Abstract
Swine vesicular disease virus (SVDV) is an Enterovirus of the family Picornaviridae that causes symptoms indistinguishable from those of foot-and-mouth disease virus. Phylogenetic studies suggest that it is a recently evolved genetic sublineage of the important human pathogen coxsackievirus B5 (CBV5), and in agreement with this, it has been shown to utilize the coxsackie and adenovirus receptor (CAR) for cell entry. The 3.0-Å crystal structure of strain UK/27/72 SVDV (highly virulent) reveals the expected similarity in core structure to those of other picornaviruses, showing most similarity to the closest available structure to CBV5, that of coxsackievirus B3 (CBV3). Features that help to cement together and rigidify the protein subunits are extended in this virus, perhaps explaining its extreme tolerance of environmental factors. Using the large number of capsid sequences available for both SVDV and CBV5, we have mapped the amino acid substitutions that may have occurred during the supposed adaptation of SVDV to a new host onto the structure of SVDV and a model of the SVDV/CAR complex generated by reference to the cryo-electron microscopy-visualized complex of CBV3 and CAR. The changes fall into three clusters as follows: one lines the fivefold pore, a second maps to the CAR-binding site and partially overlaps the site for decay accelerating factor (DAF) to bind to echovirus 7 (ECHO7), and the third lies close to the fivefold axis, where the low-density lipoprotein receptor binds to the minor group of rhinoviruses. Later changes in SVDV (post-1971) map to the first two clusters and may, by optimizing recognition of a pig CAR and/or DAF homologue, have improved the adaptation of the virus to pigs.
There are currently nine genera in the picornavirus family, including the enteroviruses, rhinoviruses, and aphthoviruses. Swine vesicular disease virus (SVDV) belongs to the Enterovirus genus. Swine vesicular disease is classified as a list A disease by the Office International des Epizooties because the lesions produced by SVDV are indistinguishable from those caused by foot-and-mouth disease virus, an aphthovirus. Like foot-and-mouth disease, swine vesicular disease is highly contagious and difficult to eradicate, and with no effective vaccine, control measures necessitate the slaughter of infected and contacted animals. SVDV is extraordinarily resistant to environmental factors and is able to resist desiccation, freezing, and the fermentation and smoking processes used to preserve food. It can survive in a pH range of 2.5 to 12 and in temperatures from 12 to −20°C for 4 to 11 months, is resistant to most common disinfectants, and must be held at 56°C for 1 h to be heat inactivated (43). SVDV isolates form a single genetic lineage within the coxsackievirus B5 (CBV5)/SVDV species in both the capsid and nonstructural coding regions, which is suggestive of a single transfer of CBV5 into pigs, and an approximate molecular clock indicates that this transfer took place between 1945 and 1965 (66).
Atomic resolution structures have been determined for representatives of four genera of the family Picornaviridae (1, 28, 42, 56), all of which are broadly similar. The nonenveloped particles are composed of four viral proteins (VPs) that make up the 30-nm-diameter capsid that surrounds approximately 7,500 nucleotides of single-stranded RNA. The capsid possesses icosahedral symmetry, being made up of 60 copies of each of the four viral coat proteins, VP1 to VP4, which are arranged with T=1 (pseudo T=3) symmetry. VP1, -2, and -3 contain on the order of 270 amino acid residues each, folded into eight-stranded antiparallel β-sheet structures with a jelly-roll topology, with the constituent strands labeled BIDG and CHEF. The loops are labeled according to the strands they connect and these form the outer surface of the virus. VP4 contains only about 70 amino acids and lies on the inside of the capsid. Apart from those on the inner surface of the capsid, the major differences among the picornavirus structures occur in the surface-exposed loop regions that define the receptor-binding and principle antigenic sites. Despite this variation, there is a remarkable conservation in the three-dimensional distribution of these sites.
The structure of human rhinovirus type 14 (HRV14) (56) revealed a canyon surrounding each fivefold axis that is wide enough to bind its cellular receptor, intercellular adhesion molecule 1 (ICAM-1), but apparently too narrow to suitably bind an antibody. This prompted the canyon hypothesis (55) proposing that the spatial seclusion of a necessarily conserved receptor-binding site might be a general strategy facilitating viral escape from immune surveillance. It was later shown that ICAM-1 does indeed bind to the canyon of HRV16 and HRV14 (35, 52) and, furthermore, that coxsackievirus A21 (CAV21), CBV3, and poliovirus (PV) (enteroviruses) bind their receptors (ICAM-1, the coxsackie and adenovirus receptor [CAR], and CD155, respectively) at an analogous site (3, 24, 57, 65). However, the structure of an HRV14-antibody complex showed that there is no strict seclusion of receptor-binding residues (59). This is certainly true of foot-and-mouth disease virus, in which the integrin attachment site is very exposed and part of the major immunogenic site (41, 62). Furthermore, the cryo-electron microscopy-visualized complex of a minor receptor group rhinovirus (HRV2) with the first three ligand-binding repeats of its receptor, the very-low-density lipoprotein receptor, reveals the binding site on the virus as a small star-shaped dome on the icosahedral fivefold axes (27). Finally, the binding site for decay accelerating factor (DAF) on echovirus 7 (ECHO7) is close to the icosahedral twofold axes (25) and is also exposed.
Classification based on pathology and serology has identified 23 CAV and 6 CBV serotypes; the virus most similar genetically to SVDV for which there is a three-dimensional structure is CBV3 (50). These viruses share 87% sequence identity for the capsid proteins. CBV3 uses two different receptors, CAR (4) and DAF (5). Experimental evidence suggests that SVDV can also utilize both CAR and DAF receptors (45). The next closest virus for which an X-ray structure is available is CAV9 (26), which shares 73% amino acid identity with SVDV for the capsid proteins. In fact, CAV9 is genetically more similar to CBVs and echoviruses than to other CAVs (9) and possesses an ∼15-amino-acid C-terminal extension of VP1 containing an arginine-glycine-aspartate (RGD) tripeptide (9), most likely an acquired integrin-binding site.
In order to be capable of disassembly and yet maintain stability outside the host, the virus must be capable of a triggered conformational change. In the case of enteroviruses and rhinoviruses, uncoating is believed to occur by the formation of A particles (58), virions with an altered structure that have been observed by electron microscopy. Three features of viral structure have been discussed as potential modulators of uncoating: first, there is a hydrophobic pocket in VP1, under the base of the canyon that can bind so-called pocket factors, stabilizing the capsid (13, 39); second, there is a myristoyl group bound to the N terminus of VP4 that may insert into the host membrane (10); and third, there are ions on the fivefold axes that may respond to the change in pH of the endocytotic vesicle and allow extrusion of the RNA through the vertex of the fivefold axes (32). Although the nature of the events leading to uncoating remains unclear, compounds that bind tightly within the VP1 pocket inhibit uncoating and represent potential therapeutic agents (60).
We report the determination of the three-dimensional structure of SVDV, which allows us to map the CAR receptor-binding site by superimposition with the CBV3/CAR complex. We then use this to analyze the sequence changes that have occurred since the presumably relatively recent transition of this virus from humans to pigs (66).
