Abstract
Transcription initiation of the copy-number control and better-than-random segregation genes of the broad-host-range and low-copy-number plasmid pSM19035 are subjected to repression by the autoregulated pSM19035-encoded ω product in Bacillus subtilis cells. The promoters of the copS (Pcop1 and Pcop2), δ (Pδ), and ω (Pω) genes have been mapped. These promoters are embedded in a set of either seven copies of a 7-bp direct repeat or in a block consisting of two 7-bp direct repeats and one 7-bp inverted repeat; the blocks are present either two or three times. The cooperative binding of ω protein to the repeats on the Pcop1, Pcop2, Pδ, and Pω promoters represses transcription initiation by a mechanism that does not exclude σARNAP from the promoters. These results indicate that ω protein regulates plasmid maintenance by controlling the copy number on the one hand and by regulating the amount of proteins required for better-than-random segregation on the other hand.
Keywords: plasmid replication, Gram-positive bacteria, protein–DNA interactions, transcriptional repressor
Naturally occurring plasmids are usually stably maintained in their bacterial hosts. This stability often must be accomplished in spite of a very low number of plasmid copies per cell. Replication-control mechanisms play a very important role here by ensuring a constant number of plasmid copies per chromosome for segregation to each daughter cell (reviewed in refs. 1–4). Additionally, several specialized plasmid stabilization modes have been identified. They can be divided into three major classes, a, b, and c. Class a groups those functions that maximize the number of plasmid units to be segregated between the daughter cells (reviewed in refs. 4–6). In class b, the plasmid encodes a toxin that kills the plasmid-free progeny and an antitoxin that neutralizes its cognate toxin or prevents its synthesis (7). Class c includes active plasmid partition systems that ensure the localization of at least one plasmid molecule at the site of future septal growth (cell-quarter site) of the daughter cells (8, 9). The class b and c systems have been described for several plasmids of Gram-negative bacteria, including P1, F, RK2, and R1 (1–4, 7). The mechanisms used by plasmids of Gram-positive bacteria to ensure that each cell receives at least one plasmid copy at cell division (better-than-random segregation) are poorly understood.
The low-copy-number and broad-host-range plasmids of the inc18 family (pIP501, pAMβ1, and pSM19035) from Gram-positive bacteria share an extensive sequence identity (>90%) in their replication (Cop, RNA III, Rep, ori; Fig. 1A) and stability (SegA and SegB) regions (10–12). pSM19035 has extraordinarily long inverted repeated sequences that comprise ≈80% of the plasmid molecule. The sequences required to ensure plasmid replication and its stable maintenance are localized within the inversely repeated segments (13, 14), which means that they are duplicated in the plasmid. Replication is controlled by two negative regulators, each of which reduces the amounts of mRNA coding for the initiation protein, Rep. One of them, Cop, represses transcription of the rep mRNA, whereas the other one is a stable antisense RNA (RNA III) that induces transcriptional attenuation within the leader region of the rep mRNA (11, 15–17). In the absence of Cop, the increased (derepressed) transcription of rep mRNA interferes, in cis, with initiation of transcription of RNA III (convergent transcription). Thus, the Cop protein limits the amounts of rep mRNA either directly (as a repressor) or indirectly, by affecting the intensity of transcription necessary to produce RNA III. The factor(s) that would control the synthesis of Cop has not been yet described.
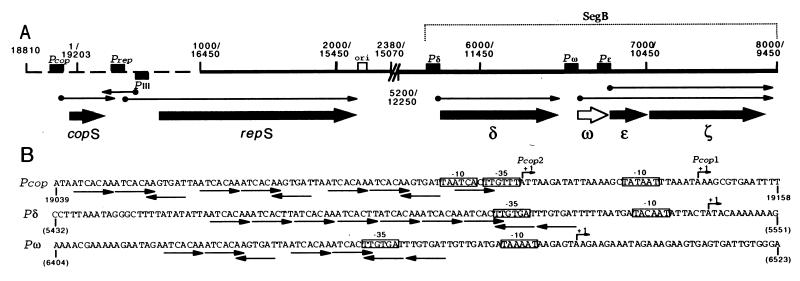
Figure 1.
Physical map of replication and better-than-random segregation genes of plasmid pDB101. (A) pDB101 has extraordinarily long inverted repeated sequences that comprise about 76% of the plasmid molecule (10). The single-copy DNA is indicated as a broken line. The duplicated sequences are denoted by a continuous line, and by double coordinates. The promoters (Pcop, Prep, PIII, Pδ, Pω, and Pɛ) and the plasmid replication origin (ori) are denoted by filled and empty boxes, respectively. RNA transcripts are indicated by thin arrows, and the gene products (copS, repS, δ, ω, ɛ, and ζ) are denoted by thick arrows. The filled arrows denote previously identified products and the open arrow, the product identified in this work. The SegB region is indicated. (B) Nucleotide sequence of the Pcop (Pcop1-Pcop2), Pδ, and Pω promoters. The transcription start sites of Pcop1, Pcop2, and Pδ and Pω, determined by primer extension assays, are denoted by bent arrows. Conserved −35 and −10 regions are indicated as open boxes. The coordinates are indicated. The heptamers and their relative orientation are indicated by thin arrows below the nucleotide sequence.
