Abstract
The cre(2C) hairpin is a cis-acting replication element in poliovirus RNA and serves as a template for the synthesis of VPgpUpU. We investigated the role of the cre(2C) hairpin on VPgpUpU synthesis and viral RNA replication in preinitiation RNA replication complexes isolated from HeLa S10 translation-RNA replication reactions. cre(2C) hairpin mutations that block VPgpUpU synthesis in reconstituted assays with purified VPg and poliovirus polymerase were also found to completely inhibit VPgpUpU synthesis in preinitiation replication complexes. Surprisingly, blocking VPgpUpU synthesis by mutating the cre(2C) hairpin had no significant effect on negative-strand synthesis but completely inhibited positive-strand synthesis. Negative-strand RNA synthesized in these reactions immunoprecipitated with anti-VPg antibody and demonstrated that it was covalently linked to VPg. This indicated that VPg was used to initiate negative-strand RNA synthesis, although the cre(2C)-dependent synthesis of VPgpUpU was inhibited. Based on these results, we concluded that the cre(2C)-dependent synthesis of VPgpUpU was required for positive- but not negative-strand RNA synthesis. These findings suggest a replication model in which negative-strand synthesis initiates with VPg uridylylated in the 3′ poly(A) tail in virion RNA and positive-strand synthesis initiates with VPgpUpU synthesized on the cre(2C) hairpin. The pool of excess VPgpUpU synthesized on the cre(2C) hairpin should support high levels of positive-strand synthesis and thereby promote the asymmetric replication of poliovirus RNA.
Poliovirus is a prototypic positive-strand RNA virus with a single-stranded genome that contains a 3′-terminal poly(A) tail and a 5′-terminal covalently linked protein, VPg (1, 15, 27). Once released into the cytoplasm of infected cells, poliovirion RNA serves as an mRNA for translation and then as a template for negative-strand synthesis. Multiple rounds of positive-strand synthesis on each molecule of negative-strand RNA result in the synthesis of excess genomic RNA. Overall, the ratio of positive- to negative-strand synthesis is greater than 30:1 (10, 24), which is essential for the production of high yields of virion RNA and progeny virus in infected cells.
cis-active replication sequences that are present at the 3′ and 5′ ends of poliovirion RNA play an important role in regulating poliovirus RNA replication. The 3′ nontranslated region (NTR) and the associated poly(A) tail are important cis-active determinants that are required for efficient negative-strand RNA synthesis (14, 21, 22, 28, 30, 36). The 5′-terminal cloverleaf structure in poliovirus RNA is a multifunctional element that is required in cis for negative-strand synthesis, RNA stability, and VPg uridylylation in membrane replication complexes (8, 14, 17, 23, 34). A new cis-acting replication element (cre) was recently identified in the genomes of several picornaviruses (9, 11, 16, 18, 20). The cre was originally identified in the P1 capsid coding region of HRV14 [cre(VP1)] (19). Mutational analysis of the cre hairpin structure suggested that the positive-sense structure was critical for function during viral RNA replication (20). Furthermore, the cre hairpin was position independent, and translation was not required for its function (20). Similar observations were made for the cre hairpins identified in other picornaviruses, including poliovirus [cre(2C)] (11, 16, 18). Therefore, multiple cis-active replication elements are required for various steps in the viral RNA replication cycle and for the specific recognition of viral RNAs in infected cells.
VPg is covalently linked to the 5′ terminus of all newly synthesized viral RNAs, and therefore it is generally assumed that VPg or a uridylylated form of VPg serves as a primer for the viral RNA polymerase, 3Dpol (1, 32, 33, 35, 37). Experimental evidence that VPg can serve as a primer for RNA synthesis was first demonstrated by Paul et al. (26), who showed that purified 3Dpol can utilize VPg and a poly(A) template to synthesize VPg-poly(U). In subsequent studies, it was shown that the cre hairpins in poliovirus and rhinovirus RNAs can function as effective templates for VPg uridylylation, especially in the presence of 3CDpro (9, 25, 29, 38). Based on this finding, it was suggested that VPgpUpU is first synthesized on the cre(2C) hairpin and then translocated to the 3′-terminal poly(A) tail to act as a primer for negative-strand synthesis (25). This model provided an explanation for the functional significance of the cre hairpins and an efficient mechanism for the synthesis of VPgpUpU.
Recently, we reported that a poliovirus RNA transcript that contained a large internal deletion in the polyprotein coding sequence was a functional template for negative-strand RNA synthesis (8). This was a surprising result, since the 2C coding sequence including the cre(2C) hairpin was deleted from this RNA. In this study, we conducted experiments to further investigate this interesting observation by examining the effect of mutations in the cre(2C) hairpin on VPg uridylylation and viral RNA replication in preinitiation replication complexes (PIRCs). The results showed that the cre(2C) hairpin was required for the synthesis of VPgpUpU and positive-strand RNA, but it was not needed for the synthesis of VPg-linked negative-strand RNA. Therefore, it appears that different mechanisms are used to uridylylate VPg during the initiation of negative- and positive-strand RNA synthesis.
MATERIALS AND METHODS
Viral cDNA clones and transcript RNAs.