MATERIALS AND METHODS
Virus purification and crystallization.
Thrice plaque-purified SVDV (strain UKG/27/72), for which the sequence was verified, was cultured in pig IB-RS-2 cells. The virus was purified by sucrose density gradient centrifugation, and the resultant material (at a concentration of 10 mg/ml) was crystallized by the sitting-drop vapor diffusion method (23) by mixing equal volumes with the reservoir solution containing 15 to 25% saturated ammonium sulfate in 100 mM phosphate buffer (pH 7.6). The “toblerone” morphology crystals (maximum, 0.1 by 0.1 by 0.2 mm3) have unit cell dimensions of a = 354.1 Å, b = 371.7 Å, c = 318.6 Å, and α = β = γ = 90° and differ from those previously reported (40).
Data collection and reduction to a space group.
A 49% complete data set, extending to 3-Å Bragg spacings and containing 406,689 unique reflections, was collected on station 14.2 at the SRS Daresbury, representing the extent of diffraction from a single unfrozen crystal. Data collection statistics are shown in Table 1. Data were recorded on an ADSC Quantum IV detector using an oscillation range of 0.5° and exposure time of 10 s per image. The data were autoindexed, integrated, and scaled using the HKL package programs DENZO and SCALEPACK (53). The symmetry of the intensity data indicated that the crystals belong to a primitive orthorhombic space group. The data set is incomplete, but the intensities of the axial reflections are consistent with space group P212121. Nevertheless, packing considerations dictate that the space group must be P21212 (or some permutation thereof), in which case the crystallographic asymmetric unit comprises half a virus particle such that there is 30-fold noncrystallographic symmetry. Inspection of the strength of different classes of reflections suggested that the position of the virus particle and its orientation conspired to render the space group pseudo-I222. Thus, Table 2 shows the relative strength of the reflections, with odd and even index sums for shells with different resolutions. While still significant, the reflections with odd index sums are far weaker than the reflections with even index sums, especially at low resolution. If the space group is assigned arbitrarily for now as P21212, then the particle must be positioned almost exactly at (0,0,1/4), with an icosahedral twofold axis aligned almost exactly along the crystallographic axis. Definitive assignment of the space group was deferred to later in the analysis.
TABLE 1.
X-ray data collection and structure refinement statistics
| Parameter (unit) | Value |
|---|---|
| Data collection and processing | |
| Wavelength (Å) | 0.979 |
| Dmin (Å) | 3.0 |
| No. of crystals | 1 |
| No. of images | 56 |
| Rotation per image (°) | 0.5 |
| No. of unique reflections | 406,689 |
| R merge (%)a | 20.7 |
| Completeness (%)b | 49.2 |
| 〈I/σ(I)〉 | 5.1 |
| Averaging correlation coefficient (20-3 Å)c | 0.95 |
| Refinement | |
| Data range (Å) | 15-3 |
| No. of unique reflections (space group I222) | 253,299 |
| Final statistics | |
| R factor (%)d | 24.5 |
| Protein atoms | 7,802 |
| Solvent | NAf |
| Nonprotein atoms | 18 Sphingosine, 15 Myristoyl |
| RMSD bond lengths (Å)e | 0.018 |
| RMSD angles (°)e | 2.2 |
| Average RMSD B (main-chain [Å2]) | 2.3 |
| Average B factor (main-chain [Å2]) | 14 |
R = 100×∑h ∑j ‖Ih, merged| − |Ih, j‖ / ∑h N|Ih, merged|, where j = 1,…, N for N data sets.
Completeness of the highest resolution shell (3.11 to 3.0 Å) was 46%.
C = 100 × ∑h (〈Fobs〉 − |Fh, obs|)(〈Fav〉 − |Fh, av|) / [∑h (〈Fobs〉 − |Fh, obs|)2 × ∑h (〈Fav〉 − |Fh,av|)2]1/2.
R = 100 × ∑h ‖Fh, obs| − |Fh, cale‖ / ∑h |Fh,obs|.
Root mean square deviation from ideal bond lengths or bond angles.
NA, not available.
TABLE 2.
Reflection intensity analysis
| Resolution range (Å) | 〈I/σ(I)〉 (h + k + l) = odd | 〈I/σ(I)〉 (h + k + l) = even |
|---|---|---|
| 20.0-8.0 | 6.0 | 15.4 |
| 8.0-6.0 | 5.9 | 12.4 |
| 6.0-4.5 | 7.5 | 11.8 |
| 4.5-3.5 | 6.8 | 9.1 |
| 3.5-3.0 | 4.5 | 5.4 |
Structure determination.
Measured diffraction intensities were converted to structure factor amplitudes using the CCP4 program TRUNCATE (15). Calculation of a self-rotation function (program CNS [8]) using data from 20- to 6.0-Å resolution, with a minimum and maximum integration radius of 20 and 280 Å, respectively, revealed that icosahedral twofold symmetry axes were, as expected, aligned with all of the crystallographic axes. Using the particle orientation derived from the rotation function assuming space group P21212 and placing a model (one-half of the CAV9 virion) at (0,0,1/4) gave a correlation coefficient between observed and calculated structure factors of 0.593 (data to 5 Å). Fine rotation and translation searches about this position gave broad peaks and did not discriminate from the initial positioning. Thirty noncrystallographic symmetry operators were calculated by use of GAP (D. I. Stuart and Jonathan Grimes, unpublished data) for the appropriately rotated CAV9 half-virion. After a few steps of positional refinement (by CNS [8]), the correlation coefficient was 0.744 (data to 3 Å). At this point a 2Fo-Fc map was calculated (Fo are observed and Fc are calculated structure factors) and used as the starting point for 10 cycles of 30-fold noncrystallographic averaging using previously described envelope procedures (16), at the end of which the R-factor and correlation coefficients in reciprocal space between observed and averaged structure factors were 21.1% and 0.95, respectively (for all data from 20- to 3-Å resolution). The final map was very clear and confirmed the sequence changes from CAV9 (Fig. 1). Iterative model building and refinement were carried out by the interactive graphics program O (30) and X-PLOR, version 3.1 (7), using positional and B-factor refinement. We established at this stage that, with the axis assignment given above, the space group is P22121, with the virus particle rotated about the a axis by 0.3°. Interestingly, we found that the most effective refinement was achieved by assuming space group I222 (with strict 15-fold noncrystallographic symmetry constraints and a bulk solvent correction). Thus, the progress to a minimum of the target function was more rapid in I222, presumably due to the lack of noise relative to P22121, for which there are a plethora of very weak (and hence poorly measured) reflections. It is interesting that interactions between local symmetry axes and crystallographic axes made space group determination problematic for these crystals, a situation encountered before in virus crystallography (14).
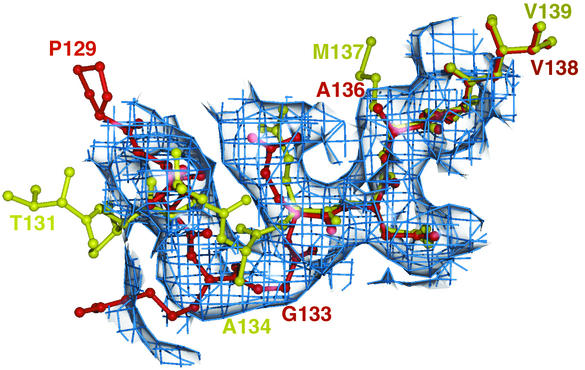
FIG. 1.