Two discrete regions, SegA and SegB, are involved in the segregation stability of pSM19035 (13). SegA is required for the conversion of oligomeric forms into monomers (class a; ref. 5). The SegB region, which assures better-than-random plasmid pSM19035 distribution, encompasses the δ, ω, ɛ, and ζ genes (refs. 13, 14; this report; Fig. 1A). The δ protein shares a high degree of homology with the family of ATPases involved in partitioning of diverse bacterial plasmids and bacterial chromosomes (class c; refs. 13, 18, 19), whereas the ɛ and ζ gene products prevent the appearance of plasmid-free segregants (class b; see above) (unpublished results).
In this report, we show that a pSM19035-borne gene ω product represents a new element involved in the regulation of pSM19035 replication. We demonstrate both in vivo and in vitro that, in addition to repressing its own synthesis, ω protein also blocks transcription from genes copS and δ. Our observations indicate that the ω protein is a negative regulator linking plasmid copy control to better-than-random segregation.
Materials and Methods
Bacterial Strains and Plasmids.
Escherichia coli strains DH5α and BL21(DE3) and Bacillus subtilis strains YB886 and YB1015 (recA4) were used (13, 20). Plasmids pDB101, pBT233, pBT291, pBT346, pBT312, and pBT347 (10, 13, 14), pBT338 (21), and pBT205 (S. Chai and J.C.A. unpublished results) were used. The plasmid-borne Pcop (pUC57, coordinates 18976 to 64 of pDB101), Pδ (pBT291, 4735 to 5635), and Pω (pCB30, 6377 to 6754) were used for promoter analysis. Plasmid pRC1 was constructed by in vitro deletions of pDB101 and is composed of the copS, repS, and erm genes (coordinates 16738-2546 and 7824–9622). The plasmid-borne δ and ω genes (pBT346–1, coordinates 4685–6754), ω and ɛ genes (pBT346–2, 5800–7081), ω gene (pBT346–3 and pT7-ω, 6312–6894), and ɛ gene (pBT346–4, 6595–7081) were constructed by in vitro deletions of pBT346. Plasmid pBT346–5, which is derived from pBT346–3, contains a STOP TAA codon at the sixth triplet of the ω gene. The 5′ noncoding region of copS (18560 to 1), δ (4695 to 5800), ω (6312 to 6522), and ɛ (6529 to 6899) genes were fused to the promoterless lacZ gene of E. coli plasmid pBT205.
Bacterial Growth, Transformation, and Plasmid Copy Number.
B. subtilis and E. coli cells were grown and made competent as described previously (20). The promoterless lacZ and its derivatives (cop:lacZ, δ:lacZ, ω:lacZ, and ɛ:lacZ fusions), present in pBT205, were integrated by a double-crossover into the amyE locus of YB886 strain. In a second step, the recA4 mutation was introduced into the background. The ω gene (6312–6727) was integrated via plasmid pBT205 into the chromosome of YB886 to construct strain YB886-ω.
YB886 and YB886-ω strains bearing plasmid pRC1 were grown to the middle exponential phase, and plasmid DNA from 5-ml cultures (OD650 = 0.8) was isolated. The relative plasmid copy number was obtained by comparing the intensity of the ethidium bromide-stained plasmid DNA bands (HindIII cleavage) with image quant (Molecular Dynamics) software.
β-Galactosidase Assays.
Aliquots of B. subtilis cells grown to OD600 = 0.5−1.0 were pelleted and resuspended in buffer Z (22) containing 0.4 mg/ml lysozyme. After 5-min incubation at 37°C, Triton X-100 (final concentration 0.08%) was added, mixed, and the lysates clarified by 5-min centrifugation at 12,000 × g. β-Galactosidase was assayed in the cell lysates as described (22).
DNA, RNA, Enzymes, and Reagents.
Covalently closed circular plasmid DNA was purified as described (20). DNA restriction and modification enzymes, ATP, RNaseA, and poly[d(I/C)] were purchased from Boehringer Mannheim. Gel-purified DNA fragments were end labeled as described (20). The concentration of DNA was determined by using molar extinction coefficients of 6,500 M−1⋅cm−1 at 260 nm and expressed as mol of DNA molecules.
The in vivo transcriptional start site of the promoter δ gene was determined by using 5′ rapid amplification of cDNA ends, Ver. 2.0 (GIBCO/BRL), according to the manufacturer.
The soluble 8-kDa ω protein (predicted molecular mass, 7,956 kDa) was purified by conventional column chromatography (phosphocellulose and Superose 12). The ω protein concentration was determined by using a molar extinction coefficient of 2,560 M−1⋅cm−1 at 280 nm and expressed in mol of ω protein tetramers. The NH2-terminal amino acid sequence of the ω protein was determined by automated Edman degradation.
Molecular Mass Determination.
Gel filtration chromatography was carried out in 50 mM Tris⋅HCl, pH 7.5/150 mM NaCl at 4°C with a flow rate of 0.5 ml/min, and the A280 was measured. Ten and twenty micrograms of ω protein (3 and 6 nmol) was applied onto the Superose 12 column (Pharmacia). A standard curve of Kav vs. log of molecular masses was determined as recommended by Pharmacia. Equilibrium centrifugation of ω protein (2, 4, and 6 μM) in 25 mM Tris⋅HCl, pH 7.5/150 mM NaCl was analyzed in a Beckman Optima XL-A (Beckman Coulter) operating at 8,000 and 15,000 rpm (An-Ti60 rotor) for 12 hr at 20°C.
Primer Extension Assays.
