Abstract
During entry into host cells, poliovirus undergoes a receptor-mediated conformational transition to form 135S particles with irreversible exposure of VP4 capsid sequences and VP1 N termini. To understand the role of VP4 during virus entry, the fate of VP4 during infection by site-specific mutants at threonine-28 of VP4 (4028T) was compared with that of the parental Mahoney type 1 virus. Three virus mutants were studied: the entry-defective, nonviable mutant 4028T.G and the viable mutants 4028T.S and 4028T.V, in which residue threonine-28 was changed to glycine, serine, and valine, respectively. We show that mutant and wild-type (WT) VP4 proteins are localized to cellular membranes after the 135S conformational transition. Both WT and viable 4028T mutant particles interact with lipid bilayers to form ion channels, whereas the entry-defective 4028T.G particles do not. In addition, the electrical properties of the channels induced by the mutant viruses are different from each other and from those of WT Mahoney and Sabin type 3 viruses. Finally, uncoating and/or cytoplasmic delivery of the viral genome is altered in the 4028T mutants: the 4028T.G lethal mutant does not release its genome into the cytoplasm, and genome delivery is slower during infection by mutant 4028T.V 135S particles than by mutant 4028T.S or WT 135S particles. The distinctive electrical characteristics of the different 4028T mutant channels indicate that VP4 sequences might form part of the channel structure. The different entry phenotypes of these VP4 mutants suggest that the ion channels may be related to VP4's role during genome uncoating and/or delivery.
Poliovirus, a member of the Picornaviridae family, encapsidates its 7,400-nucleotide positive-sensed RNA genome within an icosahedrally symmetric protein shell that is formed by 60 copies of the four capsid proteins (VP1 to VP4). VP1, VP2, and VP3 form the surface of the virion, with VP1 located at each fivefold axis and VP2 and VP3 alternately positioned around each threefold axis. VP4 in its entirety as well as the amino termini of VP1, VP2, and VP3 are buried within the interior of the capsid, lying along the inner surface of the virion shell (12).
Poliovirus entry into cells is initiated by binding to the poliovirus receptor (PVR) on the cell surface. PVR binding induces a conformational transition within the virus particle that leads to formation of altered particles (termed A particles) sedimenting at 135S (versus the 160S sedimentation value of the native particle) (references 8 and 23 and references therein). This conformational transition results in relocation of VP4 and the VP1 N termini from the particle interior to the virion exterior. The appearance of VP4 and VP1 domains on the particle surface is correlated with dramatic differences in the functional behavior of the native 160S and altered 135S particles. Functionally, these PVR-induced conformational rearrangements generate 135S particles that acquire the ability to bind to liposomes, to form ion channels in lipid bilayers, and to infect cells in a receptor-independent fashion (7, 10, 13, 27). Thus, it was hypothesized that the membrane binding properties of the 135S particle are mediated by the VP1 N termini and possibly the myristoylated VP4 protein and that these membrane interactions are involved at postreceptor stages of the viral entry pathway (4, 10, 16, 18). Consistent with this view is the genetic evidence indicating that these domains are involved in the virus entry process at stages after PVR binding and conversion to 135S particles (5, 15, 22). However the postreceptor binding stages of the viral entry and uncoating pathway are largely undefined.
Mutants 4028T.G, 4028T.V, and 4028T.S, containing, respectively, a glycine, valine, and serine substitution at threonine-28 of VP4, were previously identified from a collection of site-specific mutants originally generated to study the function of the N-terminal myristoyl modification of VP4 during viral infection (20-22). Mutants 4028T.V and 4028T.S are viable, whereas 4028T.G is nonviable. Earlier studies indicated that 4028T.G is defective at an undefined step after receptor binding (22). To date, 4028T.G is the only lethal mutant identified whose defect is in the entry pathway. Thus, we decided to study in greater detail the phenotypes of the VP4 4028T mutants and the potential role(s) of VP4 sequences during the early stages of viral entry.
We demonstrate here that VP4 is inserted into cellular membranes during the initial stages of infection by wild-type (WT) and 4028T mutant viruses. However, in contrast to infection by WT and the viable 4028T mutant viruses, cytoplasmic delivery of the 4028T.G viral genome does not occur. Analyses of the interaction of 4028T.G 160S particles with lipid bilayers demonstrate that the 4028T.G mutant virion is unable to form ion channels, whereas both 4028T.S and 4028T.V 160S particles are able to form ion channels in lipid bilayers. Thus, the ability to form ion channels is associated with the ability to successfully uncoat and deliver the viral genome into the cytoplasm. In addition, the dramatic differences in the electrical properties of the ion channels formed by 4028T.S and 4028T.V mutants suggest that the VP4 sequence contributes to the channel architecture.
MATERIALS AND METHODS
Cells and antibodies.
HeLa-S3 cells were maintained in suspension (3 × 105 to 4 × 105 cells/ml) in Joklik's modified minimal essential medium (S-MEM) supplemented with 7.5% horse serum and 1 mM MEM sodium pyruvate. For monolayer growth, cells were transferred into Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum (FCS), 50 U of penicillin/ml, 50 μg of streptomycin/ml, and 1 mM sodium pyruvate in a 5% CO2 incubator. Polyclonal antibodies against VP4 protein were generated previously in rabbits (17). Anti-3D monoclonal antibodies raised against the viral polymerase were a gift from Craig Cameron, Pennsylvania State University.
Viable viruses and plasmid cDNAs.
Poliovirus serotype 1, Mahoney strain, was the parental WT virus for all mutant studies. Viral stocks were propagated as previously described from a plaque obtained by transfection of WT poliovirus infectious cDNA (pPVM-1) into HeLa cells (25). The seed stock of the Sabin type 3 strain of poliovirus was obtained from P. Minor. Virus stocks of the viable mutant viruses (4028T.S and 4028T.V) as well as the Sabin type 3 strain of poliovirus used for these studies were generated from low-multiplicity-passage-2 (P2) stocks by a single passage in HeLa cells at high multiplicity (mulitiplicity of infection [MOI] = 10) and were purified (20). Recombinant vaccinia virus T7 stocks were generated by a low-multiplicity (MOI = 0.01) infection of HeLa cells (11). When the cytopathic effects were evident throughout the culture at 4 days postinfection (p.i.), cells were harvested and a cell lysate was generated by three cycles of freeze-thaw in phosphate-buffered saline (PBS)-AM (PBS with 10 mM MgCl2 and 0.01% bovine serum albumin) followed by sonication. Aliquoted stocks were stored at −20°C. The vaccinia virus stocks were titered in a 4-day plaque assay on HeLa cells, and typical titers were approximately 2 × 109 PFU/ml.
Production of 4028T.G mutant virus.
