Abstract
Adenovirus early proteins E4 ORF3 and E4 ORF6 have complementary functions during viral infection. Both proteins facilitate efficient viral DNA replication, late protein expression, and prevention of concatenation of viral genomes. Additionally, E4 ORF6 is involved in the shutoff of the host cell protein synthesis through its interaction with the E1B 55K protein. This complex also leads to the degradation of p53. A unique function of E4 ORF3 is the reorganization of nuclear structures known as PML oncogenic domains (PODs). The function of these domains is unclear, but PODs have been implicated in a number of important cellular processes, including transcriptional regulation, apoptosis, transformation, and response to interferon. The goal of this study was to determine the functional significance of the reorganization of PODs by E4 ORF3. Point mutations were made in the E4 ORF3 gene. These mutants were recombined into a virus lacking E4 ORF6 and expressed under the control of the natural virus E4 promoter. The panel of mutant viruses was used to investigate the role of E4 ORF3 during the course of the viral infection program. One of the mutant viruses exhibited aberrant reorganization of PODs and had a severe defect in viral DNA replication, thus leading to a dramatic decrease in virus production. A number of mutants accumulated viral DNA and infectious virus particles to wild-type levels but showed significant viral genome concatenation. These data show that E4 ORF3 is a multifunctional protein and that a specific rearrangement of nuclear PML domains is coupled to efficient viral DNA replication. This function is distinct from the role of E4 ORF3 in the regulation of virus genome concatenation via inhibition of cellular double-strand break repair.
Adenovirus (Ad) DNA replication requires a concerted effort by a number of viral as well as cellular proteins (reviewed in reference 11). The early region 4 (E4) produces a diverse set of proteins that are involved in a variety of processes crucial to virus growth. In particular, E4 open reading frame 3 (E4 ORF3) and E4 ORF6 have been shown to be important in viral DNA replication (4, 17). These two proteins have been reported to have complementary roles in efficient initiation of viral DNA replication, late Ad mRNA splicing and protein expression, and inhibition of virus genome concatemer formation (reviewed in reference 31). Also, both proteins bind another Ad early protein, E1B 55K, but for seemingly juxtaposed purposes (7, 20). E4 ORF6 has a number of unique functions, primarily facilitated by the E1B 55K interaction, such as inhibition of host cell protein synthesis and promoting the degradation of p53 (7, 8).
E4 ORF3 is necessary and sufficient to reorganize the nuclear structures known as PML oncogenic domains (PODs) (5, 10). The function of POD redistribution with respect to virus infection is still unclear. E4 ORF3 is an 11- to 14-kDa protein synthesized early in infection that is tightly associated with the nuclear matrix (28). E4 ORF3 is one of the most highly conserved Ad proteins. E4 ORF3 affects the double-strand break repair (DSBR) mechanism in cells by binding and inhibiting the function of the DNA damage-activated DNA protein kinase (DNA-PK) via its catalytic subunit (3). E4 ORF3 inhibition of DSBR could prevent the formation of viral DNA concatemers. Ad genome concatenation has been associated with decreased viral DNA replication, late protein expression, and packaging of the genome into the capsid (32). Early after infection, E4 ORF3 localizes to the nucleus in the autonomous POD structure and induces the reorganization of PODs from a punctate to a track-like appearance (10). The majority of E4 ORF3 protein is present in these track structures. However, the mechanism by which E4 ORF3 promotes this reorganization or how this benefits the virus has not been determined.
PODs are distinct nuclear structures with unknown functions. They were first recognized as electron-dense regions within the nucleus but were later analyzed by using antisera from patients with acute biliary cirrhosis (reviewed in references 2 and 9). The primary protein constituent was determined to be the promyelocytic leukemia protein PML. These structures are disrupted in the hematopoeitic disorder acute promyelocytic leukemia (APL). APL is caused when a reciprocal chromosomal translocation fuses the PML gene with the retinoic acid receptor. The resulting PML-retinoic acid receptor fusion protein is devoid of the regulatory domains of either protein, leading to constitutive activation as well as aberrant localization. Treatment of APL with all-trans retinoic acid or arsenic trioxide leads to remission of the disease, and PODs return to normal appearance, suggesting a link between the disease state and integrity of the POD complexes (24). These complexes, likely containing >20 proteins, are extremely dynamic in physical appearance and constitution. PODs are tightly bound to the nuclear matrix. PODs have been implicated in a variety of cellular functions, including transcriptional regulation, apoptosis, transformation, and response to interferon (reviewed in reference 22).
A number of DNA viruses (e.g., herpes simplex virus type 1 [HSV-1], papillomavirus, and cytomegalovirus [CMV]) have one or more proteins dedicated to localization at the POD complex, which usually results in the reorganization of the structure (1, 23; reviewed in reference 12). For example, the large viral transactivator ICP0 (or Vmw110) encoded by HSV-1 localizes to PODs, leading to their destruction through proteasome-dependent degradation of their constituents (e.g., PML and Sp100) (9, 25). Mutation of ICP0 to inhibit its ability to disrupt PODs leads to a severe defect in virus growth. Also, proteasome inhibitors block the degradation of PML and Sp100 and cause a large decrease in herpesvirus production (13).
Recently, Stracker et al. (30) provided evidence suggesting that E4 ORF3 disruption of PODs is linked to its role in the inhibition of Ad genome concatemer formation. It was also shown that E4 ORF6 acts to degrade the Mre11 DNA repair complex, which also leads to inhibition of genome concatenation. These data supported previous findings that E4 ORF3 played a role in viral DNA replication and concatemer inhibition and provide a conceptual model for the complementing roles of E4 ORF3 and E4 ORF6 in Ad replication.
Our studies focused on the ability of Ad E4 ORF3 to reorganize PODs and its effects on virus replication. Point mutations in the Ad type 5 (Ad5) E4 ORF3 gene were constructed. These mutants supplanted the E4 ORF3 gene in a virus deficient for E4 ORF6 expression. One of the mutants exhibited aberrant reorganization of PODs and had a severe defect in both viral DNA replication and virus growth. Thus, a specific reorganization of PODs is a significant step that must be achieved by the virus in order to establish a productive infection. A number of other mutants were able to efficiently accumulate viral DNA as well as produce infectious virus, while they displayed significant virus genome concatenation. These results demonstrate that E4 ORF3 is a multifunctional protein and uncouple the role of E4 ORF3 in DNA replication from the regulation of DSBR.
MATERIALS AND METHODS
Cells, viruses, and infections.
A549 and W162 (33) (gifts of G. Ketner) were maintained in Dulbecco modified Eagle medium supplemented with 10% bovine calf serum. The viruses dl309, dl355, inORF3, and dl355/inORF3 were described previously (14, 17, 19). The construction of viruses containing E4 ORF3 point mutations is described below. Virus particles were purified by two successive rounds of cesium chloride equilibrium centrifugation. All infections were carried out in an identical manner. A549 cells were infected with 200 particles/cell. After 1 h of incubation, the inoculum was removed and the cells were washed. Dulbecco modified Eagle medium supplemented with 10% bovine calf serum was added, and the cells were incubated at 37°C in 5.0% CO2.
Plasmids, mutagenesis, and cloning.
