Abstract
The design of drugs for treatment of virus infections and the exploitation of viruses as drugs for treatment of diseases could be made more successful by understanding the molecular mechanisms of virus-specific events. The process of assembly, and more specifically packaging of the genome into a capsid, is an obligatory step leading to future infections. To enhance our understanding of the molecular mechanism of packaging, it is necessary to characterize the viral components necessary for the event. In the case of adenovirus, sequences between nucleotides 200 and 400 at the left end of the genome are essential for packaging. This region contains a series of redundant bipartite sequences, termed A repeats, that function in packaging. Synthetic packaging sequences made of multimers of a single A repeat substitute for the authentic adenovirus packaging domain. A repeats are binding sites for the CCAAT displacement protein and the viral protein IVa2. Several lines of evidence implicate these proteins in the packaging process. It was not known, however, whether other cis-acting elements play a role in the packaging process as well. We utilized an in vivo approach to address the role of the inverted terminal repeats and the covalently linked terminal proteins in packaging of the adenovirus genome. Our results show that these elements are not necessary for efficient packaging of the viral genome. A significant implication of these results applicable to gene therapy vector design is that the linkage of the adenovirus packaging domain to heterologous DNA sequences should suffice for targeting to the viral capsid.
A critical step in the life cycle of any virus is the successful packaging of the nucleic acid in a capsid structure. Although a good deal is known about the molecular events of packaging for double-stranded DNA bacteriophage (e.g., φ29, T4, and P22), much less is understood for the eukaryotic virus, adenovirus (Ad). Significantly, the various double-stranded DNA bacteriophage share a similar packaging process that provides a paradigm for the packaging of double-stranded DNA viruses in general. Consistent with this idea, some of the packaging molecules of bacteriophage appear to have functional counterparts among the packaging proteins of Herpesviridae, including the portal protein, terminase, and a putative ATPase that may be part of a packaging motor (6, 13, 22, 24, 25, 28, 34, 40). Equivalent viral counterparts have not been identified for Ad.
Ad, like other bacteriophage and viruses, appears to package DNA in a polar fashion from left to right, presumably into a preformed protein capsid (reviewed in reference 39). The left-end DNA between nucleotides (nt) 200 and 400 contains a series of redundant bipartite sequences, termed A repeats (consensus: 5′-TTTGN8CGNG-3′), that are required for packaging of the genome into a capsid (9, 10, 19, 38). Synthetic packaging sequences made of multimers of a single A repeat substitute for the authentic Ad packaging domain, although with different efficiencies depending upon which A repeat is used (38). In addition, several proteins have been identified that bind to the A repeats, including a cellular activity referred to as P complex (37), which is the transcriptional regulator CCAAT displacement protein (CDP) (7), and the viral protein IVa2 (42). Several lines of evidence implicate these proteins in the packaging process. Different A repeats bind CDP with different apparent affinities in vitro, which correlates directly with how efficiently these A repeats function for packaging in vivo (7, 37). Mutations within the TTTG motif of the A-repeat consensus sequence result in loss of CDP binding in vitro, and viruses carrying similar mutations are either severely defective or are nonviable for packaging in vivo (10). Mutations that disrupt the CGNG motif of the A-repeat consensus sequence do not disrupt CDP binding (37). However, such mutations disrupt the binding of the viral protein IVa2 (42), which is involved in the packaging process (43). One possibility is that CDP and IVa2 function together in packaging. IVa2 also interacts with the viral protein L1 (15). A temperature-sensitive mutant in L1 has a significant defect in packaging (17), and virus lacking L1 coding sequences is completely defective for packaging (14). Recently, it has been shown that IVa2 may function in a subtype-specific fashion in packaging (43).
The fact that gutted Ad vectors for gene therapy are packaged when viral proteins are provided in trans by a helper virus shows that the packaging sequences, the inverted terminal repeats (ITRs), and the 5′ covalently linked terminal proteins are sufficient cis-acting elements for packaging. By necessity, gutted vectors include the ITRs and covalently linked terminal proteins for replication and the packaging sequences as the only authentic Ad sequences. The question remains as to whether the ITRs and terminal proteins are necessary for packaging. Circumstantial observations suggest a possible role for the ITRs and terminal protein. Among the more provocative observations is that CDP also binds to sequences within the ITR (37), and the ability of the packaging domain to function requires its location near the ends of the Ad genome (19). Bacteriophage φ29 has a genome structure similar to that of Ad, and its terminal protein plays a role in packaging (2, 12).
At present, there is no in vitro packaging system for Ad for facilitating the study of the packaging process. Therefore, we used an approach that would allow us to remove ITRs along with their covalently linked terminal proteins in vivo. Nicolas et al. (29) had shown that the yeast HO endonuclease cleaved at an HO site introduced into the Ad genome in vivo. Taking advantage of this system, we introduced HO endonuclease cleavage sites at key locations, resulting in genomes lacking one or both ITR(s) and associated terminal protein(s). Our results show that the packaging of truncated genomes lacking one or both ITR(s) occurs and that the packaging is an efficient process.
MATERIALS AND METHODS
Viruses and cells.
293 cells (11) were maintained as a monolayer in Dulbecco's modified Eagle's medium containing 10% bovine calf serum (HyClone) and the antibiotics penicillin and streptomycin. N52.E6 cells (35) were maintained as a monolayer in alpha modification of Eagle's medium supplemented with 10% fetal calf serum (HyClone), 2 mM glutamine, penicillin, and streptomycin. N52.E6 cells are from a helper cell line for E1a-defective viruses. Virus Ad(E1 HO site) and Ad(HO gene) were described previously (29) (the latter virus was referred to as E1 HO gene virus). Ad(ITR:HO:Ψ) was made by recombination (41) with the ClaI right-end fragment from dl309 (21) and the left end from plasmid pEBL106 (18) that had the HO endonuclease cleavage site (sequence of insert, with the HO cut indicated by the asterisk, is 5′-AGACTCGGTACCTCGAGATGGGGATCTAAATAAATTCGTTTTCAATGATTAAAATAGCATAGTCGGGTTTTTCTTTTAGTTTCAGCTTTCCGC*AACAGTATAATTTTATAAACCCTGGTTTTGGTTTTGTAGAGTGGTTCGAGATCT-3′) cloned into the unique BglII site at Ad nt 106. Ad(Double HO) has two HO sites in an inverted orientation relative to one another. Ad(Double HO) was constructed by recombination between the ClaI right-end fragment of AdE3:HO site virus (29) and a pEBL106 derivative that has the HO endonuclease cleavage site cloned into the BglII site.