The following cDNA clones of the Mahoney strain of type 1 poliovirus were used in this study. (i) DNA plasmids pT7-PV1(A)80 and pT7-PV1(A)80 ΔC869-T6011 (pT7-DJB2) were constructed as previously described and served as the parental clones for the other constructs used in this study (8). Transcripts of these plasmids were designated as PV1 RNA and PV1ΔC869-T6011 RNA, respectively. (ii) pT7-PV1(A)80 VPgY3F containing the mutations A5379T and C5380T was engineered by site-directed mutagenesis with the Transformer site-directed mutagenesis kit (Clonetech). Transcripts of this plasmid were designated PV1VPgY3F RNA. (iii) pT7-PV1(A)80 cre(2C)mut1 containing the mutations G4462A and C4465U and pT7-PV1(A)80 cre(2C)mut7 containing the mutation A4472C (29) were engineered with the QuikChange site-directed mutagenesis protocol (Stratagene). Transcripts of these plasmids were designated as PV1cre(2C)mut1 RNA and PV1cre(2C)mut7 RNA. (iv) pT7-Rz-PV1(A)80, pT7-Rz-PV1(A)80 cre(2C)mut1, and pT7-Rz-PV1(A)80 ΔC869-T6011 were identical to their respective parental plasmids, except they contained a hammerhead ribozyme (Rz) sequence between the T7 promoter and the 5′ end of the poliovirus sequence. RNA transcripts derived from these plasmids were indicated by the Rz designation. (v) pT7-PV1(A)80 ΔG7418-A7422, pT7-PV1(A)80 cre(2C)mut1ΔG7418-A7422, and pT7-PV1(A)80 cre(2C)mut7ΔG7418-A7422 were engineered as described above to contain a GUAAA deletion (ΔGUA3) in the 3′ NTR (8). All plasmids constructed were verified by sequencing. Transcripts of these plasmids were used as helper RNAs in complementation assays and were designated as PV1ΔGUA3 RNA (or wild-type helper), PV1cre(2C)mut1ΔGUA3 RNA [or cre(2C)mut1 helper], and PV1cre(2C)mut7ΔGUA3 RNA [or cre(2C)mut7 helper]. All viral RNAs were transcribed in reaction mixtures containing bacteriophage T7 RNA polymerase and 500 μM each nucleoside triphosphate (NTP) as described previously (5). RNA transcripts were stored in ethanol at −20°C and used within 2 days of transcription and purification.
RNA replication assays.
Poliovirus RNA synthesis was measured in PIRCs by method 4 as previously described (4, 6). PIRCs were resuspended in 50-μl reaction mixtures containing [α-32P]CTP and incubated at 37°C for 1 h. The resulting 32P-labeled product RNAs were analyzed by CH3HgOH-agarose gel electrophoresis. In the trans-replication assays, the PIRCs contained both a subgenomic-size template RNA that did not encode any viral replication proteins and a nonreplicating helper RNA (8). The labeled product RNAs synthesized in these reactions were isolated and characterized as described above.
VPg uridylylation assays.
The cre(2C)-dependent synthesis of VPgpUpU was assayed in reaction mixtures containing either purified polymerase and VPg (method 1) or PIRCs (method 2).
(i) Method 1.
The synthesis of VPgpUpU was measured in a 20-μl reconstituted assay that contained purified poliovirus RNA polymerase and VPg as previously described (25, 29). These reaction mixtures also contained 1 μl of a HeLa S10 translation reaction mixture, which contained 3CD but no other viral proteins. The 3CD protein included amino acid changes T181K and Q182D, which blocked normal proteolytic processing of 3CD to form 3C and 3D (2, 12). The uridylylated VPg products were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE [9 to 18% polyacrylamide]).
(ii) Method 2.
VPgpUpU was synthesized in reaction mixtures containing PIRCs and then immunoprecipitated with anti-VPg antibody. The uridylylation reactions were identical to those described above for the RNA replication assays, except the reaction mixtures contained 5 μM UTP, which was provided by the addition of 100 μCi of [α-32P]UTP (400 Ci/mmol; Amersham) and 250 μM (each) ATP, CTP, and GTP. The reactions were terminated by adding 5.6 μl of 10% SDS, resuspended in 560 μl of radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 1% sodium deoxycholate, 10 mM Tris · HCl [pH 8], 150 mM NaCl) and centrifuged at 6,000 rpm in an Eppendorf microcentrifuge (model 5415C) for 4 min. Affinity-purified anti-VPg antibody (2 μl, 0.35 mg/ml) was added to one-half of the supernatant, which was then incubated for 1.5 h on a rotary mixer at 23°C. As a control, no antibody was added to the other half of the supernatant. A 10% suspension of Pansorbin cells (Calbiochem) in RIPA buffer (100 μl) was added, and the reaction mixtures were incubated at 23°C for 45 min on a rotary mixer. The Pansorbin cells were isolated by centrifugation at 4,000 rpm for 5 min and washed once with 800 μl of RIPA buffer. VPgpUpU was released from the Pansorbin cells by resuspending the cells in 60 μl of Laemmli sample buffer and heating them at 60°C for 2 min. After centrifugation, a portion of the supernatant was analyzed by SDS-PAGE (9 to 18% polyacrylamide).
Immunoprecipitation of 32P-labeled product RNAs.
32P-labeled RNA products synthesized in PIRCs were immunoprecipitated with affinity-purified anti-VPg antibody by procedures similar to those previously described (3). Briefly, labeled product RNAs were phenol extracted six times and ethanol precipitated before immunoprecipitation. The labeled product RNAs were then incubated sequentially with affinity-purified anti-VPg antibody and then Pansorbin cells. The reaction mixture containing the immunoprecipitated labeled RNAs was adjusted to 0.4 M NaCH3CO2, phenol extracted three times, and ethanol precipitated. The immunoprecipitated RNAs were then analyzed by electrophoresis on a CH3HgOH-agarose gel.
In the proteinase K-treated control, the phenol-extracted labeled product RNA was resuspended in 200 μl of 0.5% SDS buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.5% SDS) and incubated at 37°C for 45 min with 1 μl of 20 mg of proteinase K per ml (Fisher Scientific) to digest covalently linked VPg. The RNA was then phenol extracted three times and ethanol precipitated. The proteinase K-treated RNA was then used as a negative control in the immunoprecipitation experiments described above.
RESULTS
VPg uridylylation in PIRCs.
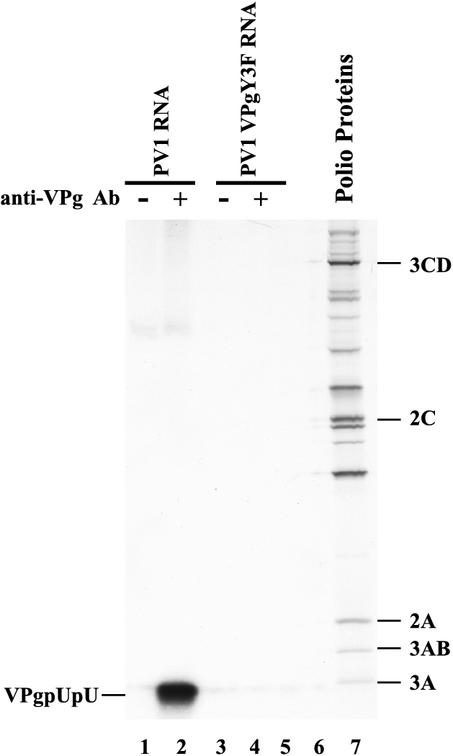
In previous studies, PIRCs isolated from HeLa S10 translation-replication reactions have proven to be very useful in investigating the molecular basis of poliovirus RNA replication (3, 4). We have further adapted this assay to examine the VPg uridylylation reaction in PIRCs. 32P-labeled VPgpUpU was immunoprecipitated with affinity-purified anti-VPg antibody from reaction mixtures containing [α-32P]UTP and PIRCs isolated from HeLa S10 translation-replication reaction mixtures containing PV1 RNA (Fig. 1, lane 2). The 32P-labeled VPgpUpU recovered in these experiments migrated to the same approximate position in the gel as poliovirus protein 3A as previously reported (32).
