Abstract
The identification of viral determinants of virulence and host determinants of susceptibility to virus-induced disease is essential for understanding the pathogenesis of infection. Obtaining this information requires infecting large numbers of animals to assay amounts of virus in a variety of organs and to observe the onset and progression of disease. As an alternative approach, we have used a murine model of viral encephalitis and an in vivo imaging system that can detect light generated by luciferase to monitor over time the extent and location of virus replication in intact, living mice. Sindbis virus causes encephalomyelitis in mice, and the outcome of infection is determined both by the strain of virus used for infection and by the strain of mouse infected. The mode of entry into the nervous system is not known. Virulent and avirulent strains of Sindbis virus were engineered to express firefly luciferase, and the Xenogen IVIS system was used to monitor the location and extent of virus replication in susceptible and resistant mice. The amount of light generated directly reflected the amount of infectious virus in the brain. This system could distinguish virulent and avirulent strains of virus and susceptible and resistant strains of mice and suggested that virus entry into the nervous system could occur by retrograde axonal transport either from neurons innervating the initial site of replication or from the olfactory epithelium after viremic spread.
Understanding the pathogenesis of a viral infection requires a knowledge of the sites of viral replication and pathways of spread throughout the body. Traditionally, this knowledge has been obtained by infecting sufficient numbers of animals to be able to evaluate several animals per time point and assaying organs for infectious virus at many times after the initiation of infection. This process requires large numbers of animals, and free virus circulating in the blood can confound the interpretation of data on the amounts of virus in a given organ. Furthermore, important sites of virus replication may be missed because appropriate samples were not taken. In vivo imaging might provide an alternative method of assaying viral infection that would eliminate these concerns.
In vivo imaging was first used to detect sites of bacterial replication in intact, living animals in 1995 (3). Both green fluorescent protein (26) and firefly luciferase (3) have been used as reporters. Luciferase offers the advantages of producing an inherently low background in animals and not accumulating (25). Luciferase can therefore be used to monitor real-time activity in living animals (6). Significant technological advances continue to be made in our ability to detect small amounts of light, and luciferase imaging has been used successfully to monitor tumors (4, 17, 21), bacterial infections (3, 19), herpes simplex virus expression (12), and viral gene expression (5, 25). However, its usefulness for studies of viral pathogenesis, host susceptibility, and virus strain differences is unclear.
Alphaviruses are an important cause of encephalitis in the Americas, causing disease in horses, birds, and humans (20). Sindbis virus (SV), a member of the alphavirus family causing primarily rash and arthritis in humans (20), causes encephalitis and paralysis in mice and therefore serves as an excellent model for studying the pathogenesis of acute viral encephalitis. Both viral and host factors play a role in determining the outcome of SV infection. SV AR339 causes fatal encephalitis in young mice but is avirulent in adult mice (10). Neuroadapted SV (NSV), which was derived by serial passage in mouse brain, is lethal for adult mice (7) and replicates to titers higher than those of AR339 (8). Both strains can spread from the periphery into the central nervous system (CNS) in young mice. Thach et al. have also shown that there are genetic determinants of resistance to fatal encephalitis in certain strains of mice (22). By comparing virulent and attenuated strains of SV, as well as susceptible and resistant strains of mice, we seek to identify the viral and host factors that lead to viral encephalitis, paralysis, and death. Here we describe the imaging of SV infections in mice and show that this imaging can quantitatively substitute for traditional methods of assaying virus replication and can reveal previously unsuspected sites of virus replication and modes of virus spread.
MATERIALS AND METHODS
Construction of recombinant viruses.
