Abstract
PB2 mutants of influenza virus were prepared by altering conserved positions in the N-terminal region of the protein that aligned with the amino acids of the eIF4E protein, involved in cap recognition. These mutant genes were used to reconstitute in vivo viral ribonucleoproteins (RNPs) whose biological activity was determined by (i) assay of viral RNA, cRNA, and mRNA accumulation in vivo, (ii) cap-dependent transcription in vitro, and (iii) cap snatching with purified recombinant RNPs. The results indicated that the W49A, F130A, and R142A mutations of PB2 reduced or abolished the capacity of mutant RNPs to synthesize RNA in vivo but did not substantially alter their ability to transcribe or carry out cap snatching in vitro. Some of the mutations (F130Y, R142A, and R142K) were rescued into infectious virus. While the F130Y mutant virus replicated faster than the wild type, mutant viruses R142A and R142K showed a delayed accumulation of cRNA and viral RNA during the infection cycle but normal kinetics of primary transcription, as determined by the accumulation of viral mRNA in cells infected in the presence of cycloheximide. These results indicate that the N-terminal region of PB2 plays a role in viral RNA replication.
Transcription and replication of influenza A virus genes is mediated by viral ribonucleoprotein complexes (vRNPs) that contain each of the eight negative-polarity, single-stranded RNA segments associated with nucleoprotein (NP) molecules and bound to the polymerase. This enzyme is a heterotrimer formed by the PB1, PB2, and PA proteins (12, 13, 27, 31). Both transcription and replication take place in the nucleus of the infected cells (24, 30) and are catalyzed by the polymerase complex. The replication of viral RNA involves the generation of a full-length RNA copy of positive polarity that is encapsidated with NP molecules and complexed with the polymerase (cRNP) and is used as an intermediate for the synthesis of viral RNA (vRNA) progeny molecules (23). However, the initiation of mRNA synthesis involves the generation of capped primers from cellular heterogeneous nuclear RNAs by a cap-stealing mechanism (34). The recognition of capped RNA requires the interaction of the polymerase with the 5′ terminus of vRNA, while the cleavage step needs binding of the polymerase to both the 5′ and 3′ termini of the template (9). The termination and polyadenylation signal consists of an oligo(U) sequence located close to the 5′ terminus of the vRNA templates (59, 61, 64). These processes require the presence of the panhandle structure (37) and the interaction of the polymerase with the conserved 5′-terminal sequences of the template (60, 62, 63).
The polymerase domains responsible for intersubunit interaction have been identified (20, 56, 73, 75), as well as the domains in the PB1 subunit that bind the vRNA template (18, 35) and the cRNA template (19). The functional contribution of each subunit to the transcription and replication processes has been studied by several approaches. The available evidence indicates that the PB1 protein harbors the polymerase activity. Thus, it can be cross-linked to the triphosphate substrate (1, 2, 8, 65) and contains amino acid motifs present in other RNA-dependent RNA polymerases (58). Mutation of the conserved residues at these motifs abolishes the transcriptional activity (5). Furthermore, extracts from baculovirus-infected cells expressing PB1 protein show some polymerase activity in vitro (33). The phenotypes of temperature-sensitive mutants affected in the PA subunit (reviewed in reference 39) suggest its involvement in vRNA synthesis.
The PA subunit is a phosphoprotein (69) that shows protease activity in vivo and in vitro (22, 68). The regions of the PA subunit responsible for this activity map to the amino-terminal third of the protein (70), close to the nuclear localization signal (47). Some mutations in PA protein affecting this proteolytic activity correlate with a decreased ability of the polymerase to synthesize cRNA from a vRNA template (55), while others do not (45). In addition to this role for the PA subunit in RNA replication, recent evidence indicates that it is also involved in transcription initiation (14).
The PB2 subunit is involved in the initiation of viral transcription. Thus, it is a cap-binding protein (6, 72, 74), and cap-primer-dependent in vitro RNA synthesis is affected by mutations in the PB2 gene (53). A series of inhibition studies in vitro also support such a role for PB2. Thus, monoclonal antibodies specific for the PB2 subunit interfere with the initiation step of mRNA-primed transcription in vitro (3), and antibodies monospecific for the C terminus of PB2 inhibit cap snatching and cap-dependent transcription in vitro but not cap binding (7). Furthermore, antibodies directed to the region from positions 300 to 550 in PB2 inhibit cap snatching and partially affect cap recognition (42). In spite of this evidence, it has recently been proposed that the enzymatic activity responsible for cap snatching resides in the PB1 subunit (36). Nevertheless, both transcription and cap-dependent endonuclease activities require the presence of the three subunits of the polymerase and the RNA template (9, 21).