Mutant 4028T.G virus stocks used for these studies were generated by using coupled vaccinia virus infection-poliovirus cDNA transfections with slight modifications (26). Vaccinia virus T7 (MOI = 10) was bound to HeLa cells (108 cells/1 ml of PBS with 10 mM MgCl2 and 0.01% bovine serum albumin) for 1 h at 37°C, and subsequently, the infection was continued in suspension by adding methionine-free S-MEM-5% FCS (5 × 106 cells/ml) for an additional hour. The cells were transfected with WT or 4028T.G mutant cDNAs (100 μg) by using 450 μg of DOTAP (Roche, Indianapolis, Ind.) and were transferred to plates (1.5 × 106 cells/ml). Actinomycin D (50 μg/ml) was added at 3.5 h posttransfection, and [35S]methionine (100 μCi/ml) was added at 4 h posttransfection. Cells were harvested at 12 h posttransfection and lysed in RSB buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 1.5 mM MgCl2, and 1% NP-40) at 4°C. The lysate was centrifuged at 1,000 × g for 10 min, and the virus was pelleted from the postnuclear supernatant by centrifugation at 50,000 rpm for 1 h at 4°C in a Beckman 70.1Ti rotor. The pellets were resuspended overnight in Tris-NaCl-EDTA-0.5% sodium dodecyl sulfate (SDS) (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, and 0.5% SDS) and were centrifuged on 15 to 30% sucrose density gradients. Fractions constituting the radiolabeled 160S virus peaks were pooled and dialyzed against PBS. To determine the yield of nonviable mutant virus, the specific infectivity of the WT virus was determined by measuring viral plaque titers on HeLa cell monolayers and normalizing the titers to the radioactivity present in the virus preparation (PFU/counts per minute). Because the WT and mutant viruses were always grown in parallel cultures under identical conditions, the specific infectivity of 4028T.G was assumed to be identical to that measured for the WT virus. Occasionally, 4028T.G preparations would be contaminated with revertant viruses (as determined by sequence analyses) generated during the vaccinia virus infection and transfection process. All 4028T.G stocks used in these studies showed no evidence of revertants as determined by plaque assays. Thus, the maximum titer of revertant viruses potentially in these mutant stocks was < 10 PFU/ml.
NR assays.
Neutral red (NR) viruses were grown by infecting 107 HeLa cells (MOI =10) with WT or 4028T mutant viruses in the dark in the presence of NR dye (10 mg/ml) (6). Cells were harvested at 8 h p.i. and were lysed in 10 mM Tris-HCl (pH 7.5)-10 mM NaCl-1.5 mM MgCl2 at 4°C. After nuclei were removed, the supernatants were titered and used as NR-160S virus stocks. NR-160S particles were converted to NR-135S particles by heating for 2 min 10 s at 51°C. In the dark, 160S or 135S particles of WT or mutant NR viruses (200 to 300 PFU) were bound for 30 min to HeLa cell monolayers on 60-mm-diameter plates and the unbound virus was removed. The infections were initiated with addition of an agar overlay (Dulbecco's modified Eagle's medium-3% FCS containing 0.8% agarose) and were placed at 37°C. The infections were exposed to white light at various times p.i. by placing the plates in triplicate on a light box for a period of 10 min. The infections were returned to the incubator and were incubated in the dark for an additional 36 to 48 h to allow plaque development.
Cell fractionation.
Radiolabeled WT or 4028T mutant viruses were bound to 108 HeLa cells (MOI = 10) at 4°C for 2 h. The cells were washed with PBS at 4°C to remove unbound virus, and infection was initiated by addition of S-MEM prewarmed to 37°C. Cells were harvested at different times by centrifugation, and the media and PBS washes were combined. The cell pellet was resuspended in hypotonic lysis buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl) and was lysed by Dounce homogenization at 4°C (20 strokes). The nuclei were removed by centrifugation at 1,000 × g for 10 min and the postnuclear supernatant was subsequently centrifuged at 100,000 × g for 30 min to obtain a microsomal membrane fraction (pellet) and a cytoplasmic fraction (supernatant). Total recovery of input-labeled virus was determined by counting an aliquot of each fraction in a liquid scintillation counter and correcting for the volume of the fraction. Samples were further analyzed on 15 to 30% linear sucrose gradients and by Western blots.
To wash the membrane fractions in high-ionic-strength buffers, the membrane pellet fractions were resuspended by vortexing vigorously in 10 mM Tris-HCl (pH 7.4) containing 10 mM, 100 mM, or 1 M NaCl (1 ml) and were then repelleted by centrifugation at 100,000 × g for 30 min at 4°C. The samples were run on SDS-polyacrylamide gels and probed for VP4 on Western blots.
Western blots.
Samples from WT- or 4028T mutant virus-infected cells were separated by SDS-12.5% polyacrylamide gel electrophoresis and were transferred to nitrocellulose or polyvinyl difluoride (PVDF) membranes. The nitrocellulose or PVDF membranes were blocked in 10 mM Tris-Cl, pH 7.5, 150 mM NaCl, and 0.1% Tween 20 (TBST) containing 5% nonfat milk or horse serum for 1 h at room temperature. Viral polymerase (3D) was detected as follows: the nitrocellulose membrane was incubated with an anti-3D monoclonal antibody (1:3,000 dilution) overnight at 4°C and subsequently with horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:3,000 dilution; Caltag) for 1 h at room temperature. The membranes were washed three times with TBST for 10 min after each antibody incubation, and the blots were developed by using Renaissance Western blot Chemiluminescence Reagent Plus (NEN, Boston, Mass.). VP4 was detected as follows: the nitrocellulose or PVDF membranes were incubated with rabbit polyclonal anti-VP4 antibody (1:2,000 dilution) and washed with TBST and subsequently incubated with alkaline phosphatase-conjugated goat anti-rabbit secondary antibody (1:7,500 dilution; Boehringer Mannheim) for 30 min each at room temperature. The blots were developed by using Nitro Blue Tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (Fisherbiotech). Images of the VP4 membrane blots were captured by using Epi II chemi darkroom transilluminator (UVP Lab Products). The intensities of the VP4 bands were quantitated by determining the integrated optical density (IOD) using the Labworks2 software (UVP laboratory products). The ratios of the IODs for salt-washed and control lanes were used to determine the relative amounts of WT and mutant VP4 that remained membrane associated.
Reverse transcriptase PCR (RT-PCR) of viral RNAs.
HeLa cells were infected with WT or 4028T.G virus (MOI = 10). Cells were harvested at different times p.i., and total RNA was extracted by using RNAzol B (Tel-Test, Friendswood, Tex.) according to the manufacturer's instructions. Briefly, RNA from 100 cell equivalents of WT- or mutant-infected cells was reverse transcribed by using random hexamers. The resultant cDNAs were PCR amplified by using primers E1 and E2 specific for the 5′ untranslated region of poliovirus (9) or by using primers specific for human β-actin (Stratagene, La Jolla, Calif.). PCR products were resolved on 1.5% agarose gels.
Lipid bilayers.