Determinants of SV virulence are present in both the structural and nonstructural regions of the genome. Figure 1 shows the construction of the recombinant viruses used in this study and the virulence of each in 4-week-old albino C57BL/6-Tyrc-2J mice and BALB/cAnNCrlBR mice (hereinafter referred to as albino B6 and BALB/c mice, respectively). TRNSV was constructed from pTRSB (14), in which the StuI/XhoI fragment (nucleotides [nt] 8572 to 11750) was replaced with that of TE12 (13), yielding a virus with the NSV glycoproteins on an AR339 consensus background. NSV7 was constructed by replacing the SacI/BglII fragment (nt 8 to 2288) and the StuI/BstEII fragment (nt 8572 to 11379) of Toto1101 with cDNA generated from NSV-infected BHK cells and by replacing the ClaI/AvrII fragment (nt 2713 to 4280) with that of pTRSB. Subsequently, the two sources of NSV structural region genes (TE12 and NSV cDNA) were found to differ at E2 glycoprotein amino acid positions 79 (threonine to glycine) and 200 (phenylalanine to serine). These constructs contain both histidine at E2 amino acid 55 and guanine at nt 5, both of which are known virulence determinants (14, 24).
FIG. 1.
Schematic representations of recombinant viruses used. The arrows indicate the subgenomic promoter. ns, nonstructural; GOI, gene of interest. Percentages of mortality of 4-week-old female albino B6 or BALB/c mice inoculated intracerebrally with 103 PFU of recombinant viruses are shown. a, P was 0.0096 when values with NSV7-Luc were compared to those with TRNSV-Luc; b, P was 0.0054 when values for albino B6 mice were compared to those for BALB/c mice (Kaplan-Meier analysis).
To construct viruses expressing luciferase as a reporter gene, we used viruses engineered to contain a second subgenomic promoter. TRNSV-Luc was constructed by removing the BsiWI/SacI fragment (nt 10381 to 13658) from an SV containing a second subgenomic promoter (11) and using it to replace the BsiWI/SacI fragment of TRNSV to create a double-subgenomic-promoter TRNSV. The luciferase gene from the pGL3 Basic vector (Promega) was amplified by PCR (primer 1, 5′GATCGGTCACCATGGAAGACG3′; primer 2, 5′GATAGGTCACCACACGGCGAT3′) and then inserted into the BstEII site downstream of the second subgenomic promoter. NSV7-Luc was constructed similarly, but the BsiWI/SacI fragment containing the subgenomic promoter was placed into the NSV7 construct. The cDNA copy was then linearized with XhoI and in vitro transcribed by using an SP6 promoter, and the resulting RNA was transfected into BHK cells with Lipofectin (Invitrogen). Approximately 24 h later, supernatant fluid containing recombinant virus was collected. Luciferase-expressing viruses were less virulent than the parent viruses (Fig. 1).
In vivo experiments.
Albino B6 (Jackson Laboratories, Bar Harbor, Maine) or BALB/c (Charles River Laboratories, Wilmington, Mass.) mice were used for all of the imaging experiments and mortality studies. All mice were inoculated intracerebrally (i.c.) or subcutaneously in the right hind foot with 103 PFU of virus. Mortality was determined in 4-week-old mice infected i.c. and monitored daily for 21 days. Prior to imaging, mice were injected intraperitoneally with 150 mg of luciferin (Xenogen Corp., Alameda, Calif.) per kg of body weight and 17 μl of a 2.5% solution of Avertin (Sigma Corp.) per g as an anesthetic. In vivo images were acquired with the IVIS charge-coupled-device camera system (Xenogen Corp.) and analyzed with the LivingImage 2.11 software package (Xenogen Corp.). For the correlation study, brains were taken from animals immediately after they were subjected to imaging for virus plaque assays and luciferase assays. For the time course study, animals were inoculated i.c. with 103 PFU of NSV7-Luc or TRNSV-Luc and subjected to imaging on the days indicated in Results. Plotted values are numbers of pixels per second as measured by the LivingImage 2.11 software. The calculated number of PFU per brain was determined from the correlation curves for either TRNSV-Luc or NSV7-Luc.
In vitro experiments.