Transcription reactions were performed in a 25-μl final volume in buffer B [25 mM Tris⋅HCl, pH 7.5/10 mM MgCl2/80 mM KCl/5 mM (NH4)2SO4/2% glycerol/12 mM NaCl] containing ATP, CTP, GTP, and UTP (200 μM of each), 2 nM of supercoiled plasmid DNA (pUC57, Pcop; pBT291, Pδ; pCB30, Pω or pBT338, Pαβ control), and 20 nM of purified B. subtilis αARNAP. The reactions were incubated for 15 min at 37°C with or without increased amounts of ω protein. The reactions were stopped by the addition of 1 μl of 0.5 M EDTA, 1 μg of carrier tRNA, 3 μl of 3 M sodium acetate, and 90 μl of ethanol to precipitate the RNA.
The primers used (5′-TACTCATTTCTAATTCT-3′ at coordinates 49 to 64, 5′-CAATTGCTTCATTTTTATTGC-3′ at coordinates 5615 to 5635, and 5′-ATTGGTGTATGAATTCGTTTTGCTTTTT-3′ at coordinates 6537 to 6564) hybridized downstream of the Pcop, Pδ, and Pω promoters, respectively. The annealing and primer extension reactions were performed as described (20). The cDNA was analyzed by electrophoresis in 6% urea-polyacrylamide gels (6% dPAGE) and detected by autoradiography.
Electrophoretic Mobility-Shift Assays (EMSA).
For EMSA experiments, the end-labeled DNA (0.2 nM) was incubated in buffer C (50 mM Tris⋅HCl, pH 7.5/10 mM MgCl2) containing 50 mM NaCl for 15 min at 37°C with different amounts of purified proteins: ω and/or σARNAP in the presence of 1 μg of poly d(I/C) as a nonspecific competitor DNA in a 20-μl final volume and immediately loaded on the gel (19). The samples were separated on a 4% nondenaturing polyacrylamide (80:1 acrylamide/bis) gel (ndPAGE). Gels were run with 1× Tris-acetate-EDTA (TAE) at 150 V at room temperature and dried before autoradiography.
DNase I Footprinting.
The reaction conditions were as for EMSA. The proteins added and their concentrations are specified in the figure legends. The 300-bp HindIII-KpnI (comprising the Pcop1 and Pcop2 promoters), the 317-bp EcoRI-SacI (Pδ), and the 426-bp HindIII-KpnI (Pω) DNA fragments were end labeled. DNaseI treatment was as previously described (23). The samples were analyzed by 6% dPAGE and dried before autoradiography.
Results
In Vivo Activity of Promoters from the Copy Control and SegB Region.
The complete nucleotide sequence of pDB101 revealed that the promoters of the copy control (copS) and of SegB share common potential regulatory sequences (ref. 10; Fig. 1B). To address which of the genes of the SegB region (except the ζ gene, which is translationally coupled to the ɛ gene), as well as the copS gene, are subjected to transcriptional control and what product is required for such a regulation, the 5′ noncoding region of these genes were fused to a promoterless lacZ gene, integrated as single copies into the amyE locus of the B. subtilis chromosome, and different combinations of plasmid-borne genes were then provided in trans. β-Galactosidase assays from the lysates of analyzed strains revealed that: (i) the fusions exhibit promoter activities with high (Pδ:lacZ), moderate (Pcop:lacZ and Pω:lacZ), and low (Pɛ:lacZ) β-galactosidase values; (ii) transcription from the promoters of the copS, δ, and ω genes is repressed by a plasmid-encoded ω gene; (iii) a point mutation in the ω gene (pBT346–5) suppresses transcriptional repression; and (vi) transcription of the promoter upstream of the ɛ gene is not directly controlled by the ω protein (Table 1).
Table 1.
Expression in vivo of the Pcop (Pcop1-Pcop2), Pδ, Pω, and Pɛ promoters fused to lacZ
| Genes provided in trans | β-galactosidase activity*
|
|||
|---|---|---|---|---|
| Pcop | Pδ | Pω | Pɛ | |
| None | 329 | 3561 | 274 | 43 |
| δωɛζ | 24 | 26 | ND | 37 |
| δω | 39 | 54 | ND | 44 |
| ωɛζ | 52 | 57 | 8 | 33 |
| ωɛ | 71 | 118 | 12 | 52 |
| ɛ | ND | 3484 | ND | 39 |
| ω | 47 | 50 | 7 | 34 |
| Δω† | 308 | 3613 | 412 | ND |
Activity specified by the tested promoters in the presence or absence of various products.
The β-galactosidase activity is expressed in Miller units.
† The sixth TTA codon of ω gene was converted into a TAA Stop triplet. ND, not done.
The ω Product and Its Putative Target Sites in pSM19035.
The ω gene is located in the SegB region between the δ and ɛ genes (Fig. 1A). It consists of 71 codons, begins with the putative “weak” GTG translation initiation triplet, and is preceded by no sequence homologous to a ribosomal loading site. The observation presented in Table 1, however, strongly suggests that ω is a bona fide gene, and that its translated product, ω protein, is most likely a transcriptional repressor.
The nucleotide sequences upstream of the copS, δ, and ω genes were compared with those of B. subtilis promoters (see refs. 24–26). Sequences showing putative −35 and −10 regions, which may be recognized by the vegetative form of B. subtilis σARNAP, were found (Fig. 1B). Features resembling a TG element are present in the Pδ and Pω promoters, and an AT-rich region is present upstream of the putative promoters (UP element) (ref. 27; Fig. 1B).
