Abstract
Homologs of the small tegument protein encoded by the UL11 gene of herpes simplex virus type 1 are conserved throughout all herpesvirus subfamilies. However, their function during viral replication has not yet been conclusively shown. Using a monospecific antiserum and an appropriate viral deletion and rescue mutant, we identified and functionally characterized the UL11 protein of the alphaherpesvirus pseudorabies virus (PrV). PrV UL11 encodes a protein with an apparent molecular mass of 10 to 13 kDa that is primarily detected at cytoplasmic membranes during viral replication. In the absence of the UL11 protein, viral titers were decreased approximately 10-fold and plaque sizes were reduced by 60% compared to wild-type virus. Intranuclear capsid maturation and nuclear egress resulting in translocation of DNA-containing capsids into the cytoplasm were not detectably affected. However, in the absence of the UL11 protein, intracytoplasmic membranes were distorted. Moreover, in PrV-ΔUL11-infected cells, capsids accumulated in the cytoplasm and were often found associated with tegument in aggregated structures such as had previously been demonstrated in cells infected with a PrV triple-mutant virus lacking glycoproteins E, I, and M (A. R. Brack, J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter, J. Virol. 73:5364-5372, 1999). Thus, the PrV UL11 protein, like glycoproteins E, I, and M, appears to be involved in secondary envelopment.
The herpesvirus virion is a complex structure that contains at least 30 different virally encoded proteins. More than 15 viral proteins constitute the tegument that surrounds the herpesvirus capsid. The tegument itself is bounded by a cell-derived lipid bilayer envelope which contains more than 10 viral (glyco)proteins (28, 35). Although the formation of capsids and the involved viral components has been delineated in some detail (41), the molecular mechanisms that drive assembly of the viral tegument and envelope during herpesvirus morphogenesis are still largely unknown. Initially, it had been postulated that herpesvirus virions are fully assembled in the nucleus, acquiring their final envelope by budding at the inner nuclear membrane. They should then be transported through the secretory pathway as complete virus particles (34). In this model, all envelope glycoproteins have to be present in the inner nuclear membrane and all tegument proteins have to be assembled onto the nascent virus particle in the nucleus. During the last few years, data have accumulated showing that capsids which bud at the inner nuclear membrane lose their primary envelope by fusion with the outer nuclear or the contiguous endoplasmic reticulum membrane and acquire a final envelope and tegument in the cytoplasm by budding into vesicles derived from the trans-Golgi network that contain mature viral glycoproteins (reviewed in reference 29). This envelopment-deenvelopment-reenvelopment mechanism revived investigations on the localization and site of addition of tegument proteins to the nascent virus particle. Thus, it was shown that several major tegument proteins are present in the cytoplasm late in infection (13, 22, 37), which is coherent with the latter model.
Homologs of the herpes simplex virus 1 (HSV-1) UL11 gene product are components of the tegument which are conserved throughout the herpesvirus family. Genes coding for homologs of HSV-1 UL11 are present, for example, in the alphaherpesviruses varicella-zoster virus (VZV) (11), equine herpesvirus 1 (42), Marek's disease virus (32), and pseudorabies virus (PrV) (12); in the betaherpesvirus human cytomegalovirus (HCMV) (8); and in the gammaherpesvirus Epstein-Barr virus (1). Although amino acid sequence identity is generally low, all UL11 homologs contain a predicted N-myristoylation signal at their N terminus as well as one or several consecutive cysteine residues which could serve as a substrate for palmitoylation (33). The conservation of these features indicates that fatty acid modification may play an important role for UL11 function. The presence of N-myristic acid has indeed been shown for the HSV-1 UL11 protein (26), the homologous VZV gene 49 product (17), and the HCMV homolog pp28 (36). When expressed in the absence of other viral proteins, pp28 was detected in a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment (36). However, in the absence of myristoylation, the protein was no longer membrane associated but was found dispersed in the cytoplasm, indicating that membrane association was disturbed. In HCMV-infected cells, pp28 was found exclusively in the cytoplasm throughout the replicative cycle and was demonstrated to accumulate in cytoplasmic vacuoles and in large membranous structures which also contained the envelope glycoproteins B, H, and gp65 (37). In HSV-1 infected cells, UL11 was detected in association with intracytoplasmic membranes (25) as well as in the perinuclear space and in sites within the nucleus (4).
Expression of the HSV-1 UL11 protein in the absence of other viral proteins also results in Golgi localization. Deletion analyses revealed that the first 49 amino acids of the protein contain the information for membrane targeting. This N-terminal part of the protein contains the myristoylation site as well as possible substrates for palmitoylation. Whereas N myristoylation is important for membrane association (although the mediated interaction is weak), subsequent palmitoylation may serve to strengthen membrane affinity (24). The importance of the cotranslational modification in intracellular targeting of the protein has been shown in an elegant study using a chimeric Rous sarcoma virus Gag protein in which the M domain, which is responsible for membrane targeting of Gag, was replaced by the complete HSV-1 UL11 protein. This hybrid protein was targeted to the Golgi apparatus instead of the plasma membrane as wild-type Gag (5). These results indicated that the HSV-1 UL11 protein possesses all necessary signals to direct the chimeric protein to the Golgi apparatus.
Although the exact function of the UL11 homologous proteins is unclear, it has been shown that HSV-1 UL11 is necessary for efficient replication. A UL11 deletion mutant formed only small plaques, and final titers were reduced 5- to 20-fold (25). A more drastic effect on viral replication, with 30- to 250-fold reduced final titers and accumulation of capsids in the nucleus at the inner lamellae of the nuclear membranes as well as an increased number of naked capsids in the cytoplasm, has been described in a study using a different HSV-1 UL11 deletion mutant (3). These authors speculated that UL11 encodes a function that facilitates nucleocapsid envelopment at the nuclear membrane and transport through the cytoplasm to the extracellular space (3).
Localization and function of the UL11 homolog of PrV have not yet been investigated. On the basis of sequence analysis, the PrV UL11 gene product is predicted to be a 63-amino-acid protein with a molecular mass of 7 kDa sharing 31% amino acid identity with HSV-1 UL11 and 40% with the corresponding protein of equine herpesvirus 1 (12). In the present study, the UL11 gene product of PrV was identified using a monospecific antiserum and its function was assessed by analysis of a UL11 deletion mutant as well as a corresponding rescuant.
MATERIALS AND METHODS
Viruses and cells.
All PrV mutants were derived from the laboratory strain Kaplan (PrV-Ka) (18). Viruses were grown on rabbit kidney (RK13) or porcine kidney cells in Eagle's minimum essential medium supplemented with 10 or 5% fetal calf serum, respectively. RK13-UL11 cells which constitutively express UL11 under control of the HCMV immediate-early promoter/enhancer were established after transfection of RK13 cells with plasmid pcDNA-UL11 (see below) followed by geneticin (Invitrogen, Karlsruhe, Germany) selection and immunofluorescence screening with the monospecific anti-UL11 antiserum (see below). RK13-gB cells (30) were used for isolation and propagation of pPrV-ΔgB (see below).