The lipids and the method used to form the planar bilayers were as previously described (27). The bilayers were made by apposition of two monolayers made from soybean lecithin (Associated Concentrate, Woodside, N.Y.) dissolved in pentane (20 mg/ml) on a small hole (60 to 80 μm in diameter) on the partition. Each compartment of the voltage clamp chamber could be stirred independently. The electrical contact was made through Ag-AgCl electrodes connected to the solution in the compartments via 3 M NaCl-3% agar bridges. The chamber was set in a sample holder of a precision temperature controller (Warner Instruments Co., Hamden, Conn.). The electrolyte solution used throughout these measurements was 300 mM NaCl and 20 mM Tris-morpholinepropanesulfonic acid, pH 7.2.
After formation of a membrane, the conductance was monitored at room temperature for a period of time varying from 20 to 40 min by applying voltage pulses of various amplitudes. Subsequently, virus was added to one compartment (under continuous stirring) and the conductance was monitored as before. This was followed by an increase in the temperature of the chamber to 31°C and monitoring of the conductance until it reached a steady state. Alternatively, the temperature in both compartments was raised to 31°C before addition of virus and the conductance was monitored (control), after which virus was added to one of the compartments (under continuous stirring) and the monitoring of the conductance continued until it reached a steady value (usually about 10 min after the temperature increase). The current traces obtained in response to applied voltages (3-min duration) were recorded by using a Dagan 3900 patch clamp amplifier (Dagan Instruments, Minneapolis, Minn.) after it was filtered at 5 kHz and digitized for further analyses either at 0.05 ms per point for channels with short open times or at 200 ms/point when the open time of the channels was very long. The single-channel conductance was calculated from the mean value of the current obtained from Gaussian fits to the distribution of the current amplitudes by using Peak Fit (Jandel Software, San Rafael, Calif.). The mean open probability was calculated by using Pclamp 8.0 software (Axon Instruments Inc., Union City, Calif.).
RESULTS
VP4 localizes to cell membranes at early stages of poliovirus infection.
Both the PVR-catalyzed and heat-induced 160S-135S conformational transitions result in irreversible externalization of the VP4 sequences from the capsid interior. During early times p.i., the majority of the particles in the viral inoculum elute from the cell surface into the media as 135S particles and these “eluted” 135S particles lack detectable VP4 protein (8). Other studies using [3H]myristate-labeled virus indicated that the radiolabel remains largely cell associated during the 1st h p.i. (29). To further define the role of VP4 during entry, the subcellular location of VP4 was determined at early times p.i. in WT- and mutant-infected cells.
Cells infected with [35S]methionine-labeled WT virus were harvested at various times over the 1st h p.i., and the cell lysates were fractionated to isolate microsomal membrane and cytoplasmic compartments. The distribution of label was analyzed in these subcellular fractions as well as the growth media (Fig. 1 and 2). At all times examined, the majority of the original input label was recovered in either the microsomal membrane or medium fractions (Fig. 1A). Initially, consistent with virus binding to PVR on the cell surface, the majority of the label was found in the membrane fraction. At later times p.i., the percentage of label detected in the membrane fraction decreased and was associated with concomitant increases of label appearing in the media. These medium and membrane fractions were analyzed further; the radiolabel present in the cytoplasmic fractions was too dilute for further analyses.
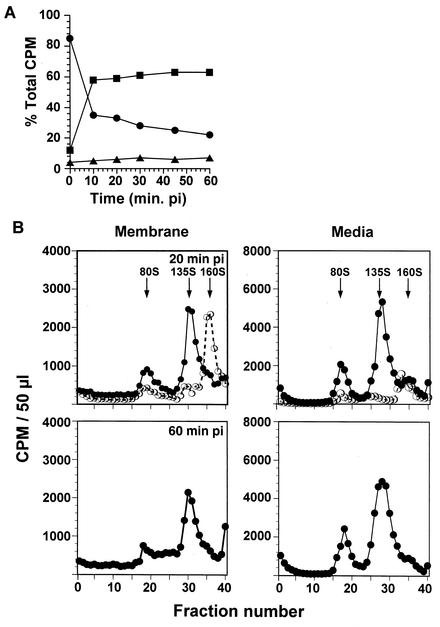
FIG. 1.
Fate of input virus particles and proteins at early times of infection. (A) Distribution of labeled virus in different compartments. Cells infected with [35S]methionine-labeled WT virus were harvested at various times p.i. and fractionated into microsomal membrane and cytosolic compartments. At each time p.i., >95% of the original input virus is recovered within these subcellular compartments and the growth media. The amount of label associated with the medium (▪), membrane (•), or cytosol (▴) fractions is expressed as a percentage of the total amount of label present in the sample at the time of harvest. (B) Sedimentation analyses of poliovirus particles. Equal volumes of the membrane and medium compartments from WT-infected cells were run on 15 to 30% linear sucrose gradients. Gradients were fractionated, and the radioactivity in each gradient fraction was determined by liquid scintillation counting. Only the samples at 0 min p.i. (○) and at 20 and 60 min p.i. (•) are shown. The positions of viral markers in the gradients are indicated.
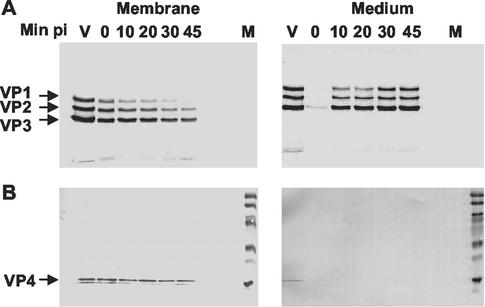
FIG. 2.
Analysis of WT viral capsid proteins in membrane and medium fractions. Equal volumes of the infected cell membrane fractions or medium at the indicated times p.i. were resolved by SDS-polyacrylamide gel electrophoresis and were transferred to nitrocellulose. The nitrocellulose filters were exposed to X-ray film (A) and were then subsequently probed with rabbit anti-VP4 antibodies to detect the VP4 protein (B). V, labeled virus; and M, prestained molecular weight markers. The relative intensities of VP1, VP2, and VP3 for each sample in the autoradiograph reflect the different methionine contents of the capsid protein: VP1, VP2, and VP3 contain, respectively, 5, 6, and 12 methionine residues.
Sedimentation analyses of the membrane fractions (Fig. 1B) indicated that the majority of the input virus was found at the start of infection as the 160S form. Consistent with the results found by previous investigators, as infection progressed over the 1st h, the membrane-associated 160S particle converted into the PVR-mediated 135S form and an increase in the “empty” 80S particle form was observed over time. In the media, the concentration of receptor-converted 135S particles in the media increased as infection progressed and was the predominant particle form (with a smaller fraction of 80S particles) that eluted back into the media from the infected cell (10). The compositions of capsid proteins found in the medium and microsomal membrane fractions were consistent with this interpretation (Fig. 2A). The major capsid proteins, VP1, VP2, and VP3, were detected initially in the membrane fractions, and the intensities decreased over time. Conversely, the intensities of the capsid proteins found in the medium samples increased over this period.