The promoter regions of the copS, δ, and ω genes are embedded in a set of two types of 7-bp conserved repeats, 5′-ATCACAA-3′ (type A) or 5′-ATCACTT-3′ (type B), and such heptameric sets of repeats in plasmid pDB101 are present only upstream of these three genes (ref. 10; Fig. 1B). Upstream of the copS and ω genes, there are 10 and 7 copies of both types of heptamers in both head-to-tail and head-to-head configurations, whereas upstream of the δ gene, there are seven copies of both types of heptamers in a head-to-tail configuration plus two head-to-head copies (Fig. 1B).
Purification and Properties of the ω Protein.
The ω protein (predicted molecular mass, 7,956 Da) of pSM19035 was purified. The ω polypeptide was more than 99% pure, as judged by SDS/PAGE and quantitative analysis of the NH2-terminal amino acid composition. The sequence of the first 10 NH2-terminal residues of the purified protein is in perfect agreement with the prediction from the nucleotide sequence of the ω gene.
The molecular mass of native ω protein was estimated by gel filtration by using a Superose 12 column. From the elution profile of the ω protein and a number of protein standards, we estimate that the Mr of the ω protein is approximately 32,000, four times that of the ω protomer. Using equilibrium centrifugation, we confirmed that the protein is largely tetrameric at high protein concentrations and largely monomeric at low protein concentrations.
ω Protein Binds Cooperatively to the Promoters of the copS, δ, and ω Genes.
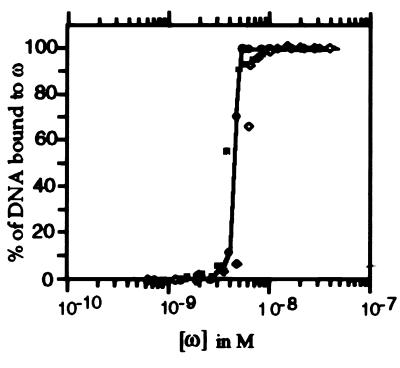
The affinity of protein ω for the Pcop, Pδ, and Pω [32P]-DNA fragments (0.2 nM) was determined by EMSA assays by following complex formation as a function of ω protein concentration. As revealed in Fig. 2, at ω protein concentrations lower than 1 nM, no binding was detected, whereas at higher protein concentrations, the amount of protein ω-DNA complex formed increased exponentially until a plateau was observed. The exponential increase in complex formation suggests that the ω protein binds to its target sequences in a cooperative manner. The apparent equilibrium constant (Kapp) of the protein ω–DNA complex formation was estimated to be similar for the three DNA segments analyzed and approximately 4 nM (Pcop1 Pcop2 3.6 nM, Pδ 4.6 nM and Pω 5.5 nM), pH 7.5, in the presence of 50 mM NaCl and at 37°C (Fig. 2).
Figure 2.
Cooperative binding of ω protein to the promoter region of copS (shaded squares), δ (filled circles), and ω (empty rhomboids). DNA fragments spanning the promoter region of copS, δ, and ω genes (0.2 nM) were radiolabeled and incubated with 1 μg of poly [d(I/C)] as nonspecific competitor DNA and an increasing concentration of the ω protein (0.2–200 nM) in buffer C containing 50 mM NaCl for 15 min at 37°C. The signals present in the protein–DNA complex and in the free DNA were determined by densitometry.
ω Protein Represses the copS, δ, and ω Promoters.
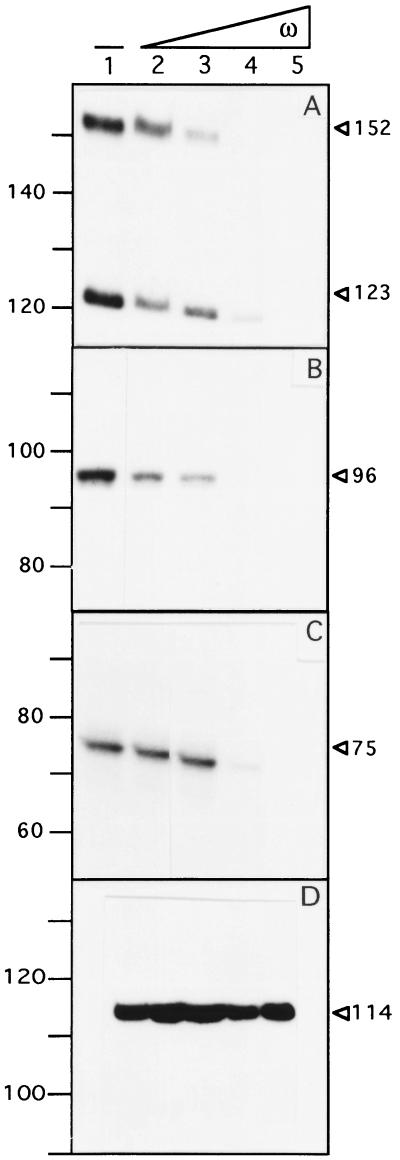
To map the transcription start points of the putative “Pcop,” “Pδ,” and “Pω” promoters in vitro and to determine whether the ω protein regulates promoter utilization, we analyzed, by primer extension, the transcripts produced in vitro by B. subtilis σARNAP holoenzyme (20 nM) on a supercoiled DNA substrate (2 nM). mRNA species of 152 nt and 123 nt (Pcop), 96 nt (Pδ), and 75 nt (Pω) in length were detected (Fig. 3 A–C). From the position of the primers, we could infer that the 123-nt, 96-nt, 75-nt, and 152-nt long transcripts start at positions corresponding to the predicted “Pcop1,” “Pδ,” and “Pω” promoters and at a new site termed “Pcop 2,” respectively (see Fig. 1B). This is in good agreement with the in vivo mapping of promoter Pcop1 (see ref. 11), Pδ, and Pω by using the 5′ rapid amplification of cDNA ends system (this work).