Whole brains from infected mice were homogenized in 2 ml of Dulbecco's modified Eagle’s medium (Gibco BRL, Grand Island, N.Y.). Twenty microliters of the homogenate was used in a luciferase expression assay (Promega, Madison, Wis.), and serial dilutions were made for standard plaque assays on BHK cells. Recombinant firefly luciferase (Promega) was used to generate a standard curve for converting relative light units from the luciferase assay to micrograms of luciferase.
Statistical analysis.
The significance of differences in rates of survival was determined by Kaplan-Meier analysis with StatView 5.0.1 (SAS Institute Inc., Cary, N.C.). Differences in levels of virus replication were determined by comparing calculated log numbers of PFU between groups by Student's t test.
RESULTS
Visualization and quantitation of virus.
The first step in developing the in vivo imaging system for SV was to determine whether luciferase-expressing SV could be detected in the brains and spinal cords of infected mice. Four-week-old albino B6 mice were inoculated i.c. with 103 PFU of NSV7-Luc and imaged 3 days postinfection (Fig. 2). Significant signal was detected in both the brain and the spinal cord. To determine whether there was a direct correlation between the light detected by the camera and the amount of virus replicating in tissue, 4-week-old albino B6 mice 1, 2, or 3 days postinfection were subjected to imaging and their tissues were harvested. The amounts of infectious virus in their brains were assayed in vitro by plaque assay, and results were compared to the amount of light quantitated in vivo by the camera (Fig. 3). The amount of infectious virus measured in vitro directly correlated with the amount of light emitted in vivo. Extrapolation of the curve in Fig. 3A showed that the limit of detection was approximately 10 PFU per g of brain. The actual limit of detection for infectious TRNSV-Luc was determined by subjecting 4-week-old albino B6 mice to imaging early after infection around the time that a signal was first apparent and then harvesting tissues for virus titration. For three mice with undetectable signal, the log10 numbers of PFU of infectious virus per gram of brain were 1.67, 3.42, and 5.44. However, the virus isolated from the brain of the animal with 5.44 log10 PFU/g was determined by IVIS imaging and plaque assay to have lost luciferase expression. Loss of luciferase expression was infrequent. Expression of luciferase by virus recovered from brains of mice sacrificed 1 to 3 days postinfection was confirmed by imaging, followed by traditional staining of the plaques. In all instances, the viruses remained luciferase positive (data not shown). Since during determination of the standard curve light was detected from animals with 3.5 and 4.4 log10 PFU/g (Fig. 3), the limit of detection is approximately 3.5 log10 PFU/g.
FIG. 2.
Imaging of 4-week-old albino B6 mice after i.c. inoculation of NSV7-Luc. Virus is detectable in the brain (A) and spinal cord (B). Animals were subjected to imaging for 3 (A) or 5 (B) min. The animal in panel B had its head covered to prevent camera saturation by the stronger signal from the brain.
FIG. 3.
Correlation between light detected by the camera and in vitro plaque assays. Light was correlated with infectious virus as measured by plaque assay. Each point represents data from a single 4-week-old albino B6 mouse studied 1 to 3 days after infection with TRNSV-Luc (A) or NSV7-Luc (B).
Comparison of virulent and avirulent strains of virus.
Virulence is usually associated with high levels of virus replication in the CNS (8, 18). To determine if in vivo imaging could be used to phenotype different strains of SV, virulent (TRNSV-Luc) and less virulent (NSV7-Luc) viruses were inoculated i.c. into 4-week-old BALB/c mice. Four animals were subjected to imaging for each virus on days 1, 3, and 5 postinfection, and the numbers of PFU per brain were calculated from the appropriate correlation curves plotting the amount of light to the number of PFU. The amount of virus in the brains of mice infected with the more virulent TRNSV-Luc, estimated by the amount of light emitted, was consistently higher than the amount in brains of mice infected with the less virulent NSV7-Luc (Fig. 4).
FIG. 4.
Detection of differences in levels of virus virulence associated with the ability of virus to replicate in the CNS. Four-week-old BALB/c mice were infected i.c. with 103 PFU TRNSV-Luc (closed symbols) or NSV7-Luc (open symbols) and subjected to imaging with the IVIS camera system on days 1, 3, and 5 after infection. The calculated number of PFU per brain was determined for each mouse at each time point from the appropriate correlation curve in Fig. 3.