The presence of VP4 proteins in the medium and membrane fractions was examined by Western blot analysis (Fig. 2B), because this protein is difficult to detect by autoradiography. VP4 was present in the membrane fractions at all times examined and was not detected in the medium fractions containing the eluted virus particles. Similarly, VP4 was not detected in the cytoplasmic fractions (data not shown). Interestingly, the amount of VP4 found in the membrane fraction appeared to remain approximately the same throughout the 1st h of infection, even though greater than 50% of the labeled input virus had eluted from the membrane. Although the VP4 at initial times p.i. could be associated with receptor-bound 160S particles, the 135S and 80S particles (where VP4 is irreversibly exposed on the particle surface) are the predominant particle forms in the membrane fractions (Fig. 1B). Thus, the VP4 detected in the membrane is either associated with the membrane-bound 135S particle or is itself associated with cellular membranes.
VP4 has been hypothesized to facilitate membrane binding of the 135S particles (16, 18). To determine the consequences of the 4028T mutations and whether the lethal defect in 4028T.G is due to the inability of the mutant VP4 protein to associate with membranes, cells infected with labeled nonviable 4028T.G or viable 4028T.S and 4028T.V mutants were harvested at 0 and 30 min p.i. and were fractionated as explained above (Fig. 3). As shown previously for the 4028T.G mutant (22), by 30 min p.i., only 135S particles were detected in the membrane fractions from mutant-infected cells and the majority of initial input virus was recovered from the medium as the 135S form. Western blot analyses demonstrated that the VP4 proteins of the 4028T.G as well as those of 4028T.S and 4028T.V mutants were also localized to membrane compartments over these early times p.i. (Fig. 3A and B). Thus, early during virus entry, both WT and 4028T mutant VP4 capsid proteins associate with cellular membranes and remain membrane associated even though a significant fraction of the 135S particles have eluted from the membrane surface.
FIG. 3.
The fate of 4028T mutant virus at early times of infection. Capsid protein analyses of WT- and 4028T mutant-infected cellular membranes. (A) 4028T.G-infected membrane fractions. (B) 4028T.S and 4028T.V mutant-infected membrane fractions. At the indicated times p.i., cells infected with radiolabeled WT or 4028T mutant viruses were fractionated and analyzed as for Fig. 2. The nitrocellulose filters were exposed to a storage phosphor screen to detect labeled viral proteins by using a Molecular Dynamics PhosphorImager (450SI) and were subsequently probed with VP4 antibodies. Only the Western and autoradiographic analyses of the membrane fractions are shown. (C) High-salt wash of WT- or 4028T mutant-infected membrane fractions. Membrane pellets from cells infected with WT or the 4028T mutants at 30 min p.i. were resuspended in buffers of increasing ionic strength. The membranes were repelleted and analyzed on Western blots for VP4. The Western blots of the membrane fractions washed in buffers containing 1 M NaCl are shown here. The IODs obtained for the VP4 bands by using Labworks2 software are indicated at the bottom of the lanes.
The VP4 proteins are N-terminally myristoylated. Other than this lipid modification, this 63-amino-acid protein does not contain significant clusters (>5 amino acids in length) of hydrophobic residues. This suggested that WT and mutant VP4 proteins might be peripherally associated with the membranes with different affinities. To characterize the VP4-membrane interaction, membrane fractions of WT- and 4028T.G-infected cells were washed with buffers of increasing strength that would remove peripherally bound proteins and the VP4 content in these membranes was reexamined on Western blots (Fig. 3C). Densitometric analysis of the VP4 proteins of WT and mutant viruses showed that, in repeat experiments, approximately 45 to 60% of VP4 protein remained associated with the membrane fraction even after washing of the membranes in buffers containing 1 M NaCl. Although the intensity of the VP4 bands decreased as buffers of increasing ionic strength were used, at any given ionic strength, there were no significant differences in the intensities of the WT and mutant VP4 proteins. Thus, based at least on their membrane retention upon exposure to different-ionic-strength buffers, there appeared to be no significant differences between the WT and 4028T mutant VP4 associations with membranes.
Interaction of 4028T mutant 135S and 160S particles with bilayers.
The localization of VP4 to cellular membranes and the defective entry phenotype of 4028T.G suggested that a direct interaction of the viral particle with cellular membranes was important for the penetration and uncoating stages of the entry process. Previous studies have shown that the WT Mahoney virus 160S particles form temperature-dependent, ion-permeable channels when they interact with lipid bilayers (27). Thus, bilayer studies were designed to determine if temperature-dependent, ion-permeable channels were formed in lipid bilayers upon interaction of other standard laboratory poliovirus strains (with sequence differences in the VP4 proteins) or the 4028T mutant particles.
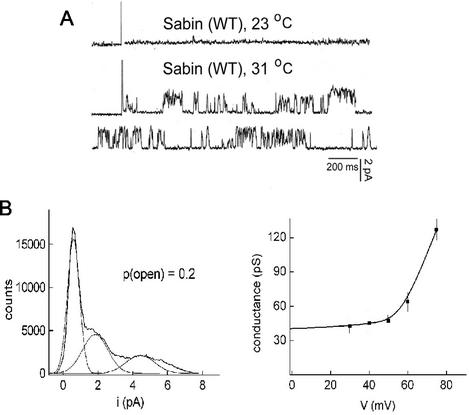
Residue 4028T (as well as the N-terminal half of the 68-amino-acid residue VP4 protein) is conserved in all standard laboratory poliovirus strains (Mahoney and Sabin type 1, Lansing and Sabin type 2, and Leon and Sabin type 3) examined. Sabin type 1 and the type 2 strains vary by only 1 amino acid residue (near the C terminus) from the Mahoney virus VP4 sequence. However, the VP4 sequences of Leon and Sabin type 3 viruses, though identical to each other, differ from the Mahoney strain at four positions throughout the C-terminal half of the protein. Residues 4034R, 4046F, 4057I, and 4067 M of Mahoney type 1 virus are replaced in the type 3 strains by residues 4034K, 4046Y, 4057L, and 4067A, respectively. Figure 4 shows the electrical characteristics of a bilayer after addition of Sabin type 3 virus 160S particles to one side of the bilayer. The figure shows that the current in response to an applied voltage of +40 mV remained constant in time at room temperature (Fig. 4A). However, as was previously shown for the Mahoney type 1 virus (27), when the temperature was raised to 31°C, the presence of channels induced by the binding of Sabin type 3 virus became evident as demonstrated by the random, sudden increases (and decreases) in the current response to an applied voltage of +40 mV (Fig. 4A, second and third traces). Analyses of the current amplitude from the complete trace yielded the amplitude distribution curve at this voltage (shown in Fig. 4B). The peaks in the amplitude distribution can be fitted by Gaussian curves to obtain the values of the current for the different states of the channel (Fig. 4B, left panel). The first Gaussian curve shown, with a center at 0.06 pA, corresponds to the closed state of the channel, the conductance of which was identical to that of the control trace (bilayers with no added virus). The time that the current dwells in this closed state (mean closed probability) corresponded to 55% of the total time that the pulse was applied. The second curve, with a center at 2.0 pA (consistent with the Gaussian curve shown), corresponds to the smallest open state, with a probability of opening of 0.2 ± 0.06. The difference between these two current values, 1.9 pA, yields the current that flowed through the channel opened at this state. Similar analyses were done at different positive and negative applied voltages, and the plot of conductance versus voltage was obtained by using the values at the peak of the distribution curve of the current amplitudes divided by the applied voltage (Fig. 4B, right panel). The curve shows that, like the channels induced by Mahoney type 1 virus, the Sabin type 3 virus channels are voltage gated, only opening at positive potentials (27). Importantly for the analyses of the 4028T mutants, the electrical properties determined for the Mahoney type 1 and Sabin type 3 virus channels are identical and indistinguishable from each other.