Infections.
Stocks of virus were prepared by lysis of infected cells by three freeze/thaw cycles and either clarified lysates were obtained by centrifugation or the particles were further purified on cesium chloride density gradients, as described previously (36). Lysates were titered by plaque assay on 293 cells and were expressed as PFU per milliliter, or purified particles were quantified by optical density at 260 nm and were expressed as particles per milliliter, as described previously (36). Single infections and coinfections in 293 cells were performed by using 150 particles/cell in single infections and 150 particles/cell of each virus in coinfections. The optimal amount of cutting by the HO endonuclease was determined empirically in coinfections in N52.E6 cells by using different amounts of the Ad(Double HO) virus and Ad(HO gene) virus relative to each other. Single infections and coinfections in N52.E6 cells were done by using 0.5 PFU of Ad(Double HO) virus/cell and 600 particles (∼30 PFU/cell) of Ad(HO gene) virus/cell. Cells were harvested 24 h postinfection. 293 cells were used in the infections, unless indicated otherwise in the figure legends.
Packaging assays.
High-molecular-weight DNA was prepared from a fraction of the infected cells (∼2 × 106 infected cells), and viral particles were purified from the remaining fraction (∼5 × 108 to 1 × 109 infected cells). Isolation of high-molecular-weight DNA was performed by first preparing nuclei followed by protease digestion and phenol-chloroform extraction, as described previously (9). Viral particles were isolated as previously described for stock preparations and were purified on density gradients, and DNA was prepared by protease digestion and phenol-chloroform extraction as described previously (30). A fluorescent-based Southern blot filter hybridization assay was used to quantify the relative amounts of HO-cut and -uncut DNA in the coinfection experiments, as described previously (30). Restriction endonucleases used for digestion of DNAs are indicated in the figure legends. For analysis of cuts at the left end of the Ad genome, a probe of 649 bp was used that included Ad nt 180 to 828. For the analysis of the left-end ITR, a probe from Ad nt 1 to 194 was used. This probe had an additional 514 bp derived from plasmid sequences in order to increase the specific activity of the probe. For the analysis of HO cutting at the right end of the Ad genome, a 503-bp probe that included Ad nt 28667 to 29169 was used.
Calculations for Tables 1 to 3.
TABLE 1.
Ad genomes lacking the left ITR are packaged efficientlya
| Expt | Nuclear DNA
|
Viral DNA
|
Relative ratio
|
Relative packaging efficiency | |||
|---|---|---|---|---|---|---|---|
| % Cut | % Intact | % Cut | % Intact | Cut | Intact | ||
| 1 | 21 | 79 | 24 | 76 | 1.1 | 1.0 | 1.1 |
| 2 | 28 | 72 | 24 | 76 | 0.9 | 1.1 | 0.8 |
Shown are the results of determining relative quantities of cut and intact genomes from two experiments (Experiments 1 and 2). Ad(ITR:HO:Ψ) and Ad(HO gene) were coinfected, and nuclear and viral DNA was analyzed by Southern blot hybridization as shown in Fig. 2. Nuclear DNA is the relative level of replicated genomes that was intact (% intact) or lacking the left ITR (% cut). Viral DNA is the relative level of cut and intact genomes packaged into viral particles. The relative ratio equals the percent packaged DNA divided by the percent nuclear DNA. Calculations used to define the relative packaging efficiency are described in Materials and Methods. A value of 1 indicates that the cut and intact genomes were packaged with the same efficiency.
TABLE 3.
Ad genomes lacking the left and right ITRs are packageda
| Expt | Peak fraction
|
Nuclear DNA
|
Packaged genomes
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Right end
|
Left end
|
Right end
|
Left end
|
% Left end cut with right end intact | % Left end cut with right end cut | |||||
| % Cut | % Intact | % Cut | % Intact | % Cut | % Intact | % Cut | % Intact | |||
| 1 | 93 | 7 | 32 | 68 | 17 | 83 | 24 | 76 | 1.7 | 30 |
| 2 | 91 | 9 | 30 | 70 | 24 | 76 | 28 | 72 | 2.5 | 28 |
| 3 | 87 | 13 | 26 | 74 | 19 | 81 | 17 | 83 | 2.2 | 24 |
Columns labeled Peak fraction or Nuclear DNA show the percent of genomes that were cut at either the left-end or the right-end HO site or that were not cleaved by HO endonuclease (% Intact). Peak fraction refers to the most enriched fractions from the CsCl gradient of viral DNA with genomes cut at the E3 HO site, as shown in Fig. 6. Experiments 1 and 2 represent results obtained by using virus particles purified through two rounds of CsCl equilibribrium centrifugation; experiment 3 was performed by using virus particles from one round of gradient purification. The calculations for determination of % packaged genomes are described in Materials and Methods.
Quantification of signals from probes hybridized in Southern analyses was done by detection of the intensity of the signal by scanning filters on the Storm 860 PhosphorImager (Molecular Dynamics) and quantifying the results by using Imagequant software version 1.2 (Molecular Dynamics), as described previously (30). For results shown in Tables 1 to 3, percent cut and percent intact were determined by dividing the volume of the signal from either the cut or intact hybridized band by the volume of the total signal (cut plus intact), multiplied by 100. This gives the relative percentage of the cut and intact genomes. The relative packaging efficiency takes into account the relative amount packaged to the relative amount of the nuclear pool of DNA. This relative ratio is calculated from the percentage found in the viral particle and the percentage replicated (nuclear DNA). These relative ratios are shown in Tables 1 and 2 and are calculated by dividing the percent cut or the percent intact in viral particles by the percent cut or the percent intact in the nuclear DNA, respectively. By dividing the ratio of cut DNA by the ratio of intact DNA, the relative packaging efficiency was determined. A number close or equal to 1 means that the HO-cut Ad genome packaged with an efficiency similar to that of the intact Ad genome. For Table 3, the percent left-end cut with right-end intact was calculated by multiplying the percent right-end intact in the peak viral fraction by the percent left-end cut in the nuclear fraction. This was done because the percent right-end intact in the peak fraction included both Ad genomes with cut left ends and intact left ends. We wanted to determine the relative amount of cut left ends coming from molecules with the right ends intact. The percent left-end cut in the nuclear fraction is equal to the percent left-end cut in the packaged fraction, since we showed that left-end-cut Ad genomes package at a relative ratio of 1 (Table 1). Percent left-end cut with right-end cut is calculated by subtracting the percent left-end cut with right-end intact from the percent left-end cut from the peak fraction.