Preparation of monospecific anti-UL11, anti-gM, and anti-gB sera.
For generation of an UL11-specific antiserum, the complete UL11 open reading frame (ORF) of PrV was amplified by PCR using Platinum Pfx DNA polymerase (Invitrogen), primers UL11FOR (5′-CACAGAATTCATGGGACAGTGTTGCTGCCG-3′) and UL11REV (5′-CACAGCGGCCGCGCAGTCAGTACGCCCGCGAG-3′) (nucleotides [nt] 1733 to 1753 and nt 1910 to 1929, respectively; GenBank accession no. X97257) (12), and cloned genomic BamHI fragment 3 as the template. UL11 start and stop codons are shown in boldface, and EcoRI and NotI sites introduced for convenient cloning are underlined. After cleavage, the 196-bp amplification product was inserted into the EcoRI/NotI-digested vector pcDNA3 (Invitrogen), giving rise to pcDNA-UL11, and into the vector pGEX-4T-1 (Amersham Biosciences, Freiburg, Germany). The obtained plasmid pcDNA-UL11 was used for generation of a complementing cell line (RK13-UL11), and plasmid pGEX-UL11 was used for UL11 expression in Escherichia coli. The 33-kDa glutathione S-transferase (GST)-UL11 fusion protein was isolated from sodium dodecyl sulfate (SDS)-10% polyacrylamide gels and used for rabbit immunization as described previously (20). Serum obtained after the fifth immunization was used in this study.
For generation of a polyclonal anti-glycoprotein B (gB) serum, gC-negative PrV (PrV-8411) (19) was purified by sedimentation through a 40% sucrose cushion. Glycoproteins were then solubilized in concanavalin A buffer (10 mM Tris, 0.5 mM MgCl2, 0.5 mM MnCl2, 0.5% Zwittergent 3-12) and purified by standard concanavalin A affinity chromatography. The eluate containing the glycoproteins was dialyzed, freeze-dried, resuspended in buffer A (25 mM MES [pH 6.1], 0.5% Zwittergent 3-12), and applied to a 1-ml Heparin HiTrap column (Amersham Biosciences). Of the solubilized PrV glycoproteins, only gB and gC have been shown to bind to heparin (38). Unbound protein was removed by extensive washing with buffer A. Heparin-bound gB was then eluted with 1 M NaCl in buffer A. gB was recovered after dialysis and lyophilization. Purity was checked by SDS-polyacrylamide gel electrophoresis. One rabbit was immunized three times with approximately 100 μg of purified gB at 4-week intervals. Serum obtained after the third immunization was used for this study.
For generation of a gM-specific antiserum, the 3′ part of the UL10 ORF was amplified by PCR using primers PgM-TF, 5′-CAGAATTCCGCCTGGTCCGCGCTG-3′, and pM-end, 5′-CACAGAATTCTTATTCAAAGCCGAGGTTCTCGTACAC-3′, corresponding to nt 2594 to 2610 and nt 2404 to 2430, respectively, of the sequence available at GenBank accession no. X97257 (12), and cloned BamHI fragment 3 as the template. The resulting PCR fragment encompassed codons 326 to 393 of the UL10 gene. Both primers contained EcoRI restriction sites (underlined) which were used for cloning of the PCR product into prokaryotic expression vector pGEX-4T-1. After transformation of E. coli, the C terminus of gM was expressed as a GST fusion protein and used for immunization of a rabbit (20). Serum obtained after the fourth immunization was used in the study.
Generation of pPrV-ΔgB.
For the present study, a PrV gB deletion mutant was cloned as a bacterial artificial chromosome (BAC). To this end, the mini-F plasmid vector pMBO131 (31) was digested with SalI, blunt ended by Klenow treatment, and inserted into an EcoRI-digested plasmid construct which already contained an expression cassette for enhanced green fluorescent protein (EGFP) flanked by PrV DNA fragments originating from upstream and downstream of the gB gene (P021) (30). The resulting plasmid (pΔgB-GFP/MBO) and virion DNA of PrV-Ka were used for calcium phosphate-mediated cotransfection (16) of RK13-gB cells. A GFP-expressing PrV recombinant was plaque purified from the progeny obtained. Circular viral DNA was prepared from infected cells and used for electroshock transformation of E. coli as described previously (14). Bacteria carrying pMBO131 derivatives were selected on agar plates containing 30 μg of chloramphenicol/ml, and full-length clones of the PrV genome were identified by restriction analyses of plasmid DNA and by transfection of RK13-gB cells. One infectious clone, named pPrV-ΔgB, was further propagated and used for mutation of UL11 (see below). pPrV-ΔgB and second site mutants derived from it have to be propagated in gB-expressing cells. However, they can be easily rescued by cotransfection of noncomplementing cells with plasmids containing the authentic PrV gB gene. The removal of bacterial vector and EGFP expression cassette sequences from the PrV genome precludes unwanted effects of these foreign DNA sequences on viral gene expression and replication. Thus, the in vitro growth properties of a gB rescuant of pPrV-ΔgB were indistinguishable from those of the parental PrV-Ka (data not shown).
Deletion of the PrV UL11 gene.
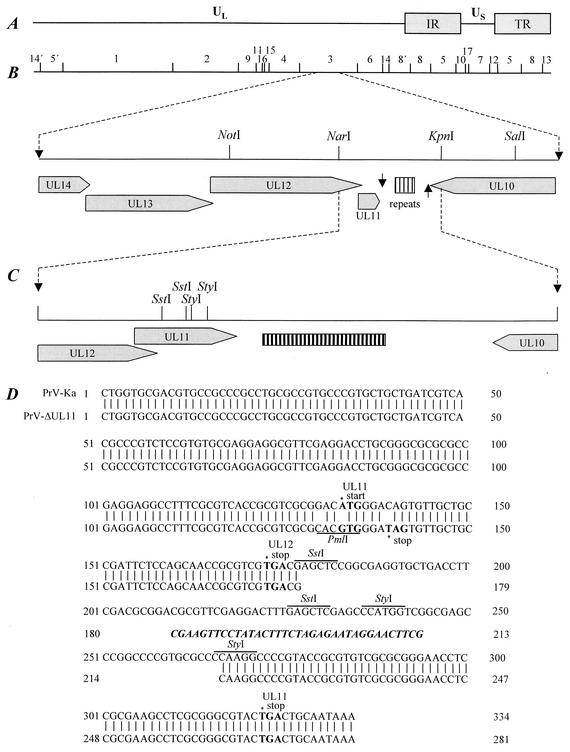
For mutation of the UL11 ORF, a genomic 2,703-bp NotI/SalI fragment (Fig. 1) of PrV strain Ka was cloned into phagemid vector pBluescript SK(+) (Stratagene, Amsterdam, The Netherlands) from which the unique SstI site had been removed. The resulting plasmid pBl-B3NS was subsequently shortened to 968 bp by cleavage with KpnI and religation and by double digestion with NotI and NarI (Fig. 1B), Klenow polymerase treatment, and religation. The obtained plasmid pBl-B3NK was used for site-specific mutagenesis (23) of the UL11 ORF with the synthetic oligonucleotide UL11-MUT (5′-GGCAGCAACACTaTCCCAcGTgCGCGACGCGGTG-3′), which is complementary to nt 1719 to 1752 of the PrV UL12-to-UL6 sequence (GenBank accession number X97257) (12). It contains three base exchanges (shown in lower case) to mutate the UL11 start codon, to generate a stop codon at amino acid position 3, and to introduce a PmlI marker site (underlined) (Fig. 1D).