Sample region of an Fo-Fc omit map (omitting residues 130 to 137 of VP1). The electron density is shown as a blue mesh. SVDV residues and CAV9 residues from the same loop are shown as red and yellow ball-and-stick diagrams, respectively, and are labeled in the corresponding colors. The electron density clearly shows that the SVDV phases have no residual bias from the CAV9 model used to produce them. Figures were generated with a version of MOLSCRIPT (36) modified by R. Esnouf (11, 12) and with Raster3D (46).
Protein sequence accession numbers.
The coordinates and structure factors have been deposited with Brookhaven Protein Data Bank (accession number 100P).
RESULTS
Overall structure.
The structure was solved by molecular replacement and refined by use of strict noncrystallographic constraints (see Materials and Methods and Table 1). The electron density maps were of good quality and consistent with the sequence. A sample region of an omit Fo-Fc map (omitting residues 130 to 137 of VP1) is shown in Fig. 1, together with the SVDV and CAV9 coordinates. The final R factor for the model was 24.5%. Eighty-four percent of the residues lie in the most favored region of the Ramachandran plot and 1.6% lie in disallowed regions (38). In light of the excellent ratio of X-ray observations to parameters, the good stereochemistry of the model, and the satisfactory backbone torsion angles, the model can be considered essentially refined and reliable (Table 1). The overall structure is shown in Fig. 2A.
FIG.2.
Structure of SVDV. (A) Depicted centrally is an SVDV capsid comprising 60 copies of the proteins VP1, -2, -3, and -4. The proteins are shown as “worms” and are color coded (VP1 is blue, VP2 is green, VP3 is red, and VP4 is yellow). A protomeric unit (one copy each of VP1 to -4) is omitted. Around the periphery of the capsid a copy of each of the SVDV proteins is shown as a worm in red. Superimposed on each is a copy of the similar protein from CBV3 (green), CAV9 (yellow), and PV type 3 (blue). The pocket factor is depicted in aquamarine within the VP1 β-barrel. (B) Chain traces of SVDV (red) superimposed on CBV3 (green) around a fivefold vertex. Myristoyl groups are shown as ball-and-stick representations and colored green or red, respectively. Residue Arg 132 is shown as a blue ball-and-stick representation. The ions on the fivefold axis are shown in lilac.
The secondary structures of CAV9 (26), CBV3 (50), and SVDV are compared in Fig. 2A and 3.
FIG. 3.
Structure-based alignment (61) of the SVDV capsid proteins with those of CBV3 and CAV9. Conserved residues are shown boxed with a black background. Helices and strands assigned by definition of secondary structure of proteins (DSSP) (31) are labeled corresponding to standard picornaviral nomenclature and are represented by coils and arrows, respectively. Solvent accessibility (acc) within a pentamer was calculated by DSSP and is shown by the graduations in the bar below the sequence. Black graduations, relative accessibility of >40%; gray graduations, relative accessibility of >10%. This figure was produced using Espript (20).
SVDV is, as expected, more similar to CBV3 than to CAV9. This is borne out by superimposition of the coordinates (61), matching 806 residues (root mean square deviation [RMSD] = 0.45 Å) between SVDV and CBV3 compared to 800 residues (RMSD = 0.56 Å) between SVDV and CAV9 and 776 residues (RMSD = 0.83 Å) between SVDV and PV type 3 (13).
VP1.
This protein is often the most variable among picornaviruses, contributing the greatest fraction of the outer surface area of the virus capsid. It associates to form the fivefold vertices of the icosahedral particle, which in the entero-, rhino-, and cardioviruses protrude from the rest of the capsid (Fig. 2). In SVDV, VP1 comprises 283 residues, all but the first 12 of which have been built into the electron density map. The core of the β-barrel structure is well conserved, while the loops connecting the strands, with fewer structural constraints, vary in length and position. The three loops that cluster at the fivefold axes, the BC, DE, and HI loops (residues 82 to 88, 127 to 139, and 223 to 230 [Fig. 2A and 3]), vary most in structure from those of CBV3 and CAV9. The DE and BC loops in particular deviate from those of CAV9. The BC loop has a three-residue insertion in CAV9 relative to CBV3. The DE loop in SVDV is similar in conformation to that of CBV3 but, strikingly, bears arginine residue 132, which forms a crown at the apex of the fivefold axes (Fig. 2B).
The N-terminal 12 amino acids cannot be accurately modeled (as was the case for CBV3); however, there is weak electron density corresponding to the position of the N terminus modeled for CAV9 and tentatively modeled for PV (13), suggestive of a similar structure. In CAV9, the terminus forms a finger that points towards the base of the fivefold axes, partly following the same course as the VP4 N terminus such that a short region of parallel β-sheet may be formed. This structure contrasts with the N-terminal helix of HRV16 VP1 and may represent a general difference between rhino- and enteroviruses.
The location of the C terminus is closely similar to that for CBV3, while the corresponding residues in CAV9 diverge, presumably as a result of its C-terminal extension containing an RGD receptor recognition motif. VP1 has a rather loosely packed core compared to the other major capsid proteins and accommodates a fatty acid molecule. The electron density for this is very clear.
The pocket factor.
The rhino- and enteroviruses possess a hydrophobic pocket under the base of the canyon of VP1 that extends from a pore on the virus surface through the interior of the β-barrel until it is closed off by loops of VP1 (Fig. 2A). This pocket is normally occupied by a natural lipid pocket factor, which can be replaced by one or two molecules of a suitable antiviral compound. In the case of HRV14 and HRV3, the pocket is empty (56, 69).
In CBV3, the VP1 β-barrel accommodates a molecule of fatty acid classified as palmitic acid (C16); however, in SVDV, the electron density is commensurate with the binding of a longer molecule (Fig. 4). Overall, the height of the electron density within the pocket is 75% of the surrounding protein, which is consistent with full occupation of the pocket given the lipid nature of the pocket factor. However, the electron density is weaker towards the mouth of the pocket, which is suggestive of flexibility in this region or the binding of pocket factors of heterogeneous length. In the CAV9 structure (26), the electron density observed in the pocket seemed to indicate that two molecules of the antiviral compound WIN 51711 were bound (the virus was crystallized in the presence of this compound). These molecules appeared to be bound head-to-head (with the second blocking the mouth of the pocket) and suggested binding of a longer natural pocket factor. In SVDV, the electron density for the fatty acid molecule overlaps the molecule of palmitic acid modeled into CBV3 (50) in its entirety but extends further towards the mouth of the pocket, in the vicinity of which the electron density exhibits asymmetric protrusions and becomes less well defined. Thus, it would appear that the polar head group is towards the exterior or mouth of the pocket while the aliphatic chain extends the full length towards the nearby fivefold axis. The head group does not extend as far towards the exterior as that observed in CAV9 (26), probably due to the blocking effect of Tyr 178 (VP3), which is a leucine in CAV9. In the absence of a definitive chemical identification for the molecule in SVDV, we have modeled sphingosine (C18) into the electron density. Although this does not fully account for all the less well-connected density towards the mouth of the pocket, the oxygen and nitrogen of the head group hydrogen bond in a satisfactory way to the nitrogen and carbonyl oxygen of Ser 192 (VP1). Sphingosine has been previously identified binding into the pocket of PV types 1 and 3 (13).