TABLE 2.
Ad genomes lacking the right ITR are packaged efficientlya
| Expt | Nuclear DNA
|
Viral DNA
|
Relative ratio
|
Relative packaging efficiency | |||
|---|---|---|---|---|---|---|---|
| % Cut | % Intact | % Cut | % Intact | Cut | Intact | ||
| 1 | 25 | 75 | 19 | 81 | 0.8 | 1.1 | 0.7 |
| 2 | 11 | 89 | 13 | 87 | 1.2 | 1.0 | 1.2 |
Shown are the results of determining relative quantities of cut and intact genomes from two experiments (Experiments 1 and 2). Ad(double HO) and Ad(HO gene) were coinfected, and nuclear and viral DNA was analyzed by Southern blot hybridization as shown in Fig. 4. Calculations of the relative levels of nuclear, viral, and packaged DNAs were made as described for Table 1.
RESULTS
Because of the lack of an in vitro system to study Ad packaging, it was necessary to devise a system that allowed us to examine the potential role of the ITRs and the covalently linked terminal proteins for packaging in vivo. Previously, Nicolas et al. (29) had engineered an Ad containing a 181-bp DNA sequence that included a cleavage site for the Saccharomyces cerevisiae HO endonuclease. HO cleavage sites were placed within either the E1 or the E3 regions of the Ad genome. Cleavage at these sites was observed upon coinfection of either HO site virus with an Ad that was engineered to express the S. cerevisiae HO endonuclease gene. Taking advantage of this system, we introduced an HO cleavage site at the left end of the Ad genome between the ITR and packaging sequences. Due to the close proximity of the left ITR to the packaging domain, we anticipated that if either of the ITRs had any role in packaging, it would likely be the left ITR. A schematic diagram of the structure of this viral genome is shown in Fig. 1 and is referred to as Ad(ITR:HO:Ψ). Cleavage at this site by HO endonuclease should result in the production of two fragments. The resulting large fragment would be composed of approximately 99% of the Ad genome, including the packaging sequences and all sequences to the right, which includes the right-end ITR. The smaller fragment that includes the left ITR and the covalently linked terminal protein would be less than 1% of the Ad genome. The results of Southern blot analysis of DNAs isolated following a coinfection of Ad(ITR:HO:Ψ) and the virus that expresses the HO endonuclease, Ad(HO gene) (Fig. 1), are shown in Fig. 2. DNA was isolated from nuclei of infected cells and formed the pool of replicated viral DNA in the cell. Distinct diagnostic left-end fragments were generated by digestion of either Ad(ITR:HO:Ψ) or Ad(HO gene) with XbaI followed by hybridization with a probe that recognized Ad nt 180 to 828 (Fig. 2, nuclear DNA, ITR:HO:Ψ, and HO gene). DNAs from the coinfection of the two viruses yielded not only the diagnostic fragments from each virus but also an additional fragment unique to the coinfection (Fig. 2, Nuclear DNA, Coinfection; the unique fragment is indicated by an asterisk). This XbaI fragment is of the predicted size produced upon cleavage at the HO endonuclease site at the left end of the Ad(ITR:HO:Ψ) genome. Determination of the quantities of cut and intact genomes from the results of four independent experiments showed that 17 to 30% of the Ad genomes were cleaved by HO endonuclease in vivo. The amount of cutting was similar to what had been observed previously (29).
FIG. 1.
Schematic diagram of the structures of the Ad used in this study. Strands of DNA are indicated by double black lines. Grey boxes indicate the ITRs, ovals indicate the covalently linked terminal protein, and the white boxes indicate the packaging sequences (ψ). The curved lines indicate that the diagram is not drawn to scale. Ad(ITR:HO:ψ) has a 135-bp fragment containing a cleavage site for S. cerevisiae HO endonuclease inserted at nt 106. Ad(Double HO) contains the HO site at nt 106 as well as a second HO site in the E3 region inserted at nt 29602. The HO gene virus has a cassette containing the cytomegalovirus enhancer/promoter sequences, a synthetic splice site, and the S. cerevisiae HO gene sequences inserted in place of Ad5 nt 355 to 2965.
FIG. 2.
Ad genomes lacking the left ITR package efficiently. (A) 293 cells were infected with the Ad(ITR:HO:Ψ) virus or the Ad(HO gene) virus or were coinfected with both viruses. Forty-eight hours postinfection a fraction of the cells was used to isolate nuclear DNA. Viral particles were isolated from the remaining fraction, and viral DNA was prepared. Shown are the results of a Southern blot analysis of the DNAs digested with XbaI and detected by using a probe corresponding to Ad nt 180 to 828. The lane labeled Marker indicates the molecular size standards. XbaI digestion of the Ad(ITR:HO:Ψ) genome gave rise to a DNA fragment of 1,480 bp. Upon cleavage with HO endonuclease, this fragment was reduced to two fragments of 1,320 (indicated by an asterisk) and 190 bp. XbaI digestion of the Ad(HO gene) genome gave rise to a DNA fragment of ∼1,000 bp. (B) The structures of the Ad(ITR:HO:Ψ) and Ad(HO gene) viral genomes. The HO cleavage site, relevant XbaI sites, and the probe positions are indicated.