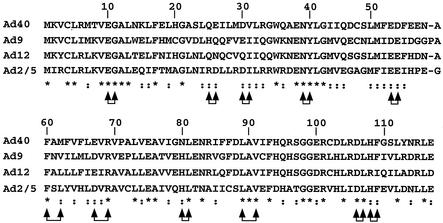
Plasmid pAd-CMV-HA contains Ad5 left-end sequences (nucleotides [nt] 1 to 450), the CMV promoter, a hemagglutinin (HA) epitope, and adjacent cloning sites, followed by Ad5 sequences from nt 3330 to 5780. Ad5, Ad9, and Ad12 E4 ORF3 reading frames were amplified by PCR and cloned downstream of the HA epitope. Plasmid pBS-E4 was constructed by introducing Ad5 right-end sequences from nt 33593 to 35935 into pBluescript (Stratagene). Double alanine substitution mutations were introduced into the E4 ORF3 gene in pBS-E4 by site-directed mutagenesis with complementary oligonucleotide pairs (Quick-Change Mutagenesis; Stratagene) (Fig. 1). Double alanine substitution mutations were introduced in place of conserved and/or charged residues at 10- to 15-amino acid intervals in E4 ORF3 (Fig. 2). The mutants are designated pmXnXn, where X is the amino acid mutated and n is the residue number. The mutants were E10G11, R25D26, D30I31, N39Y40, E52E53, F59L61, D66R68, H79L80, L88V90, D105L106, and H107F108.
FIG. 1.
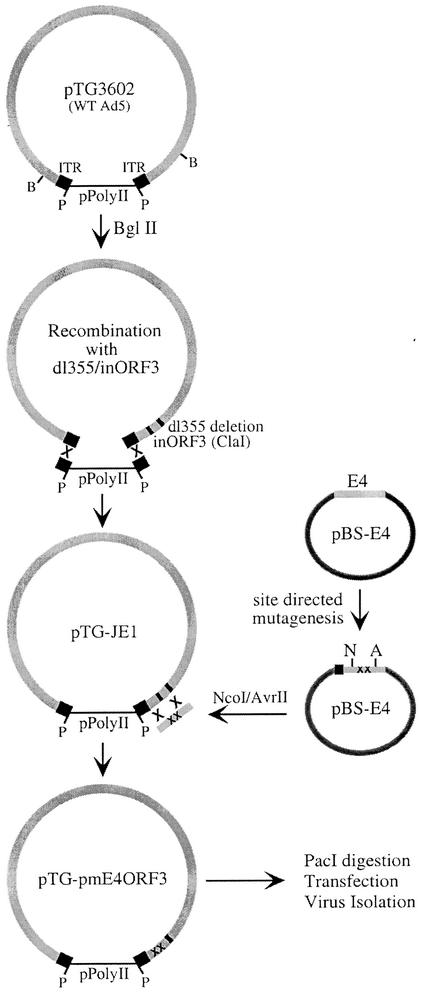
Construction of viruses containing E4 ORF3 point mutants. Mutations in the E4 ORF3 reading frame initially were constructed in plasmid pBS-E4, which contains the Ad5 E4-coding region without ORF6 or ORF7. Double alanine point mutations were introduced into the E4 ORF3 gene by site-directed mutagenesis with complementary primer pairs. An E4 fragment containing the point mutants was released from the vector by NcoI and AvrII cleavage. The recombinant plasmid pTG-JE1 contains entire genome of mutant virus dl355/inORF3. A ClaI site in the E4 ORF3 reading frame was used to linearize the plasmid for bacterial recombination with overlapping fragments containing E4 ORF point mutations. Mutagenized viral genomes were excised from the plasmid by PacI digestion and used for transfection of the E4-complementing cell line W162. Viruses were isolated and propagated by using W162 cells. WT, wild type; ITR, inverted terminal repeat.
FIG. 2.
Alignment of E4 ORF3 proteins and point mutations. The sequences of E4 ORF3 proteins from multiple Ad serotypes were identified in the National Center for Biotechnology Information database and analyzed with the Multi-Align program (http://dot.imgen.bcm.tmc.edu). The asterisks below the sequences indicate identical amino acids. Double dots below the sequences indicate conserved amino acids. The joined arrow pairs indicate amino acids replaced by double alanine residues.
A plasmid containing the intact, wild-type Ad5 genome, pTG-3602, was obtained from Transgene, S.I. (Strasbourg, France) (6) (Fig. 1). The majority of the Ad5 sequences were removed by digestion with BglII, resulting in a linear bacterial plasmid with minimal sequences from the left and right ends of the Ad5 genome. Recombination of the genome of mutant dl355/inORF3 (17) with BglII-digested pTG3602 was carried out in Escherichia coli BJ5183 bacteria as previously described (6), resulting in formation of the plasmid pTG-JE1 (Fig. 1). The viral DNA contains a 14-bp deletion in E4 ORF6 (dl355 mutation) (14) and an 8-bp ClaI insertion in E4 ORF3 (inORF3 mutation) (17). The presence of the dl355/inORF3 genome was confirmed by restriction endonuclease analysis and DNA sequencing.
AvrII-NcoI DNA fragments were excised from pBS-E4 plasmids containing different E4 ORF3 point mutations. Plasmid pTG-JE1 was linearized within E4 ORF3 with ClaI and treated with calf intestinal phosphatase. Recombination of the E4 AvrII-NcoI fragments into the linearized pTG-JE1 plasmid was performed as previously described (6). Briefly, 500 ng of the AvrII-NcoI E4 fragment and 100 ng of linearized pTG-JE1 plasmid were electroporated into E. coli BJ5183 bacteria. Recombinants were selected in the presence of ampicillin and streptomycin. The resulting plasmids were referred to as pTG-pmXnXn, designating the E4 ORF3 mutations listed above. These plasmids were propagated in E. coli XL-1 bacteria. Mutations were verified by DNA sequencing.
Construction of recombinant Ads carrying E4 ORF3 point mutations.
Mutated Ad genomes were excised from the pTG-pmXnXn plasmids by PacI digestion. Four micrograms of viral DNA was used to transfect complementing W162 cells by using Fugene 6 transfection reagent according to the instructions of the manufacturer (Roche). After 8 days, the cells were harvested and viruses were isolated by plaque purification on W162 cells. The viruses were propagated in W162 cells, purified by two successive bandings on cesium gradients, and stored at −20°C. Virus sequences were confirmed by DNA sequencing.
Construction of E1 replacement viruses.
E1 replacement viruses were constructed by overlapping homologous recombination. Briefly, a fragment containing Ad5 sequences from nt 918 to 35935 with a deletion of E4 ORF1, ORF2, and ORF3 (E4-dlORF1-3) (17) was generated by digestion with ClaI and isolated on a sucrose gradient. pAd-CMV-HA plasmids, with wild-type and mutated E4 ORF3 sequences, were linearized with EcoRI and used to cotransfect E1-complementing 293 cells with the ClaI fragment of E4-dlORF1-3. The mutants used were E10G11, R25D26, N39Y40, E52E53, D66R68, H79L80, L88V90, and D105L106. Overlapping recombination resulted in viruses lacking E1A and most of E1B as well as E4 ORF1 to ORF3 while possessing CMV-driven HA-E4 ORF3 wild-type and mutant genes at the left end. Recombinant viruses were plaque purified, amplified on 293 cells, and confirmed by DNA sequencing.
Virus growth and DNA replication.