Comparison of susceptible and resistant strains of mice.
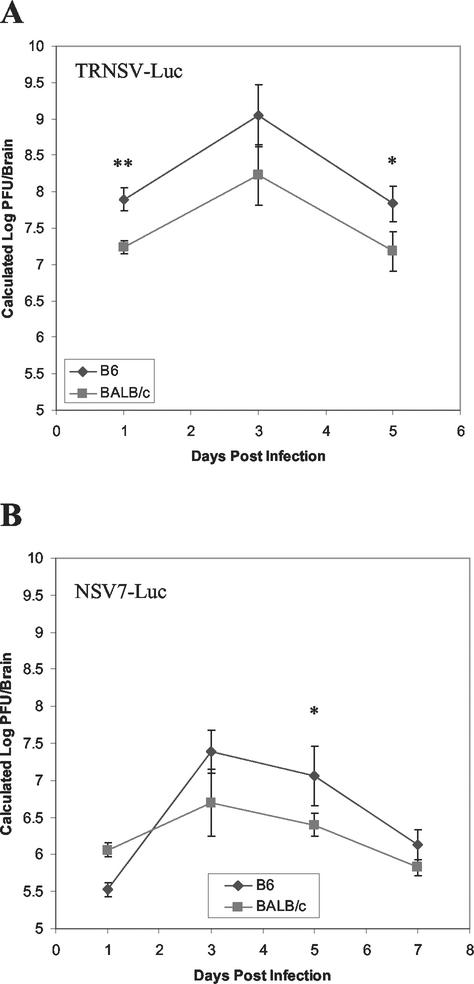
Strains of mice vary in their susceptibilities to fatal encephalitis, and this correlates with the efficiency of virus replication in the CNS (22). To determine whether this in vivo imaging system could also be used for phenotyping the susceptibilities of different mouse strains, TRNSV-Luc was inoculated into susceptible (albino B6) and resistant (BALB/c) strains of mice (Fig. 5A). The amount of light detected by the in vivo imaging of the brains of susceptible mice was consistently higher than the amount of light detected from the brains of resistant mice. To corroborate the ability of the luciferase imaging system to differentiate between susceptible and resistant mouse strains, we infected the same two strains of mice with the less virulent NSV7-Luc recombinant SV (Fig. 5B). Again, the amount of light emitted from the infected mice tended to be greater in albino B6 mice.
FIG. 5.
Detection of differences in levels of virus replication in susceptible and resistant strains of mice. Four-week-old susceptible albino B6 or resistant BALB/c mice were infected with TRNSV-Luc (A) or NSV7-Luc (B) and subjected to imaging with the IVIS system. Values plotted are averages of results for four mice, and error bars are ± standard errors. *, P < 0.10; **, P < 0.05 (Student's t test).
Sites of peripheral replication and CNS invasion.
To determine whether invasion of the CNS from peripheral sites of replication could be visualized, 10 11-day-old albino B6 mice were inoculated subcutaneously in the right rear foot with TRNSV-Luc and subjected to imaging daily for 6 days to monitor the spread of virus from the periphery to the CNS. All mice had detectable virus in the right foot at 8 h postinoculation (Fig. 6A). We detected virus in the noses of 6 of 10 mice at 1 day postinfection (Fig. 6B). Four of 10 mice had detectable virus in their lower spinal cords at day 3 (Fig. 6C). The source of this light was confirmed to be the spinal cord by postmortem dissection and imaging (data not shown). Three of six of the mice with nasal replication had detectable virus in the brain 1 to 2 days later (Fig. 6D), while only one of four of the mice with spinal cord replication had detectable virus in their brains at later time points. All animals were subjected to imaging until death, 5 or 6 days postinfection.
FIG. 6.