FIG. 4.
Ligplot (63) representations of the VP1 hydrophobic pockets of SVDV and CBV3.
As can be seen in Fig. 4, the binding site is hydrophobic except for a few polar interactions in the mouth region. The dimensions of the pocket are very similar for the CAV9, SVDV, and CBV3 structures, the major exception being a threonine at residue 96 (VP1) in SVDV (Thr 97 [VP1] in CAV9) which is a proline in CBV3. Ten residues are involved in hydrophobic interactions with the backbone of the pocket factor in SVDV, six of the analogous residues in CBV3 are ligands, and a slightly different subset of six are ligands in CAV9. There are more differences in ligands in the mouth region between CBV3 and SVDV.
VP2.
VP2 alternates with VP3 around the threefold axes (Fig. 2A) of the virion and is relatively exposed at the capsid surface. Of the 263 residues of VP2, all but the N-terminal 9 residues have been built into the model. As for VP1, weak density allowed us to visualize a path for these residues, although they cannot be accurately modeled. The N terminus starts on the inside of the capsid towards an icosahedral threefold axis and finishes exposed to the surface close to a twofold axis. The least conserved area of the structure with respect to CBV3 and CAV9 is the puff, a region implicated in receptor binding of PV (47). The puff is made up of two sequential loops (residues 135 to 141 and 160 to 166, between β-sheets E and F) (Fig. 3), with the second loop being more exposed on the virus surface and in CBV3 carrying a two-residue insertion relative to CAV9 and SVDV.
Pro 83 (perfectly conserved among picornaviruses [37]) adopts the cis conformation that appears to be a general feature of picornavirus structures. Isomerization of this residue has been suggested to play a role in structural rearrangements associated with interpentamer interactions (37).
Electron density corresponding to an RNA nucleotide making a ring-stacking interaction with Trp 38 is another feature reported for several other picornaviruses (2, 13, 22, 50) that is conserved in SVDV. The base is observed adjacent to an indent in the inner surface at the junction of the three proteins in the biological protomer.
VP3.
VP3 contains 238 residues, all of which are modeled into the structure. The conserved β-barrel is relatively compact (Fig. 2A), and an extended N-terminal tail begins close to a fivefold axis, stacked above the N termini of VP1 and VP4, which together with the other fivefold-related copies form a β-cylinder that is important for pentamer stability. The VP3 C terminus is external. The most prominent and variable surface protrusion (known as the “knob”) comprises residues 58 to 69 and occupies a position adjacent to the VP1 C terminus (Fig. 3). This loop is similar in SVDV and CBV3, but differs in CAV9. There is variation to a lesser degree in the BC and EF loops (comprising residues 74 to 79 and 137 to 141, respectively), with CAV9 again adopting a conformation distinct from those of CBV3 and SVDV.
VP4.
VP4 is the most conserved of all the viral proteins and lies on the interior of the capsid. Of the 69 amino acids, a central portion (residues 15 to 24) corresponding to the most variable sequence is disordered and cannot be modeled. The protein has little secondary structure (Fig. 2A and 3), beginning close to an icosahedral fivefold axis and snaking towards the nearest threefold axis such that the C terminus of VP4 is in the vicinity of the N terminus of VP2. Since the nine residues at the start of VP2 are not visible, it is not possible to define the precise relationship between these two termini that are covalently linked prior to the cleavage of VP0, the final step in the maturation of the virion.
The N-terminal glycine of VP4 is covalently attached to a myristic acid moiety by an amide linkage, and the myristoyl groups cluster around the fivefold axes underneath the VP3 β-cylinder. The carboxylate group of the myristoyl forms a hydrogen bond with Thr 28 of VP4. This interaction is conserved in picornaviruses, and mutagenesis studies have shown that it is important for normal viral assembly and infectivity in PVs (48).
Ions.
On the fivefold axes there are both discrete spheres of density and more continuous areas of density indicative of ions, the latter apparently interacting with each other and the surrounding protein via rings of water molecules. These are detailed in Table 3 and depicted in Fig. 2B. The two most peripheral binding sites appear to be unique to SVDV and seem to be coordinated by Arg 132 of VP1, which is on the very apex of the fivefold axis and has been implicated as a determinant of pathogenicity and plaque phenotype (34). The third ion binds adjacent to the DE loop of VP1 and corresponds to a putative ion-binding site found in CBV3, human rhinoviruses, and Mengo virus (42, 50, 68). In HRV16, it has been putatively classified as a zinc ion because of coordination by histidine residues (22, 68). A ring of water molecules lies between the fourth and fifth site and a further ring of water molecules links to the sixth site, similar to a site in CAV9. Below this putative ion there is a further ring of water molecules, possibly coordinated by Asn 11 (VP3) and Ser 6 (VP3) (the latter is a replacement in SVDV for an Asn in CBV3). None of the ion-binding sites on this axis appear to be fully occupied since the density at any given site is 30 to 70% of the height of the surrounding protein backbone density.
TABLE 3.
Putative ion-binding sites
| Distance from particle center (Å) | Corresponding density in other picornavirus structures | Putative identity | Likely coordination |
|---|---|---|---|
| Threefold axis | |||
| 148 | HRV14, CAV9, CBV3 | Ca2+ | Asp 203 (VP3) OD1 |
| Fivefold axis | |||
| 164 | Arg 132 (VP1) NE, Gly 133 (VP1)N | ||
| 160 | Arg 132 (VP1) NE, Gly 133 (VP1)N | ||
| 152 | HRV14, MENGO, HRV16, CBV3, CAV9 | Zn2+ | Gln 134 (VP1) O, Asp 135 (VP1)OD1 |
| 148 | CBV3, CAV9 | Asp 135 (VP1) OD1, Ala 136 (VP1)O | |
| 140 | Val 138 (VP1) CG2 | ||
| 135 | CAV9 | Gly 174 (VP1) O | |
| 124 | CAV9, HRV14 | CI− | Ser 6 (VP3) OG, Thr 4 (VP3) OG1 |
| 113 | CAV9 | Myristoyl C8 |
There is an area of density clearly resembling an ion on the icosahedral threefold axes (radius, 148 Å) at approximately 120% of the height of the surrounding protein density (Table 2). The electron density clearly shows the ion to interact with the side chain of Asp 203 of VP3. This corresponds to a Ca2+ binding site established for HRV14. This site was also reported for CBV3 (50) and CAV9 (26), although in the latter it is coordinated by Pro 202 (VP3).
Surface characteristics and receptor binding.