In order to examine the structure of the packaged genomes, DNA was purified from viral particles from single infections with either Ad(ITR:HO:Ψ) or Ad(HO gene) and from coinfections of the two viruses and were analyzed in the same fashion as that used for the replicated DNA (Fig. 2, Viral DNA). Single bands were observed in the CsCl density gradients in the purification of viral particles from coinfections of Ad(ITR:HO:Ψ) and Ad(HO gene) (data not shown). Loss of the left end due to cutting at the left-end HO site had an undetectable influence on the buoyant density of the particles, consistent with the fact that less than 1% of the genome was removed upon cutting at the left-end HO site. As was seen with the replicated DNA, single infections with either Ad(ITR:HO:Ψ) or Ad(HO gene) yielded the single diagnostic fragment for each virus. Analysis of the DNA from the coinfection showed the diagnostic fragments for each of the viruses and, in addition, the fragment that was unique to the coinfection. The relative quantities of cleaved and uncleaved genomes in the nuclear DNAs and in the viral particle DNAs are shown in Table 1 along with the relative packaging efficiencies of each. These results show that the genomes of Ad(ITR:HO:Ψ) which were cleaved by HO endonuclease were competent substrates for packaging. Importantly, the efficiency of packaging a genome lacking the left ITR and its covalently linked terminal protein was the same as the efficiency of packaging the intact genome.
As a control to show that packaging of the right-end fragment lacking the left-end ITR was dependent upon the packaging sequences and to test if HO cleavage resulted in efficient release of the cleaved DNA fragment from the remainder of the viral genome, we examined the viral genomes packaged in a coinfection of Ad(E1 HO site) and Ad(HO gene) (schematics of the viral genomes are shown in Fig. 3B). Ad(E1 HO site) has an HO cleavage site located at the left end of the Ad genome situated on the right side of the packaging domain. Cleavage of Ad(E1 HO site) by HO endonuclease yields a left-end DNA fragment of 450 bp containing the left ITR and packaging domain and a larger right-end fragment that includes the remaining 99% of the genome. Analysis of the nuclear DNA isolated from cells from the coinfection showed that cleavage of the HO site occurred (Fig. 3A, Nuclear DNA), consistent with previous results (29). Analysis of the genomes packaged in the coinfection showed that the right-end fragment that lacks the left-end ITR and adjacent packaging domain was not packaged (Fig. 3A, Viral DNA). This result is consistent with the requirement for the cis-acting packaging signal to be linked to the viral genome to direct DNA packaging. Furthermore, this result demonstrates that the presence of the HO enzyme does not play a role in regulating the packaging of Ad DNA within our experimental system, for example, by bridging a cleaved left-end fragment to the remainder of the viral genome.
FIG. 3.
Elimination of the packaging domain in addition to the left ITR results in the loss of packaging of the right-end fragment. (A) 293 cells were infected with either Ad(HO gene) or Ad(E1 HO site) or were coinfected with both viruses. Forty-eight hours postinfection nuclear DNA (labeled Nuclear DNA) was isolated from cells from the three different infections. Viral particles were isolated from cells coinfected with both viruses, and viral DNA was purified (labeled Viral DNA). Shown are the results of a Southern blot of DNAs digested with SmaI and detected by using a probe corresponding to Ad nt 2982 to 3655. SmaI digestion of the Ad(E1 HO site) virus DNA yields a left-end fragment of 1,850 nt. Upon cleavage with HO endonuclease, this fragment was reduced to two fragments of 1,400 and 450 bp. The uncut fragment of 1,850 bp and the 1,400-bp HO-cut fragment (indicated by an asterisk) hybridize to the probe. The 450-bp left end was not recognized by the probe used. (B) The structures of the genomes of the Ad(HO gene) and Ad(E1 HO site) viral genomes. The HO cleavage site, relevant SmaI sites, and the probe position are indicated. Both viruses are derived from an E1 deletion of nt 356 to 2965 and carry different-size inserts, hence the relative difference in the sizes of the left ends.
The left-end ITR fragment of Ad(ITR:HO:Ψ) genomes (nt 1 to 106) cleaved by HO endonuclease should not package due to the lack of packaging sequences. However, it was formally possible that the left-end ITR fragment could be packaged along with the right-end fragment by virtue of protein-protein interactions, e.g., CDP binding to the ITR and the packaging sequences or by interactions between the terminal protein and other viral proteins. We analyzed the DNAs found in viral particles to determine if the small left-end fragment was packaged. A plasmid standard was included in the Southern blot analysis with a titration of a plasmid that contains the left end of Ad with an HO site (Fig. 4, molecules of standard). The probe hybridized to Ad nt 1 to 195. Analysis of the nuclear DNA isolated from cells infected with either Ad(ITR:HO:Ψ) or Ad(HO gene) or coinfected with both viruses showed a band unique to the coinfections (Fig. 4, Nuclear DNA, Coinfection) that was not seen in the single infections [Ad(HO gene) and Ad(ITR:HO:Ψ)]. This fragment of approximately 200 nt is of the predicted size to correspond to the left-end ITR fragment released by HO endonuclease cleavage of Ad(ITR:HO:Ψ) genomes. However, no corresponding 200-nt fragment was observed in the DNA isolated from viral particles from a coinfection (Fig. 4, Viral DNA, Coinfection). The number of molecules of cleaved Ad DNA loaded on the gel for the nuclear sample and the particle sample corresponded to 9 × 109 and 1 × 1010, respectively. These amounts were within the detectable range of the assay as seen in the titration of the plasmid standard (Fig. 4, molecules of standard). Therefore, the left-end ITR and its covalently linked terminal protein are not needed for efficient packaging of Ad DNA and are not packaged into virus particles when separated from the remainder of the viral genome.
FIG. 4.
The left-end 160-bp fragment from the HO-cleaved Ad(ITR:HO:Ψ) genome is not packaged. 293 cells were infected with Ad(HO gene) or Ad(ITR:HO:Ψ) or were coinfected with both viruses. Shown are the results of a Southern blot analysis of nuclear DNAs and viral particle DNAs prepared after virus infection. Also shown is a titration of a plasmid standard. The plasmid used for the standard was pEB-HO (see Materials and Methods), a plasmid that contains Ad nt 1 to 1340 with the HO site inserted at nt 106. Digestion of pEB-HO with EcoRI and BglII yielded three fragments due to partial cleavage at the BglII site. A fragment of 249 bp corresponds to Ad sequences 1 to 106 plus the 135-bp HO site insertion; the fragment of 108 bp includes Ad nt 1 to 106. The 700-bp probe used was derived from pEB-HO and includes Ad nt 1 to 194 and 506 nt of adjacent plasmid sequences. The largest fragment corresponds to the remaining plasmid and Ad sequences. The number of molecules of plasmid loaded in each lane of the standard is indicated. Molecules (∼9 × 109) of HO-cleaved DNA were loaded in the nuclear DNA (labeled Coinfection). Molecules (∼1 × 1010) of HO-cleaved DNA were loaded in the viral particle lane (labeled Coinfection). Shown below is a longer exposure of the lower region of the Southern blot encompassing the smaller fragments of 249 and 108 bp. The asterisk indicates the 160-nt fragment unique to the coinfection, i.e., the HO endonuclease-cleaved DNA fragment.