Single-step growth curves were used to measure virus growth. At 4, 20, 32, and 44 h postinfection, cells were harvested and lysed by four cycles of freeze-thawing. Virus growth was determined by plaque assay in the E4-complementing cell line, W162. Viral DNA replication was determined by Southern blot analysis. Total nuclear DNA from 6-, 12-, 18-, and 24-h infections was isolated as previously described (29). These samples were diluted in Tris-EDTA at 1:10, 1:30, 1:100, and 1:200, respectively. The DNA was denatured and prepared for hybridization as previously described (17). The DNA was bound to Hybond-N membrane (Amersham) by slot blotting, followed by UV cross-linking. The probe corresponded to a PCR fragment from Ad5 nt 1 to 194 and was fluorescently labeled by using the Atto-Phos system (Promega). The probe was hybridized to the membrane and detected according to the manufacturer's instructions. The signal was visualized and quantified with a Molecular Dynamics Storm 860 phosphorimager and ImageQuant software.
Detection of Ad proteins.
Infected A549 cells were harvested at 9 and 18 h postinfection and washed three times with phosphate-buffered saline (PBS). Cell pellets were resuspended in 3 volumes of radioimmunoprecipitation assay (RIPA) lysis buffer (150 mM NaCl, 1.0% NP-40, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris [pH 8.0]) supplemented with a protease inhibitor cocktail. The cells were vortexed and then lysed by a freeze-thaw cycle. Cellular debris was precipitated at 25,000 × g, and the soluble protein fraction was collected. Twenty micrograms of total protein was separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a Hybond-P (Amersham) membrane, and probed with the appropriate antibody. The 9-h samples were analyzed for DNA binding protein (DBP), and the 18 h samples were analyzed for penton. Mouse monoclonal anti-DBP antibodies (A1-6 and B6-8) were gifts from A. Levine and were used as a mixture at a 1:50 dilution each. The rabbit polyclonal antipenton antibody was a gift from C. Anderson and was used at a 1:1,000 dilution.
Immunofluorescence.
A549 cells were seeded on coverslips and infected as described above. At 4, 14, and 24 h postinfection, cells were fixed with −20°C methanol for 5 min. The cells were blocked in 10% goat serum in PBS for 1 h. The antibodies were diluted in PBS containing 1.5% goat serum and incubated with the cell monolayers for 1 h at room temperature. The dilutions were 1:300 anti-PML (PG-M3) mouse monoclonal antibody (Santa Cruz Biotechnology), 1:5 anti-E4ORF3 (6A11) rat monoclonal antibody (a gift from T. Dobner), and 1:600 anti-DBP rabbit polyclonal antibody (a gift from C. Anderson), respectively. For Rad50, A549 cells were infected for 18 h and a 1:250 dilution of anti-Rad50 (2C6) mouse monoclonal antibody (Novus Biologicals) was used. The coverslips were then washed three times (5 min each) with PBS. The cells were then incubated with a 1:300 dilution of fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (IgG) (Zymed), Texas red isothiocyanate-conjugated goat anti-rat IgG (Zymed), or Alexa 350 goat anti-rabbit IgG (Molecular Probes) for 45 min at room temperature in the dark and then washed three times with PBS. The coverslips were mounted on slides in Fluoromount G (Southern Biotechnology Associates). Immunofluorescence was viewed and photographed with a Zeiss microscope with Axiovision software.
Pulsed-field gel electrophoresis.
Infected A549 cells were harvested at 36 h postinfection and prepared for pulsed-field gel electrophoresis as previously described (3). Briefly, cells were harvested by trypsinization, washed once with PBS, and then resuspended in 100 μl of PBS with 125 mM EDTA. Next, 100 μl of 1.25% low-melting-point agarose in 50 mM Tris (pH 7.4) and 125 mM EDTA was added to the cells. The solidified plugs were treated with 1.2% SDS, 0.125 mM EDTA, and 1 mg of proteinase K per ml overnight at 50°C. The plugs were washed three times over 8 h with 50 mM EDTA. A 1.0% agarose gel was loaded with 30 μl of melted plug. The gel was run at 6 V/cm with a switching time of 6 s for 20 h. The DNA was transferred to Hybond-N. The DNA was probed with a 32P-labeled Ad genome probe as previously described (17).
Immunoprecipitation analyses.
A549 or 293 cells were infected with E1 replacement viruses expressing HA-tagged E4 ORF3 wild-type and mutant proteins for 12 h. Infected cells were harvested and lysed in RIPA buffer. A control experiment using 35S-labeled Ad-infected cells, and immunoprecipitation demonstrated that RIPA buffer extracted ∼90% of E4 ORF3 protein present in the cell. The extracts were immunoprecipitated with 3 μg of anti-E1B 55K rat monoclonal antibody (Calbiochem), anti-p300 (N-15) rabbit polyclonal antibody (Santa Cruz Biotechnology), anti-CREB binding protein (CBP) (A-22) rabbit polyclonal antibody (Santa Cruz Biotechnology), and anti-DNA-PK (catalytic subunit) mouse monoclonal antibody (Upstate Biotechnology). Immune complexes were separated by SDS-PAGE and transferred to Hybond-P (Amersham) by standard methods. Membranes were probed with anti-HA mouse monoclonal antibody (Roche). Proteins were visualized with ECL reagents (Amersham).
RESULTS
Construction and propagation of viruses containing point mutations in E4 ORF3.
In order to fully investigate the role of E4 ORF3 in Ad infection, viruses that expressed wild-type and mutant E4 ORF3 proteins at physiological levels in the absence of E4 ORF6 were constructed. Mutation of E4 ORF6 was necessary to eliminate the complementation of E4 ORF3 defects by E4 ORF6 expression. Figure 1 illustrates the approach taken for the production of the panel of mutant viruses. Utilizing the bacterial recombination system described by Chartier et al. (6), an infectious Ad5 clone that contains the E4 ORF3 and ORF6 mutations of mutant virus E4-dl355/inORF3 (17) was constructed. This mutant virus has a 14-bp deletion in the E4 ORF6 gene, thus eliminating this protein without affecting the expression of the splice product E4 ORF6/7. The dl355/inORF3 virus also has an 8-bp ClaI linker insertion in E4 ORF3, disrupting the gene and providing a target site for recombination in bacteria to introduce mutations into the ORF3 reading frame in the natural E4 context.
Plasmid pTG-3602 contains the intact, wild-type Ad5 genome in a pPolyII background (Fig. 1). A majority of the Ad5 genome was removed by BglII digestion. Taking advantage of the remaining homologous Ad5 sequences, the viral genome of dl355/inORF3 was introduced into the plasmid by recombination in E. coli, as described previously (6). The new infectious clone, named pTG-JE1, was used to introduce individual E4 ORF3 point mutations into the E4 region. A transfer plasmid (pBS-E4 [Fig. 1]) that contains all of the Ad5 E4 region rightward of the ORF6 deletion in dl355 was constructed. This plasmid was used for site-directed mutagenesis of the E4 ORF3 gene. The mutagenesis scheme was to substitute neighboring conserved and/or charged residues with alanine pairs at 10- to 15-amino-acid intervals (ORF3 mutants depicted in Fig. 2). Individual mutations were introduced into pTG-JE1 by homologous recombination in E. coli. Mutant viral genomes were excised from the plasmid by PacI digestion and transfected into the E4-complementing cell line W162 (33). Mutant viruses were plaque purified and propagated. These mutants contained a deletion in E4 ORF6 and double alanine point mutations in E4 ORF3.