FIG. 1.
Construction of PrV-ΔUL11. (A) A schematic map of the PrV genome shows the unique long (UL) and unique short (US) regions, the inverted repeat sequences (IR and TR), and the positions of BamHI restriction sites. Numbers indicate BamHI restriction fragments. (B) Enlargement of the PrV UL11 gene region. The UL14, UL13, UL12, and UL11 genes are transcribed into 3′-coterminal mRNAs which share a common polyadenylation signal (arrow pointing down). The UL11 gene is followed by a region containing direct repeat elements. Glycoprotein gM encoded by the UL10 gene is transcribed in anti-parallel orientation followed by a polyadenylation signal (arrow pointing up). (C and D) Construction of PrV-ΔUL11 (C) and sequence comparison to wild-type PrV-Ka (D) are shown. Start and stop codons of the UL11 ORF and the stop codon of the UL12 ORF are shown in bold, the SstI and StyI sites used for deletion and the introduced PmlI site are underlined, and the 36 nucleotides of the remaining FRT site are shown in boldface italics.
For site-directed mutagenesis, uracil-containing single-stranded DNA of pBl-B3NK was produced after transformation of dUTPase and uracil-DNA-glycosylase-negative bacteria (E. coli CJ236; New England Biolabs, Frankfurt, Germany) and infection with helper phage (VCSM13; Stratagene) according to the protocols of the distributors. After hybridization with UL11-MUT, the second DNA strand was completed in vitro using Klenow polymerase and T4 DNA ligase and bacteria (E. coli XL1Blue MRF′; Stratagene) were transformed. By double digestion of the resulting plasmid with SstI and StyI (Fig. 1C), three small DNA fragments (10, 30, and 49 bp) representing UL11 codons 16 to 45 were removed and, after Klenow polymerase treatment, replaced by a 1,258-bp BstBI fragment of plasmid pKD13 (10), which contains a kanamycin resistance gene flanked by Flp recombinase recognition target (FRT) sites, giving rise to plasmid pB1-ΔUL11KF. The resulting 2,137-bp insert pBl-ΔUL11KF fragment was amplified by PCR with vector-specific M13 (−47) and M13 reverse (−48) primers (New England Biolabs) and Pfx DNA polymerase (Invitrogen).
The resulting product was used for mutagenesis of pPrV-ΔgB in E. coli, utilizing the Red recombinase of bacteriophage λ (10). To this end, bacteria containing pPrV-ΔgB were transformed with the helper plasmid pKD46, which carries the Red recombinase under control of an arabinose-inducible promoter. After induction, the cells were transformed by electroshock with the pBl-ΔUL11KF PCR product and recombinant clones were selected on agar plates containing 30 μg of chloramphenicol/ml and 50 μg of kanamycin/ml. Subsequently, the kanamycin resistance gene was excised from the BAC after transformation with helper plasmid pCP20 (9) expressing the FRT site-specific Flp recombinase. After recombination, both helper plasmids could be easily eliminated, since they possess temperature-sensitive replication origins. Finally, the mini-F plasmid vector and the adjacent EGFP expression cassette were also removed from the PrV genome after cotransfection (16) of RK13 cells with BAC DNA and plasmid pUC-B1BclI, which contains the authentic gB gene of strain PrV-Ka within a 6,971-bp BclI fragment. The resulting PrV-ΔUL11 virus mutant was characterized by restriction analysis and Southern blot hybridization. The presence of the expected point mutations and deletions within the UL11 gene region was confirmed by PCR amplification and DNA sequencing (Fig. 1).
A PrV-ΔUL11 rescue mutant was isolated after cotransfection of PrV-ΔUL11 DNA and a 2.0-kb NotI/KpnI fragment comprising the wild-type UL11 ORF (Fig. 1). Transfection progeny was screened for expression of UL11 by indirect immunofluorescence. One plaque isolate, PrV-ΔUL11R, was further analyzed.
Virus purification and immunoblotting.
For virus purification, porcine kidney cells were infected with PrV-Ka, PrV-ΔUL11, and PrV-ΔUL11R and incubated until a complete cytopathic effect developed. Remaining intact cells were lysed by freezing (−70°C) and thawing (37°C), and cellular debris was removed by low-speed centrifugation. Virions were sedimented from the supernatant by centrifugation for 1 h at 22,000 rpm (TST-28 rotor; Kontron). The pellet was resuspended in TBSal (200 mM NaCl, 2.6 mM KCl, 10 mM Tris-HCl [pH 7.5], 20 mM MgCl2, 1.8 mM CaCl2), layered onto a discontinuous sucrose gradient (30, 40, and 50% sucrose), and centrifuged for 2 h at 20,000 rpm in a TST-28 rotor (Kontron). Virions accumulating at the boundary between 40 and 50% sucrose were harvested by aspiration, pelleted, and resuspended in TBSal. Purified virions (3 mg per lane) were separated by SDS-10 or -15% polyacrylamide gel electrophoresis, electrotransferred onto nitrocellulose membranes, and reacted for 1 h at room temperature with monospecific antisera against UL11 (dilution, 1:20,000; this study), UL49 (dilution, 1:100,000) (6), UL37 (dilution, 1:100,000) (20), US3 (dilution, 1:100,000) (21), or gM (dilution, 1:100,000; this study) and with monoclonal antibodies against glycoproteins gB (b43-b5; dilution, 1:500) (30), gC (B16-c8; dilution, 1:100) (21), and gE (A9-b15; dilution, 1:100) (30). Binding of peroxidase-conjugated secondary antibodies (Dianova, Hamburg, Germany) was detected by chemiluminescence (SuperSignal; Pierce, Bonn, Germany) and recorded on X-ray film.