It was feasible that a single ITR, independent of its relative location to the packaging sequences, was both necessary and adequate for packaging. In the experiments described thus far, the right-end ITR and its covalently linked terminal protein were still present in the Ad(ITR:HO:Ψ) genomes cleaved at the left end by HO endonuclease. A viral genome containing two HO sites was constructed in order to remove both ITRs in a coinfection. A schematic diagram of this viral genome, Ad(Double HO), is shown in Fig. 1. Ad(Double HO) was constructed by recombination between a left-end fragment from Ad(ITR:HO:Ψ) and a right-end fragment from the AdE3:HO site (29). It had the left-end HO cleavage site between the left ITR and the packaging sequences as well as a second HO cleavage site in the E3 region. Cleavage at the E3 HO site or at both the left-end site and the E3 site resulted in genomic fragments containing the packaging sequences that are approximately 82 and 81.5% of the full length, respectively. These truncated genomes are of a sufficient size so as not to be perturbed for stable packaging by size alone (31). Results of Southern blot analysis of the nuclear DNAs from single infections and coinfection of Ad(Double HO) and Ad(HO gene) showed that both the left-end and right-end HO sites were cleaved by HO endonuclease (Fig. 5A). Because cleavage was not 100% efficient, multiple genomic structures were generated. A schematic diagram of the four possible structures generated by HO cleavage of the Ad(Double HO) genome that could be found in the pool of nuclear DNA from the coinfection is shown in Fig. 6B. Examination of the structures of the genomes packaged into viral particles from a coinfection of Ad(Double HO) and Ad(HO gene) showed that genomes cleaved at either the left end or the right end were packaged (Fig. 6A). Packaging of the cleaved fragment containing the right 18% of the genome, including the right-end ITR and covalently linked terminal protein, was not observed as expected, since this fragment lacks the packaging domain (data not shown). This is consistent with results reported by Nicolas et al. that were seen for the virus with a single HO site in E3 (29). The efficiency of packaging genomes cleaved at the right end (including genomes with both cut and intact left ends) was similar to the efficiency of packaging genomes with intact right ends (Table 2). Taken together, the results showed that both the left-end HO site and the E3 HO site are accessible to HO endonuclease in genomes that contain both sites. In addition, genomes lacking right-end and left-end ITRs are packaged. Furthermore, as was seen for the genomes lacking the left ITR (Fig. 2, Table 1), the packaging of genomes lacking the right-end ITR was as efficient as packaging of genomes with intact right ends.
FIG. 5.
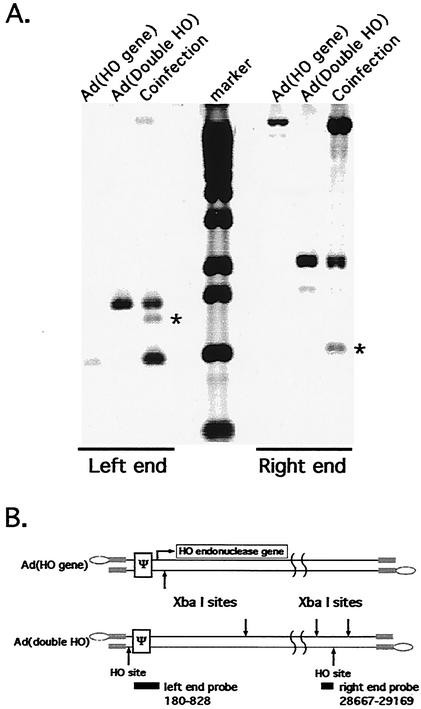
Left- and right-end HO sites in the Ad(Double HO) genome are cleaved by HO endonuclease. (A) Nuclear DNAs were isolated 24 h postinfection from infections of N52.E6 cells with either Ad(HO gene), Ad(Double HO), or a coinfection of both viruses. Results of Southern blot analysis of XbaI-digested DNAs hybridized with either a left-end probe (Ad nt 180 to 828) or right-end probe (Ad nt 28667 to 29169) are shown. XbaI digestion of the Ad(Double HO) genome gave rise to a DNA fragment of 2,067 bp. Upon cleavage by HO endonuclease, two DNA fragments of 1,016 and 1,051 bp were generated. The asterisks indicate the XbaI fragments that are unique to the coinfection and recognized by the probe. (B) Illustrated are the structures of the Ad(Double HO) and Ad(HO gene) virus genomes. The HO-cut sites, relevant XbaI sites, and the probe's positions are indicated.
FIG. 6.
Both right-end-cut and left-end-cut genomes are found in viral particles from a coinfection with Ad(Double HO) and Ad(HO gene). (A) The results of Southern blot analysis of DNA isolated from infected cell nuclei (labeled Nuclear DNA) and from CsCl gradient purified viral particles (labeled Viral DNA) are shown. DNAs were digested with XbaI and were hybridized with either a probe of Ad nt 180 to 828 (labeled Left end) or Ad nt 28667 to 29169 (labeled Right end). The bands in the hybridization that are unique to the coinfection are indicated by asterisks. (B) Schematic diagrams for the four different genomic structures of Ad(Double HO) that could be generated by cleavage with HO endonuclease at either or both of the HO sites.
The question remained what fraction, if any, of the Ad genomes that were cut at both the right and left ends was packaged. These genomes would lack both ITRs and their covalently linked terminal proteins. To answer this question, viral particles containing genomes cleaved at the E3 HO site were purified away from particles with intact right ends, and then the structures of the left ends were determined. It was feasible to separate particles with genomes cut at the E3 HO site from particles with intact genomes based on their different buoyant densities in a cesium chloride gradient, since genomes cleaved at the E3 HO site are ∼82% the length of the intact genome. Purification of viral particles from a coinfection of Ad(Double HO) and Ad(HO gene) resulted in the appearance of two bands on a cesium chloride equilibrium gradient (data not shown). The denser band corresponded to particles of Ad(Double HO), Ad(HO gene), and particles with genomes cleaved at the left-end HO site. The second, less dense band floated at a density of approximately 1.32 gm/ml, which was lighter than the wild-type particle density of 1.34 gm/ml.