Figure 3.
Primer extension assay of the Pcop1-Pcop2, Pδ, Pω, and Pαβ in the absence or presence of the ω protein. The supercoiled plasmid DNA (2 nM) was incubated with 20 nM of σARNAP in the absence or presence of increasing concentrations of ω protein (15, 30, 60, and 120 nM) and subjected to in vitro transcription followed by primer extension. In A, Pcop1-Pcop2; in B, Pδ; in C, Pω; and in D, Pαβ as a nonspecific control. The lengths of the cDNAs obtained in the presence or absence of ω are indicated. The length of the standards in the region of interest are indicated. −, absence of protein ω.
As shown in Fig. 3 A–C, a regulatory effect of increased concentrations of the ω protein (15, 30, 60, and 120 nM) on transcription from the in vitro Pcop1, Pcop2, Pδ, and Pω promoters in the purified system by using the vegetative B. subtilis σARNAP (20 nM) and supercoiled pUC57 (Pcop1 and Pcop2), pBT291 (Pδ), and pCB30 (Pω) DNA template was observed. The addition of 7.5 and 15 ω tetramers per plasmid molecule reduced by about half the transcription initiation of the Pδ and the Pcop2 promoters, respectively (Fig. 3 A and B). Transcription initiation of the Pcop1 and the Pω is repressed in the presence of 30 ω tetramers (Fig. 3 A and C). Concentrations of ω protein equal to or higher than those required to repress the promoters of the copS, δ, and ω genes did not affect the expression of an unrelated promoter (Pαβ of the SegA region; ref. 21) (Fig. 3D). From the results presented in Fig. 3D, we can rule out that a contaminant RNase or any other nonspecific effect could be responsible for the lack of RNA synthesis at higher ω concentrations.
ω Protein Does Not Affect the Binding of σARNAP to the Pcop, Pδ, and Pω Promoter Regions.
The potential transcriptional repression mechanisms can be divided into three general types (reviewed in ref. 28). Transcriptional repressors could act: (i) at the binding stage by inhibiting the binding of RNAP to the promoter region (steric hindrance); (ii) at post-RNAP binding; here the repressor binds to the promoter region together with RNAP in a way that hinders the formation of either the open complex or promoter clearance by RNAP (protein–protein interaction); or (iii) by influencing the DNA structure immediately adjacent to the promoter. To address the above hypotheses, we analyzed initially the complexes formed by σARNAP and ω to the Pcop1 Pcop2, Pδ, and Pω promoter by EMSA experiments. Similar coexistence of both ω protein and σARNAP on DNA were obtained for all three promoters, hence only the results of the best in vivo- and in vitro-characterized Pcop1-Pcop2 promoter (see refs. 11, 13) are shown (Fig. 4).
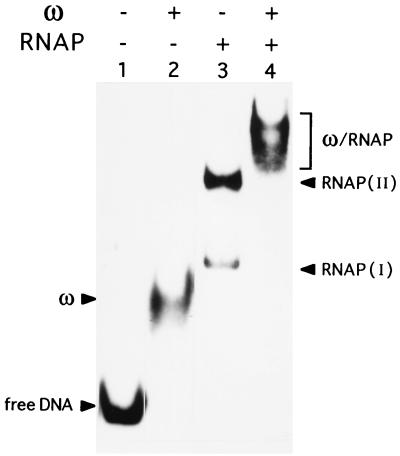
Figure 4.
The ω protein does not displace σARNAP from the Pcop1-Pcop2 DNA. [α32P]-Pcop1-Pcop2 DNA (2 nM) incubated with 60 nM ω protein (lane 2) or 60 nM σARNAP (lane 3) or with both the ω (60 nM) and σARNAP (60 nM) (lane 4) and the complexes analyzed by ndPAGE. The free DNA, ω, and σARNAP complexes RNAP (I) and (II) are indicated. + and − indicate the presence or absence of the denoted protein, respectively.
We incubated the [32P]-Pcop1-Pcop2 DNA (2 nM) with 60 nM of ω, 60 nM of σARNAP, or 60 nM of both proteins, and the reaction mixture was subjected to ndPAGE. In the presence of ω protein, all the DNA is complexed (Fig. 4, lane 2). In the presence of σARNAP, two complexes (indicated as I and II) are observed (Fig. 4, lane 3). When both ω and σARNAP proteins were present in the reaction mixture, a new diffuse and slowly moving complex was observed (Fig. 4, lane 4).