FIG. 1.
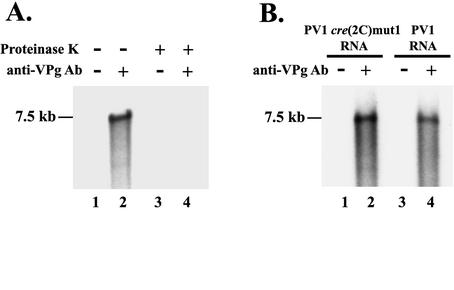
Synthesis of uridylylated VPg in PIRCs. HeLa S10 translation-RNA replication reaction mixtures containing 2 mM guanidine HCl were prepared with PV1 RNA and PV1VPgY3F RNA. VPg uridylylation was measured in PIRCs resuspended in reaction mixtures containing [α-32P]UTP. The labeled VPgpUpU synthesized in these reactions was immunoprecipitated with anti-VPg antibody (Ab) as described in Materials and Methods and resolved by SDS-PAGE. [35S]methionine-labeled poliovirus proteins were used as markers (lane 7).
VPg is covalently linked to UMP by a phosphodiester bond between the 5′ phosphate in UMP and the single tyrosine residue in VPg (1, 31). To confirm that [32P]UMP in the immunoprecipitated VPg was linked to tyrosine, we used viral transcript RNA that contained a VPg(Y3F) mutation. VPg uridylylation was completely blocked in PIRCs containing PV1 VPg(Y3F) RNA (Fig. 1, compare lanes 2 and 4). Therefore, the synthesis of VPgpUpU was dependent on the tyrosine residue of VPg, indicating that authentic VPgpUpU was synthesized in PIRCs. Additionally, VPg uridylylation in PIRCs was completely inhibited by 2 mM guanidine HCl (data not shown), consistent with the findings of Lyons et al. (17).
Effect of cre(2C)mut1 on VPgpUpU synthesis.
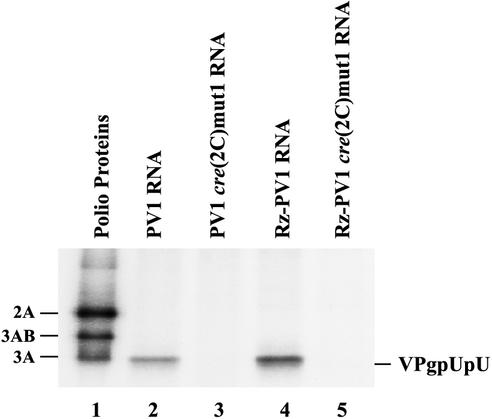
Several mutations that disrupt the structure of the cre(2C) hairpin have been characterized in previous studies (29). cre(2C)mut1 contains two silent nucleotide changes that disrupt the upper stem of the cre(2C) hairpin structure and was previously shown to inhibit virus formation in transfected cells and block VPg uridylylation in vitro (29). We engineered the cre(2C)mut1 changes into PV1 RNA and Rz-PV1 RNA. Both mutant RNAs had a lethal phenotype in transfected cells (data not shown). By using the previously described reconstituted assay that contained synthetic VPg and purified 3Dpol (25), we confirmed that 32P-labeled VPgpUpU was synthesized in reaction mixtures containing the wild-type RNAs (Fig. 2, lanes 2 and 4), but not in reaction mixtures containing the PV1cre(2C)mut1 RNAs (Fig. 2, lanes 3 and 5).
FIG. 2.
Effect of cre(2C)mut1 on VPg uridylylation in reconstituted reactions. VPg uridylylation was measured in reconstituted reaction mixtures containing 3Dpol, VPg, [α-32P]UTP, the indicated RNAs, and 3CD as described in Materials and Methods (lanes 2 to 5). 32P-labeled VPgpUpU synthesized in these reactions was resolved by SDS-PAGE (9 to 18% polyacrylamide). [35S]methionine-labeled poliovirus proteins were used as markers and are shown in lane 1.
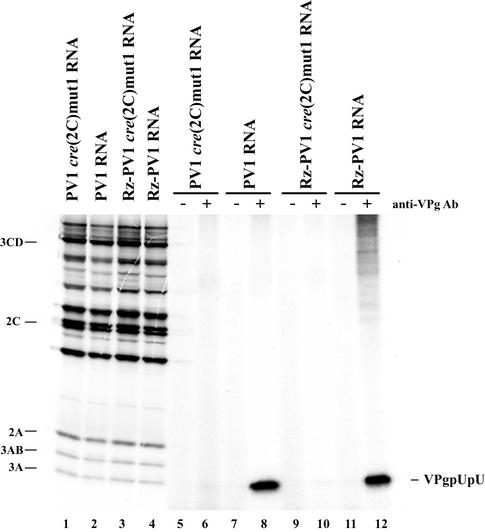
To ensure that similar results would also be observed in membrane-associated replication complexes, we assayed for VPgpUpU synthesis in PIRCs isolated from HeLa S10 translation-replication reaction mixtures containing either wild-type RNA or PV1cre(2C)mut1 RNA. No significant effect on viral protein synthesis or polyprotein processing was observed in the reaction mixtures containing the cre(2C)mut1 RNAs or wild-type RNAs (Fig. 3, lanes 1 to 4). The synthesis of 32P-labeled VPgpUpU, however, was completely inhibited in the reaction mixtures containing the PV1cre(2C)mut1 RNA (Fig. 3, compare lanes 6 and 10 with lanes 8 and 12). Therefore, cre(2C)mut1 inhibited VPgpUpU synthesis below detectable levels in both the reconstituted reactions and in the membrane-associated PIRCs.