Fractions of viral particles produced in the coinfection collected either from a single cesium chloride equilibrium gradient or the pool of lighter density particles were run on a second gradient to further enrich for the lighter-density particles. Density measurements confirmed that the fractions contained particles that floated at a density lighter than that of wild-type virus particles. DNA prepared from each fraction was analyzed by Southern blot hybridization. The results with particles purified by a second round of CsCl equilibrium centrifugation are shown in Fig. 7. Lane N shows the signals obtained from the analysis of the nuclear fraction from the coinfection of Ad(Double HO) and Ad(HO gene) examining cleavage at the E3 HO site. Note that the intensity of the signal from the intact Ad(Double HO) genome is significantly stronger than that from the genome cut at the E3 HO site. Lanes 1 to 12 show the relative signals from the intact and E3-cut genomes from particles for each fraction taken from the top of the gradient. Fractions 5 to 8 contain particles that are substantially enriched for genomes lacking the right ITR due to cutting at the E3 HO site. The relative intensities of the signals for intact and E3-cut genomes in fractions 5 to 8 are inverted compared to those of the nuclear fraction. The relative quantities of the intact and E3-cut genomes in the peak fraction from three experiments are shown in Table 3. Enrichment of the particles with E3-cut genomes to levels between 87 and 93% was achieved. The percentage of genomes that had intact right ends in these peak fractions was between 7 and 13%. If none of the genomes cleaved at both the right and left ends were packaged, then the percentage of left-end-cut genomes in the peak fraction would have been between 1.7 and 2.5% in these experiments (Table 3) due to the contaminating intact right-end genomes in the enriched fractions. Contrary to this supposition, the percentage of left-end-cut genomes in the enriched fractions was between 26 and 32% (Table 3). Taking into account the percent contribution from the contaminating genomes with intact right ends but cleaved left ends, between 24 and 30% of the genomes isolated in the enriched fractions lack both the left and right ITRs (Table 3). These results demonstrate that genomes lacking both the left and right ITRs are packaged. The percentages of genomes cleaved at both the right and left ITRs are similar to the percentage of total genomes with left-end cuts (Table 3, Nuclear DNA). These results show that an Ad genome lacking both ITRs and their covalently linked terminal proteins are efficiently packaged.
FIG. 7.
Viral particles with genomes lacking the right end are enriched from particles with full-length genomes on a CsCl equilibrium gradient. 293 cells were coinfected with Ad(Double HO) and Ad(HO gene) viruses. Viral particles were enriched for the lighter-density fraction on a CsCl equilibrium gradient and were repurified by using a second gradient (see Materials and Methods). Fractions were collected from the top of the gradient, and DNA was purified from the particles in each fraction. The XbaI-digested DNAs were hybridized with the right-end probe, Ad nt 28667 to 29169, as described in the legend to Fig. 4.
DISCUSSION
Both designing drugs for the treatment of virus infections and the exploitation of viruses as drugs for treatment of diseases can be made more successful by understanding the molecular mechanisms of virus-specific events. The process of assembly of the virus, and more specifically packaging of the viral genome into a protein capsid, is an obligatory step leading to future infections. To enhance our understanding of the molecular mechanism of packaging, it is necessary to characterize the viral components necessary for the event.
In the case of Ad, it is clear that sequences between ∼200 to 400 nt at the left end of the genome are essential for packaging. However, it was not known whether other cis elements were playing a role in the packaging process as well. Exploitation of Ad for gene therapy in treating both genetic disorders and cancers has led to the development of various vectors. The helper-dependent vectors contain only Ad packaging sequences and the ITRs with covalently linked terminal proteins (reviewed in reference 26). The ITRs and covalently linked terminal proteins are required for replication of the virus, but it was unclear whether they contributed to the packaging process as well. In this report, we demonstrate that removal of the ITRs and their covalently linked terminal proteins does not disable the remaining Ad genome for efficient packaging.
We employed an in vivo method to remove the ITRs from the Ad genome. It had been previously shown that HO endonuclease sites in the Ad genome could be cut by HO endonuclease in vivo (29). We surmised that if the ITRs were contributing to packaging, then the left ITR would be the more critical element because of its close location to the packaging domain. This seemed consistent with the fact that the genome is packaged in a polar fashion from left to right and that the packaging sequences are inactive when moved more than 600 bp from the left end (reviewed in reference 39). Furthermore, the cellular protein CDP, which is implicated in packaging, binds not only to the packaging sequences but also to sequences within the ITR (7, 37). Our results show that viral genomes lacking either the left-end ITR, the right-end ITR, or both ITRs are efficiently packaged. These results suggest that the packaging sequences (nt 200 to 400) are the only sequences necessary for Ad packaging. Calculation of the packaging efficiency of genomes lacking both ITRs depends on the distribution of left-end-cut molecules within the fractions enriched for right-end-cut viral genomes (Fig. 6 and Table 3). Viral DNAs in the enriched fractions that had intact right ends may have left-end cuts equal to the percentage found in the nuclear DNA pool, or viral DNAs with intact right ends may have been enriched for left-end-cut molecules. If the population of uncut right-end fragments in the peak fractions in Table 3 exhibited left-end cuts equivalent to that found in the nuclear pool of DNA, then genomes lacking both ITRs were packaged as efficiently as wild-type DNA (experiment 1, 125%; experiment 2, 98%; experiment 3, 140%). If, however, the population of uncut right-end fragments in the peak fractions in Table 3 were completely enriched for left-end-cut genomes, then the packaging efficiencies of genomes cut at both ends was somewhat reduced (experiment 1, 104%; experiment 2, 75%; experiment 3, 76%). In both cases, it is clear that genomes lacking both ITRs are packaged at or close to wild-type efficiency. We believe that the former possibility is the most probable, since there is no reason to believe that genomes with intact right ends would be entirely enriched for left-end-cut molecules, since the differences in size are minimal (only 145-bp difference in size with molecules ∼29,500 bp in length). A previous report suggested that sequences within the first 400 nt at the right end of the Ad genome contributed to efficient packaging in the context of a helper-dependent (gutted) Ad vector backbone (32). While these sequences may improve the packaging of a gutted Ad vector genome by three- to fourfold, it is clear from our work that right-end sequences play no role in viral DNA packaging with an otherwise wild-type virus.