Indirect immunofluorescence and confocal microscopy.
RK13 cells grown on coverslips were inoculated with PrV-Ka at a multiplicity of infection (MOI) of ca. 0.001. After 20 h, cells were fixed with 90% acetone for 20 min at −20°C or with 3% paraformaldehyde (PFA) for 20 min at ambient temperature with or without subsequent permeabilization with 3% PFA-0.3% Triton X-100. For blocking, fixed cells were incubated with phosphate-buffered saline (PBS)-10% fetal calf serum for 30 min. After repeated washing with PBS, the monolayer was incubated for 1 h with anti-UL11 serum (dilution, 1:100; this study) and with Alexa 488-conjugated secondary antibodies (Molecular Probes, Leiden, The Netherlands). Slides were washed repeatedly with PBS after each step. Fluorescence was preserved with a 9:1 mixture of glycerol and PBS containing 25 mg of 1,4-diazabicyclooctane per ml and 1 μg of propidium iodide per ml for chromatin counterstaining. The slides were analyzed in a confocal laser scan microscope (LSM 510; Zeiss, Oberkochen, Germany).
Determination of plaque size.
PrV-infected RK13 and RK13-UL11 cells were incubated for 2 days at 37°C in methylcellulose medium. For each virus mutant, 50 plaques were analyzed microscopically and the average plaque diameters were determined. Values were calculated in comparison to those of PrV-Ka, which was set at 100%. Average percentages and standard deviations were determined from three independent experiments.
One-step growth analysis.
To monitor one-step growth, RK13 cells were infected with PrV-Ka, PrV-ΔUL11, and PrV-ΔUL11R at a MOI of 5 for 1 h at 4°C. Thereafter, the inoculum was replaced by prewarmed medium and virus was allowed to penetrate for 1 h at 37°C. Remaining extracellular virus was subsequently inactivated by low-pH treatment. Cells and supernatants were harvested separately immediately and after 4, 8, 12, 24, and 36 h of incubation at 37°C. Virus progeny was titrated on RK13 cells. Since no significant differences were observed in extra- and intracellular virus titers, the two were added and average values and standard deviations of three independent experiments were calculated.
Electron microscopy.
For ultrathin sectioning, RK13 or RK13-UL11 cells were infected at a MOI of 1 and fixed at 12 h postinfection (p.i.). Fixation, dehydration, and embedding for routine microscopy and for intracellular immunolabeling of viral proteins were performed as described previously (20). Reactivity of monospecific antisera against UL11 and gB proteins was visualized with 10-nm gold-tagged secondary anti-rabbit antibodies (GAR10; British Biocell International). The counterstained ultrathin sections were analyzed with an electron microscope (Tecnai 12; Philips, Eindhoven, The Netherlands).
RESULTS
Identification of the PrV UL11 gene product.
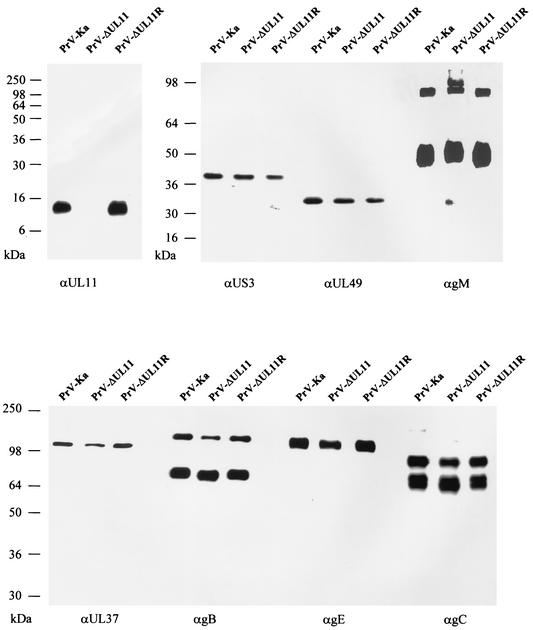
For identification of the PrV UL11 gene product, the complete ORF was cloned into the prokaryotic expression vector pGEX-4T-1, expressed in E. coli as a GST fusion protein, and used for immunization of a rabbit. In Western blot analyses of purified PrV-Ka and PrV-ΔUL11R virions, the obtained antiserum specifically detected a 10- to 13-kDa protein which was absent from PrV-ΔUL11 (Fig. 2), identifying the PrV UL11 protein as a structural component of extracellular virus particles. For control, parallel blots were probed with sera specific for tegument components (US3, UL49, and UL37) or with sera or monoclonal antibodies against viral envelope glycoproteins (gM, gB, gC, and gE). All of these proteins appeared identical in the tested mutants (Fig. 2).
FIG. 2.
Identification of the PrV UL11 protein. Purified PrV-Ka, PrV-ΔUL11, and PrV-ΔUL11R virions were analyzed by Western blotting using monospecific antisera against the UL11, US3, UL49, and UL37 proteins as well as gM or monoclonal antibodies against gB, gE, and gC. Proteins were separated on an SDS-15% polyacrylamide gel for detection of the PrV UL11 gene product, and SDS-10% polyacrylamide gels were used for detection of the other tegument or envelope proteins. Locations of molecular mass markers are shown on the left.
Localization of PrV UL11 in infected cells and in virus particles.
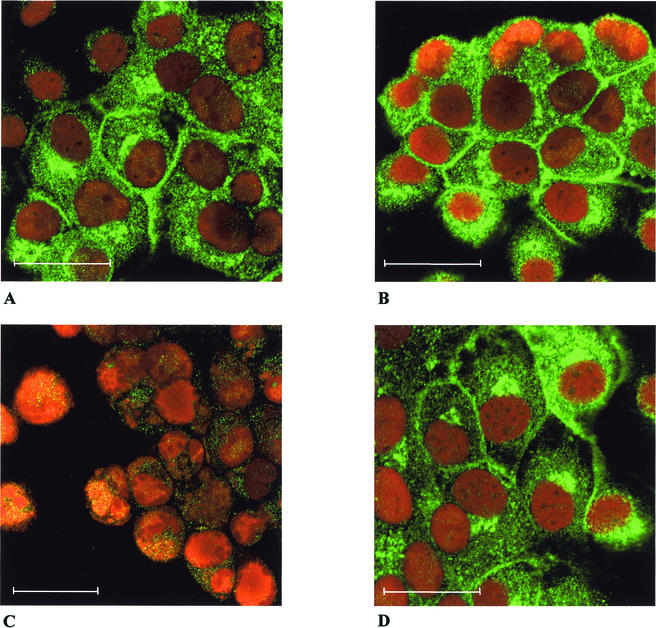
To analyze the intracellular localization of the PrV UL11 protein, confocal laser scan immunofluorescence microscopy of virus-infected cells under plaque assay conditions was performed. As shown in Fig. 3A, in RK13 cells infected with PrV-Ka, UL11-specific fluorescence was found predominantly in large speckles in the cytoplasm as well as lining the plasma membrane. A similar picture was observed after infection of RK13-UL11 cells (Fig. 3B). No UL11-specific fluorescence was detectable on nonpermeabilized cells (data not shown), indicating that UL11 is localized to the inner face of the plasma membrane. Specificity of the reaction was demonstrated by the absence of staining in RK-13 cells infected with PrV-ΔUL11 (Fig. 3C), whereas infection of RK13-UL11 cells with PrV-ΔUL11 produced typical UL11-specific staining (Fig. 3D).