Tracking the spread of virus from the periphery to the CNS in 11-day-old albino B6 mice inoculated s.c. with 103 PFU of TRNSV-Luc in the right hind footpad. Representative images of viral replication at the site of inoculation 8 h postinfection (A), the nose on day 1 (B), the lower spinal cord on day 3 (C), and the brain on day 4 (D). All animals were subjected to imaging for 5 min.
DISCUSSION
Traditionally, the study of viral pathogenesis has involved infecting large numbers of animals and sacrificing several animals per time point to determine viral titers in different organs. Separate groups of animals need to be infected and observed to acquire morbidity and mortality data. Indirect correlations can then be drawn between the amount of virus in a particular tissue and manifestations of disease. However, to phenotype mouse strains and characterize recombinant strains of virus, it would be advantageous to be able to draw direct correlations between viral load and outcome of infection. These studies have shown that the IVIS camera system, combined with viruses engineered to express luciferase, can be used to quantitatively visualize viral infections in living mice. Not only does this allow the viral infection to be monitored over time in a single animal, it also allows disease progression to be directly linked to virus replication. Additionally, experiments can now be carried out with fewer animals because interanimal variation is reduced due to the availability of longitudinal data for a single animal and both viral replication in multiple organs at many times and mortality can be determined for the same animal. Data acquisition is also more rapid because assaying organs for infectious virus by traditional methods requires a minimum of 2 days to read the assay results whereas images can be quantitated within minutes.
The limit of detection determined in these experiments is approximately 103 PFU/g, which is higher than the limit of detection of virus in tissue for the standard plaque assay, approximately 102 PFU/g, but lower than that determined for luciferase-expressing adenovirus in muscle gene transfer experiments (106 PFU) (25). The ability to monitor animals longitudinally compensates for the loss in sensitivity compared to that of the plaque assay. The limit of detection will vary between different strains of luciferase-expressing viruses due to the varying strength of the promoter used to drive luciferase expression and the amount of mRNA produced (compare Fig. 3A and B). For this reason, separate correlation curves must be generated for each virus studied.
For alphaviruses, as well as other viruses found in plasma, the spread of virus from the periphery to the CNS has been difficult to study due to the confounding effects of free virus in the blood. Even if the animal is perfused, some blood will remain in the organs and precise determinations of the amount of virus in organs is not possible. Documentation of replication in a specific tissue requires identification of infected cells by immunohistochemistry or in situ hybridization. Because luciferase requires ATP for generating light from luciferin, the IVIS system measures only virus actively replicating in cells; plasma viremia is not measured, and any signal acquired is from the organ, not the blood. This method can be combined with nonlethal methods of determining plasma viremia (e.g., retro-orbital bleeding) to provide an even more complete picture of viral replication in the whole animal.
Early studies suggested that SV enters the brain by replicating in cerebral capillary endothelial cells (9), but in subsequent studies these cells have not been observed to be infected (8). Other viruses that cause encephalitis can enter the CNS by retrograde axonal transport from the olfactory epithelium (15) or peripheral sites of replication (2, 15). For SV, the question has been difficult to answer because of the confounding problem of the simultaneous presence of a high titer of virus in blood at the time of CNS entry. By imaging after peripheral inoculation, significant replication was seen in the lower spinal cords, noses, and brains of different animals. In all cases, replication in either the nose or the lower spinal cord preceded detection of virus replication in the brain. We also detected diffuse light from the abdominal region in some animals (data not shown), consistent with previously observed limited replication in a variety of peripheral sites (23). Our data support the hypothesis that virus entered the CNS by replicating first in the nasal neuroepithelium, as has previously been described for St. Louis encephalitis virus (15) and Venezuelan equine encephalitis virus, an alphavirus known to be infectious by the respiratory route (1). This route of entry requires spread through the blood to the olfactory epithelium prior to retrograde transport to the brain. Strains of SV differ in their abilities to produce a sustained viremia, and this presumably affects efficient entry by this route. In some animals SV appears to have entered the CNS by retrograde axonal transport from a peripheral site of replication by direct transport to the spinal cord as has been demonstrated for rabies virus (2) and polio virus (16). This variability among animals in mode of entry was not recognized in previous studies and may account for some of the difficulty in determining how SV entered the CNS from the periphery.