FIG. 3.
Effect of cre(2C)mut1 on VPg uridylylation in PIRCs. Viral protein synthesis was measured in HeLa S10 translation-replication reaction mixtures containing the indicated RNAs and [35S]methionine (1.2 mCi/ml) (lanes 1 to 4). VPg uridylylation was measured in PIRCs isolated from identical HeLa S10 translation-replication reaction mixtures containing each of the indicated RNAs. Reaction mixtures containing the PIRCs and [α-32P]UTP were incubated at 37°C for 1 h. Labeled VPgpUpU was immunoprecipitated with anti-VPg antibody (Ab) as described in Materials and Methods (lanes 5 to 12). The labeled viral proteins and immunoprecipitated products were resolved by SDS-PAGE (9 to 18% polyacrylamide).
The effect of cre(2C)mut1 on viral RNA replication.
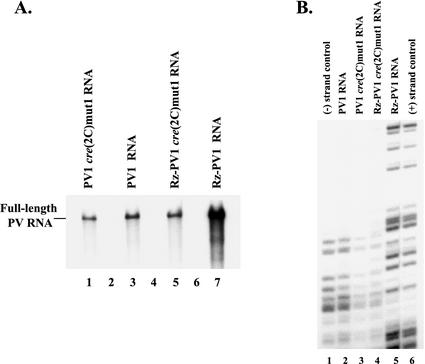
In previous studies, we showed that synthesis of both negative- and positive-strand RNAs occurs in PIRCs isolated from HeLa S10 translation-replication reaction mixtures containing poliovirion RNA (3, 4, 7). Viral RNA synthesis initiates with the synthesis of negative-strand RNA, which is followed by the asymmetric synthesis of excess positive-strand RNA and infectious virus (3, 4). One-dimensional RNase T1 fingerprints were used to confirm the polarity of the product RNAs synthesized in these reactions (see Fig. 4 and 7 in reference 4). In contrast, only negative-strand RNA is synthesized in PIRCs containing poliovirus transcript RNA (PV1 RNA). PV1 RNA contains two nonviral 5′-terminal G nucleotides that have no effect on negative-strand synthesis but inhibit the synthesis of labeled positive-strand RNA below detectable levels in PIRCs (5, 6, 13). Therefore, only labeled negative-strand RNA is synthesized in PIRCs containing PV1 transcript RNA (Fig. 4A, lane 3). The negative polarity of the labeled RNA synthesized on PV1 RNA was again confirmed with a one-dimensional RNase T1 fingerprint analysis (Fig. 4B, compare lanes 1 and 2).
FIG. 4.
Effect of cre(2C)mut1 on negative- and positive-strand RNA synthesis. (A) PIRCs containing the indicated viral RNAs were prepared as described in Materials and Methods, resuspended in reaction mixtures containing [α-32P]CTP, and incubated at 37°C for 1 h. The 32P-labeled product RNAs synthesized in these reactions were characterized by CH3HgOH-agarose gel electrophoresis. (B) 32P-labeled product RNAs were isolated from reactions identical to those described in panel A above, digested with RNase T1, and then characterized by electrophoresis in a 20% polyacrylamide-7 M urea gel as previously described (4). Because of the increased amount of 32P-labeled product RNA from the Rz-PV1 RNA reaction, only 20% of the digested RNA from this reaction was loaded in lane 5. RNase T1 digests of [32P]CMP-labeled poliovirus negative-strand (lane 1) and positive-strand (lane 6) transcript RNAs were used as strand-specific oligonucleotide markers. The 32P-labeled oligonucleotides were detected by autoradiography.
FIG. 7.
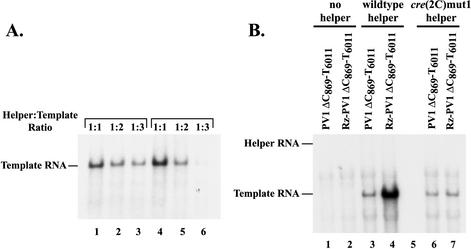
Effect of cre(2C)mut1 on negative- and positive-strand RNA synthesis in trans-replication assays. (A) PIRCs were isolated from HeLa-S10 translation replication reaction mixtures that contained a nonreplicating helper and template RNAs at the indicated molar ratios. In lanes 1 to 3, the helper and template RNA concentrations were varied, while the total RNA concentration was kept constant at 50 μg/ml. In lanes 4 to 6, the helper RNA concentration was kept constant at 38 μg/ml, while the template RNA concentration was varied. PIRCs were resuspended in reaction mixtures that contained [α-32P]CTP and were incubated for 1 h at 37°C. The 32P-labeled product RNAs were analyzed by CH3HgOH-agarose gel electrophoresis. (B) HeLa S10 translation-replication reactions that contained 2 mM guanidine HCl and the indicated template and helper RNAs were incubated at 34°C for 4 h. The overall RNA concentration in each reaction mixture was 50 μg/ml with equal molar amounts of the helper and template RNAs. PIRCs were isolated from these reaction mixtures and resuspended in a reaction mixture containing [α-32P]CTP and then incubated for 1 h at 37°C. The 32P-labeled product RNAs were analyzed by CH3HgOH-agarose gel electrophoresis. The positions of the template and helper RNAs in the gel are indicated above. Control experiments showed that equivalent amounts of the viral replication proteins were synthesized in each reaction (data not shown).