Growth of mutant viruses.
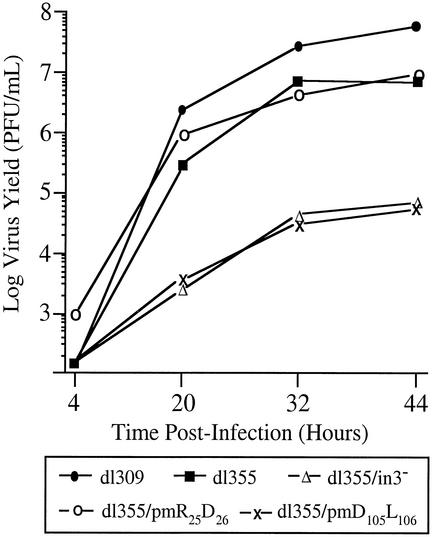
Single-step growth curves for noncomplementing A549 cells were used to evaluate the effects of the E4 ORF3 mutations on overall viral growth properties. Mutation of E4 ORF6 resulted in an ∼10-fold reduction in virus growth in A549 cells, and the additional deletion of E4 ORF3 further reduced virus growth ∼100-fold (Fig. 3), as previously described (4, 17). One of the E4 ORF3 mutant viruses, dl355/pmD105L106, was severely defective for growth in A549 cells (Fig. 3); this mutant was as defective for growth as dl355/inORF3, which does not express functional E4 ORF3, demonstrating a significant deficiency in E4 ORF3 activity. With one exception, the remainder of the E4 ORF3 mutant viruses grew to levels within ∼3-fold of dl355, which expresses wild-type E4 ORF3; the exception was mutant dl355/pmE10G11, which was reduced ∼15-fold in virus yield (Table 1). Due to the significant growth defect observed with mutant dl355/pmD105L106, this virus was used to further characterize the function of E4 ORF3 in virus growth.
FIG. 3.
Viral growth kinetics in A549 cells. A549 cells were infected with wild-type or mutant viruses at a multiplicity of 200 particles per cell. Virus yields in cellular lysates were measured at the times indicated by plaque assay on W162 cells. The results represent the averages from three independent experiments.
TABLE 1.
Growth properties of mutant virusesa
| Virus | Designation | Virus yield (PFU/ml) at:
|
||
|---|---|---|---|---|
| 4 h | 20 h | 32 h | ||
| dl309 | dl309 | 2.5 × 102 | 2.4 × 106 | 2.25 × 107 |
| dl355 | dl355 | 2.5 × 102 | 2.8 × 105 | 7.75 × 106 |
| dl355/inORF3 | dl355/in3 | 2.5 × 102 | 2.3 × 103 | 4.35 × 104 |
| dl355/pmE10G11 | pmEG | 5.0 × 102 | 2.0 × 104 | 5.25 × 105 |
| dl355/pmR25D26 | pmRD | 1.0 × 103 | 8.35 × 105 | 4.15 × 106 |
| dl355/pmD30I31 | pmDI | 1.0 × 103 | 5.0 × 106 | 5.0 × 106 |
| dl355/pmN39Y40 | pmNY | 2.5 × 102 | 7.5 × 105 | 2.0 × 107 |
| dl355/pmE52E53 | pmEE | 2.5 × 102 | 1.8 × 105 | 5.0 × 106 |
| dl355/pmF59L61 | pmFL | 2.5 × 102 | 6.0 × 105 | 2.5 × 106 |
| dl355/pmD66R68 | pmDR | 5.0 × 102 | 4.0 × 105 | 2.25 × 107 |
| dl355/pmH79L80 | pmHL | 2.5 × 102 | 1.9 × 106 | 7.5 × 106 |
| dl355/pmL88V90 | pmLV | 2.5 × 102 | 1.5 × 105 | 2.3 × 106 |
| dl355/pmD105L106 | pmDL | 2.5 × 102 | 4.0 × 103 | 3.25 × 104 |
| dl355/pmH107F108 | pmHF | 2.5 × 102 | 3.25 × 105 | 1.5 × 106 |
A549 cells were infected with the wild type and mutant viruses at a multiplicity of infection of 200 particles/cell. Virus yields in cellular lysates were measured at the times indicated by plaque assay on W162 cells. The full names of the mutant viruses and the abbreviated designations used in the figures are shown.
Viral DNA replication.
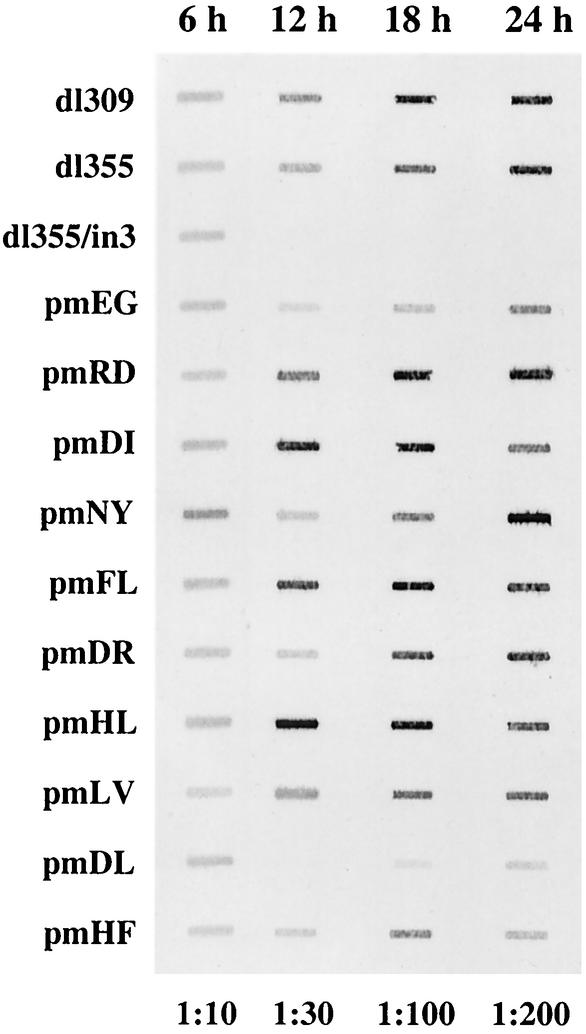
The phenotype observed with the mutant virus dl355/pmD105L106 could be attributed to a number of possible defects in the infection program. Viral DNA replication of the mutant viruses was analyzed (Fig. 4). The level of viral DNA at 6 h postinfection is reflective of input DNA, since this is before the onset of viral DNA replication. These levels were within 1.5-fold of one another for all of the viruses examined. The levels of viral DNA accumulation observed with mutants at later times after infection were entirely consistent with the data obtained in the single-step growth curves (Fig. 4 and Table 1). With the majority of the mutant viruses, DNA replication was comparable to that of wild-type Ad (dl309), whereas mutants dl355/inORF3 and dl355/pmD105L106 were significantly impaired in viral DNA accumulation. The decreases in viral DNA production for dl355/pmD105L106 were 47.4-fold at 12 h, 9.1-fold at 18 h, and 5-fold at 24 h postinfection (Fig. 4). These results suggest that the deficiency in virus production with mutant dl355/pmD105L106 likely reflects inefficient establishment of DNA replication, although dl355/pm D105L106 was able to accumulate DNA at later time points. Several of the mutant viruses (pmD30I31, pmF59L61, and pmH79L80) displayed modestly increased levels of viral DNA replication at the 12- and 18-h time points. This effect was reproducible, although we do not understand the basis for this result.
