoco, Y. Kasuya, F. X. Claret, D. R. Green, and M. Karin. 1999. Withdrawal of survival factors results in activation of the JNK pathway in neuronal cells leading to Fas ligand induction and cell death. Mol. Cell. Biol. 19:751-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, C. K., A. P. Hope, and W. J. Atwood. 1998. The human polyomavirus, JCV, does not share receptor specificity with SV40 on human glial cells. J. Neurovirol. 4:49-58. [DOI] [PubMed] [Google Scholar]
- 32.Lynch, K. J., and R. J. Frisque. 1991. Factors contributing to the restricted DNA replicating activity of JC virus. Virology 180:306-317. [DOI] [PubMed] [Google Scholar]
- 33.Lynch, K. J., and R. J. Frisque. 1990. Identification of critical elements within the JC virus DNA replication origin. J. Virol. 64:5812-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch, K. J., S. Haggerty, and R. J. Frisque. 1994. DNA replication of chimeric JC virus-simian virus 40 genomes. Virology 204:819-822. [DOI] [PubMed] [Google Scholar]
- 35.Monaco, M. C., B. F. Sabath, L. C. Durham, and E. O. Major. 2001. JC virus multiplication in human hematopoietic progenitor cells requires the NF-1 class D transcription factor. J. Virol. 75:9687-9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pho, M. T., A. Ashok, and W. J. Atwood. 2000. JC virus enters human glial cells by clathrin-dependent receptor-mediated endocytosis. J. Virol. 74:2288-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raj, G. V., J. Gordon, T. J. Logan, D. J. Hall, A. Deluca, A. Giordano, and K. Khalili. 1995. Characterization of glioma cells derived from polyomavirus-induced brain tumors in hamsters. Int. J. Oncol. 7:801-808. [DOI] [PubMed] [Google Scholar]
- 38.Raj, G. V., and K. Khalili. 1994. Identification and characterization of a novel GGA/C-binding protein, GBP-i, that is rapidly inducible by cytokines. Mol. Cell. Biol. 14:7770-7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranganathan, P. N., and K. Khalili. 1993. The transcriptional enhancer element, kappa B, regulates promoter activity of the human neurotropic virus, JCV, in cells derived from the CNS. Nucleic Acids Res. 21:1959-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rencic, A., J. Gordon, J. Otte, M. Curtis, A. Kovatich, P. Zoltick, K. Khalili, and D. Andrews. 1996. Detection of JC virus DNA sequence and expression of the viral oncoprotein, tumor antigen, in brain of immunocompetent patient with oligoastrocytoma. Proc. Natl. Acad. Sci. USA 93:7352-7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renner, K., H. Leger, and M. Wegner. 1994. The POU domain protein Tst-1 and papovaviral large tumor antigen function synergistically to stimulate glia-specific gene expression of JC virus. Proc. Natl. Acad. Sci. USA 91:6433-6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadowska, B., R. Barucco, K. Khalili, and M. Safak. 2003. Regulation of human polyomavirus JC virus gene transcription by AP-1 in glial cells. J. Virol. 77:665-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Safak, M., R. Barrucco, A. Darbinyan, Y. Okada, K. Nagashima, and K. Khalili. 2001. Interaction of JC virus agno protein with T antigen modulates transcription and replication of the viral genome in glial cells. J. Virol. 75:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Safak, M., G. L. Gallia, S. A. Ansari, and K. Khalili. 1999. Physical and functional interaction between the Y-box binding protein YB-1 and human polyomavirus JC virus large T antigen. J. Virol. 73:10146-10157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Safak, M., G. L. Gallia, and K. Khalili. 1999. A 23-bp sequence element from human neurotropic JC virus is responsive to NF-kappa B subunits. Virology 262:178-189. [DOI] [PubMed] [Google Scholar]
- 46.Safak, M., G. L. Gallia, and K. Khalili. 1999. Reciprocal interaction between two cellular proteins, Puralpha and YB-1, modulates transcriptional activity of JCVCY in glial cells. Mol. Cell. Biol. 19:2712-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Safak, M., and K. Khalili. 2001. Physical and functional interaction between viral and cellular proteins modulate JCV gene transcription. J. Neurovirol. 7:288-292. [DOI] [PubMed] [Google Scholar]
- 48.Safak, M., B. Sadowska, R. Barrucco, and K. Khalili. 2002. Functional interaction between JC virus late regulatory agnoprotein and cellular Y-box binding transcription factor, YB-1. J. Virol. 76:3828-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with “mini-extracts”, prepared from a small number of cells. Nucleic Acids Res. 17:6419.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390-2400. [DOI] [PubMed] [Google Scholar]
- 51.Shaulian, E., M. Schreiber, F. Piu, M. Beeche, E. F. Wagner, and M. Karin. 2000. The mammalian UV response: c-Jun induction is required for exit from p53-imposed growth arrest. Cell 103:897-907. [DOI] [PubMed] [Google Scholar]
- 52.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 53.Small, J. A., G. Khoury, G. Jay, P. M. Howley, and G. A. Scangos. 1986. Early regions of JC virus and BK virus induce distinct and tissue-specific tumors in transgenic mice. Proc. Natl. Acad. Sci. USA 83:8288-8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tavis, J. E., and R. J. Frisque. 1991. Altered DNA binding and replication activities of JC virus T-antigen mutants. Virology 183:239-250. [DOI] [PubMed] [Google Scholar]
- 55.Trowbridge, P. W., and R. J. Frisque.1995. Identification of three new JC virus proteins generated by alternative splicing of the early viral mRNA. J. Neurovirol. 1:195-206. [DOI] [PubMed] [Google Scholar]
- 56.van Straaten, F., R. Muller, T. Curran, C. Van Beveren, and I. M. Verma. 1983. Complete nucleotide sequence of a human c-onc gene: deduced amino acid sequence of the human c-fos protein. Proc. Natl. Acad. Sci. USA 80:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varakis, J., G. M. ZuRhein, B. L. Padgett, and D. L. Walker. 1978. Induction of peripheral neuroblastomas in Syrian hamsters after injection as neonates with JC virus, a human polyoma virus. Cancer Res. 38:1718-1722. [PubMed] [Google Scholar]
- 58.Vogt, P. 2001. Jun, the oncogene. Oncogene 20:2365-2377. [DOI] [PubMed] [Google Scholar]
- 59.Walker, D. L., B. L. Padgett, G. M. ZuRhein, A. E. Albert, and R. F. Marsh. 1973. Human papovavirus (JC): induction of brain tumors in hamsters. Science 181:674-676. [DOI] [PubMed] [Google Scholar]
- 60.Wegner, M., D. W. Drolet, and M. G. Rosenfeld. 1993. Regulation of JC virus by the POU-domain transcription factor Tst-1: implications for progressive multifocal leukoencephalopathy. Proc. Natl. Acad. Sci. USA 90:4743-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei, G., C. K. Liu, and W. J. Atwood. 2000. JC virus binds to primary human glial cells, tonsillar stromal cells, and B-lymphocytes, but not to T lymphocytes. J. Neurovirol. 6:127-136. [DOI] [PubMed] [Google Scholar]
- 62.Wisdom, R. 1992. AP-1: one switch for many signals. Exp. Cell Res. 253:180-185. [DOI] [PubMed] [Google Scholar]
- 63.Zullo, J., C. D. Stiles, and R. L. Garcea. 1987. Regulation of c-myc and c-fos mRNA levels by polyomavirus: distinct roles for the capsid protein VP1 and the viral early proteins. Proc. Natl. Acad. Sci. USA 84:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]