The surface profile is essentially identical to that observed for the coxsackieviruses (26, 50). Around each fivefold protrusion, there are five distinct depressions of about 15 Å deep rather than the continuous circular canyon observed in the rhino- and polioviruses. The structure forming a ridge between each depression is composed of the C terminus of VP3, the first loop of the puff of VP2, and residues of the C-terminal loop (around residue 262) of VP1. A further depression on the icosahedral twofold axes, also approximately 15 Å deep, is enclosed by four walls made up of symmetrically related pairs of VP3 and VP2.
The relative exposure of capsid residues (Fig. 3) correlates with their antigenicity. Antigenic characterization of SVDV has been carried out by analysis of monoclonal antibody-resistant mutants and by peptide scanning.
Five neutralization sites were defined by Kanno et al. (33) that are analogous to PV neutralization sites 1, 2, 3A, 3B, and 3C. Two more sites not previously recognized as areas of the capsid involved in neutralization in picornaviruses were identified by Nijhar et al. (51), corresponding to residue 261 at the C terminus of VP1 and residue 234 at the C terminus of VP3. Both of these sites (approximate separation, 17 Å) are located on the ridge that divides the canyon depression into five distinct depressions. The structure of the VP3 C terminus is highly conserved among CAV9, CBV3, and SVDV, and residue 234 is certainly accessible (Fig. 3). The analogous residues are Glu 234 in CBV3 and Asp 234 in CAV9. The monoclonal antibody escape mutation (D234E) in SVDV thus corresponds to a reversion to the same amino acid as that in CBV3. The conformation of the VP1 C terminus is also conserved among the three viruses, with residue 261 being accessible. The analogous residues are Phe 262 in CAV9 and Lys 259 in CBV3, and the escape mutation in SVDV (G261R) reverts the residue to the same charge as that in CBV3. Since most of the virus surface is likely to be antigenic, it is surprising that these neutralization sites have not been recognized in other viruses, especially since the structure is conserved and they do not appear to be involved in capsid stabilizing interactions. Of a further seven immunodominant regions identified by peptide mapping, only two are exposed on the mature capsid, one of which overlaps site 3A; however, homologous regions were found in CAV9 (the N termini of VP1 and VP3, the C terminus of VP1, and the VP3 knob and VP2 puff regions) (Fig. 3). This suggests that the VP3 N termini might transiently externalize along with the VP1 and VP4 N termini upon binding to the cellular receptor or that protein precursors and dissociated subunits may induce antibodies during infection. All of the major epitopes of SVDV appear to be conformational rather than linear, which has implications for the development of assays for the virus.
Surprisingly, the necessity for the conservation of receptor-binding residues and the contrary variation observed at antigenic sites do not require a strict segregation of antigenic and receptor-binding sites (in fact, many receptor-binding sites are exposed and overlap antigenic sites [17, 62]). Nevertheless, the depressions known as the canyon or pit in picornaviruses are frequently used as sites of receptor binding (3, 24, 35, 65). CAV9 binds an integrin via the RGD motif inserted into the C terminus of VP1 and exposed on the virus capsid (26) but is also able to use another receptor of unknown identity, and the marked sequence variation compared to CBV3 and SVDV suggests that its receptor usage is different from that of these viruses. It has been shown that the six CBV serotypes (CBV1 to CBV6), together with SVDV (45) and many adenoviruses, share a common receptor on human cells known as CAR.
CAR has two immunoglobulin-like extracellular domains (D1 and D2), a transmembrane domain, and a cytoplasmic domain. For CBVs, CAR mediates both attachment and infection. Cryo-electron microscopy has been used to visualize CAR bound to CBV3 (24). CAR binds radially within the canyon. The distal end of D1 binds to the north rim and floor of the canyon and the A and G β-strands bind to the south rim of the canyon. The majority of the interactions of CAR with CBV3 involve VP1 residues, although VP2 and VP3 also participate in binding and charge complementarity seems to be important. The residues involved in the binding interface appear to be conserved among the six CBV serotypes and SVDV (24). We have performed a simple superimposition of these coordinates (PDB code 1JEW) (6) by use of the structure homology program (61) which enabled us to compare the footprint of the receptor in CBV3 with the analogous region in SVDV (Fig. 5). However, since the coordinates (PDB code 1JEW) show a steric clash at the bottom of the pocket in CBV3, the model is unlikely to be sufficiently accurate to allow a discussion of the role of individual residues. The most obvious difference in the binding site is in the second loop of the puff of VP2 residues 161 to 164 in which there is a two-residue insertion in CBV3. The altered conformation of this loop hints at altered binding specificity which may be attributable to differences between the porcine and human CAR. The importance of this VP2 loop in determining receptor specificity is further substantiated by a mutation, N165D, in CBV3 which attenuates the myocarditic phenotype and is postulated to alter the electrostatic interaction between Lys 166 and CAR residue Glu 28 or Glu 29 (24).
FIG. 5.
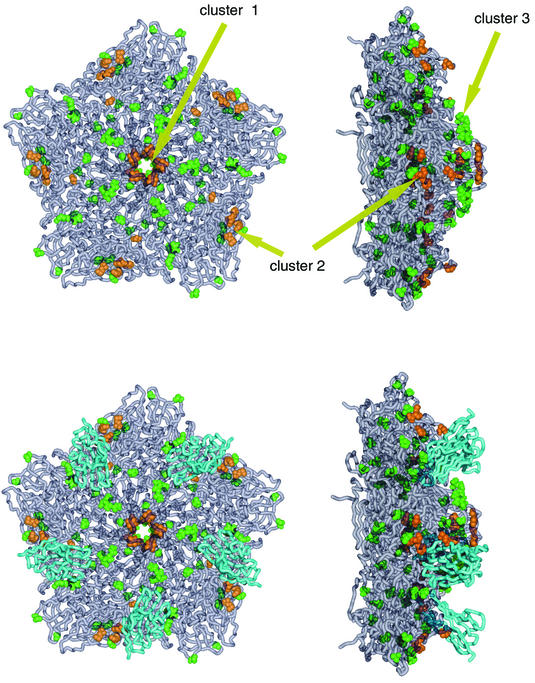
Sequence variation and receptor binding. (Top) Orthogonal views of an SVDV pentamer are shown with the proteins shown as worms in gray. Amino acid substitutions between CBV5 and SVDV are shown in green and substitutions that have occurred in SVDV since 1971 are depicted in orange. Residues in cluster 2 also correspond to the site of DAF binding in ECHO7 (25). (Bottom) Orthogonal views of an SVDV/CAR complex modeled based on the CBV3/CAR complex coordinates (PDB code 1JEW) (6). The CAR receptor is shown in aquamarine.
Mapping adaptive substitutions onto the capsid structure.
Using an alignment based on two early strains of CBV5 (Faulkner/KY/52 and 1954/UK/85) and 10 SVDV isolates spanning 1966 to 1993, we identified the amino acid substitutions between the two viruses and then classified them chronologically, placing those occurring post-1971 in a separate subset (Table 4). We have mapped these onto the three-dimensional structure together with the superimposed CAR receptor coordinates (30, 61) (Fig. 5). Overall, the substitutions cluster into the following three sites: I, on the icosahedral fivefold axis; II, at the CAR interaction site; and III, adjacent to the fivefold axes. Later changes map to sites I and II. One of the site II substitutions corresponds to residue Glu 163 of VP2 located in the loop that adopts different conformations in CBV3 and SVDV. Three of the site II mutations (Lys 148, Met 154, and Glu 163 of VP2) also correspond, by structural alignment of the SVDV coordinates with those of the ECHO7/DAF cryo-electron microscopy complex (1M11 [6, 25]), with residues in the DAF-binding footprint.