FIG. 4.
Electrical characteristics of bilayers exposed to Sabin type 3 virus. Planar bilayers were formed either at room temperature, and their conductances were checked for a minimum of 30 min. Viral particles were added before the increase in temperature to 31°C. (A) The panel shows the current-versus-time traces at 23°C obtained after addition of Sabin type 3 virus (8 × 104 particles/ml). The next two traces correspond to the same membrane after the temperature was increased to 31°C. Voltage = +40 mV. (B, left panel) The current amplitude distribution curve for a +40-mV pulse (3-min duration) is shown together with the fit to the curve by using Gaussian curves. The first peak corresponds to the closed state of the channel, whereas the second peak corresponds to the open state of the channel. The single-channel conductance can be determined from the mean value of current distribution. The ratio of the area under the curves reflects the probability of opening. (B, right panel) The conductance-versus-voltage curve. Each point represents the mean value of the conductance (and standard deviation) obtained at each of the voltages indicated from at least three different bilayers.
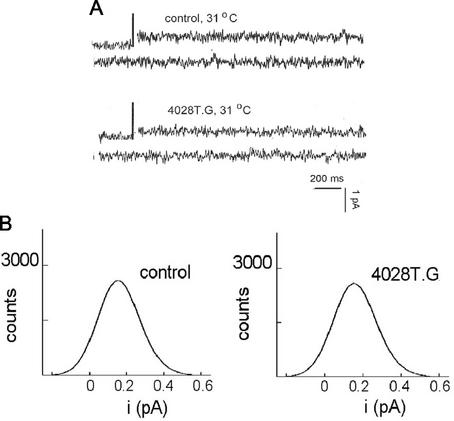
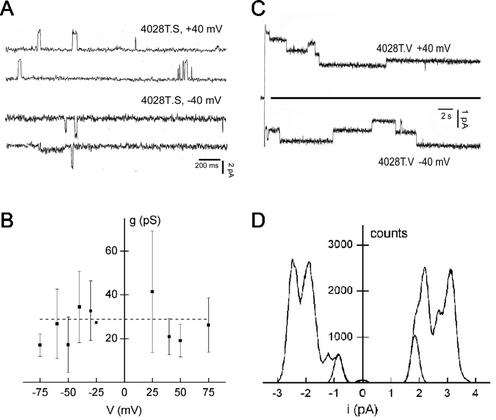
The abilities of the 4028T mutant particles to interact with lipid bilayers were examined at various applied voltages (Fig. 5 and 6). Again as observed for the Sabin type 3 and Mahoney type 1 viruses, no channel openings were detected at room temperature for any of the mutant viruses (data not shown). However, unexpectedly, when the nonviable 4028T.G 160S particles were added to a planar lipid bilayer, either at 23°C (and then raised to 31°C; not shown) or at 31°C (Fig. 5A), no channels were observed. This suggests that the glycine-for-threonine substitution in this mutant prevents the virus from interacting with lipid bilayers in a channel-productive manner. The inability to form channels of 4028T.G may be due to the threonine residue being essential to channel structure or to the fact that any substitution at residue 4028T would prevent the 160S particles from productively interacting with lipid bilayers. Thus, mutants 4028T.S and 4028T.V were also examined (Fig. 6A and C). Ion channel openings were observed for both viable 4028T mutants at 31°C. However, a comparison of the traces of current as a function of time clearly showed that the 4028T.S and 4028T.V mutant channels are very different from each other and from the ones made by the WT virus.
FIG. 5.
Electrical characteristics of bilayers exposed to 4028T.G virus particles. Planar bilayers were formed at 31°C and their conductances were checked for a minimum of 30 min. Voltage = +40 mV. Viral particles were added. (A) Current-versus-time traces of a bilayer in the absence of virus (first two traces, control) and the same bilayer after addition of 4028T.G virus (107 particles/ml), both at 31°C. (B) The resultant current amplitude distributions of the bilayers in the absence of virus (left panel) and in the presence of 4028T.G virus (right panel) are shown.
FIG. 6.
Electrical characteristics of bilayers exposed to 4028T.S and 4028T.V particles. (A) The panel shows the current-versus-time traces at 31°C in the presence of 4028T.S (∼5 × 108 particles/ml) in response to voltages of +40 mV and −40 mV. The virus was added at room temperature, and after 40 min, the temperature was increased to 31°C. (B) Conductance-versus-voltage plot obtained from the current amplitude histograms as described in Materials and Methods and legend of Fig. 4. (C) Current-versus-time plot for a bilayer exposed to 4028T.V (∼2 × 109 particles/ml) at 31°C. The current amplitude distributions corresponding to voltages of +40 mV and −40 mV for one membrane (5-min pulse duration) are shown in panel D, together with the Gaussian fit to the minimum current amplitude obtained. See text for further details.
The channels made by 4028T.S (Fig. 6A) have a short lifetime and open sparingly (mean open probability ≈ 0.02). Furthermore, when opened, the channels showed fast closings and openings with a frequency that was too high for the current amplitude to be properly resolved (not shown). This was reflected in the large standard deviations for the single-channel conductances as shown in the plot of conductance versus voltage (Fig. 6B). The mean value of conductance of the 4028T.S channels at the lower voltages, 28 ± 10 pS, is lower than the 47-pS value determined for the Mahoney WT parent (or Sabin type 3) virus channels. The other important difference between the WT and 4028T.S mutant channels is that the mutant channels open both at positive and at negative potentials, whereas the WT channels open only at positive potentials (Fig. 6A and B). Even more striking are the channels induced by mutant 4028T.V (Fig. 6C and D). The four traces of current as function of time showed that these channels remained open for long periods of time (>0.5 s). In addition, the channels did not close to the value of current that was obtained for bilayers in the absence of virus (indicated by the solid line [0.06 to 1 pA at 40 mV]), as was the case for the channels induced by WT, Sabin type 3, and 4028T.S viruses. Rather, the 4028T.V channels closed at a higher current amplitude (1.2 to 2.5 pA at 40 mV) that was invariant with the voltage amplitude (Fig. 6D) (current amplitude distribution). The channels induced by the 4028T.V virus open both at positive and at negative potentials and have a conductance of 15 pS that is independent of voltage.