FIG. 3.
Intracellular localization of the PrV UL11 protein. RK13 (A and C) or RK13-UL11 (B and D) cells were infected under plaque assay conditions with PrV-Ka (A and B) or PrV-ΔUL11 (C and D), and immunofluorescence analysis was performed by confocal laser scan microscopy using the monospecific serum against the UL11 protein and FITC-conjugated secondary antibodies (green). Chromatin was counterstained with propidium iodide (red). Bar, 25 μm.
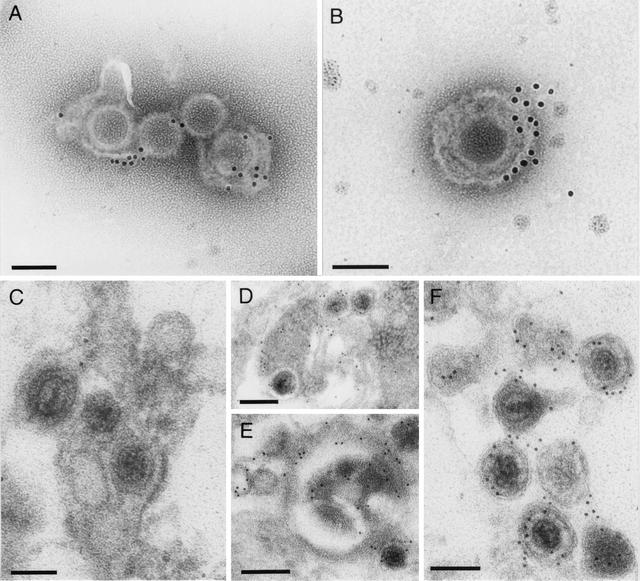
To locate the UL11 protein in virus particles, immunostaining was performed on purified virions and examined by electron microscopy. The UL11 antiserum labeled virus particles only when the envelope was partly disrupted (Fig. 4A and B), thereby allowing access of the antibody to the inner part of the virion. The gold particles were found in close proximity to the envelope, which corroborates the assumption that the PrV UL11 protein, like its HSV-1 counterpart, is a membrane-associated tegument protein. In ultrathin sections of PrV-Ka-infected RK13 cells, the monospecific α-UL11 serum labeled intracytoplasmic enveloped virions (Fig. 4D) and intracellular membranes which are presumably Golgi-derived (Fig. 4E) as well as extracellular virus particles (Fig. 4F). Primary enveloped virus particles in the perinuclear space and in the nuclear membranes were free of specific gold labeling (Fig. 4C), indicating that UL11 is added to nascent virions in the cytoplasm.
FIG. 4.
Virion localization of the PrV UL11 protein. Purified extracellular PrV virions (A and B) as well as RK13 cells infected at an MOI of 1 for 12 h with PrV-Ka (C to F) were prepared for electron microscopy. Ultrathin sections (C to F) and purified virus particles (A and B) were subsequently incubated with anti-UL11 serum and 10-nm gold-tagged secondary anti-rabbit antibodies and counterstained. PrV UL11 was detected in partly disrupted, purified virus particles (A and B) as well as in enveloped intracytoplasmic (D and E) and extracellular virions (F) but was absent from primary enveloped virus particles in the perinuclear space (C). Bars, 100 nm in panels A to C and F and 200 nm in panels D and E.
Deletion of the UL11 gene from the PrV genome.
For the present study, a novel BAC clone of the PrV genome, pPrV-ΔgB, was constructed which contains a mini-F plasmid vector and an EGFP expression cassette at the essential gB gene locus. Due to deletion of a major part of the gB gene, pPrV-ΔgB is not infectious in noncomplementing eukaryotic cells and is therefore safer than the previously described full-length clones of PrV (14, 39, 40). PrV-ΔUL11 was generated by mutagenesis of pPrV-ΔgB in E. coli by deletion of UL11 ORF codons 16 to 45, which were replaced by an FRT site consisting of 36 bp of foreign noncoding DNA (Fig. 1D). The remaining UL11 sequences do not contain possible start codons. Expression of the N-terminal part of the protein containing the predicted N-myristoylation site was also excluded by site-specific mutagenesis of the ATG initiation codon into GTG. Moreover, the CAG sequence at the third codon was replaced by the TAG stop codon. None of these alterations affected the predicted amino acid sequence of the overlapping UL12 ORF (Fig. 1). The gB defect of the resulting pPrV-ΔgBΔUL11 BAC was repaired after transfection with a plasmid comprising the PrV gB gene, giving rise to PrV-ΔUL11. DNA of this mutant virus was used for isolation of a UL11 rescue mutant after cotransfection with a plasmid containing UL11-specific sequences.
Virions of the UL11 deletion and rescue mutants were purified by sucrose gradient centrifugation and analyzed by Western blotting. As expected, the UL11 protein was absent from PrV-ΔUL11 virions and present in PrV-ΔUL11R virus particles (Fig. 2). All other tested tegument components and envelope glycoproteins were incorporated into virus particles in similar amounts in the absence or presence of UL11 (Fig. 2).
In vitro growth properties of PrV-ΔUL11.
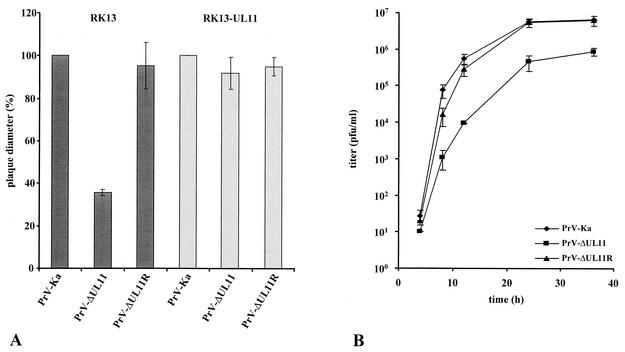
Successful isolation of PrV-ΔUL11 on noncomplementing cells had already demonstrated that the PrV UL11 gene product is not required for virus replication in cell culture. To analyze growth properties in more detail, RK13 and complementing RK13-UL11 cells were infected with PrV-Ka, PrV-ΔUL11, and PrV-ΔUL11R under plaque assay conditions and plaque diameters were measured microscopically at 2 days p.i. (Fig. 5A). Whereas the plaques formed on UL11-expressing cells were comparable in size and morphology for all viruses tested, the plaque diameters of PrV-ΔUL11 were reduced by approximately 60% on noncomplementing RK13 cells. In one-step growth analyses, PrV-ΔUL11 exhibited decreased titers at all time points, with final titers ca. 10-fold reduced compared to PrV-Ka (Fig. 5B). The replication defect of PrV-ΔUL11 was complemented in RK13-UL11 cells (data not shown) and corrected in PrV-ΔUL11R (Fig. 5), indicating that no other fortuitous mutations contribute to the observed phenotype. These data show that UL11 is not essential for virus growth in cell culture but is required for efficient viral replication and spread.