FIG. 4.
Viral DNA accumulation in A549 cells. A549 cells were infected with the wild type or mutant viruses at a multiplicity of 200 particles per cell. Total nuclear DNA was isolated at the times indicated above the blot, diluted as indicated below the blot, and applied to nylon membrane by using a slot blot apparatus. The blots were hybridized with a fluorescently labeled oligonucleotide probe. The signal was detected and quantified with a Molecular Dynamics Storm 860 phosphorimager and ImageQuant software.
Viral protein expression.
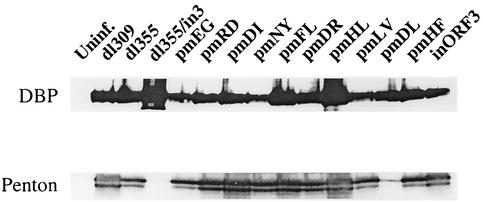
Infected-cell extracts were analyzed for early and late Ad protein expression. Extracts prepared early after infection (9 h) were probed for the expression of DBP (Fig. 5, top panel), an early protein necessary for Ad DNA replication. All of the viruses, including mutants dl355/inORF3 and dl355/pmD105L106, expressed DBP at similar levels. The same results were found when E1A protein expression was analyzed (data not shown), indicating that the decrease in Ad genome replication with mutants dl355/inORF3 and dl355/pmD105L106 was not due to insufficient early gene expression. Also, the presence of normal amounts of DBP suggests that the Ad replication machinery is available. Extracts prepared late after infection (18 h) were probed for the expression of penton (Fig. 5, bottom panel), a late Ad protein that is a key component of the virus capsid. A majority of the viruses produced penton to high levels. However, mutants dl355/inORF3 and dl355/pmD105L106 were substantially reduced for penton expression. The decrease in penton expression is in direct relation to the level of virus DNA replication observed with these mutants.
FIG. 5.
Viral protein synthesis in A549 cells. A549 cells were infected with wild-type or mutant viruses at a multiplicity of 200 particles per cell. Cells were infected for 9 h (top) or 18 h (bottom) and lysed in RIPA lysis buffer. Proteins were separated by SDS-12.5% PAGE, transferred to Hybond-P, and probed with the antibodies indicated on the left. The proteins were visualized with ECL reagents.
These data show that viruses containing point mutations in E4 ORF3 are capable of producing the early proteins necessary for virus DNA replication regardless of E4 ORF3 status. Conversely, expression of late Ad proteins is dependent on the ability of E4 ORF3 to facilitate virus DNA replication. Viral DNA replication with the mutant virus dl355/pm D105L106 was within 5-fold of that with dl355 at late times after infection, yet this mutant had >100-fold-decreased virus production. It appears that the defect in growth of dl355/pm D105L106 may be a synergistic one, coupling the decreases in viral DNA accumulation and late protein expression.
Reorganization of PML nuclear structures.
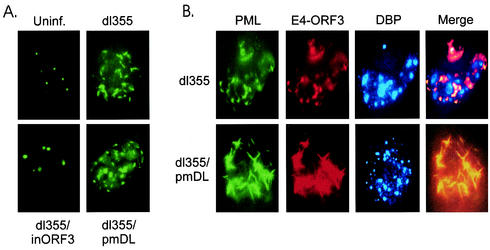
Ads from serotypes 2, 3, 5, 7, 9, and 12 were all found to redistribute the POD complex (data not shown), suggesting an evolutionary conservation of function. It was suggested previously that viral replication is linked to the dynamic reorganization of POD structures by E4 ORF3 (5, 10). However, this relationship has been poorly defined with respect to the mechanism by which E4 ORF3 redistributes POD components and to what end this serves the virus replication program. The reorganization of POD structures was analyzed with the wild type and the entire panel of mutant viruses at various times throughout the infection cycle by indirect immunofluorescence, using antibodies directed against PML, E4 ORF3, and DBP (Fig. 6). All of the point mutant viruses exhibited staining patterns similar to that of dl355, and thus they are described as having wild-type staining and dl355 is shown as the representative.
FIG. 6.
Localization of PML and viral proteins. A549 cells were infected with wild-type or mutant viruses at a multiplicity of 200 particles per cell. (A) Cells were infected for 4 h, and the POD structure was visualized with anti-PML antibody and fluorescein isothiocyanate-labeled secondary antibody. Uninf., uninfected. (B) Cells were infected for 14 h, and the POD structure, E4 ORF3 protein, and DBP-containing virus replication centers were visualized with anti-PML, anti-E4 ORF3, and anti-DBP, as described in Materials and Methods.
At 4 h postinfection, the redistribution of PML-containing structures into the previously described “track” appearance was observed in cells infected with viruses that express wild-type E4 ORF3 protein (dl355 is shown in Fig. 6A; data not shown for wild-type Ad5 and the remainder of the panel of point mutant viruses) as well as the pmD105L106 mutant protein (Fig. 6A). All the viruses tested expressed E4 ORF3 proteins to equivalent levels, which colocalized with PML (data not shown). Virus dl355/inORF3 does not express E4 ORF3, and thus PODs were not reorganized and E4 ORF3 was not present in cells infected with this mutant (Fig. 6A). It had been shown previously that DBP is weakly expressed at 4 h postinfection, and thus no DBP staining was evident in these samples.
At 14 h postinfection, expression of wild-type E4 ORF3 reorganized PML-containing structures into a more circular appearance (Fig. 6B, dl355). The PML/E4 ORF3 staining was completely coincident. DBP-containing Ad replication centers were readily evident, and the PML/E4 ORF3 signals appeared to be excluded from or only overlapped at the edges of the Ad DNA replication factories (Fig. 6B, dl355, Merge). It appears that the PML/E4 ORF3 structures may act to sequester DBP-positive replication centers to certain centers within the nucleus, perhaps to areas that are appropriate for virus DNA replication. The dl355/pmD105L106 virus continued to express high levels of E4 ORF3 protein at 14 h postinfection (Fig. 6B, dl355/pmDL). However, this virus maintained the track-like reorganization of PODs, failing to manipulate PML to encircle the DBP replication centers. The PML tracks seen in dl355/pmD105L106-infected cells were exaggerated in length compared to those seen in cells infected with a virus expressing wild-type E4 ORF3. DBP was easily observed in cells infected with dl355/pmD105L106, but it was present in the nucleus in a large number of small, dispersed points, in contrast to the organized centers observed in cells expressing wild-type ORF3. These results correlate the inefficiency of DNA replication observed with mutant dl355/pmD105L106 with an aberrant reorganization of PML structures and an inability to organize Ad DNA replication centers in a typical manner.
Roles of E4 ORF3 in viral DNA replication and inhibition of genome concatenation are genetically separable.