Viral RNAs that have the correct 5′-terminal sequence can be prepared by using transcripts that contain a 5′-terminal hammerhead ribozyme (Rz-PV1 RNA). As expected, Rz-PV1 RNA functions like poliovirion RNA in cell-free reactions and in transfected cells (7, 13). Previous studies showed that full-length positive-strand RNA is synthesized in reaction mixtures containing Rz-PV1 RNA and virion RNA but not in reaction mixtures containing PV1 RNA (7, 13). In addition, equivalent titers of infectious virus are produced in HeLa S10 translation-replication reaction mixtures containing either Rz-PV1 RNA or poliovirion RNA when translation is normalized to equivalent levels (data not shown). Therefore, Rz-PV1 RNA allows for both negative- and positive-strand RNA synthesis (Fig. 4A, lane 7). The increase in labeled product RNA synthesized in reaction mixtures containing the Rz-PV1 RNA represents positive-strand RNA that is synthesized in addition to negative-strand RNA (Fig. 4A, compare lanes 3 and 7) (7, 13). The polarity of the labeled RNA synthesized in reaction mixtures containing Rz-PV1 RNA was confirmed by RNase T1 fingerprint analysis (Fig. 4B, lane 5). As expected, positive-strand RNA was the predominant RNA synthesized in this reaction (Fig. 4B, compare lanes 5 and 6). Therefore, by using both PV1 RNA and Rz-PV1 RNA, it is possible to specifically measure negative- and positive-strand RNA synthesis.
In reactions with PV1cre(2C)mut1 and Rz-PV1cre(2C)mut1 RNAs, we determined how cre(2C)mut1 specifically affected negative-and positive-strand RNA synthesis. Equivalent amounts of labeled RNA were synthesized in PIRCs containing either PV1cre(2C)mut1 RNA or PV1 RNA (Fig. 4A, lanes 1 and 3). RNase T1 fingerprints verified that labeled negative-strand RNA was synthesized in both reactions (Fig. 4B, lanes 2 and 3). These results indicated that inhibition of the cre(2C)-dependent synthesis of VPgpUpU had no significant effect on the synthesis of negative-strand RNA. In contrast, experiments with Rz-PV1cre(2C)mut1 RNA indicated that positive-strand synthesis was strongly inhibited by cre(2C)mut1. No increase in labeled RNA synthesis was observed with Rz-PV1cre(2C)mut1 RNA compared to that observed with PV1cre(2C)mut1 RNA or PV1 RNA (Fig. 4A, compare lane 5 with lanes 1 and 3). This was in contrast to the marked increase in positive-strand RNA synthesis observed with the wild-type ribozyme transcript (Rz-PV1 RNA) (Fig. 4A, lanes 5 and 7). Since no increase in labeled RNA synthesis was observed with Rz-PV1cre(2C)mut1 RNA above that observed with PV1cre(2C)mut1 RNA, we concluded that cre(2C)mut1 specifically inhibited positive-strand synthesis without affecting negative-strand synthesis. The negative polarity of the RNA synthesized in reaction mixtures containing either Rz-PV1cre(2C)mut1 RNA or PV1cre(2C)mut1 RNA was confirmed by RNase T1 fingerprint analysis (Fig. 4B, lanes 3 and 4).
VPg is linked to negative-strand product RNA.
The results presented above suggested that negative-strand RNA was efficiently synthesized in reactions in which the cre(2C)-dependent synthesis of VPgpUpU was inhibited. Since cre(2C)-dependent uridylylation of VPg was only required for the initiation of positive-strand RNA synthesis, we determined if the negative-strand RNA synthesized in the cre(2C)mut1 reactions was linked to VPg. RNA replication reactions were performed with PIRCs that contained either PV1 RNA or PV1cre(2C)mut1 RNA. Labeled product RNA was isolated from both reactions and phenol extracted to remove noncovalently linked proteins. The labeled product RNA from the PV1 RNA reaction immunoprecipitated with anti-VPg antibody, indicating that it was linked to VPg (Fig. 5A, lane 2). As expected, pretreatment of this RNA with proteinase K to remove VPg blocked its immunoprecipitation with anti-VPg antibody (Fig. 5A, lane 4). Labeled RNA isolated from the PV1cre(2C)mut1 RNA reaction was then tested for its ability to immunoprecipitate with anti-VPg antibody. This RNA immunoprecipitated with anti-VPg antibody at levels equivalent to those observed with labeled RNA from the PV1 RNA reaction (Fig. 5B, compare lanes 2 and 4). These results showed that equivalent amounts of VPg-linked negative-strand RNA were synthesized on both PV1 RNA and PV1cre(2C)mut1 RNA.
FIG. 5.
Negative-strand product RNA is linked to VPg. (A) PIRCs formed with PV1 RNA were resuspended in reaction mixtures containing [α-32P]CTP and were incubated for 1 h at 37°C. The 32P-labeled product RNA was phenol extracted and ethanol precipitated. Half of the product RNA was treated with proteinase K to remove VPg as described in Materials and Methods. The labeled product RNAs were then immunoprecipitated with anti-VPg antibody (Ab) and analyzed by CH3HgOH-agarose gel electrophoresis. (B) PIRCs formed with PV1cre(2C)mut1 RNA or PV1 RNA were resuspended in reaction reactions containing [α-32P]CTP and incubated at 37°C for 1 h. The 32P-labeled product RNAs were phenol extracted, ethanol precipitated, and immunoprecipitated with anti-VPg antibody. The immunoprecipitated products were analyzed by CH3HgOH-agarose gel electrophoresis.
Effect of cre(2C) mutations on RNA synthesis in trans-replication assays.
To further investigate the requirement for the cre(2C)-dependent synthesis of VPgpUpU, we performed trans-replication assays. In these assays, the replication proteins were provided in trans to copy template RNAs that contained a large internal deletion, which included the cre(2C) hairpin (PV1ΔC869-T6011 RNA and Rz-PV1ΔC869-T6011 RNA) (Fig. 6). RNA replication was measured in PIRCs that were isolated from HeLa S10 translation-replication reaction mixtures that contained a template RNA and either wild-type helper RNA or cre(2C)mut1 helper RNA. All of the helper RNAs used in this study contained a 3′ NTR deletion mutation that inhibited their replication so that RNA synthesis was only observed on the template RNAs. To optimize these reactions, RNA synthesis was measured on PV1ΔC869-T6011 RNA in reactions in which the ratio of wild-type helper RNA to template RNA was varied from 1:1 to 1:3. Maximum amounts of labeled RNA synthesis were observed in reaction mixtures containing equivalent amounts of the helper and template RNAs (Fig. 7A, compare lanes 1 to 3 and 4 to 6). As expected, labeled RNA synthesis was not observed on the template RNAs in the absence of the helper RNA (Fig. 7B, lanes 1 and 2).