FIG. 5.
Growth properties of PrV-ΔUL11. (A) Analysis of plaque size. RK13 (dark bars) or RK13-UL11 cells (light bars) were infected with PrV-Ka, PrV-ΔUL11, or PrV-ΔUL11R under plaque assay conditions, and plaque diameters were measured microscopically at 48 h p.i. The mean plaque size of PrV-Ka was set at 100%, and relative plaque sizes as well as standard deviations were calculated from three independent experiments. (B) One-step growth kinetics. RK13 cells were infected with PrV-Ka, PrV-ΔUL11, or PrV-ΔUL11R at an MOI of 5 and harvested at 0, 4, 8, 12, 24, and 36 h p.i. Total virus titers and standard deviations for three independent experiments are shown.
Absence of UL11 impairs secondary envelopment in the cytoplasm.
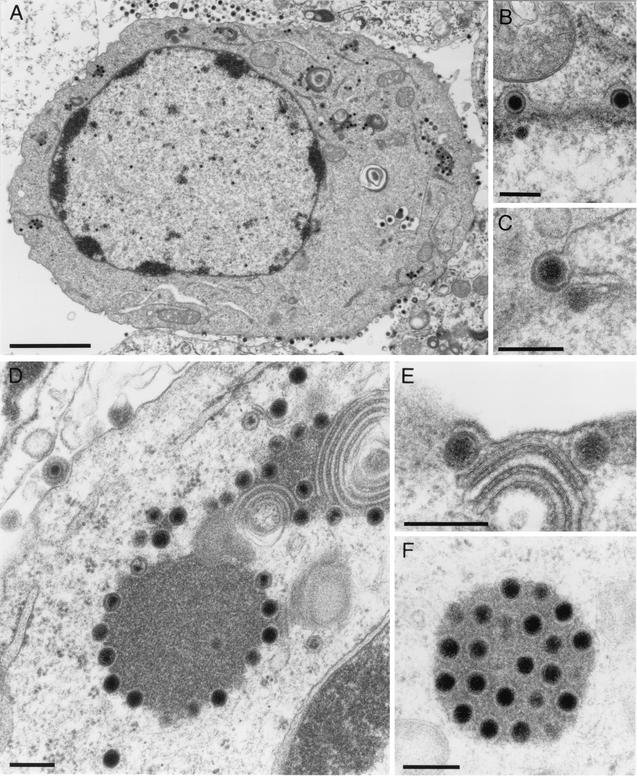
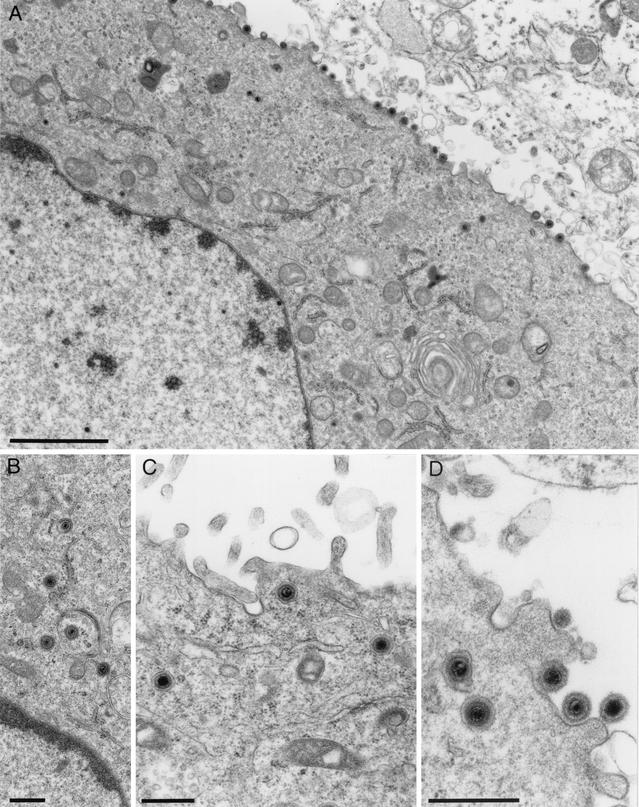
To investigate in detail the role of UL11 during virus replication, RK13 cells were infected with PrV-ΔUL11 and examined by electron microscopy at 12 h after infection. As shown in Fig. 6, cells infected with PrV-ΔUL11 showed normal capsid morphogenesis in the nucleus with no obvious accumulation of capsids at the inner lamella of the nuclear membrane (Fig. 6A) and apparently unimpaired nuclear egress (Fig. 6B and C). However, in the absence of the PrV UL11 protein, intracytoplasmic membranes (in particular those in the vicinity of or encompassing the Golgi apparatus) were distorted. The observed membrane stacks were bent and seemed to be tightly connected, possibly impeding efficient access of tegument proteins or capsids to the site of secondary envelopment (Fig. 6E). Surprisingly, in most but not all PrV-ΔUL11-infected cells, intracytoplasmic capsids associated with tegument aggregated in structures that had previously been demonstrated in cells infected with a PrV triple mutant lacking glycoproteins E, I, and M (PrV-gEIM−; Fig. 6D and F) (7). This suggests that in the absence of UL11, morphogenesis is impaired before secondary envelopment occurs. However, in contrast to the almost complete block of secondary envelopment in PrV-gEIM−-infected cells, complete virions are formed and released in the absence of UL11, which correlates with the observed production of infectious virus progeny (Fig. 5B). None of these defects was observed on complementing cells infected with PrV-ΔUL11 (Fig. 7A), and secondary envelopment (Fig. 7B), as well as transport of enveloped virus particles to (Fig. 7C) and release at (Fig. 7D) the cell surface, appeared unimpaired.
FIG. 6.