Limitations of this system include the light-absorbing properties of dark hair and skin pigment. The albino B6 mice used in this study maintain the genetic background of the commonly studied B6 mouse without the loss of signal due to dark hair and skin. The stability of luciferase gene expression is also essential. During the generation of the correlation curves, the determination of the limit of detection, and the monitoring of the course of infection longitudinally, we encountered only 1 animal out of over 40 evaluated in which the replicating virus had lost luciferase expression. Addition of the luciferase gene also attenuates SV (Fig. 1), but it attenuates all strains to similar degrees in susceptible strains of mice; therefore, useful comparisons between strains can still be made. It is also likely that the correlation curves between light emitted and infectious virus will be different early and late in the infection, since the relative levels of viral RNA and the ability to detect infectious virus are altered late in infection by the appearance of the immune response, particularly neutralizing antibody. However, at any given point in time, more light equates to more replicating virus, and the use of this technique will be applicable to in vivo study of any virus that can be engineered to express luciferase.
Acknowledgments
These studies were supported by the National Institutes of Health through research grant NS18597 (D.E.G.) and training grant AI07417 (S.H.C.).
We thank Darlene Jenkins, Bo Nelson, and William Anderson at Xenogen Corp. for generous assistance with the equipment, software, and analysis.
REFERENCES
- 1.Charles, P. C., E. Walters, F. Margolis, and R. E. Johnston. 1995. Mechanism of neuroinvasion of Venezuelan equine encephalitis virus in the mouse. Virology 208:662-671. [DOI] [PubMed] [Google Scholar]
- 2.Charlton, K. M. 1994. The pathogenesis of rabies and other lyssaviral infections: recent studies. Curr. Top. Microbiol. Immunol. 187:95-119. [DOI] [PubMed] [Google Scholar]
- 3.Contag, C. H., P. R. Contag, J. I. Mullins, S. D. Spilman, D. K. Stevenson, and D. A. Benaron. 1995. Photonic detection of bacterial pathogens in living hosts. Mol. Microbiol. 18:593-603. [DOI] [PubMed] [Google Scholar]
- 4.Contag, C. H., D. Jenkins, P. R. Contag, and R. S. Negrin. 2000. Use of reporter genes for optical measurements of neoplastic disease in vivo. Neoplasia 2:41-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contag, C. H., S. D. Spilman, P. R. Contag, M. Oshiro, B. Eames, P. Dennery, D. K. Stevenson, and D. A. Benaron. 1997. Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem. Photobiol. 66:523-531. [DOI] [PubMed] [Google Scholar]
- 6.Contag, P. R., I. N. Olomu, D. K. Stevenson, and C. H. Contag. 1998. Bioluminescent indicators in living mammals. Nat. Med. 4:245-247. [DOI] [PubMed] [Google Scholar]
- 7.Griffin, D. E., and R. T. Johnson. 1977. Role of the immune response in recovery from Sindbis virus encephalitis in mice. J. Immunol. 118:1070-1075. [PubMed] [Google Scholar]
- 8.Jackson, A. C., T. R. Moench, B. D. Trapp, and D. E. Griffin. 1988. Basis of neurovirulence in Sindbis virus encephalomyelitis of mice. Lab. Investig. 58:503-509. [PubMed] [Google Scholar]
- 9.Johnson, R. T. 1965. Virus invasion of the central nervous system. Am. J. Pathol. 46:929-943. [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, R. T., H. F. McFarland, and S. E. Levy. 1972. Age-dependent resistance to viral encephalitis: studies of infections due to Sindbis virus in mice. J. Infect. Dis 125: 257-262. [DOI] [PubMed] [Google Scholar]