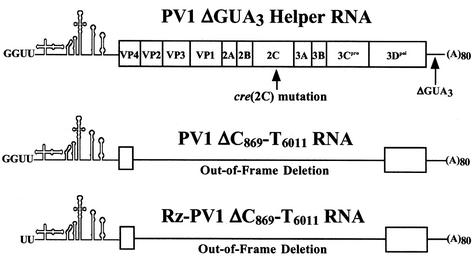
FIG. 6.
Diagram of the RNAs used in the trans-replication experiments. PV1ΔGUA3 RNA, a nonreplicating helper RNA, is full length and contains either a wild-type 2C sequence (wild-type helper) or a cre(2C) mutation [cre(2C)mut1 helper or cre(2C)mut7 helper]. The PV1ΔC869-T6011 and Rz-PV1ΔC869-T6011 template RNAs contain a large out-of-frame deletion and do not encode any viral proteins.
Wild-type helper RNA supported negative-strand synthesis on PV1ΔC869-T6011 RNA (Fig. 7B, lane 3) and both negative- and positive-strand syntheses on Rz-PV1ΔC869-T6011 RNA (Fig. 7B, lane 4). In contrast, cre(2C)mut1 helper RNA supported equivalent amounts of labeled RNA synthesis on both template RNAs (Fig. 7B, compare lanes 6 and 7). Since no increase in labeled product RNA was observed with the Rz template RNA, it appears that only negative-strand synthesis was supported by the cre(2C)mut1 helper RNA. This confirmed our previous observations that the cre(2C)-dependent synthesis of VPgpUpU was only required for positive-strand RNA synthesis.
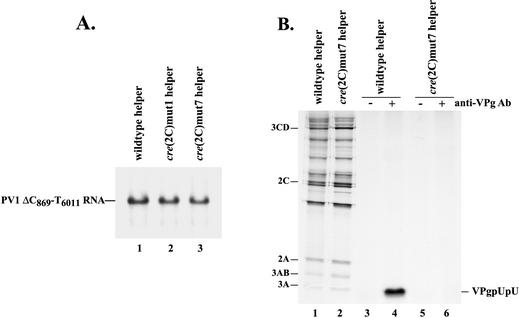
A second mutation in the cre(2C) hairpin that was previously shown to block VPg uridylylation is cre(2C)mut7 (29). This mutation alters the A1 nucleotide in the A1A2A3CA motif in the loop portion of the cre(2C) hairpin and completely blocks the VPg uridylylation reaction (see Discussion) (29). To further demonstrate that the cre(2C)-dependent synthesis of VPgpUpU is not required for negative-strand synthesis, cre(2C)mut7 helper RNA was used in trans-replication assays. PIRCs were formed in HeLa S10 translation-replication reactions with PV1ΔC869-T6011 RNA as the template and either a wild-type, cre(2C)mut1, or cre(2C)mut7 helper RNA. Negative-strand synthesis in PIRCs containing the cre(2C)mut7 helper RNA was similar to that observed in reactions with either the wild-type or cre(2C)mut1 helper RNA (Fig. 8A, lanes 1 to 3). As expected, cre(2C)mut7 had no effect on protein synthesis or polyprotein processing in the translation reactions (Fig. 8B, lanes 1 and 2) and completely inhibited VPgpUpU synthesis in the PIRCs (Fig. 8B, compare lanes 4 and 6). These results confirmed that the cre(2C)-dependent synthesis of VPgpUpU was not required for negative-strand synthesis.
FIG. 8.
Effect of cre(2C)mut7 on negative-strand RNA synthesis and VPg uridylylation. (A) The effect of cre(2C)mut1 and cre(2C)mut7 on negative-strand RNA synthesis was measured in trans-replication assays like those described in the legend to Fig. 7. PIRCs were isolated from HeLa S10 trans-replication reaction mixtures that contained a PV1ΔC869-T6011 RNA template and the indicated nonreplicating helper RNAs. The PIRCs were resuspended in reaction mixtures containing [α-32P]CTP and then incubated at 37°C for 1 h. The 32P-labeled product RNAs were then analyzed by CH3HgOH-agarose gel electrophoresis. (B) Viral protein synthesis was measured in HeLa S10 translation-replication reaction mixtures containing either wild-type helper RNA or PV1cre(2C)mut7 helper RNA and [35S]methionine (1.2 mCi/ml) (lanes 1 and 2). VPg uridylylation was measured in PIRCs isolated from identical HeLa S10 translation-replication reaction mixtures containing the indicated RNAs. Reaction mixtures containing the PIRCs and [α-32P]UTP were incubated at 37°C for 1 h. 32P-labeled VPgpUpU was immunoprecipitated with anti-VPg antibody (Ab) as described in Materials and Methods (lanes 3 to 6). The labeled viral proteins and immunoprecipitated products were resolved by SDS-PAGE (9 to 18% polyacrylamide).
DISCUSSION
In this study, we show that mutations in the cre(2C) hairpin in poliovirus RNA completely blocked the synthesis of VPgpUpU in PIRCs isolated from HeLa S10 translation-replication reactions. Inhibition of the cre(2C)-dependent synthesis of VPgpUpU in PIRCs blocked positive-strand RNA synthesis but had no significant effect on the synthesis of VPg-linked negative-strand RNA. These results suggest a model for poliovirus RNA replication in which the 3′ poly(A) tail in virion RNA is used as the template for VPg uridylylation and the initiation of negative-strand RNA synthesis. In contrast, the cre(2C) hairpin is used as the template for the synthesis of VPgpUpU, which is then used as a primer for positive-strand synthesis at the 3′ end of negative-strand RNA templates.
VPg uridylylation in PIRCs.
One of the first steps in the initiation of poliovirus RNA replication involves the formation of uridylylated VPg, which then serves as a primer for the poliovirus RNA polymerase (10, 33). Previous studies showed that uridylylated VPg and VPg-linked poly(U) are synthesized in reconstituted in vitro assays containing purified polymerase (3Dpol), UTP, synthetic VPg, and a poly(A) template (26). Subsequent studies showed that VPg is also uridylylated in reconstituted reaction mixtures that contain viral protein 3CD and poliovirus RNA. In this case, the cre(2C) hairpin in poliovirus RNA functions as the primary template for VPg uridylylation (25). The poliovirus cre(2C) hairpin consists of a stem-loop structure with a conserved A1A2A3CA motif in the loop (29). The results of previous studies showed that the duplex structure of the stem region of the cre(2C) hairpin is required for uridylylation activity (11, 29). In addition, the A1 residue in the A1A2A3CA loop sequence is proposed to function as the template for the addition of both U residues in VPgpUpU as part of a “slide-back” mechanism by the viral polymerase (9).