TABLE 4.
Sequence comparison of SVDV and CBV5
| Protein | Amino acid substitutions between SVDV and CBV5 proteins | Amino acid substitutions between SVDV and CBV5 proteins (post-1971) |
|---|---|---|
| VP1 | 45, 83, 84, 186, 227, 281 | 6, 7, 131, 136, 210 |
| VP2 | 108, 151, 223 | 143, 148, 154, 163 |
| VP3 | 6, 35, 76, 92, 108, 204, 235 | 5 |
DISCUSSION
SVDV is antigenically and phylogenetically closely related to CBV5 and is believed to have arisen within the past century from a single transfer of CBV5 into pigs (21, 66, 67). The close structural similarity to CBV3 that we observe agrees with this. Furthermore, SVDV shares with CBVs the ability to interact with the coxsackie and adenovirus receptor CAR, and it has been suggested that along with some CBVs, SVDV interacts with the coreceptor CD55 (DAF) (45). While porcine CAR transcripts have been detected in pig liver tissue, it is not known whether a homolog of human DAF is expressed in pigs.
The distribution of sequence changes between CBV5 and SVDV clusters into three sites (Fig. 5), one on the fivefold axes, one betwixt the CAR-binding site (by analogy with CBV3) and the DAF-binding site (by analogy with ECHO7, close to the icosahedral twofold axes [25]), and one close to the fivefold axes corresponding to the low-density lipoprotein receptor-binding site (27) in minor receptor group human rhinoviruses. The tendency of picornaviruses to utilize either the canyon (or pit) (57) or the twofold or fivefold axes as receptor-binding sites suggests that the cluster of changes close to the fivefold axes may also be connected with the binding of a second receptor (45). Substitutions that have occurred in later SVDV isolates (post-1971) seem to map to the first two sites, suggesting that these changes may be attributable to the better adaptation of the virus to a porcine CAR receptor and/or the putative DAF coreceptor. The second loop of the VP2 puff, residues 160 to 165, seems to be important for determining receptor specificity. If the low-density lipoprotein site does bind a second receptor (possibly a novel DAF binding site), then the absence of changes in this region in later SVDV isolates might suggest that further refinement of this binding in SVDV confers no selective advantage on the virus. Overall, the pattern of surface changes between CBV and SVDV is in line with the idea that viruses use antigenic variation to mediate the evolution of cell tropism (44).
In order to determine which regions of the genome are associated with virulence, pigs have been infected with recombinant SVDV (34). Unlike in PV, the 5′ noncoding region does not contain sequences that are essential for virulence; however, residue 132 of VP1 and residue 20 of the 2A protease have been implicated as determinants of pathogenicity and plaque phenotype (34). Residue 132 of VP1, an arginine in strain UK/27/72, projects from the tip of the DE loop, adjacent to the fivefold axis, forming a crown around the axis. The analogous residue in CBV3 is Gln 130 and in CAV9 is Ala 134. An arginine at this position certainly alters the overall charge of this loop. It has been reported (34) that mutation of this residue in SVDV to a glutamine (i.e., reversion to the same amino acid as in CBV3), in combination with an arginine at residue 20 of the 2A protease, correlates with a large-plaque phenotype and virulence in pigs, irrespective of the remainder of the genome. This does not agree with the fact that strain UK/27/72 is highly virulent. However, residues within the VP1 DE loop are major determinants of pathogenicity in other enteroviruses, e.g., Thr 129 in CBV4 and Ile 143 in PV type 2. In SVDV, Arg 132 appears to coordinate a density on the fivefold axis that is reminiscent of an ion, and thus it may affect the extrusion of the N terminus of VP1 and VP4 via the fivefold axis upon cell attachment. Alternatively, it may have a role in mediating receptor interactions directly via charge complementarity or indirectly via interactions with the BC loop as suggested for PV (49, 54). Although this would represent an economical way of engineering a heparin-binding site via a single mutation, there is no evidence for this, and the Arg residue does not arise from tissue culture adaptation, since most passaged isolates have a Glu at this position.
The hydrophobic pocket inside the β-barrel of picornavirus VP1 is often occupied by fatty-acid-like electron density belonging to a so-called pocket factor. It is thought that a kinetic equilibrium exists between occupied and unoccupied pocket states and that the presence of a pocket factor or drug inhibits uncoating (19). The inhibition could be caused by prevention of the collapse of the hydrophobic pocket or by blocking the flow of ions into the virus interior by increasing the rigidity of ion ligands on the fivefold symmetry axes. Removal of the pocket factor then allows the virus structure to “breathe,” presumably allowing conformational changes that are necessary for uncoating. Rhino- and enteroviruses have an extended β-sheet spanning the pentamer interfaces, and rearrangements in this sheet may be part of the process of uncoating (64). For PVs such rearrangements are proposed to be initiated by receptor binding and the displacement of the pocket factor. In contrast, the VP1 β-barrels of representatives of the aphthovirus and cardiovirus genera for which structures are available could not accommodate a pocket factor. These viruses have no role for one since they have a different mechanism for uncoating, dissociating via pentamers instead of A particles (18). For SVDV we find a substantial molecule in the pocket, an observation that may provide a route to an antiviral agent effective against SVDV. This adds further weight to the theory that longer pocket factors are found in enteroviruses than in rhinoviruses, possibly reflecting greater conformational changes on receptor binding. Certainly, enteroviruses have greater acid stability than rhinoviruses (which become inactivated below pH 6), being able to survive transit through the stomach. This stability may originate in part from the presence of a large pocket factor of cellular origin binding within the VP1 β-barrel.
The clustering of amino acid changes between SVDV and CBV3 along the fivefold axes of the virion is indicative of some functional change. It is known that the axis is important for disassembly; thus, environmentally induced release of ions may initiate a process by which the myristoyl group, VP4, and N terminus of VP1 are extruded (19). There is evidence (29) that in SVDV, the N terminus of VP1 is exposed upon cell attachment and that this region is involved in virus entry. The substitutions between SVDV and CBV3 do not seem to alter the overall charge distribution within the channel but rather increase its diameter and thus its permeability. In agreement with this, there seem to be more ions strung like beads along the length of the axis than have been reported for other picornaviruses. The increased number of ions on the fivefold axes, together with the large pocket factor, may provide some explanation of the robustness of SVDV in the face of extreme environmental factors.
Acknowledgments
We thank Aldo Dekker (Central Institute for Animal Disease Control, Lelystad, The Netherlands) for supplying sequence data prior to publication. We also thank Liz Duke for help with data collection; Robert Esnouf, Jonathan Diprose, and Jun Dong for computer support; and Linda Vincent for help with manuscript preparation.
This work was funded by the BBSRC.