RNA delivery from 4028T.G particles is defective.
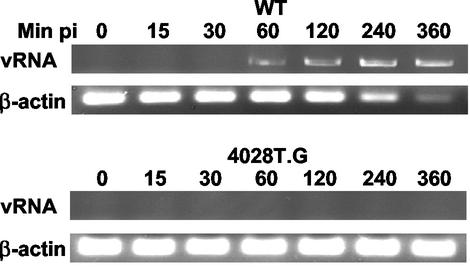
Previous studies showed that transfection of the 4028T.G genome into cells led to synthesis and assembly of mutant 160S particles. Subsequent binding of these particles to the PVR led to formation of 135S particles, demonstrating that these mutant particles are able to bind receptor and to undergo the receptor-dependent, conformational changes at 37°C (22). These data indirectly suggested that the 4028T.G mutant was defective during the penetration and uncoating stages of the entry process. To confirm that cytoplasmic delivery of the viral genome was defective in 4028T.G virus-infected cells, RNA was extracted from mutant- and WT virus-infected cells and analyzed by RT-PCR (Fig. 7). In WT virus-infected cells, viral RNA was detected within 60 min p.i. and the signal continued to increase as genome replication and transcription occurred over the 6-h infection period. At all times examined, no viral RNA sequences were detected in 4028T.G virus-infected cells. The sensitivity of this RT-PCR assay for viral RNAs increases at later times p.i. due to the increased numbers of viral replicons. Because mutant 4028T.G was replication competent when the viral genome was transfected into cells (22), the absence of detectable viral RNA sequences in mutant-infected cells even at late times p.i. indicated that the virus failed to successfully deliver the viral genome into the cytoplasm to initiate viral replication. In addition, the absence of viral replication and translation in cells infected with the 4028T.G mutant was indirectly corroborated by the levels of β-actin RNA seen in WT virus- and mutant virus-infected cells. The decreases in β-actin signal observed at late times p.i. in WT virus-infected cells were consistent with inhibition of host transcription due to cleavage of host transcription factors by the 3C viral protease (31, 32). In contrast, there was no change in the β-actin mRNA levels in 4028T.G-infected cells over time. In addition, no viral protein expression was detected in 4028T.G-infected cells by Western blot analyses (data not shown). These results support the interpretation of previous studies that the glycine substitution for threonine-28 in VP4 results in a virus that is unable to uncoat and successfully deliver its RNA genome into the cell.
FIG. 7.
Analyses of viral RNA (vRNA) synthesis in 4028T.G-infected cells. RNAs were extracted at the indicated times p.i. from either WT- or 4028T.G-infected cells. RNA samples from equivalent numbers of infected cells were analyzed by RT-PCR to detect viral RNAs or β-actin mRNAs. The products were resolved on a 1.5% agarose gel.
Entry kinetics of viable 4028T mutants.
The observation that mutant 4028T.G was defective in genome uncoating and/or delivery and unable to form channels suggests that the ability to form channels and to uncoat the viral genome might be related. If so, then the differences in the electrical properties of the 4028T.S and 4028T.V ion channels suggest that genome uncoating and delivery might also be altered in these mutants as well. Previous studies had shown that infections by poliovirus particles grown in NR dye (NR virus) are photosensitive (6, 30) and that infections with NR-160S or NR-135S particles become photoresistant within approximately 30 to 40 min p.i., with half-life (t[1/2]) for 160S equal to 16 to 19 min p.i. and t[1/2] for 135S equal to 14 to 17 min p.i. (13). In addition, the period of photosensitivity is lengthened in infections by viral capsid mutants displaying uncoating defects (15). Thus, the length of time that the viral infection remains photosensitive is an indirect measure of the overall kinetics of virus entry and reflects the combined rates of the 160S-to-135S conformational transition and the other stages leading to genome uncoating and delivery.
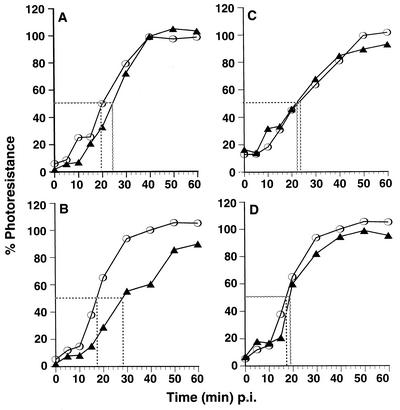
The lengths of the photosensitive periods for infections by the viable NR-4028T mutant viruses were compared with those of the NR-WT virus (Fig. 8). The period of photosensitivity for infections by 4028T.V 160S particles was slightly longer (approximately 2 to 4 min) than that for the WT virus and would normally be of marginal significance (Fig. 8A). However, this difference was more apparent when infections were initiated with 4028T.V 135S virions (Fig. 8B). When cells are infected with 135S particles, the period of 4028T.V photosensitivity was prolonged (4028T.V t[1/2] for 135S = 25 to 28 min, whereas WT t[1/2] for 135S = 15 to 18 min, ranging over several experiments) and, as evidenced by the shallower slope, the rate that 4028T.V infections became photoresistant was slower than that measured in the parallel WT virus infection. When infections were initiated with 4028T.S 160S virions, both the kinetics of photoresistance as well as the overall periods of photosensitivity (as indicated by the t[1/2] values) were indistinguishable from those of the WT viruses (Fig. 8C). In addition, no consistent differences were detected in infections with 4028T.S versus WT NR-135S particles (Fig. 8D).
FIG. 8.
Kinetics of NR resistance. NR-labeled WT (open symbol) or NR-4028T mutant (filled symbol) viruses were bound to HeLa cell monolayers. Infection was initiated by addition of an agar overlay. At the indicated times p.i., the infection was exposed to white light for 10 min. Plaques were allowed to develop over 36 to 48 h in the dark and were subsequently stained and counted. Titers from plates kept entirely in the dark were considered 100%. (A and B) Infection with 4028T.V 160S and 135S particles, respectively. (C and D) Infection with 4028T.S 160S and 135S particles, respectively.
It is interesting that the mean open times of 4028T.V particles in the lipid bilayers was significantly longer than those for the WT Mahoney, Sabin type 3, and 4028T.S viruses. Although a larger number of mutants need to be examined, this suggests that a long mean open time of the channels may influence the uncoating kinetics. Mechanistically, the open channels may locally alter the electrical properties of the membrane environment and thus alter the events leading to uncoating.