As part of this study, we showed that two previously characterized mutants that completely inhibit VPg uridylylation in the reconstituted assays, cre(2C)mut1 and cre(2C)mut7, also inhibited VPgpUpU synthesis in PIRCs. In cre(2C)mut1, the duplex structure of the cre(2C) hairpin is disrupted by mutating two nucleotides in the stem region, and in cre(2C)mut7, the uridylylation template nucleotide (A1) in the A1A2A3CA loop sequence is altered (29). Therefore, all of the labeled VPgpUpU that was synthesized in PIRCs was dependent on the cre(2C) hairpin. Being able to inhibit VPgpUpU synthesis in these reactions allowed us to investigate the relationship between the cre(2C)-dependent uridylylation of VPg and viral RNA replication.
cre(2C)-dependent VPg uridylylation and negative-strand synthesis.
Recent studies from our laboratory demonstrated that a subgenomic poliovirus RNA that did not contain the cre(2C) sequence (i.e., PV1ΔC869-T6011 RNA [or DJB2]), was a functional template for negative-strand synthesis in a trans-replication assay containing a wild-type helper RNA (8). This observation suggested that either the cre(2C) hairpin was not required for negative-strand synthesis, or it was able to function in trans as part of the wild-type helper RNA. In the present study, we showed that negative-strand RNA synthesis on PV1ΔC869-T6011 RNA was not inhibited in trans-replication assays containing a cre(2C)mut1 helper RNA. In addition, negative-strand synthesis was not inhibited on full-length cre(2C)mut1 RNA. These results indicate that the cre(2C)-dependent synthesis of VPgpUpU is not required for negative-strand synthesis.
In a previous study, however, it was shown that cre(2C)mut1 RNA, upon transfection, produced revertants after five blind passages (29). This result suggests that extremely low levels of VPgpUpU synthesis and RNA replication may occur in cells transfected with cre(2C)mut1 RNA. In contrast, revertants were never observed after repeated passages with cre(2C)mut7 RNA (29). This was consistent with the proposal that the A1 nucleotide in the A1A2A3CA loop sequence serves as the template for the VPg uridylylation reaction as previously discussed. cre(2C)mut7 creates an absolute block in VPgpUpU synthesis. Finding that negative-strand synthesis was not inhibited in assays containing cre(2C)mut7 confirmed the results of the experiments with cre(2C)mut1. Therefore, we concluded that the cre(2C)-dependent synthesis of VPgpUpU is not required for negative-strand synthesis. This raises a question about the mechanism used to synthesize negative-strand RNA. Is VPg required to initiate negative-strand synthesis? Immunoprecipitation of labeled negative-strand RNA with anti-VPg antibody showed that negative-strand RNA synthesized in reactions containing cre(2C)mut1 RNA was covalently linked to VPg. Thus, VPg is used to initiate negative-strand synthesis, even though VPgpUpU synthesized on the cre(2C) hairpin is not required for this reaction.
cre(2C)-dependent VPg uridylylation and positive-strand synthesis.
In marked contrast to negative-strand synthesis, positive-strand synthesis was totally dependent on the synthesis of VPgpUpU on the cre(2C) hairpin. Positive-strand synthesis was inhibited in PIRCs containing cre(2C)mut1 RNA and in trans-replication assays containing cre(2C)mut1 helper RNA. Therefore, the cre(2C) hairpin is required for the synthesis of VPgpUpU, which is then used as a primer to initiate positive-strand synthesis. The cre hairpins in the coding region of picornavirus genomes were initially described as cis-active replication elements that are required for viral RNA replication, apparently at the level of negative-strand synthesis (11, 16, 20). However, the measurement of negative-strand RNA levels in transfected cells is complicated by the fact that the synthesis of the vast majority of negative-strand RNA is dependent on the synthesis of positive-strand RNA, which serves as a template for negative-strand synthesis in subsequent rounds of amplification. Hence, a complete block in positive-strand synthesis will severely inhibit the accumulation of negative-strand RNA. As shown in this study, the poliovirus cre(2C) mutants completely inhibited positive-strand RNA synthesis, suggesting that this could have been a factor limiting the detection of negative-strand synthesis in the previous studies. Since optimal amounts of input viral RNA are added to the HeLa-S10 translation-replication reaction mixtures to maximize viral protein synthesis and RNA replication, negative-strand synthesis can be directly measured in PIRCs isolated from these reactions. Therefore, this system offers a distinct advantage in terms of measuring negative- and positive-strand RNA synthesis in the context of membrane-associated RNA replication complexes that support authentic viral RNA replication and virus formation.
Models for VPg uridylylation and asymmetric viral RNA replication.
Two models have been proposed to explain the mechanisms involved in VPg-primed negative-strand RNA synthesis. The first model suggests that VPg is uridylylated on the 3′ poly(A) tail in poliovirus RNA and then used as a primer for negative-strand synthesis (26). The second model proposes that VPgpUpU synthesized on the cre(2C) hairpin is translocated to the 3′ end of the poly(A) tail, where it acts as a primer for negative-strand synthesis (25). The results of this study clearly support a model in which VPg is uridylylated on the 3′ poly(A) tail and then elongated into negative-strand RNA. It appears that VPg itself is used to initiate negative-strand synthesis, and that in this case, the uridylylated form of VPg is only a transient intermediate that is elongated into full-length negative-strand RNA. Our results also indicate that VPgpUpU, which is synthesized on the cre(2C) hairpin, is used during positive-strand synthesis. The cre(2C)-dependent uridylylation of VPg appears to generate a pool of VPgpUpU, which functions as a primer to initiate positive-strand synthesis by using the AA sequence at the 3′ end of negative-strand RNA templates. This would provide an efficient mechanism for the initiation of multiple rounds of positive-strand RNA synthesis.