These results are in contrast to what is seen with the bacteriophage φ29. The φ29 genome has a structure similar to that of Ad. It contains a linear, double-stranded DNA genome and a covalently linked protein at the termini that functions in replication of the phage (reviewed in reference 24). In addition, this protein in conjunction with the DNA plays a role in the recognition of the portal vortex made up of the connector, ATPase, and pRNA complex (12) to initiate DNA packaging. Similarly, the bacteriophage PRD1 has covalently linked terminal proteins that functions in replication. At present, however, it is not known whether PRD1 terminal proteins plays a role in packaging. A notable difference between Ad and φ29 is their host—a eukaryotic versus bacterial cell, respectively. The molecular organization of the host cells is quite different. It is recognized that the nucleus of a eukaryotic cell is highly organized (4, 5). Not only are the chromosomes ordered in the nucleus, but subdomains also have been identified for transcriptionally active and inactive DNA, replicating DNA, and other nuclear functions, such as splicing. Several different domains are seen within an Ad-infected nucleus (3, 20). Significantly, Ad proteins involved in replication localize to subdomains that are distinct from domains marked by the presence of the Ad assembly protein L1 (16). These latter domains may in fact be assembly sites for the virus. Consistent with this idea, terminal protein of Ad has been shown biochemically to be associated with the nuclear matrix fraction. Mutations in terminal protein that result in a reduction in association with the nuclear matrix fraction also show reduced early gene transcription (33). Altogether, these results suggest the tantalizing possibility that terminal protein maintains the Ad genome at a nuclear site for transcription and replication and that this site is distinct from the virus assembly site. Our results are consistent with this model. Also consistent with this model is the observation that among CDP DNA binding sites are matrix attachment sites (MARs) (1, 8, 27). MARs have been implicated in gene regulation. CDP is a transcriptional repressor (1, 8, 27), and one mechanism of CDP repression of gene expression may be the exclusion of other MAR binding proteins from their target sites. For example, CDP binding at MAR elements was found to compete for the interaction of matrix attachment proteins SATB1 and Bright from binding to the nuclear matrix, thus preventing activation of gene expression (1, 8, 27). Repression by CDP occurs both by competition for sites bound by activators and by CDP recruitment of the chromatin remodeling component histone deacetylase-1 (23). Taken together, such interactions could contribute to changes in localization and/or the chromatin structure of the adjacent DNA. CDP binding to the packaging sequences of Ad could lead to a relocalization of the Ad genome in the nucleus and/or the alteration of the chromatin of the Ad genome resulting in a packaging competent genome that is recognized by viral proteins. In this scenario, the absence of the ITRs and terminal proteins would not affect the packaging of the Ad genome.
Our results suggest that the only sequences necessary for the packaging of DNA into an Ad capsid are the packaging sequences located between nt ∼200 and 400 in the Ad5 genome. It seems unlikely that nucleotides between nt 106 (where the HO endonuclease site was located) and 200 are necessary, since mutants of Ad with different deletions that encompass these sequences replicate and package their DNA at wild-type or nearly wild-type levels in coinfection experiments (38). At present, the helper-dependent Ad vectors used for gene therapy include both the Ad ITRs and packaging sequences in order to amplify the genomic DNA and then package it, respectively. A helper virus is required for gutted vector production. Our results suggest that a new generation of vectors could be amplified independent of Ad and later packaged in an Ad capsid. For example, if an Ad in vitro packaging system was developed, then the simple linkage of a 200-bp Ad packaging domain to any DNA sequence would direct virus assembly. Such an approach may obviate the need for helper viruses to allow the production of gutted Ad vectors that are entirely pure, a major obstacle in the development of gutted Ad vector for human gene therapy.
Acknowledgments
We thank Hamish Young for generously providing E1 HO site virus, E1 HO gene virus, and E3 HO site virus. We thank all the members of our laboratory for many helpful discussions and Jihong Yang for excellent technical assistance.
This work was supported by Public Health Service grant AI41636.
REFERENCES
- 1.Banan, M., I. C. Rojas, W. H. Lee, H. L. King, J. V. Harriss, R. Kobayashi, C. F. Webb, and P. D. Gottlieb. 1997. Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8a gene. J. Biol. Chem. 272:18440-18452. [DOI] [PubMed] [Google Scholar]
- 2.Bjornsti, M. A., B. E. Reilly, and D. L. Anderson. 1983. Morphogenesis of bacteriophage phi29 of Bacillus subtilis: oriented and quantized in vitro packaging of DNA protein gp3. J. Virol. 45:383-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bridge, E., S. Medghalchi, S. Ubol, M. Leesong, and G. Ketner. 1993. Adenovirus early region 4 and viral DNA synthesis. Virology 193:794-801. [DOI] [PubMed] [Google Scholar]
- 4.Carmo-Fonseca, M. 2002. The contribution of nuclear compartmentalization to gene regulation. Cell 108:513-521. [DOI] [PubMed] [Google Scholar]
- 5.Cremer, T., and C. Cremer. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292-301. [DOI] [PubMed] [Google Scholar]
- 6.Duffy, C., and M. Feiss. 2002. The large subunit of bacteriophage lambda's terminase plays a role in DNA translocation and packaging termination. J. Mol. Biol. 316:547-561. [DOI] [PubMed] [Google Scholar]
- 7.Erturk, E., P. Ostapchuk, S. Wells, K. Gregg, A. Nepveu, J. P. Dudley, and P. Hearing. Binding of CCAAT Displacement Protein, CDP, to adenovirus packaging sequences. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 8.Goebel, P., A. Montalbano, N. Ayers, E. Kompfner, L. Dickinson, C. F. Webb, and A. J. Feeney. 2002. High frequency of matrix attachment regions and cut-like protein x/CCAAT-displacement protein and B cell regulator of IgH transcription binding sites flanking Ig V region genes. J. Immunol. 169:2477-2487. [DOI] [PubMed] [Google Scholar]