DISCUSSION
The data here show that, during poliovirus entry into cells, receptor-triggered conformational changes lead to extrusion of VP4 sequences from the virion interior and subsequent insertion of this capsid protein into cellular membranes. The collective phenotypes of the 4028T mutants indicate that the VP4 protein affects the ability of the virus to form ion channels and is likely to contribute to the channel structure. The failure of 4028T.G to deliver its genome and the slower delivery of 4028T.V mutant genome suggest that the ability to form functional ion channels and the electrical properties of these channels might be related to the penetration and/or uncoating process of poliovirus entry.
Architecture of the ion channel.
The domains of the virus that form the ion channels are unknown. The N-terminal domain of VP1 has previously been postulated to form an ion channel (10). These domains can be modeled as amphipathic helices, which could insert into membranes to form channels. In addition, proteolytic removal of the VP1 N termini abolishes the ability of 135S particles to bind to liposomes, suggesting that liposome attachment is mediated through the VP1 N termini. Because VP4 is N-terminally modified with myristate, it was hypothesized that this lipid modification coupled with VP4's interactions with VP1 N termini in the virion was important to facilitate membrane insertion of VP1 during poliovirus entry (10). The data here strongly indicate that VP4's role during entry is more than just to facilitate VP1 insertion into membranes. The absence of ion channels in 4028T.G as well as the differences in the electrical characteristics of the 4028T.S and 4028T.V mutant ion channels result from relatively conservative substitutions of WT Mahoney virus VP4 threonine-28. In contrast, the ion channels induced by Sabin type 3 virus (whose VP4 protein contains 4-amino-acid-residue differences from the Mahoney strain but maintains threonine-28) are electrically indistinguishable from those induced by the Mahoney strain. Although possible, it is difficult to imagine how such striking differences in electrical properties could be achieved with single-amino-acid substitutions if VP4 was not one of the domains forming the ion channel. Thus, the data suggest that VP4 sequences contribute to the structure of the ion channels and are involved in the gating of these channels. The contributions of the VP1 N-terminal domains to the architecture of the ion channel remain to be defined.
Role of the ion channels and poliovirus entry.
Genetic studies have established the importance of the VP1 N-terminal domains during virus entry. Viruses with small deletions or missense mutations in the N terminus of VP1 show delayed kinetics of viral RNA release into the cytoplasm (15) or enhanced uncoating kinetics (5). Similarly, the entry-defective phenotypes of the 4028T.G and 4028T.V mutants here show that VP4 is also important during virus penetration and/or uncoating. Within the structure of the 160S virion, VP1 N-terminal domain and VP4 are in physical contact. The combined uncoating and RNA delivery phenotypes of the VP4 and VP1 N-terminal mutants suggest that these domains not only interact structurally within the particle but may be also functionally linked in the uncoating process. Thus, the relocation of VP4 and VP1 domains from the particle interior, triggered during infection upon PVR binding, may not be independent events but rather the coordinate externalization of a structurally defined unit (such as an ion channel) that is composed of both VP4 and VP1 N termini. The resultant insertion of this unit into membranes could be important for the uncoating process.
Two general models have been proposed as to the role of the ion channels in poliovirus entry (2, 14, 24). One model is that the channels provide a pathway for transport of the RNA genomes across the cell membrane into the cytosol. The second alternative model assumes that poliovirus can utilize an endosomal route of entry. In this situation, the channels may be important for release of the viral particle into the cytosol via lysis of the endosomal membrane. The data here do not differentiate between these models, and the specific role(s) of these ion channels during poliovirus uncoating remains to be defined.
Infectivity of the eluted 135S or A particle.
During infection, the majority of the input virus elutes from the cell surface as a 135S particle that is deficient in VP4. Detection of the majority of VP4 in cellular membrane compartments even after 135S particles have eluted from the membrane fractions now provides an explanation for the presence of the “VP4-minus” 135S eluted particle in the media. PVR-mediated formation of the 135S particles leads to the coordinate insertion of VP4 along with the VP1 N-terminal domains. If the interaction of myristoylated VP4 with membranes is stronger than its interactions with the 135S particle, then, when the 135S particle elutes back into the medium, the majority of the VP4 would remain associated with the membrane.
Previously, it was demonstrated that the eluted 135S virus particle, like the purified in vitro-converted 135S particle, is infectious in a PVR-independent manner (7). This infectivity can be neutralized by anti-VP4 antibody (data not shown). Because the Western blots here as well as data from many investigators indicate that VP4 is not detected in medium samples containing the eluted 135S particle, these observations would appear to be contradictory. It is possible that small amounts of VP4 (≤10%) remain associated with the eluted 135S particle. When a synthetic VP4 peptide of known concentration is used, standardization of the Western blots shown here indicates that the threshold of VP4 detection is between 3 and 6 ng of VP4 protein or approximately five or six copies of VP4/particle (data not shown). Thus, if less than 10% of the VP4 protein remained associated with the 135S particle in the media, it would not have been detected and might be sufficient to mediate infection by the 135S particle. Alternatively, the VP4 capsid precursor, VP0, is also found in the in vitro converted and eluted 135S particles. It is possible that it is the VP4 sequences in the three or four copies of VP0 protein present in the 135S particle, rather than the VP4 capsid protein per se, that are required for infection by poliovirus.
VP4 threonine-28 mutants.
Characterization of a number of site-directed capsid protein mutants has demonstrated that many of these mutations have multiple and often unexpected phenotypes (reference 3 and references therein). In this regard, the 4028T mutants are no different from other previously characterized capsid mutants, displaying perturbations in both virion assembly and virus entry pathways. Previous comparisons of the 4028T mutant viruses with the WT virus had shown that these mutants accumulated assembly-inefficient or incompetent assembly intermediates, leading to decreased yields and stabilities of 160S particles (20). Consistent with the decreased stability of the 160S particles, the rates of conversion from 160S to 135S particles are higher for 4028T mutants than for WT virus (data not shown) and suggest that the activation energy required for this conversion in the mutant particles is less than in WT Mahoney virus particles (28). Decreased stability of the 4028T mutant assembly intermediates and 160S particles is explained by the observed stabilizing interactions that VP4 threonine-28 forms within the resolved structures of the 160S virion (12) and the empty capsid assembly intermediate (1) and the presumed structure of the 15S pentamer assembly intermediate. Interestingly, assembly of the nonviable 4028T.G mutant is two to five times more efficient than for the viable mutants, suggesting that the effects of a glycine substitution on the stability of the assembly intermediates and assembly of 160S particles are not as pronounced as when a serine or valine replaces the WT threonine residue at this position.