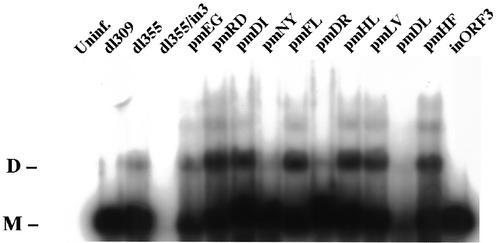
Viral genome concatenation was analyzed with the panel of mutant viruses by pulsed-field gel electrophoresis (Fig. 7). Monomer-length viral DNA was observed in cells infected with wild-type Ad5 (dl309) and a mutant that does not express E4 ORF3 (inORF3). Monomer- and dimer-length genomes were seen in cells infected with dl355, the mutant that does not express E4 ORF6 protein. Increased levels of dimeric and higher-order genomes, indicative of augmented genome concatentaion, were observed in cells infected with a number of the E4 ORF3 point mutants in comparison to dl355 (Fig. 7, pmRD, pmDI, pmFL, pmHL, pmLV, and pmHF). The levels of genome concatenation observed with these mutants are comparable with those observed with E4 mutants in previous studies using different cell lines (3, 30, 32). Very low levels of viral DNA were evident in cells infected with dl355/inORF3 or dl355/pmD105L106, as expected, and thus genome catenation was difficult to quantify. However, dl355/pmD105L106 did not inhibit concatemer formation, as seen faintly in Fig. 7 and upon a longer exposure of the blot (data not shown). It has been suggested that E4 ORF3 ensures efficient virus DNA replication and late protein expression by blocking the inhibitory effects of Ad genome concatenation (3, 30, 32). Our data indicate otherwise. Significant concatemer formation was observed with a number of the E4 ORF3 point mutants (Fig. 7) that did not interfere with viral DNA replication (Fig. 4). These results demonstrate that the functions of E4 ORF3 in viral DNA replication and inhibition of genome concatenation are genetically separable.
FIG. 7.
Ad genome concatemer formation. A549 cells were infected with wild-type or mutant viruses at a multiplicity of 200 particles per cell. Cells were harvested 36 h after infection, solidified in agarose plugs, and treated with SDS-proteinase K solution. The DNA were separated on 1.0% agarose by pulsed-field gel electrophoresis and transferred to Hybond-N. Viral unit-length genome and multimers were hybridized with 32P-labeled Ad genomic probe. Uninf., uninfected; D, dimer; M, monomer.
E4 ORF3 functions do not correlate with association of known binding partners.
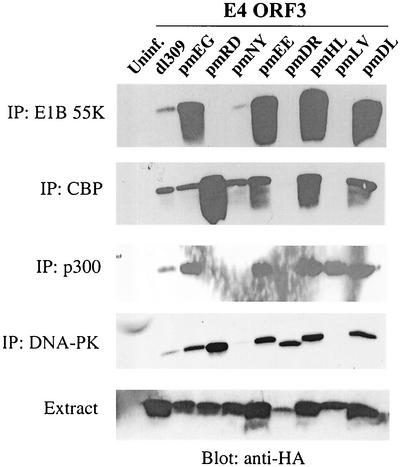
The E4 ORF3 protein has been shown to interact with the Ad E1B 55K protein during viral infection and in transformed cells (21, 26), as well as with the cellular protein kinase DNA-PK, by using transient-expression assays (3). The latter interaction has been linked to Ad inhibition of DSBR. The E4 ORF3 protein effects the nuclear reorganization of the histone acetyltransferase CBP in a manner analogous to E4 ORF3-induced PML redistribution (34). We wanted to test the interaction of the panel of E4 ORF3 mutant proteins with known protein binding partners, as well as potential binding partners, based on published results. The panel of E4 ORF3 point mutants was built into recombinant Ad vectors that were defective for expression of early region 1 proteins and E4 ORF1, ORF2, and ORF3 (to eliminate expression of wild-type E4 ORF3 protein). The E4 ORF3 proteins were tagged at their amino termini with an HA epitope. The ability of the wild-type and mutant E4 ORF3 proteins to interact with cellular and viral polypeptides was analyzed by immunoprecipitation following infection of A549 cells. Antibodies directed against E1B 55K, CBP, p300, and DNA-PK were used for immunoprecipitation, and coprecipitation of E4 ORF3 was analyzed by Western blotting with anti-HA antibody. The immunoprecipitation data are presented in Fig. 8, and the results are summarized in Table 2. Wild-type E4 ORF3 protein was found reproducibly to coprecipitate with all four of the proteins examined. However, the E4 ORF3 mutant proteins exhibited a complex interaction profile, with no clear pattern for binding evident. For example, mutant proteins pmR25D26 and pmN39Y40 bound to CBP, but they did not bind the closely related protein p300. Conversely, mutant protein pmL88V90 bound to p300 but not CBP. Importantly, the pmD105L106 mutant protein was capable of binding all four of the different proteins examined, yet this mutant had the most severe defect in growth properties of the mutants under study. Further, no correlation was evident between the binding of E4 ORF3 to DNA-PK and the ability of the corresponding mutant viruses to inhibit genome concatenation. The primary constituent of PODs, PML, was unable to interact with E4 ORF3 when anti-PML antibody was used for immunoprecipitation (data not shown). This finding indicates that PML may not be the direct target of E4 ORF3. It also served as a control immunoprecipitation with an isotype-matched antibody to the monoclonal antibody used for DNA-PK. In addition, a nonspecific rabbit antibody did not immunoprecipitate E4 ORF3 as a control for the CBP and p300 immunoprecipitations. In summary, no direct correlation was observed between the binding of E4 ORF3 to different viral and cellular proteins and the efficiency of virus DNA replication, POD rearrangement, inhibition of genome concatenation, or virus growth. Thus, the relevance of these protein-protein interactions to virus replication remains unclear. However, the fact that certain of the E4 ORF3 point mutations disrupt the interaction with one or more different binding partners while other mutations do not affect such binding demonstrates the specificity of these interactions and suggests that they may play a role in a function(s) of E4 ORF3 that is not yet understood.
FIG. 8.
Interaction of E4 ORF3 point mutants with cellular and viral proteins. A549 cells (for CBP, p300, and DNA-PK) or 293 cells (for E1B 55K) were infected with E1 replacement viruses expressing HA-tagged wild-type or mutant E4 ORF3 proteins at a multiplicity of 200 particles per cell. Cells were infected for 12 h, and RIPA lysates were prepared. Extracts were immunoprecipitated (IP) with the antibody indicated at the left. Proteins were separated by SDS-20% PAGE and transferred to Hybond-P. Membranes were probed with anti-HA mouse monoclonal antibody. Proteins were visualized with ECL reagents. Uninf., uninfected.
TABLE 2.
Binding properties of mutant E4 ORF3 proteins
| E4 ORF3 | Bindinga to:
|
|||
|---|---|---|---|---|
| E1B55K | DNA-PK | p300 | CBP | |
| Wild type | + | + | + | + |
| pmEG | + | + | + | + |
| pmRD | − | + | − | + |
| pmNY | +/− | − | − | + |
| pmEE | + | + | + | + |
| pmDR | − | + | − | − |
| pmHL | + | + | + | + |
| pmLV | − | − | + | − |
| pmDL | + | + | + | + |
Results of coimmunoprecipitation analyses.