REFERENCES
- 1.Acharya, R., E. Fry, D. I. Stuart, G. Fox, D. Rowlands, and F. Brown. 1989. The three-dimensional structure of foot-and-mouth disease virus at 2.9A resolution. Nature 337:709-716. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, E., and M. G. Rossmann. 1990. Analysis of the structure of a common cold virus, human rhinovirus 14, refined at a resolution of 3.0A. J. Mol. Biol. 211:763-801. [DOI] [PubMed] [Google Scholar]
- 3.Belnap, D. M., B. M. McDermott, Jr., D. J. Filman, N. Cheng, B. L. Trus, J. Z. Harmon, V. R. Racaniello, J. M. Hogle, and A. C. Steven. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. USA 97:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 5.Bergelson, J. M., J. G. Mohanty, R. L. Crowell, N. F. St. John, D. M. Lublin, and R. W. Finberg. 1995. Coxsackievirus B3 adapted to growth in RD cells binds to decay-accelerating factor (CD55). J. Virol. 69:1903-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The protein data bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunger, A. T. 1992. X-PLOR version 3.1: a system for X-ray crystallography and NMR. Yale University Press, New Haven, Conn.
- 8.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 9.Chang, K. H., P. Auvinen, T. Hyppia, and G. Stanway. 1989. The nucleotide sequence of coxsackievirus A9: implications for receptor binding and enterovirus classification. J. Gen. Virol. 70:3269-3280. [DOI] [PubMed] [Google Scholar]
- 10.Chow, M., J. F. E. Newman, D. J. Filman, J. M. Hogle, D. Rowlands, and F. Brown. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327:482-486. [DOI] [PubMed] [Google Scholar]
- 11.Esnouf, R. M. 1997. An extensively modified version of molscript which includes greatly enhanced colouring capacities. J. Mol. Graph. 15:133-138. [DOI] [PubMed] [Google Scholar]
- 12.Esnouf, R. M. 1999. Further additions to Molscript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D 55:938-940. [DOI] [PubMed] [Google Scholar]
- 13.Filman, D. J., R. Syed, M. Chow, A. J. Macadam, P. D. Minor, and J. M. Hogle. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8:1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filman, D. J., M. W. Wien, J. A. Cunningham, J. M. Bergelson, and J. M. Hogle. 1998. Structure determination of echovirus 1. Acta Crystallogr. D 54:1261-1272. [DOI] [PubMed] [Google Scholar]
- 15.French, S., and K. Wilson. 1978. On the treatment of negative intensity observations. Acta Crystallogr. A 34:517-525. [Google Scholar]
- 16.Fry, E., R. Acharya, and D. Stuart. 1993. Methods used in the structure determination of foot-and-mouth disease virus. Acta Crystallogr. A 49:45-55. [DOI] [PubMed] [Google Scholar]
- 17.Fry, E., S. M. Lea, T. Jackson, J. W. I. Newman, F. M. Ellard, W. E. Blakemore, R. Abu-Ghazaleh, A. Samuel, A. M. Q. King, and D. I. Stuart. 1999. The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. EMBO J. 18:543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fry, E., D. Logan, R. Abu-Ghazaleh, W. Blakemore, S. Curry, T. Jackson, S. Lea, R. Lewis, J. Newman, N. Parry, D. Rowlands, A. King, and D. I. Stuart. 1992. Molecular studies on the structure of foot and mouth disease virus, p. 71-80. In P. Goodenough (ed.), Protein engineering. CPL Press, Berkshire, United Kingdom.
- 19.Giranda, V. L., B. A. Heinz, M. A. Oliveira, I. Minor, K. H. Kim, P. R. Kolatkar, M. G. Rossmann, and R. R. Rueckert. 1992. Acid-induced structural changes in human rhinovirus 14: possible role in uncoating. Proc. Natl. Acad. Sci. USA 89:10213-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: multiple sequence alignments in Postscript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 21.Graves, J. H. 1973. Serological relationships of swine vesicular disease virus and coxsackie B5 virus. Nature 245:314-415. [DOI] [PubMed] [Google Scholar]
- 22.Hadfield, A. T., W. Lee, R. Zhao, M. A. Oliveira, I. Minor, R. R. Rueckert, and M. G. Rossmann. 1997. The refined structure of human rhinovirus 16 at 2.15A resolution: implications for the viral life cycle. Structure 5:427-441. [DOI] [PubMed] [Google Scholar]
- 23.Harlos, K. 1992. Micro-bridges for sitting-drop crystallizations. J. Appl. Crystallogr. 25:536-538. [Google Scholar]
- 24.He, Y., P. R. Chipman, J. Howitt, C. M. Bator, M. A. Whitt, T. S. Baker, R. J. Kuhn, C. W. Anderson, P. Freimuth, and M. G. Rossmann. 2001. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat. Struct. Biol. 8:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He, Y., F. Lin, P. R. Chipman, C. M. Bator, T. S. Baker, M. Shoham, R. J. Kuhn, M. E. Medof, and M. G. Rossmann. 2002. Structure of decay-accelerating factor bound to echovirus 7: a virus-receptor complex. Proc. Natl. Acad. Sci. USA 99:10325-10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendry, E., H. Hatanaka, E. Fry, M. Smyth, J. Tate, G. Stanway, S. Santti, M. Maaronen, T. Hyppia, and D. Stuart. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 7:1527-1538. [DOI] [PubMed] [Google Scholar]
- 27.Hewat, E. A., E. Neumann, J. F. Conway, R. Moser, B. Ronacher, T. C. Marlovits, and D. Blaas. 2000. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. EMBO J. 19:6317-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9A resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 29.Jimenez-Clavero, M. A., E. Escribano-Romero, A. Douglas, and V. Ley. 2001. The N-terminal region of the VP1 protein of swine vesicular disease virus contains a neutralization site that arises upon cell attachment and is involved in viral entry. J. Virol. 75:1044-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577-2637. [DOI] [PubMed] [Google Scholar]
- 32.Kalko, S. G., R. E. Cachau, and A. M. Silva. 1992. Ion channels in icosahedral viruses: a comparative analysis of the structures and binding sites at their fivefold axes. Biophys. J. 63:1133-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanno, T., T. Inoue, Y. Wang, A. Sarai, and S. Yamaguchi. 1995. Identification of the location of antigenic sites of swine vesicular disease virus with neutralization-resistant mutants. J. Gen. Virol. 76:3099-3106. [DOI] [PubMed] [Google Scholar]
- 34.Kanno, T., D. Mackay, T. Inoue, G. Wilsden, M. Yamakawa, R. Yamazoe, S. Yamaguchi, J. Shirai, P. Kitching, and K. Murakami. 1999. Mapping the genetic determinants of pathogenicity and plaque phenotype in swine vesicular disease virus. J. Virol. 73:2710-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolatkar, P. R., J. Bella, N. H. Olson, C. M. Bator, T. S. Baker, and M. G. Rossmann. 1999. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 18:6249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 37.Krishnaswamy, S., and M. G. Rossmann. 1990. Structural refinement and analysis of Mengo virus. J. Mol. Biol. 211:803-844. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 39.Lewis, J. K., B. Bothner, T. J. Smith, and G. Siuzdak. 1998. Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. USA 95:6774-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, W., Q. Zhao, M. Bartlam, Y. Ding, D. Li, Y. Chen, H. Bao, Q. Xie, and Z. Rao. 2002. Purification, crystallization and preliminary X-ray analysis of swine vesicular disease virus. Acta Crystallogr. 58:1056-1058. [DOI] [PubMed] [Google Scholar]
- 41.Logan, D., R. Abu-Ghazaleh, W. Blakemore, S. Curry, T. Jackson, A. King, S. Lea, R. Lewis, J. F. E. Newman, N. Parry, D. Rowlands, D. Stuart, and E. Fry. 1993. Structure of a major immunogenic site on foot-and-mouth disease virus. Nature 362:566-568. [DOI] [PubMed] [Google Scholar]
- 42.Luo, M., G. Vriend, I. Kamer, E. Minor, E. Arnold, M. G. Rossmann, U. Boege, D. G. Scraba, G. M. Duke, and A. C. Palmenberg. 1987. The atomic structure of Mengo virus at 3.0A resolution. Science 235:182-191. [DOI] [PubMed] [Google Scholar]
- 43.Mann, J. A. 1981. Swine vesicular disease, p. 365-381. In E. P. J. Gibbs (ed.), Virus diseases of farm animals, vol. 2. Academic Press, London, United Kingdom.