Virion morphogenesis of PrV-ΔUL11. RK13 cells were infected with PrV-ΔUL11 at a MOI of 1 and processed for electron microscopy at 12 h p.i. (A) Overview of an infected cell with intranuclear capsids and extracellular virus particles. (B and C) Unimpaired primary envelopment is shown. (D to F) Distorted Golgi or Golgi-derived membranes (D and E) as well as aggregations of cytoplasmic nucleocapsids with tegument (D and F) are shown. Bars: 3 μm (A), 250 nm (B to F).
FIG. 7.
Electron microscopy of PrV-ΔUL11-infected RK13-UL11 cells. Transcomplementing RK13-UL11 cells were infected with PrV-ΔUL11 and fixed at 12 h p.i. Electron microscopic examination reveals apparently unaffected virus morphogenesis (A), including secondary envelopment (B) and transport to (C) and release at (D) the plasma membrane as well as Golgi membranes of normal appearance (A). Bars: 2 μm (A), 500 nm (B to D).
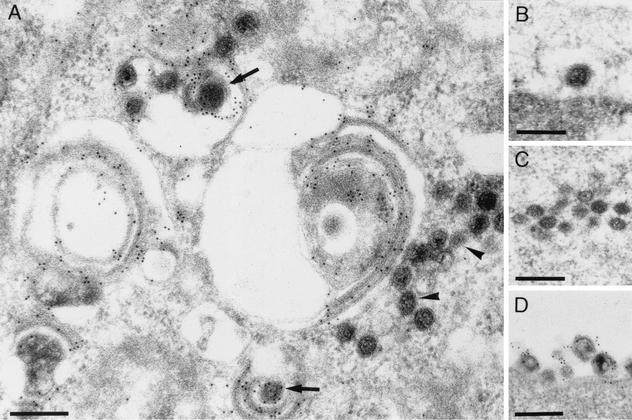
The tightly connected membranes observed in PrV-ΔUL11-infected RK13 cells possess the fuzzy appearance of viral envelopes (Fig. 6E). To test whether they contain viral glycoproteins, ultrathin sections were labeled with a gB-specific serum and analyzed by immunoelectron microscopy. As shown in Fig. 8A, these membrane stacks exhibited heavy labeling with the antiserum, indicating that they contain at least gB. As expected, extracellular virions were also labeled by the anti-gB serum (Fig. 8D), whereas primary enveloped particles in the perinuclear space (Fig. 8B) as well as naked capsids in the cytoplasm (Fig. 8A and C) were not labeled.
FIG. 8.
Immunoelectron microscopy. Using a gB-specific serum, distorted membrane stacks formed in the absence of UL11 were analyzed by immunoelectron microscopy. The distorted membranes were labeled heavily (A), as were enveloped intracytoplasmic (arrows in panel A) and extracellular (D) virus particles. Primary enveloped virions (B) and cytoplasmic nucleocapsids (A [arrowheads] and C) were free of specific labeling. Bar, 250 nm.
DISCUSSION
The PrV UL11 protein belongs to a group of highly conserved herpesvirus tegument proteins whose functions are still unclear. Using a rabbit antiserum raised against the complete UL11 protein expressed as a GST fusion protein in E. coli, we were able to identify the PrV UL11 gene product as a constituent of PrV virions localized in the interior of the virus particle but associated with the viral envelope. In infected cells, PrV UL11-specific labeling was predominantly detectable in speckles in the cytoplasm indicative of Golgi or Golgi-derived membranes and at the plasma membrane. No fluorescence was found in nonpermeabilized cells (data not shown), indicating that the UL11 protein is oriented towards the cytoplasmic face of the plasma membrane, which parallels its orientation in virus particles. In contrast to the HSV-1 UL11 protein (4), the PrV homolog could not to be detected at the nuclear membrane and primary enveloped virions in the perinuclear space were also free of UL11-specific labeling. In contrast, enveloped intracytoplasmic and extracellular virus particles were decorated, indicating that the PrV UL11 protein is added onto nascent virus particles after nuclear egress. Although we cannot strictly exclude the possibility that the PrV UL11 protein is present at the nuclear membrane beneath the detection limit or that it is present in a form which is not recognized by the anti-UL11 serum, absence of the PrV UL11 protein from the nuclear membrane correlates with the observation that neither virus assembly in the nucleus nor nuclear egress of PrV is detectably affected in PrV-ΔUL11-infected cells. This result contrasts with findings with a HSV-1 UL11 deletion mutant, which exhibits an accumulation of capsids at the inner nuclear membrane (4), suggesting that the HSV-1 UL11 protein may facilitate nuclear egress. The reason for these different phenotypes is unclear at present.
In contrast, general in vitro growth properties of UL11-negative PrV and HSV-1 are similar. Neither the PrV nor the HSV-1 UL11 protein is essential for viral replication in cell culture. However, deletion of the UL11 gene from the PrV genome (this study) or from HSV-1 (3, 25) resulted in a significantly decreased plaque size, indicative of an impairment in cell-to-cell spread, and reduced virus titers, showing that UL11 plays an important role in viral replication. The defects in PrV-ΔUL11 could be rescued on complementing cells, which provide only the UL11 protein in trans, and in a rescue mutant, which clearly demonstrates that the observed phenotypes are solely due to the lack of UL11. Since the UL12 and UL11 ORFs partly overlap and genes coding for UL14 to UL11 are transcribed into 3′-coterminal mRNAs, we took special care not to impair expression of the neighboring genes but deleted most of UL11-specific sequences. By the use of BAC technology and flp recombinase-mediated deletion of the introduced antibiotic resistance gene required for selection, only 36 nucleotides of foreign DNA were finally present at the former UL11 locus, which should have minimized any possible unwanted negative effects on adjacent genes.
The most striking phenotype for PrV-ΔUL11 was found in electron microscopic examinations of infected RK13 cells. Here, membranes in the vicinity of or encompassing the Golgi apparatus were observed to form tightly packed stacks with round or ovoid appearance, which were not found in cells infected with PrV-ΔUL11R or with the parental PrV-Ka. The distorted membranes are characterized by the presence of fuzzy surface projections, which resemble the projections seen on mature virus particles, indicating that viral envelope glycoproteins are targeted to these membranes. In immunolabeling experiments using a gB specific polyclonal antiserum, these membranous structures were heavily labeled, indicating that at least gB is present in these membranes.
In PrV-ΔUL11-infected cells, moreover, unenveloped capsids accumulated in the cytoplasm, sometimes in association with electron-dense material which presumably represents tegument. Similar clusters containing capsids and associated tegument were also observed in cells infected by another mutant, PrV-gEIM−, which lacks glycoproteins gE, gI, and gM (7), or with mutant PrV-107 gM−, which lacks only the cytoplasmic tail of gE in combination with gM (6). In these mutants, secondary envelopment is almost completely blocked, large capsid-tegument clusters are observed, and only very few infectious particles are formed (6, 7). We hypothesized that gE and gM mediate a similar step in virion morphogenesis in a synergistic manner. Since the carboxy termini of gE and gM have been shown to bind the major viral tegument protein UL49, these glycoproteins are probably involved in directing tegumented capsids to the budding site through their presence in the vesicle membrane and their interaction with tegument proteins (15). Apparently, the PrV UL11 protein is involved in a similar step of virion morphogenesis leading to secondary envelopment.