The asymmetric replication of poliovirus RNA is a fundamental property of the viral RNA replication cycle. The process of limiting negative-strand synthesis and promoting positive-strand synthesis establishes asymmetry during the replication cycle. Negative-strand initiation is believed to require the formation of a circular RNP complex between the 5′ and 3′ ends of the viral genome (8, 14, 34). It is possible that the formation of this circular complex plays an important role in facilitating the uridylylation of VPg on the 3′ poly(A) sequence and the initiation of negative-strand synthesis. The elongation of the VPg-linked nascent negative strand by the viral polymerase would disrupt this circular complex and restrict additional rounds of negative-strand initiation (8). In addition, having a relatively inefficient mechanism for VPg uridylylation that involves the use of the 3′ poly(A) tail would further limit negative-strand synthesis. In contrast, the availability of a pool of VPgpUpU preformed on the cre(2C) hairpin could potentially drive the initiation of positive-strand synthesis and explain the rapid formation of multiple nascent positive strands on individual negative-strand templates. Overall, employing two different mechanisms to uridylylate VPg during negative- and positive-strand synthesis could result in an asymmetric mode of replication that favors positive-strand RNA synthesis. This model helps explain the molecular basis for the asymmetric replication of poliovirus and other picornaviruses and perhaps additional positive-strand RNA viruses with VPg-linked genomes.
Acknowledgments
This work was supported by Public Health Service grants AI15539 and AI32123 from the National Institute of Allergy and Infectious Diseases.
We thank Brian O'Donnell for excellent technical assistance and Sushma Ogram for critically reading the manuscript.
REFERENCES
- 1.Ambros, V., and D. Baltimore. 1978. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J. Biol. Chem. 253:5263-5266. [PubMed] [Google Scholar]
- 2.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, D. J., and J. B. Flanegan. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71:8482-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1996. Assays for poliovirus polymerase, 3Dpol, and authentic RNA replication in HeLa S10 extracts. Methods Enzymol. 275:35-57. [DOI] [PubMed] [Google Scholar]
- 6.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton, D. J., B. J. Morasco, L. E. Smerage, and J. B. Flanegan. 2002. Poliovirus RNA replication and genetic complementation in cell-free reactions, p. 461-469. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 8.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerber, K., E. Wimmer, and A. V. Paul. 2001. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of 2Apro. J. Virol. 75:10979-10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giachetti, C., and B. L. Semler. 1991. Role of a viral membrane polypeptide in strand-specific initiation of poliovirus RNA synthesis. J. Virol. 65:2647-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris, K. S., S. R. Reddigari, M. J. H. Nicklin, T. Hämmerle, and E. Wimmer. 1992. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J. Virol. 66:7481-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, Y. F., A. Nomoto, B. M. Detjen, and E. Wimmer. 1977. A protein covalently linked to poliovirus genome RNA. Proc. Natl. Acad. Sci. USA 74:59-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobert, P. E., N. Escriou, J. Ruelle, and T. Michiels. 1999. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. USA 96:11560-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 75:10696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mason, P. W., S. V. Bezborodova, and T. M. Henry. 2002. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 76:9686-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKnight, K. L., and S. M. Lemon. 1996. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 70:1941-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melchers, W. J. G., J. G. J. Hoenderop, H. J. Bruins Slot, C. W. A. Pleij, E. V. Pilipenko, V. I. Agol, and J. M. D. Galama. 1997. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J. Virol. 71:686-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirmomeni, M. H., P. J. Hughes, and G. Stanway. 1997. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J. Virol. 71:2363-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray, K. E., A. W. Roberts, and D. J. Barton. 2001. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA 7:1126-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novak, J. E., and K. Kirkegaard. 1991. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J. Virol. 65:3384-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul, A. V., J. H. van Boom, D. Fillipov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 27.Pettersson, R. F., J. B. Flanegan, J. K. Rose, and D. Baltimore. 1977. 5′-terminal nucleotide sequences of polio virus polyribosomal RNA and virion RNA are identical. Nature 268:270-272. [DOI] [PubMed] [Google Scholar]
- 28.Pilipenko, E. V., K. Poperechny, S. V. Maslova, W. J. G. Melchers, H. J. Bruins Slot, and V. I. Agol. 1996. cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary ('kissing') interactions. EMBO J. 15:5428-5436. [PMC free article] [PubMed] [Google Scholar]
- 29.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohll, J. B., D. H. Moon, D. J. Evans, and J. W. Almond. 1995. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol. 69:7835-7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothberg, P. G., T. J. Harris, A. Nomoto, and E. Wimmer. 1978. The genome-linked protein of picornaviruses. V. O4-(5′-uridylyl)tyrosine is the bond between the genome-linked protein and the RNA of poliovirus. Proc. Natl. Acad. Sci. USA 75:4868-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeda, N., R. J. Kuhn, C.-F. Yang, T. Takegami, and E. Wimmer. 1986. Initiation of poliovirus plus-strand RNA synthesis in a membrane complex of infected HeLa cells. J. Virol. 60:43-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takegami, T., B. L. Semler, C. W. Anderson, and E. Wimmer. 1983. Membrane fractions active in poliovirus RNA replication contain VPg precursor polypeptides. Virology 128:33-47. [DOI] [PubMed] [Google Scholar]
- 34.Teterina, N. L., D. Egger, K. Bienz, D. M. Brown, B. L. Semler, and E. Ehrenfeld. 2001. Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J. Virol. 75:3841-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toyoda, H., C.-F. Yang, N. Takeda, A. Nomoto, and E. Wimmer. 1987. Analysis of RNA synthesis of type 1 poliovirus by using an in vitro molecular genetic approach. J. Virol. 61:2816-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, J., J. M. Bakkers, J. M. Galama, H. J. Bruins Slot, E. V. Pilipenko, V. I. Agol, and W. J. Melchers. 1999. Structural requirements of the higher order RNA kissing element in the enteroviral 3′UTR. Nucleic Acids Res. 27:485-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wimmer, E. 1982. Genome-linked proteins of viruses. Cell 28:199-201. [DOI] [PubMed] [Google Scholar]
- 38.Yang, Y., R. Rijnbrand, K. L. McKnight, E. Wimmer, A. Paul, A. Martin, and S. M. Lemon. 2002. Sequence requirements for viral RNA replication and VPg uridylylation directed by the internal cis-acting replication element (cre) of human rhinovirus type 141. J. Virol. 76:7485-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]