- 44.Martinez, M. A., N. Verdaguer, M. G. Mateu, and E. Domingo. 1997. Evolution subverting essentiality: dispensability of the cell attachment Arg-Gly-Asp motif in multiply passaged foot-and-mouth disease virus. Proc. Natl. Acad. Sci. USA 94:6798-6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martino, T. A., M. Petric, H. Weingartl, J. M. Bergelson, M. A. Opavsky, C. D. Richardson, J. F. Modlin, R. W. Finberg, K. C. Kain, N. Willis, C. J. Gauntt, and P. P. Liu. 2000. The coxsackie-adenovirus receptor (CAR) is used by reference strains and clinical isolates representing all six serotypes of coxsackievirus group b and by swine vesicular disease virus. Virology 271:99-108. [DOI] [PubMed] [Google Scholar]
- 46.Merrit, E. A., and M. E. P. Murphy. 1994. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50:869-873. [DOI] [PubMed] [Google Scholar]
- 47.Minor, P. D., M. Ferguson, D. M. A. Evans, J. W. Arnold, and J. P. Icenogle. 1986. Antigenic structure of polioviruses of serotypes 1, 2 and 3. J. Gen. Virol. 67:1283-1291. [DOI] [PubMed] [Google Scholar]
- 48.Moscufo, N., and M. Chow. 1992. Myristate-protein interactions in poliovirus: interactions of VP4 threonine 28 contribute to the structural conformation of assembly intermediates and the stability of assembled virions. J. Virol. 66:6849-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moss, E. G., and V. R. Racaniello. 1991. Host range determinants located on the interior of the poliovirus capsid. EMBO J. 10:1067-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muckelbauer, J. K., M. Kremer, I. Minor, G. Diana, F. J. Dutko, J. Groarke, D. C. Pevear, and M. G. Rossmann. 1995. The structure of coxsackievirus B3 at 3.5A resolution. Structure 3:653-667. [DOI] [PubMed] [Google Scholar]
- 51.Nijhar, S. K., D. Mackay, E. Brocchi, N. P. Ferris, P. Kitching, and N. J. Knowles. 1999. Identification of neutralizing epitopes on a European strain of swine vesicular disease virus. J. Gen. Virol. 80:277-282. [DOI] [PubMed] [Google Scholar]
- 52.Olson, N. H., P. R. Kolatkar, M. A. Oliveira, R. H. Cheng, J. M. Greve, A. McClelland, T. S. Baker, and M. G. Rossmann. 1993. Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. USA 90:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 54.Ren, R., E. G. Moss, and V. R. Racaniello. 1991. Identification of two determinants that attenuate vaccine-related type 2 poliovirus. J. Virol. 65:1377-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rossmann, M. G. 1989. The canyon hypothesis. Hiding the host cell receptor attachment site on a viral surface from immune surveillance. J. Biol. Chem. 264:14587-14590. [PubMed] [Google Scholar]
- 56.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, R. R. Rueckert, B. Sherry, and G. Vriend. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 57.Rossmann, M. G., J. Bella, P. R. Kolatkar, Y. He, E. Wimmer, R. J. Kuhn, and T. S. Baker. 2000. Cell recognition and entry by rhino- and enteroviruses. Virology 269:239-247. [DOI] [PubMed] [Google Scholar]
- 58.Rueckert, R. R. 1996. Picornaviridae: the viruses and their replication., p. 609-654. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 59.Smith, T. J., E. S. Chase, T. J. Schmidt, N. H. Olson, and T. S. Baker. 1996. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature 383:350-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith, T. J., M. J. Kremer, M. Luo, G. Vriend, E. Arnold, G. Kamer, M. G. Rossmann, M. A. McKinlay, G. D. Diana, and M. J. Otto. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 19:1286-1293. [DOI] [PubMed] [Google Scholar]
- 61.Stuart, D. I., M. Levine, H. Muirhead, and D. K. Stammers. 1979. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6A. J. Mol. Biol. 134:109-142. [DOI] [PubMed] [Google Scholar]
- 62.Verdaguer, N., M. G. Mateu, D. Andreu, E. Giralt, E. Domingo, and I. Fita. 1995. Structure of the major antigenic loop of foot-and-mouth disease virus complexed with a neutralizing antibody: direct involvement of the Arg-Gly-Asp motif in the interaction. EMBO J. 14:1690-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wallace, A. C., R. A. Lakowski, and J. M. Thornton. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Prot. Eng. 8:127-134. [DOI] [PubMed] [Google Scholar]
- 64.Warwicker, J. 1992. Model for the differential stabilities of rhinovirus and poliovirus to mild acidic pH, based on electrostatics calculations. J. Mol. Biol. 223:247-257. [DOI] [PubMed] [Google Scholar]
- 65.Xiao, C., C. M. Bator, V. D. Bowman, E. Rieder, Y. He, B. Herbert, J. Bella, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2001. Interaction of coxsackievirus A21 with its cellular receptor, ICAM-1. J. Virol. 75:2444-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, G., D. T. Haydon, N. J. Knowles, and J. W. McCauley. 1999. Molecular evolution of swine vesicular disease virus. J. Gen. Virol. 80:639-651. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, G., G. Wilsden, N. J. Knowles, and J. W. McCauley. 1993. Complete nucleotide sequence of a coxsackie B5 virus and its relationship to swine vesicular disease virus. J. Gen. Virol. 74:845-853. [DOI] [PubMed] [Google Scholar]
- 68.Zhao, R., A. T. Hadfield, M. J. Kremer, and M. G. Rossmann. 1997. Cations in human rhinoviruses. Virology 227:13-23. [DOI] [PubMed] [Google Scholar]
- 69.Zhao, R., D. C. Pevear, M. J. Kremer, V. L. Giranda, J. A. Kofron, R. J. Kuhn, and M. G. Rossmann. 1996. Human rhinovirus 3 at 3.0A. Structure 4:1205-1220. [DOI] [PubMed] [Google Scholar]