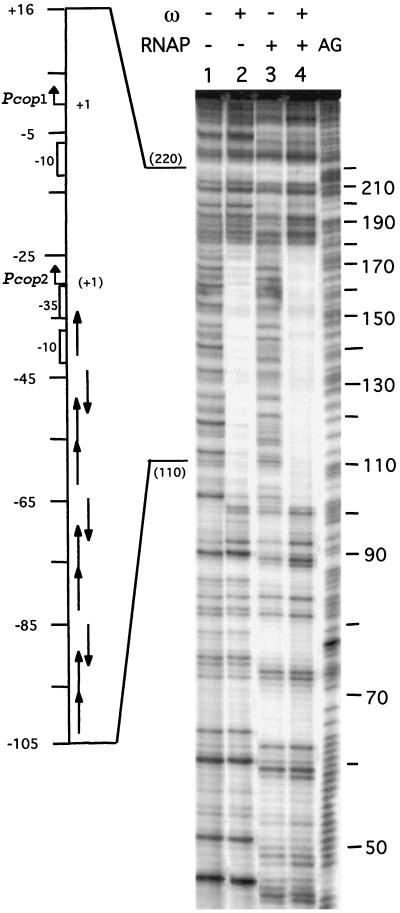
To localize the sequences recognized by protein ω, σARNAP, and both proteins, the protein–DNA complexes were analyzed by DNase I footprints. As shown in Fig. 5, lane 2, protein ω protected a contiguous region on Pcop1 Pcop2 DNA from the nuclease attack. This binding site was ≈78 bp (at positions −32 to −109) in length for the top strand, and ≈79-bp for the bottom strand (positions −33 to −109) (see Fig. 5, lane 2, coordinates 102–184). It is likely, therefore, that ω protein oligomerizes on the Pcop1-Pcop2 promoter region. σARNAP protection from nuclease attack matches the location of the Pcop1-Pcop2 promoter regions and also makes an extensive contact with the upstream region (positions +20 to −172) (see Fig. 5, lane 3). The presence of ω protein does not occlude the binding of σARNAP, because the presence of both proteins protects a contiguous region characteristic of the ω protein from nuclease attack in addition to the σARNAP protected region. The same general conclusion can be extended to the Pδ and Pω promoters.
Figure 5.
Interaction of σARNAP and the ω protein with Pcop1-Pcop2 DNA. [α32P]-Pcop1-Pcop2 DNA (2 nM) incubated with 60 nM ω protein (lane 2) or 60 nM σARNAP (lane 3) or with both ω (60 nM) and σARNAP (60 nM) proteins (lane 4). The location of the promoters and heptamers and their orientation and the transcription start sites are indicated at the side of the gel. The position +1 is the transcription start site of the Pcop1 promoter and the +1 position of promoter Pcop2 is denoted between parenthesis. The symbols + and − indicate the presence or absence of the denoted protein, respectively. AG, the A + G reaction loaded in parallel with the samples.
ω Protein Represses Pcop1-Pcop2 and Indirectly Increases Plasmid Copy Number.
We have demonstrated that a plasmid-encoded ω protein represses transcription of the negative copy-control element, CopS, about 7-fold (Table 1). A decrease in CopS synthesis predicts an increase in the number of plasmid copies to be segregated (see ref. 11). To address this hypothesis, a pBD101 minireplicon (pRC1, containing the copS, rnaIII, repS genes, and ori; Fig. 1A), which occurs at about three to five copies per cell, was introduced into the ω-free (YB886) and the chromosomal-borne ω gene (YB886-ω) and the number of plasmid copies in midexponential phase measured. In YB886-ω, a 5.5-fold increase in the number of copies of the plasmid pRC1 (22 ± 5 copies per cell) is observed when compared with the wild-type YB886 strain. Hence, if we consider that each daughter cell receives half the number of plasmid copies present at cell division and the frequency of formation of plasmid-free cells (L) as a function of the copy number (n) of the plasmid appear with a given probability [L = (0.5)2n] (see refs. 1–4), we have to consider that the expression of the chromosomally based ω gene may render the plasmid stably inherited in practice, because L values below 10−6 cannot readily be measured (13).
Discussion
A global control of vital plasmid functions such as replication, transfer, and stable maintenance has been reported only for the IncP plasmids from Gram-negative bacteria (4, 31). We have demonstrated in this communication that such global control also is present in the inc18 family from Gram-positive bacteria. However, here it functions in a different context, because a pSM19035-encoded ω product acts as a regulator of plasmid genes for copy-number control and stable inheritance. The ω protein represses the transcription of the negative copy-control element, CopS, about 7-fold. Replication in plasmids of the inc18 family is controlled by two elements (Cop and the stable antisense RIII), each of which reduces the amount of the rep mRNA (15, 17). The long-lived antisense RNA III and the constitutively expressed Cop protein, however, should be poor regulators, especially during the early phase of plasmid establishment or during situations when plasmid copy number falls below the wild-type level (11, 17). Unlike plasmids R1 (29) and pMV158(pLS1) (30), which carry both a short-lived antisense RNA and a repressor protein as copy-number control genes, in the inc18 plasmids the rep mRNA cannot be corrected by an active mechanism through the action of an upstream promoter. This can be achieved by gene ω, which maps outside of the minimal replicon (coordinates 18810 to 2380 in Fig. 1A) and is able to correct downward fluctuations in plasmid copy number because ω-mediated decreased synthesis of CopS will derepress transcription of the rep mRNA and indirectly decreases transcription of the antisense RNA III. We show that, indeed, repression of CopS synthesis by the ω protein correlates with an increase in plasmid copy number and indirectly ensures stable plasmid maintenance. It is likely that the interplay of RNA III, CopS, and ω protein is part of a negative-feedback control system of the basic replicon of inc18 plasmids.
The δ protein shares a significant degree of identity with the family of ATPases involved in plasmid partitioning (18). Unlike the ParA and SopA proteins that regulate expression of the P1 par and F sop operon, respectively, or IncA of plasmid RK2 that is indirectly involved in its regulation (1, 3, 31), the δ protein does not regulate its own synthesis. Transcription initiation of the δ gene is decreased about 70-fold by the presence of the ω product in trans. If δ protein is the functional counterpart of ParA, Sop, or IncA, we could hypothesize that an excess of δ destabilizes pSM19035 partition, as has been shown for ParA, SopA, and IncA (1–4).