In the absence of gE, gM or the UL11 protein, secondary envelopment still occurs and extracellular virions are produced, albeit in smaller amounts. However, in cells infected with UL11 (this study) or gM deletion mutants (6), tegumented nucleocapsids accumulate, indicating that UL11 and gM have a modulating function which can at least partly be executed by other viral proteins. We are currently trying to isolate a UL11/gM double mutant to analyze in more detail a possible interplay between these two viral proteins.
Although in PrV-gM−-infected cells aggregation of tegumented capsids occurred as in PrV-ΔUL11 infection, formation of the described distorted membrane stacks was not observed (6). Thus, this feature appears to be specific for deletion of UL11. It can be speculated that association of UL11 with the vesicle membrane may allow efficient addition of other tegument proteins and finally of nucleocapsids onto the site of secondary envelopment, thus preventing spontaneous interaction of the glycoprotein-containing membranes resulting in the observed stacks.
According to DNA sequence analysis, the deduced PrV UL11 protein should comprise 63 amino acids with a molecular mass of 7 kDa (12). In Western blot experiments, the anti-UL11 serum reacted specifically with an approximately 10- to 13-kDa protein of purified wild-type PrV virions and with a protein of similar size in lysates of infected cells (data not shown). Based on the broad protein band, it is likely that the signal comprises several UL11 protein species. Different species have also been described for the HSV-1 UL11 protein (4, 26) which most probably are a result of different protein modifications. For the HSV-1 (25), VZV (17), and HCMV UL11 homologs (36), myristoylation and phosphorylation have been demonstrated. In addition, the HSV-1 UL11 protein have recently been shown to be palmitoylated (24). Conservation of the N-myristoylation signal as well as one or several consecutive cysteine residues in the deduced amino acid sequences of all UL11 homologs identified so far makes it highly likely that fatty acid modifications are present on all UL11 homologs, including the PrV UL11 protein. Myristoylation and/or palmitoylation occurs on a wide variety of viral and cellular proteins, and many myristylated proteins are bound to membranes (33). However, for stable membrane association, a second modification (e.g., palmitoylation) is required (33). HSV-1 UL11 has been shown to be doubly acylated and has been detected in tight association with membranes (4, 5, 24, 26). Mutation of the three cysteine residues suspected to be the sites for addition of palmitate led to a weakened membrane binding capacity, at least in the absence of other viral gene products in transient transfection assays (24), indicating that both modifications are necessary for the observed membrane affinity. We are currently testing whether the PrV UL11 protein is also myristoylated and/or palmitoylated and whether these modifications are required for membrane association and function of UL11. Myristoylation and palmitoylation may provide not only membrane binding but also membrane targeting function (27, 33). Doubly acylated proteins are often associated with specialized regions in membranes designated as lipid rafts. Lipid raft association takes place in the Golgi complex (2), and it might be speculated that the herpesvirus envelopes are formed by modified lipid rafts in which the viral membrane glycoproteins are concentrated and from which cellular proteins are excluded. Experiments to test for lipid raft association of viral glycoproteins and UL11 are under way.
Acknowledgments
We thank G. A. Smith and B. L. Wanner for providing plasmids and E. coli strains for BAC cloning and mutagenesis, Charlotte Ehrlich, Uta Hartwig, Nadine Müller, and Petra Meyer for excellent technical assistance, and H. Stephan and E. Zorn for photographic help.
REFERENCES
- 1.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. F. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature (London) 310:207-211. [DOI] [PubMed] [Google Scholar]
- 2.Bagnat, M., and K. Simons. 2002. Lipid rafts in protein sorting and cell polarity in budding yeast Saccharomyces cerevisiae. Biol. Chem. 383:1475-1480. [DOI] [PubMed] [Google Scholar]
- 3.Baines, J., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines, J., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowzard, J. B., R. J. Visalli, C. B. Wilson, J. S. Loomis, E. M. Callahan, R. J. Courtney, and J. W. Wills. 2000. Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J. Virol. 74:8692-8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchinson, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 12.Dijkstra, J. M., W. Fuchs, T. C. Mettenleiter, and B. G. Klupp. 1997. Identification and transcriptional analysis of pseudorabies virus UL6 to UL12 genes. Arch. Virol. 142:17-35. [DOI] [PubMed] [Google Scholar]
- 13.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 17.Harper, D. R., and H. O. Kangro. 1990. Lipoproteins of varicella-zoster virus. J. Gen. Virol. 71:459-463. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 19.Karger, A., and T. C. Mettenleiter. 1995. Cell surface proteoglycans are not essential for infection by pseudorabies virus. J. Virol. 69:3482-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 22.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacLean, C. A., A. Dolan, F. E. Jamieson, and D. J. McGeoch. 1992. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J. Gen. Virol. 73:539-547. [DOI] [PubMed] [Google Scholar]
- 26.MacLean, C. A., B. Clark, and D. J. McGeoch. 1989. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J. Gen. Virol. 70:3147-3157. [DOI] [PubMed] [Google Scholar]
- 27.McCabe, J. B., and L. G. Berthiaume. 1999. Functional roles for fatty acylated amino-terminal domains in subcellular localization. Mol. Biol. Cell 10:3771-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 29.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Connor, M., M. Peifer, and W. Bender. 1989. Construction of large DNA segments in Escherichia coli. Science 244:1307-1312. [DOI] [PubMed] [Google Scholar]
- 32.Osterrieder, N. 1999. Sequence and initial characterization of the UL10 (glycoprotein M) and UL11 homologous genes of serotype 1 Marek's disease virus. Arch. Virol. 144:1853-1863. [DOI] [PubMed] [Google Scholar]
- 33.Resh, M. D. 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451:1-16. [DOI] [PubMed] [Google Scholar]
- 34.Roizman, B., and D. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 35.Roizman, B., and P. Pellet. 2001. The family herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Virology, 46th ed. Lippincott-Raven, Philadelphia, Pa.
- 36.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawitzky, D., A. Voigt, H. Zeichhardt, and K.-O. Habermehl. 1996. Glycoprotein B (gB) of pseudorabies virus interacts specifically with the glycosaminoglycan heparin. Virus Res. 41:101-108. [DOI] [PubMed] [Google Scholar]
- 39.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 42.Telford, E. A. R., M. S. Watson, D. McBirde, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus 1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]