- 9.Graeble, M., and P. Hearing. 1990. Adenovirus type 5 packaging domain is composed of a repeated element that is functionally redundant. J. Virol. 64:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graeble, M., and P. Hearing. 1992. cis and trans requirements for the selective packaging of adenovirus type 5 DNA. J. Virol. 66:723-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham, F. L., J. SMiley, W. C. Russell, and R. Nairu. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-72. [DOI] [PubMed] [Google Scholar]
- 12.Grimes, S., and D. Anderson. 1989. In vitro packaging of bacteriophage phi29 DNA restriction fragments and the role of the terminal protein gp3. J. Mol. Biol. 209:91-100. [DOI] [PubMed] [Google Scholar]
- 13.Guasch, A., J. Pous, B. Ibarra, F. X. Gomis-Ruth, J. M. Valpuesta, N. Sousa, J. L. Carrascosa, and M. Coll. 2002. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J. Mol. Biol. 315:663-676. [DOI] [PubMed] [Google Scholar]
- 14.Gustin, K. E., and M. J. Imperiale. 1998. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72:7860-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gustin, K. E., P. Lutz, and M. J. Imperiale. 1996. Interaction of the adenovirus L1 52/55-kilodalton protein with the IVa2 gene product during infection. J. Virol. 70:6463-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasson, T. B., D. A. Ornelles, and T. Shenk. 1992. Adenovirus L1 52- and 55-kilodalton proteins are present within assembling virions and colocalize with nuclear structures distinct from replication centers. J. Virol. 66:6133-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasson, T. B., P. D. Soloway, D. A. Ornelles, W. Doerfler, and T. Shenk. 1989. Adenovirus L1 52- and 55-kilodalton proteins are required for assembly of virions. J. Virol. 63:3612-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatfield, L., and P. Hearing. 1993. The NFIII/OCT-1 binding site stimulates adenovirus DNA replication in vivo and is functionally redundant with adjacent sequences. J. Virol. 67:3931-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hearing, P., R. J. Samulski, W. L. Wishart, and T. Shenk. 1987. Identification of a repeated sequence element required for efficient encapsidation of the adenovirus type 5 chromosome. J. Virol. 61:2555-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 22.Leffers, G., and V. B. Rao. 2000. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J. Biol. Chem. 275:37127-37136. [DOI] [PubMed] [Google Scholar]
- 23.Li, S., L. Moy, N. Pittman, G. Shue, B. Aufiero, E. J. Neufeld, N. S. LeLeiko, and M. J. Walsh. 1999. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J. Biol. Chem. 274:7803-7815. [DOI] [PubMed] [Google Scholar]
- 24.Meijer, W. J., J. A. Horcajadas, and M. Salas. 2001. Phi 29 family of phages. Microbiol. Mol. Biol. Rev. 65:261-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore, M., N. Horikoshi, and T. Shenk. 1996. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. USA 93:11295-11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morsy, M. A., and C. T. Caskey. 1999. Expanded-capacity adenoviral vectors-the helper-dependent vectors. Mol. Med. Today. 5:18-24. [DOI] [PubMed] [Google Scholar]
- 27.Nepveu, A. 2001. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 270:1-15. [DOI] [PubMed] [Google Scholar]
- 28.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolas, A. L., P. L. Munz, E. Falck-Pedersen, and C. S. Young. 2000. Creation and repair of specific DNA double-strand breaks in vivo following infection with adenovirus vectors expressing Saccharomyces cerevisiae HO endonuclease. Virology 266:211-224. [DOI] [PubMed] [Google Scholar]
- 30.Ostapchuk, P., and P. Hearing. 2001. Pseudopackaging of adenovirus type 5 genomes into capsids containing the hexon proteins of adenovirus serotypes B, D, or E. J. Virol. 75:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parks, R. J., and F. L. Graham. 1997. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J. Virol. 71:3293-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandig, V., R. Youil, A. J. Bett, L. L. Franlin, M. Oshima, D. Maione, F. Wang, M. L. Metzker, R. Savino, and C. T. Caskey. 2000. Optimization of the helper-dependent adenovirus system for production and potency in vivo. Proc. Natl. Acad. Sci. USA 97:1002-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaack, J., W. Y. Ho, P. Freimuth, and T. Shenk. 1990. Adenovirus terminal protein mediates both nuclear matrix association and efficient transcription of adenovirus DNA. Genes Dev. 4:1197-1208. [DOI] [PubMed] [Google Scholar]
- 34.Scheffczik, H., C. G. Savva, A. Holzenburg, L. Kolesnikova, and E. Bogner. 2002. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res. 30:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schiedner, G., S. Hertel, and S. Kochanek. 2000. Efficient transformation of primary human amniocytes by E1 functions of Ad5: generation of new cell lines for adenoviral vector production. Hum. Gene Ther. 11:2105-2116. [DOI] [PubMed] [Google Scholar]
- 36.Schmid, S., and P. Hearing. 1999. Adenovirus assembly, p. 85-90. In P. Seth (ed.), Adenoviruses: basic biology to gene therapy. R. G. Landes Company, Austin, Tex.
- 37.Schmid, S., and P. Hearing. 1998. Cellular components interact with adenovirus type 5 minimal DNA packaging domains. J. Virol. 72:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid, S. I., and P. Hearing. 1997. Bipartite structure and functional independence of adenovirus type 5 packaging elements. J. Virol. 71:3375-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmid, S. I., and P. Hearing. 1995. Selective encapsidation of adenovirus DNA. Curr. Top. Microbiol. Immunol. 199:67-80. [DOI] [PubMed] [Google Scholar]
- 40.Simpson, A. A., Y. Tao, P. G. Leiman, M. O. Badasso, Y. He, P. J. Jardine, N. H. Olson, M. C. Morais, S. Grimes, D. L. Anderson, T. S. Baker, and M. G. Rossmann. 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stow, N. D. 1981. Cloning a DNA fragment from the left-hand terminus of the adenovirus type 2 genome and its use in site-directed mutagenesis. J. Virol. 37:171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, W., and M. J. Imperiale. 2000. Interaction of the adenovirus IVa2 protein with viral packaging sequences. J. Virol. 74:2687-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, W., J. A. Low, J. B. Christensen, and M. J. Imperiale. 2001. Role for the adenovirus IVa2 protein in packaging of viral DNA. J. Virol. 75:10446-10454. [DOI] [PMC free article] [PubMed] [Google Scholar]