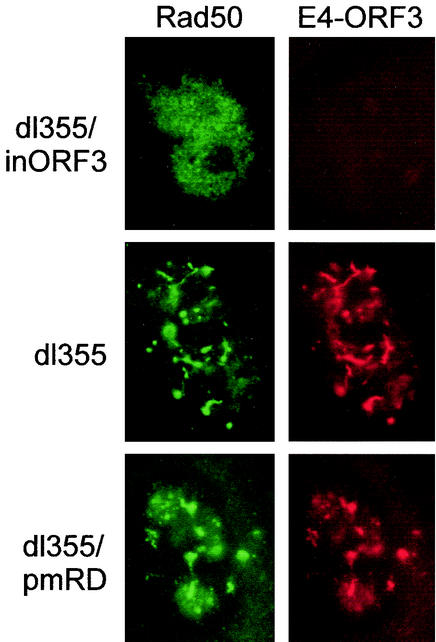
Relocalization of the MRE11/NBS1/Rad50 complex does not inhibit concatenation.
The presence of concatenated genomes with a number of the E4 ORF3 mutant viruses suggested that the mutant E4 ORF3 proteins were defective for inhibition of the DSBR machinery. Recent work has shown that E4 ORF3 inhibits DNA-PK (3) as well as relocalizes components of the MRE11/NBS1/Rad50 DNA repair complex (30). We analyzed localization of Rad50 by indirect immunofluorescence in cells infected with viruses that lack E4 ORF6 and that express wild-type E4 ORF3 (dl355), no E4 ORF3 (dl355/inORF3), and a mutant E4 ORF3 that did not inhibit Ad genome concatenation (dl355/pmR25D26). These results are shown in Fig. 9. Rad50 had a diffuse nuclear staining pattern in cells infected with dl355/inORF3 (Fig. 9) and in uninfected cells (data not shown), with occasional punctate staining. In cells that expressed wild-type E4 ORF3 (Fig. 9, dl355), Rad50 was redistributed to dispersed nuclear foci that were largely coincident with the pattern of E4 ORF3 localization. This staining was similar to previously published data for cells infected with Ad (30). The same relocalization of Rad50 was observed in cells infected with mutant virus dl355/pmR25D26 (Fig. 9). Rad50 foci localized to the periphery of DBP-containing structures, and very similar relocalization patterns were observed for MRE11 and NBS1 (data not shown). We conclude that relocalization of the Rad50 component of the MRE11/NBS1/Rad50 DNA repair complex in Ad infection is not sufficient to inhibit DSBR.
FIG. 9.
Relocalization of Rad50 by E4 ORF3. A549 cells on coverslips were infected with wild-type or mutant viruses at multiplicity of 200 particles per cell. Cells were infected for 18 h, and the Rad50 and E4 ORF3 proteins were visualized by using anti-Rad50 and anti-E4 ORF3, as described in Materials and Methods.
DISCUSSION
The reorganization of PML nuclear domains is a common feature associated with infection by many DNA viruses. Early after infection with papovaviruses (e.g., simian virus 40 and human papillomavirus), Ads, and herpesviruses (e.g., herpes simplex virus type 1 and CMV), the viral genome is localized adjacent to POD structures (reviewed in reference 22). POD reorganization appears to be a prerequisite for viral DNA replication, but not early gene expression, to commence. In order to analyze the effect of E4 ORF3 on virus replication and PML nuclear domains, an infectious clone that facilitated the expression of mutant E4 ORF3 proteins from the natural virus promoter was developed. Our data show that the redistribution of PODs by the Ad E4 ORF3 protein alone is not sufficient to initiate the DNA replication program. Rather, a specific reorganization of PML nuclear bodies appears to be required for efficient Ad DNA replication. Very early after infection (4 h), the mutant virus dl355/pmD105L106 disrupted PODs in a manner similar to that of wild-type Ad5, yet at later times, aberrant PML redistribution was evident with this mutant (Fig. 6). Mutant dl355/pmD105L106 exhibited a severe defect in DNA replication in noncomplementing cells (Fig. 4) at a time when aberrant POD structures were evident. Wild-type E4 ORF3 caused PML to encircle the DBP-containing viral replication centers (Fig. 6B, dl355). This is reminiscent of cellular DNA replication factories being adjacent to, but not colocalized with, PML nuclear domains (16). The mutant virus dl355/pmD105L106 was unable to reorganize PODs in this manner. Instead, exaggerated track-like structures were formed with this mutant virus, and DBP was localized to numerous, punctate structures in the nucleus (Fig. 6B, dl355/pmD105L106). Thus, a specific pattern of POD reorganization correlates directly with the organization and function of Ad DNA replication centers.
The highly conserved sequence of the E4 ORF3 protein among different Ad serotypes and the ability of evolutionarily diverse Ads to modify the appearance of POD structures attest to the important role that the reorganization of PML nuclear bodies plays in establishing virus infection. What is not apparent is if the reorganization of PODs to specific nuclear structures recruits Ad replication proteins to particular sites in the nucleus that are compatible with viral DNA replication or if viral replication proteins target nuclear regions that are devoid of PML to establish replication factories. However, it is clear that Ad replication centers form within newly reorganized PML structures in the nucleus, consistent with an earlier report (18). It is possible that the newly synthesized viral DNA itself displaces the contents of the nucleus, including the PML/E4 ORF3 track structures. However, this seems unlikely, since other cellular proteins in the nucleus remain unaffected and the nuclear boundaries are extended to only minimal degrees. The nature of the PML/E4 ORF3 track structures and their cause-effect relationship with viral replication centers remain an interesting and open area for study.
It is well established that the E4 ORF3 and E4 ORF6 proteins have compensatory functions in viral infection and are important for efficient viral DNA replication, late gene expression, and virus production (4, 17). A recent report linked the ability of both proteins to regulate the MRE11 DNA repair complex with the overlapping activities of these proteins in viral infection (30). The E4 ORF3 protein disrupts the organization of the MRE11 DNA repair complex in PML nuclear domains, and the E4 ORF6 protein targets the MRE11 component of this complex for ubiquitin-dependent degradation. Our results are consistent with that report but also demonstrate that the role of these E4 proteins in the establishment of Ad DNA replication precedes their role in the inhibition of cellular DSBR. The function of E4 ORF3 in Ad DNA replication is genetically separable from its inhibition of genome concatenation, since a number of mutant viruses were capable of efficient DNA replication and virus production yet were unable to inhibit concatenation of viral genomes (Fig. 7). Also, a virus that could not block genome multimerization (dl355/pmR25D26) was able to relocalize Rad50 DNA into the same pattern as a virus that can inhibit most concatenation (dl355) (Fig. 9). These data indicate that the relocalization of Rad50, and likely that of the entire MRE11/NBS1/Rad50 complex, is not sufficient to inhibit DSBR. The primary defect observed with the mutant virus dl355/pmD105L106 was a severe deficiency in the initiation of viral DNA replication. This defect correlated directly with a significant decrease in virus growth exhibited by this mutant. Thus, while Ad genome concatenation is a process that the virus has evolved several proteins to circumvent, genome multimerization does not have a major impact on virus yield in cultured cells. These results are contrary to previously accepted notions that E4 ORF3 and E4 ORF6 are redundant in their roles in replication and inhibition of genome concatenation. It is likely that the profound defect observed in overall virus yield with the inhibition of both E4 ORF3 and E4 ORF6 functions (mutants dl355/inORF3 and dl355/pmD105L106) (Fig. 3 and Table 1) is a synergistic effect caused by the lack of viral DNA replication combined with significantly reduced late gene expression (Fig. 5).