We have demonstrated that the ω protein decreases it own transcription by about 40-fold. Because ω, ɛ, and ζ genes form an operon with two promoters, Pω reading the three genes and Pɛ only the ɛ and ζ genes (I.S. and P.C., unpublished results), and the expression of the ɛ and ζ genes prevents the appearance of plasmid-free segregants, the ω protein regulates most likely the expression of the two better-than-random segregation systems of pSM19035 (class b and c systems; see Introduction).
In vitro repression of transcription initiation at the Pcop2 and Pδ promoters requires an occupancy of about half of the ω recognition heptamers per plasmid molecule, because the amount of ω protein needed to repress transcription of both promoters is similar (about 15–30 tetramers) to the Kapp (18–22 ω molecules). Transcription initiation of the Pcop1 and Pω promoters is repressed in the presence of about 30–60 ω tetramers per DNA substrate (Fig. 3).
The purified ω protein binds with high cooperativity to the promoter region of the copS, δ, and ω genes with a similar affinity. A close look at the DNase I protected region revealed that the −32 to −109 of Pcop1-Pcop2 (Fig. 5), −20 to −85 of Pδ, and −20 to −70 of Pω regions (data not shown) are occupied by the ω protein, and an extended region is occupied by σARNAP. The extent of the overlap in the binding sites and the distribution of the heptamer repeat varies for each of the promoters. There are 10 and 7 copies of both types of heptamers upstream of the copS and ω genes (positions −34 to −103, with +1 being the start of Pcop1 and −22 to −68, with +1 being the start of Pω), whereas in the case of the Pδ promoter, there are nine copies of both types of heptamers (positions −22 to −82, with +1 being the start of Pδ). Our preliminary data suggest that the affinity of ω for its cognate site seems to be proportional to the number of copies of the direct repeat heptamer (A.d.l.H. and J.C.A., unpublished results).
We have shown that the 32-kDa ω tetramer binds with similar affinity to the asymmetric and directly repeated heptamers of the Pδ promoter and to the contiguous array of two heptamers in a direct repeat (head to tail) and one overlapping heptamer in an inverted repeat (head to head), present three times in the Pcop1 (see Fig. 1B). Each heptamer might constitute a unit for ω protein interaction, but the effective binding of ω protein to one or two heptamers is insufficient under EMSA for the formation of a cooperative nucleoprotein complex (unpublished results). We could envisage that binding of the ω tetramer requires at least three properly oriented heptamers. Such organization of binding sites has been well documented for eukaryotic transcription factors that are active as monomers [e.g., TFIIIA (32)], dimers [e.g., STAT family of proteins (33)], dimers or tetramers [e.g., TRE, VDR, RAR, RXR (retinoic acid, thyroid hormones, and vitamin D receptors) proteins (34–36)], or trimers [e.g., HSF protein (37)] that bind cooperatively to tandemly repeated units. Recognition of arrays of short DNA repeats by a specific transcriptional regulator, instead of a dimer or a tetramer binding to the two halves of a palindromic sequence, is rare in bacteria. The 40.6-kDa (dimeric) Fur repressor that binds to three to four 6-bp units (40), the 12.8-kDa ParR repressor-segregation protein that binds to two sets of 11-bp units (38), the 32-kDa (monomeric) RepA repressor-replication protein that binds to five 19-bp units (39), and the 32-kDa (tetrameric) ω repressor that binds to seven to nine 7-bp units (this work) are among the few described examples. In the case of the Fur repressor, it has been documented that it inhibits transcription initiation by blocking the entry of RNAP to the promoter (40). Here, we demonstrated that the ω tetramer does not occlude the binding of σARNAP to the promoter region.
Acknowledgments
We thank Margarita Salas and José M. Lazaro (Centro de Biología Molecular “Severo Ochoa,” Madrid, Spain) for the gift of B. subtilis σARNAP, H. Gaenze for performing the ω protein sequencing, Marta Lages for analytical ultracentrifugation of ω protein, and F. Studier (Brookhaven National Laboratory, New York) for the gift of BL21(DE3) strain. A.B.d.l.H. was the recipient of a Fellowship of the Gobierno Vasco, and S.F. and S.A., of the Comunidad de Madrid. This work was partially supported by Grants PB 96-0817 and BIO4-CT98-0250 to J.C.A. and Grant 6 P04A 027 09 from the Polish State Committee for Research to P.C.