- 11.Levine, B., J. E. Goldman, H. H. Jiang, D. E. Griffin, and J. M. Hardwick. 1996. Bc1-2 protects mice against fatal alphavirus encephalitis. Proc. Natl. Acad. Sci. USA 93:4810-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luker, G. D., J. P. Bardill, J. L. Prior, C. M. Pica, D. Piwnica-Worms, and D. A. Leib. 2002. Noninvasive bioluminescence imaging of herpes simplex virus type 1 infection and therapy in living mice. J. Virol. 76:12149-12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lustig, S., A. C. Jackson, C. S. Hahn, D. E. Griffin, E. G. Strauss, and J. H. Strauss. 1988. Molecular basis of Sindbis virus neurovirulence in mice. J. Virol. 62:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKnight, K. L., D. A. Simpson, S. C. Lin, T. A. Knott, J. M. Polo, D. F. Pence, D. B. Johannsen, H. W. Heidner, N. L. Davis, and R. E. Johnston. 1996. Deduced consensus sequence of Sindbis virus strain AR339: mutations contained in laboratory strains which affect cell culture and in vivo phenotypes. J. Virol. 70:1981-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monath, T. P., C. B. Cropp, and A. K. Harrison. 1983. Mode of entry of a neurotropic arbovirus into the central nervous system. Reinvestigation of an old controversy. Lab. Investig. 48:399-410. [PubMed] [Google Scholar]
- 16.Ohka, S., W. X. Yang, E. Terada, K. Iwasaki, and A. Nomoto. 1998. Retrograde transport of intact poliovirus through the axon via the fast transport system. Virology 250:67-75. [DOI] [PubMed] [Google Scholar]
- 17.Rehemtulla, A., L. D. Stegman, S. J. Cardozo, S. Gupta, D. E. Hall, C. H. Contag, and B. D. Ross. 2000. Rapid and quantitative assessment of cancer treatment response using in vivo bioluminescence imaging. Neoplasia 2:491-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherman, L. A., and D. E. Griffin. 1990. Pathogenesis of encephalitis induced in newborn mice by virulent and avirulent strains of Sindbis virus. J. Virol. 64:2041-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siragusa, G. R., K. Nawotka, S. D. Spilman, P. R. Contag, and C. H. Contag. 1999. Real-time monitoring of Escherichia coli O157:H7 adherence to beef carcass surface tissues with a bioluminescent reporter. Appl. Environ. Microbiol. 65:1738-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. (Erratum, 58:806.) [DOI] [PMC free article] [PubMed]
- 21.Sweeney, T. J., V. Mailander, A. A. Tucker, A. B. Olomu, W. Zhang, Y. Cao, R. S. Negrin, and C. H. Contag. 1999. Visualizing the kinetics of tumor-cell clearance in living animals. Proc. Natl. Acad. Sci. USA 96:12044-12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thach, D. C., T. Kimura, and D. E. Griffin. 2000. Differences between C57BL/6 and BALB/cBy mice in mortality and virus replication after intranasal infection with neuroadapted Sindbis virus. J. Virol. 74:6156-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trgovcich, J., J. F. Aronson, and R. E. Johnston. 1996. Fatal Sindbis virus infection of neonatal mice in the absence of encephalitis. Virology 224:73-83. [DOI] [PubMed] [Google Scholar]
- 24.Tucker, P. C., and D. E. Griffin. 1991. Mechanism of altered Sindbis virus neurovirulence associated with a single-amino-acid change in the E2 glycoprotein. J. Virol. 65:1551-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu, J. C., G. Sundaresan, M. Iyer, and S. S. Gambhir. 2001. Noninvasive optical imaging of firefly luciferase reporter gene expression in skeletal muscles of living mice. Mol. Ther. 4:297-306. [DOI] [PubMed] [Google Scholar]
- 26.Yang, M., E. Baranov, A. R. Moossa, S. Penman, and R. M. Hoffman. 2000. Visualizing gene expression by whole-body fluorescence imaging. Proc. Natl. Acad. Sci. USA 97:12278-12282. [DOI] [PMC free article] [PubMed] [Google Scholar]