However, the interactions formed by threonine-28 within the 160S particles are unlikely to explain the uncoating differences seen here during virus entry. The channel-forming abilities and the ion channel characteristics of the 4028T mutants as well as the kinetic differences in photosensitivity of infections with NR mutant 135S particles occur after the VP4 sequences are relocated from the particle interior and inserted into cellular membranes. Previous studies have demonstrated that the myristoyl modification and 9 residues at the VP4 N terminus are sufficient to target VP4 via intracellular trafficking pathways to detergent-insoluble membrane microdomains (19). It is possible that, during entry, VP4 sequences may be similarly targeted to detergent-insoluble membrane microdomains or lipid rafts after receptor binding on the extracellular surface of the plasma membrane. Thus, although the environment surrounding threonine-28 within the structure of the 135S particle and in membranes is unknown, it is likely to be different from that observed in the 160S structure. Consequently, the pleiotropic phenotypes for this set of mutants (stability of 160S particles and assembly intermediates versus NR-induced photosensitivity and ion channel characteristics) likely reflect the different chemical environments surrounding threonine-28 and the potentially different functions of the VP4 domains in the assembly pathway of 160S virions and in the virus entry pathway after formation of the 135S particle.
Acknowledgments
This work was supported by Public Health Service grant AI42390 from the National Institute of Allergy and Infectious Diseases and by a grant from the Dana Foundation.
We thank D. C. Tosteson and J. M. Hogle for helpful discussions and comments.
REFERENCES
- 1.Basavappa, R., R. Syed, O. Flore, J. P. Icenogle, D. J. Filman, and J. M. Hogle. 1994. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 A resolution. Protein Sci. 3:1651-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belnap, D. M., D. J. Filman, B. L. Trus, N. Cheng, F. P. Booy, J. F. Conway, S. Curry, C. N. Hiremath, S. K. Tsang, A. C. Steven, and J. M. Hogle. 2000. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 74:1342-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow, M., R. Basavappa, and J. M. Hogle. 1997. The role of conformational transitions in poliovirus pathogenesis, p. 157-186. In W. Chiu, R. Garcea, and R. Burnette (ed.), Structural biology of viruses. Oxford University Press, Oxford, United Kingdom.
- 4.Chow, M., J. F. Newman, D. Filman, J. M. Hogle, D. J. Rowlands, and F. Brown. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327:482-486. [DOI] [PubMed] [Google Scholar]
- 5.Couderc, T., F. Delpeyroux, H. Le Blay, and B. Blondel. 1996. Mouse adaptation determinants of poliovirus type 1 enhance viral uncoating. J. Virol. 70:305-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowther, D., and J. L. Melnick. 1962. The incorporation of neutral red and acridine orange into developing poliovirus particles making them photosensitive. Virology 14:11-21. [DOI] [PubMed] [Google Scholar]
- 7.Curry, S., M. Chow, and J. M. Hogle. 1996. The poliovirus 135S particle is infectious. J. Virol. 70:7125-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Sena, J., and B. Mandel. 1977. Studies on the in vitro uncoating of poliovirus. II. Characteristics of the membrane-modified particle. Virology 78:554-566. [DOI] [PubMed] [Google Scholar]
- 9.Egger, D., L. Pasamontes, M. Ostermayer, and K. Bienz. 1995. Reverse transcription multiplex PCR for differentiation between polio- and enteroviruses from clinical and environmental samples. J. Clin. Microbiol. 33:1442-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fricks, C. E., and J. M. Hogle. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 A resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 13.Huang, Y., J. M. Hogle, and M. Chow. 2000. Is the 135S poliovirus particle an intermediate during cell entry? J. Virol. 74:8757-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalko, S. G., R. E. Cachau, and A. M. Silva. 1992. Ion channels in icosahedral virus: a comparative analysis of the structures and binding sites at their fivefold axes. Biophys. J. 63:1133-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirkegaard, K. 1990. Mutations in VP1 of poliovirus specifically affect both encapsidation and release of viral RNA. J. Virol. 64:195-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kräusslich, H.-G., C. Hölscher, Q. Reuer, J. Harber, and E. Wimmer. 1990. Myristoylation of the poliovirus polyprotein is required for proteolytic processing of the capsid and for viral infectivity. J. Virol. 64:2433-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Q., A. G. Yafal, Y. M.-H. Lee, J. Hogle, and M. Chow. 1994. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of these sequences at physiological temperature. J. Virol. 68:3965-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marc, D., G. Masson, M. Girard, and S. van der Werf. 1990. Lack of myristoylation of poliovirus capsid polypeptide VP0 prevents the formation of virions or results in the assembly of noninfectious virus particles. J. Virol. 64:4099-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin-Belmonte, F., J. A. Lopez-Guerrero, L. Carrasco, and M. A. Alonso. 2000. The amino-terminal nine amino acid sequence of poliovirus capsid VP4 protein is sufficient to confer N-myristoylation and targeting to detergent-insoluble membranes. Biochemistry 39:1083-1090. [DOI] [PubMed] [Google Scholar]
- 20.Moscufo, N., and M. Chow. 1992. Myristate-protein interactions in poliovirus: interactions of VP4 threonine 28 contribute to the structural conformation of assembly intermediates and the stability of assembled virions. J. Virol. 66:6849-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moscufo, N., J. Simons, and M. Chow. 1991. Myristoylation is important at multiple stages in poliovirus assembly. J. Virol. 65:2372-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moscufo, N., A. G. Yafal, A. Rogove, J. Hogle, and M. Chow. 1993. A mutation in VP4 defines a new step in the late stages of cell entry by poliovirus. J. Virol. 67:5075-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, New York, N.Y.
- 24.Rueckert, R. R. 1990. Picornaviruses and their replication, p. 507-548. In B. N. Fields and D. M. Knipe (ed.), Virology, 2nd ed. Raven Press, New York, N.Y.
- 25.Sarnow, P. 1989. Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J. Virol. 63:467-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simons, J., A. Rogove, N. Moscufo, C. Reynolds, and M. Chow. 1993. Efficient analysis of nonviable poliovirus capsid mutants. J. Virol. 67:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tosteson, M. T., and M. Chow. 1997. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J. Virol. 71:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsang, S. K., P. Danthi, M. Chow, and J. M. Hogle. 2000. Stabilization of poliovirus by capsid-binding antiviral drugs is due to entropic effects. J. Mol. Biol. 296:335-340. [DOI] [PubMed] [Google Scholar]
- 29.Wetz, K., and T. Kucinski. 1991. Influence of different ionic and pH environments on structural alterations of poliovirus and their possible relation to virus uncoating. J. Gen. Virol. 72:2541-2544. [DOI] [PubMed] [Google Scholar]
- 30.Wilson, J. N., and P. D. Cooper. 1963. Aspects of the growth of poliovirus as revealed by the photodynamic effects of neutral red and acridine orange. Virology 21:135-145. [DOI] [PubMed] [Google Scholar]
- 31.Yalamanchili, P., U. Datta, and A. Dasgupta. 1997. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J. Virol. 71:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yalamanchili, P., K. Weidman, and A. Dasgupta. 1997. Cleavage of transcriptional activator Oct-1 by poliovirus encoded protease 3Cpro. Virology 239:176-185. [DOI] [PubMed] [Google Scholar]