Abbreviation
- EMSA
electrophoretic mobility-shift assay
References
- 1.Nordström K, Austin A. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- 2.Hiraga S. Annu Rev Biochem. 1992;61:283–306. doi: 10.1146/annurev.bi.61.070192.001435. [DOI] [PubMed] [Google Scholar]
- 3.Willians D R, Thomas C M. J Gen Microbiol. 1992;138:1–16. doi: 10.1099/00221287-138-1-1. [DOI] [PubMed] [Google Scholar]
- 4.Helinski D R, Toukdarian A E, Novick R P. In: Escherichia coli and Salmonella, Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, editor. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2295–2324. [Google Scholar]
- 5.Alonso J C, Ayora S, Canosa I, Weise F, Rojo F. FEMS Microbiol Lett. 1996;142:1–10. doi: 10.1111/j.1574-6968.1996.tb08399.x. [DOI] [PubMed] [Google Scholar]
- 6.Summers D. Mol Microbiol. 1998;29:1137–1145. doi: 10.1046/j.1365-2958.1998.01012.x. [DOI] [PubMed] [Google Scholar]
- 7.Jensen R B, Gerdes K. Mol Microbiol. 1995;11:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 8.Gordon G S, Sitnikov D, Webb C D, Teleman A, Straight A, Losick R, Murray A W, Wright A. Cell. 1997;99:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 9.Niki H, Hiraga S. Cell. 1997;90:951–957. doi: 10.1016/s0092-8674(00)80359-1. [DOI] [PubMed] [Google Scholar]
- 10.Ceglowski P, Alonso J C. Gene. 1994;145:33–39. doi: 10.1016/0378-1119(94)90319-0. [DOI] [PubMed] [Google Scholar]
- 11.Brantl S. Mol Microbiol. 1994;14:473–483. doi: 10.1111/j.1365-2958.1994.tb02182.x. [DOI] [PubMed] [Google Scholar]
- 12.Pujol C, Chédin F, Ehrlich S D, Janniére L. Mol Microbiol. 1998;29:709–718. doi: 10.1046/j.1365-2958.1998.00940.x. [DOI] [PubMed] [Google Scholar]
- 13.Ceglowski P, Boitsov A, Karamyan N, Chai S, Alonso J C. Mol Gen Genet. 1993;241:579–585. doi: 10.1007/BF00279900. [DOI] [PubMed] [Google Scholar]
- 14.Ceglowski P, Boitsov A, Chai S, Alonso J C. Gene. 1993;136:1–12. doi: 10.1016/0378-1119(93)90441-5. [DOI] [PubMed] [Google Scholar]
- 15.Brantl S, Behnke D. J Bacteriol. 1993;175:4052–4061. doi: 10.1128/jb.175.13.4052-4061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Chatelier E, Ehrlich S D, Janniére L. Mol Microbiol. 1994;14:463–471. doi: 10.1111/j.1365-2958.1994.tb02181.x. [DOI] [PubMed] [Google Scholar]
- 17.Brantl S, Wagner G H. J Bacteriol. 1997;179:7016–7024. doi: 10.1128/jb.179.22.7016-7024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motallebi-Veshareh M, Rouch D A, Thomas C M. Mol Microbiol. 1990;4:1455–1463. doi: 10.1111/j.1365-2958.1990.tb02056.x. [DOI] [PubMed] [Google Scholar]
- 19.Lin D C, Grossman A D. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Maniatis T, Fritsch E F. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Rojo F, Alonso J C. Nucleic Acids Res. 1994;20:1855–1860. doi: 10.1093/nar/22.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 23.Chai S, Lurz R, Alonso J C. J Mol Biol. 1995;252:386–398. doi: 10.1006/jmbi.1995.0505. [DOI] [PubMed] [Google Scholar]
- 24.Gross C A, Lonetto M, Losick R. In: Transcriptional Regulation. Yamamoto K, McKnight S, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 129–176. [Google Scholar]
- 25.Helmann J D. Nucleic Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monsalve M, Calles B, Mencía M, Salas M, Rojo F. Mol Cell. 1997;1:99–107. doi: 10.1016/s1097-2765(00)80011-8. [DOI] [PubMed] [Google Scholar]
- 27.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 28.Roy S, Garges S, Adhya S. J Biol Chem. 1998;273:14059–14062. doi: 10.1074/jbc.273.23.14059. [DOI] [PubMed] [Google Scholar]
- 29.Nordström K, Wagner G. Trends Biochem Sci. 1994;19:294–300. doi: 10.1016/0968-0004(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 30.del Solar G, Espinosa M. Mol Microbiol. 1992;6:83–94. doi: 10.1111/j.1365-2958.1992.tb00840.x. [DOI] [PubMed] [Google Scholar]
- 31.Thorsted P B, Macartney D P, Akhtar P, Haines A S, Ali N, Davidson P, Stafford P, Pocklington M J, Pansegrau W, Wilkins B M, et al. J Mol Biol. 1998;282:969–990. doi: 10.1006/jmbi.1998.2060. [DOI] [PubMed] [Google Scholar]
- 32.Rhodes D, Klug A. Cell. 1986;46:123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- 33.Xu X, Sun Y L, Hoey T. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 34.Umesono K, Murakami K K, Thompson C C, Evans R M. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Näär A M, Boutin J M, Lipkin S M, Yu V C, Holloway J M, Glass C K, Rosenfeld M G. Cell. 1991;65:1267–1279. doi: 10.1016/0092-8674(91)90021-p. [DOI] [PubMed] [Google Scholar]
- 36.Lin B C, Wong C W, Chen H W, Privalsky M L. J Biol Chem. 1997;272:9860–9867. doi: 10.1074/jbc.272.15.9860. [DOI] [PubMed] [Google Scholar]
- 37.Xiao H, Perisic O, Lis J T. Cell. 1991;64:585–593. doi: 10.1016/0092-8674(91)90242-q. [DOI] [PubMed] [Google Scholar]
- 38.Jensen R B, Dam M, Gerdes K. J Mol Biol. 1994;236:1299–1309. doi: 10.1016/0022-2836(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 39.Chattoraj D K, Mason R, Wickner S H. Cell. 1988;52:551–557. doi: 10.1016/0092-8674(88)90468-0. [DOI] [PubMed] [Google Scholar]
- 40.Escolar L, Pérez-Martin J, de Lorenzo V. J Mol Biol. 1998;283:537–547. doi: 10.1006/jmbi.1998.2119. [DOI] [PubMed] [Google Scholar]