In this work, we studied the biological properties of point mutations engineered in the PB2 subunit at positions conserved among type A, B, and C influenza viruses that could be aligned with residues of the eIF4E family of cap-binding proteins. Surprisingly, these mutations reduced the capacity of RNPs reconstituted from cloned cDNAs to replicate in vivo but did not substantially alter their ability to transcribe in vitro. Some of these mutations could be rescued into infectious virus. The mutant viruses showed delayed kinetics of cRNA and vRNA accumulation, but their primary transcription rate was indistinguishable from that of wild-type virus.
MATERIALS AND METHODS
Biological materials.
The COS-1 cell line (17) was provided by Y. Gluzman and was cultivated as described previously (50). The recombinant vaccinia virus vTF7-3 (16) was a gift of B. Moss. The origin of plasmids pGPB1, pGPB2, pGPA, and pGNPpolyA has been described previously (53, 54). The pIVACAT1-S plasmid (57) was kindly provided by P. Palese. Mutant plasmid pGPB2Δ124-759 has been described previously (53). The preparation and characterization of rabbit serum and mouse monoclonal antibodies specific for the PB2 polymerase subunit have been described (3). Construction of plasmid pT7NSCAT-RT, able to transcribe the cat gene flanked by the NS gene termini in negative polarity, and of pT7NSΔCAT-RT has been described (49). Plasmid pT7cNSΔCAT-RT, which expresses a deleted cat RNA of positive polarity, has been described previously (55). Plasmid pGPB2His, containing a His tag at the C terminus of PB2, will be described in detail elsewhere (E. Area et al., unpublished data).
Plasmid pPG177, expressing a glutathione S-transferase (GST)-VP39 (O-methyltransferase) fusion protein (71), was provided by P. Gershon. The plasmids containing the cDNAs of WSN virus RNAs under control of the polymerase I promoter as well as the expression plasmids encoding the WSN virus polymerase and NP genes (46) were a gift of Y. Kawaoka. Plasmid vector pHH21 (46) was kindly provided by G. Hobom.
Mutant construction.
For mutant construction, two PCR products were obtained from PB2 cDNA with two complementary mutagenic oligonucleotides and one appropriate external oligonucleotide. A second PCR was performed with the previous PCR products and the two external oligonucleotides. Finally, it was digested with two enzymes with unique sites (Asp718I and PflMI) and cloned into plasmid pGPB2 previously digested with the same enzymes. The mutation introduced and the sequence corresponding to the PCR product were checked for each mutant plasmid. The sequence of the mutagenic oligonucleotides used is available upon request. Standard conditions were used for DNA restriction, DNA isolation, ligation, and Escherichia coli transformation (67). The generation of rescue plasmids derived from vector pHH21 containing the A/Victoria/3/75 genes (A. Falcón et al., unpublished data) will be described elsewhere. The mutations present in pGPB2 plasmids were transferred to pGPB2His or pHHVicPB2 by exchange of restriction fragment PflMI-NdeI.
Transfection.
Cultures of COS-1 cells were infected with vTF7-3 virus at a multiplicity of infection of 5 to 10 PFU per cell. After virus adsorption for 1 h at 37°C, the cultures were washed with Dulbecco's modified Eagle's medium (DMEM) and transfected with a mixture of plasmids containing (for 35-mm dishes) pGPB1 (500 ng), pGPA (100 ng), pGNPpolyA (2 μg), pGPB2 (or mutants thereof) (500 ng), and either pT7NSCAT-RT, pT7NSΔCAT-RT, or pT7cNSΔCAT-RT (2 μg). The total concentration of DNA in the mixture was kept constant by adding the required amount of plasmid pGEM3. In some experiments, transfection of plasmid pT7NSCAT-RT was omitted and NSCAT RNA was transfected at 5 h postinfection, as described previously (43). The DNA mixtures were diluted to 100 μl of DMEM, and in a separate tube, cationic liposomes (2 μl per μg of DNA) were diluted to 100 μl in DMEM. The contents of both tubes were mixed, kept at room temperature for 15 min, and added to the culture plates containing 1 ml of DMEM. Transfections of cultures in 60- or 100-mm dishes were scaled up accordingly. Cationic liposomes were prepared as described previously (66).
Virus rescue.
HEK 293T cells were cotransfected with a mixture of 12 plasmid DNAs (100 ng each) as described previously (46). They included eight genomic plasmids encoding the viral segment cDNAs under the control of the polymerase I promoter and the four expression plasmids encoding the three polymerase subunits and the NP. Transfection was carried out with Lipofectamine Plus (Gibco) under the conditions recommended by the manufacturer. At 16 h posttransfection, the cells were plated onto an excess of MDCK cells. When cytopathic effect was apparent, the supernatant medium was used for a plaque assay, and a well-isolated plaque was picked and used to produce a virus stock at a low multiplicity of infection. The identity of rescued mutant viruses was ascertained by sequencing of DNAs derived from PB2 RNA segment by reverse transcription (RT)-PCR amplification.
RNA probe synthesis.