Previously, it had been shown that E4 ORF3 interacts with both viral (E1B 55K) and cellular (DNA-PK) proteins and also regulates the subnuclear localization of the transcriptional regulator CBP (3, 21, 26, 34). The association of E4 ORF3 with these proteins has been linked to functions important for virus replication or transformation. In addition to E1B 55K and DNA-PK, the wild-type E4 ORF protein was found to interact with histone acetyltransferases CBP and p300 (Fig. 8). The panel of mutant E4 ORF3 proteins, however, exhibited a complex pattern of association with these viral and cellular proteins. It was clear from these analyses that the binding of different proteins by E4 ORF3 does not correlate with virus DNA replication, POD rearrangement, inhibition of genome concatenation, or virus growth. In each case, a viral mutant that could not bind one or more E4 ORF3-associated proteins but showed wild-type growth properties was identified (Table 2). Most strikingly, point mutant D105L106 was the most defective virus identified, yet the D105L106 mutant protein was able to bind all of the associating proteins as well as or better than wild-type E4 ORF3 protein. These data indicate that the primary functional binding target of E4 ORF3 has not been determined. The interaction of E4 ORF3 with the DSBR protein DNA-PK was implicated in the mechanism of inhibition of concatenation of viral genomes (3). However, our data demonstrate that the interaction of E4 ORF3 with DNA-PK may not be essential for inhibition of concatemer formation, nor is concatenation wholly detrimental to virus replication.
The increase in binding of certain E4 ORF3 mutant proteins to some binding partners (Fig. 7) suggests that there is competition in vivo for interaction with limiting E4 ORF3 protein. For example, mutant pmRD did not bind E1B 55K or p300 but showed enhanced binding to CBP. That this does not represent a global misfolding of mutant E4 ORF3 proteins is supported by the fact that all mutants grew as well as the wild type, with the exception of pmDL. PML, the primary component of PODs, was unable to interact with E4 ORF3, indicating that POD reorganization is carried out in a manner not dependent on the direct interaction of PML and E4 ORF3.
The specific mechanism by which E4 ORF3 achieves POD reorganization and DNA replication initiation remains unclear. It also is not clear how the E4 ORF6 protein accomplishes the same goal without any discernible rearrangement of PML nuclear bodies. E4 ORF6 recently was shown to interact with an E3 ubiquitin ligase complex to direct the degradation of specific cellular proteins such as p53 and MRE11 (15, 27). It is possible that E4 ORF6 achieves a similar goal of establishing Ad DNA replication centers by directing the degradation of a cellular protein(s) that inhibits Ad replication early after infection, but without affecting overall POD architecture. This idea would indicate that the adjacent localization of Ad DBP with E4 ORF3-reorganized PML structures observed in wild-type Ad infection reflects the establishment of Ad replication centers at sites that lack PML, rather than PML recruitment of Ad replication proteins to specific nuclear domains. Otherwise, it would be difficult to reconcile how E4 ORF6 allows the establishment of Ad replication centers in regions that lack PML.
The analysis of E4 ORF3 mutants expressed from the natural E4 promoter and without E4 ORF6 has provided new information about the function of this Ad protein. Our data show that E4 ORF3 is a multifunctional protein that temporally regulates the viral infection program. Virus DNA replication is controlled by the coordinated reorganization of POD complexes into a specific structure as manipulated by E4 ORF3. This function of E4 ORF3 appears to induce an initiating event for DNA replication. Later, the E4 ORF3 protein regulates viral genomic concatenation by interfering with the induction of cellular DSBR (3, 30). These two functions complement one another to allow for the maximal yield of viral genomes available for virus assembly.
Acknowledgments
We gratefully acknowledge Thomas Dobner, Carl W. Anderson, and Arnold Levine for making antibody reagents available to us. We also thank Gary Ketner for providing technical advice for the pulsed-field gel electrophoresis and Wadie Bahou and Jean Wainer for advice and use of their equipment. We thank all of the members of our laboratory for many helpful discussions; Michael Hayman, Dafna Bar-Sagi, Kenneth Marcu, Carl Anderson, and David Spector for expert advice; and Jihong Yang for excellent technical assistance.
This work was supported by Public Health Service grant CA 28146 to P.H. J.D.E. was supported by NIH training grant CA09176.
REFERENCES
- 1.Ahn, J. H., and G. S. Hayward. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71:4599-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borden, K. L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase Virology 263:307-312. [DOI] [PubMed] [Google Scholar]
- 4.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carvalho, T., J. S. Seeler, K. Ohman, P. Jordan, U. Pettersson, G. Akusjarvi, M. Carmo-Fonseca, and A. Dejean. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobner, T., N. Horikoshi, S. Rubenwolf, and T. Shenk. 1996. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 272:1470-1473. [DOI] [PubMed] [Google Scholar]
- 9.Doucas, V. 2000. The promyelocytic (PML) nuclear compartment and transcription control. Biochem. Pharmacol. 60:1197-1201. [DOI] [PubMed] [Google Scholar]
- 10.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 11.Evans, J. D., and P. Hearing. 2002. Adenovirus Replication, p. 39-70. In D. T. Curiel and J. T. Douglass (ed.), Adenoviral vectors for gene therapy. Elsevier, New York, N.Y.
- 12.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 13.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada, J. N., A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hozak, P., D. A. Jackson, and P. R. Cook. 1994. Replication factories and nuclear bodies: the ultrastructural characterization of replication sites during the cell cycle. J. Cell Sci. 107:2191-2202. [DOI] [PubMed] [Google Scholar]
- 17.Huang, M. M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 20.Konig, C., J. Roth, and M. Dobbelstein. 1999. Adenovirus type 5 E4orf3 protein relieves p53 inhibition by E1B-55-kilodalton protein. J. Virol. 73:2253-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leppard, K. N., and R. D. Everett. 1999. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J. Gen. Virol. 80:997-1008. [DOI] [PubMed] [Google Scholar]
- 22.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660-667. [DOI] [PubMed] [Google Scholar]
- 23.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 24.Melnick, A., and J. D. Licht. 1999. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood 93:3167-3215. [PubMed] [Google Scholar]
- 25.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nevels, M., B. Tauber, E. Kremmer, T. Spruss, H. Wolf, and T. Dobner. 1999. Transforming potential of the adenovirus type 5 E4orf3 protein. J. Virol. 73:1591-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarnow, P., P. Hearing, C. W. Anderson, N. Reich, and A. J. Levine. 1982. Identification and characterization of an immunologically conserved adenovirus early region 11,000 Mr protein and its association with the nuclear matrix. J. Mol. Biol. 162:565-583. [DOI] [PubMed] [Google Scholar]
- 29.Schmid, S., and P. Hearing. 1999. Adenovirus DNA packaging, p. 49-59. In W. S. Wold (ed.), Adenovirus methods and protocols. Humana Press, Totowa, N.J.
- 30.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 31.Tauber, B., and T. Dobner. 2001. Molecular regulation and biological function of adenovirus early genes: the E4 ORFs. Gene 278:1-23. [DOI] [PubMed] [Google Scholar]
- 32.Weiden, M. D., and H. Ginsberg. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. USA 91:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinberg, D. H., and G. Ketner. 1983. A cell line that supports the growth of a defective early region 4 deletion mutant of human adenovirus type 2. Proc. Natl. Acad. Sci. USA 80:5383-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wienzek, S., and M. Dobbelstein. 2001. Viral and cellular factors that target the promyelocytic leukemia oncogenic domains strongly activate a glucocorticoid-responsive promoter. J. Virol. 75:5391-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]