The synthesis of NSCAT RNA, which contains the cat gene in negative polarity with the termini of the NS segment, was carried out as described previously (53). For synthesis of the cap-snatching probe, plasmid pGEM7Zf(+) was digested with SmaI and transcribed with Sp6 polymerase in the presence of 1 mM cap analog 7mGpppG, 0.2 mM GTP, 0.5 mM each UTP and CTP, and 50 μM [α-32P]ATP (1.2 Ci/mmol). The capped oligonucleotide product was further methylated with vaccinia virus O-methyltransferase as described previously (71). To that aim, a recombinant GST-O-methyltransferase fusion protein was expressed and purified from E. coli as described previously (71). The probes for RNA protection have been described previously (54, 55).
RNA assays.
The in vivo accumulation of vRNA, cRNA, and mRNA after reconstitution of RNPs from cDNAs was determined as described previously (54, 55). In brief, cultures of COS-1 cells were infected with vTF7-3 virus and transfected with pGEM plasmids encoding the polymerase subunits and the NP as described above. The template RNA was provided by transfection of plasmid pT7NSΔCAT-RT or pT7cNSΔCAT-RT. Total cell RNA was isolated, fractionated into polyadenylated and nonpolyadenylated RNAs, and the presence of viral RNAs was determined by an RNase protection assay, as described previously (54, 55).
The accumulation of vRNA, cRNA, and mRNA in cells infected with wild-type or mutant recombinant viruses was performed by real-time RT-PCR with oligonucleotide primers specific for the NP segment, whose sequences are available upon request. To detect mRNA specifically, the RT primer contained an oligo(dT) stretch fused to the sequence complementary to the 3′ terminus of NP mRNA. Likewise, the specificity of cRNA detection was obtained by using an RT primer containing the sequence of the 5′ terminus of NP vRNA, not represented in the mRNA. Appropriate dilutions of the samples were boiled for 5 min and rapidly cooled to 0°C. The RT step was carried out for 10 min at 50°C in the presence of the RT primer. After inactivation for 10 min at 95°C, the PCR step was performed in the presence of both primers. Control experiments in which the RT primer was omitted in the RT step verified that no fluorescence could be obtained. Quantitation was carried out with the Sybr Green RT-PCR kit from Applied Biosystems as recommended by the manufacturer. A standard curve was obtained with RNA isolated at late times after high-multiplicity infection with influenza virus. Only determinations that led to a standard curve with a fit better than 0.99 were considered. Denaturation curves and agarose gel electrophoresis of each reaction product verified that it contained a single amplification fragment of the expected melting point and length.
In vitro transcription and cap snatching.
For in vitro assays, recombinant RNPs containing either wild-type or mutant PB2 proteins were reconstituted in vivo and purified by two successive step glycerol gradients in 150 mM NaCl-50 mM Tris. HCl, pH 7.8 (51) for 8 h at 55,000 rpm and 4°C in an SW55 rotor. The fractions of the gradient were assayed for transcriptional activity and Western blotting with anti-PB1, -PB2, and -PA antibodies. Active fractions were pooled, and the amount of RNPs was determined by Western blot with anti-PA monoclonal antibodies. As a control, RNPs were reconstituted in the absence of RNA template and purified in parallel. In vitro synthesis of viral RNAs was carried out as described previously (53), with purified recombinant RNPs as the enzyme source.
To assay cap snatching, the labeled capped probe (2.5 × 105 cpm; 0.25 pmol) was incubated with purified recombinant RNPs under the conditions described for in vitro synthesis, except that no ribonucleoside triphosphates were added. After incubation, the nucleic acids were isolated by incubation for 30 min with proteinase K (50 μg/ml) in TNE buffer containing 1% sodium dodecyl sulfate and phenol extraction. Finally, the material was analyzed on 18% polyacrylamide-urea RNA gels.
Protein analyses.
Western blot was carried out as described previously (41). In brief, cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Immobilon filters, and the membranes were saturated with 3% bovine serum albumin for 1 h at room temperature. The filters were incubated with a 1:200 dilution of a mixture of anti-PB2 or anti-PA monoclonal antibodies or a 1:10,000 dilution of anti-PB1 rabbit serum for 1 h at room temperature. The filters were washed two times for 30 min with phosphate-buffered saline containing 0.25% Tween 20 and incubated with a 1:3,000 dilution of goat anti-mouse or goat anti-rabbit IgG conjugated to horseradish peroxidase. Finally, they were washed two times for 30 min as above and developed by enhanced chemiluminescence. Protein labeling in vivo was carried out as described previously (76). After high-multiplicity infection, the cultures were incubated in DMEM containing 1/10th the normal concentration of methionine and cysteine and labeled with 35S-labeled methionine and cysteine to a final concentration of 200 μCi/ml. At various times after infection, total cell extracts were prepared in Laemmli sample buffer and analyzed by polyacrylamide gel electrophoresis and autoradiography. The conditions for binding of His-tagged polymerase to Ni2+-nitrilotriacetic acid resin will be described elsewhere (E. Area et al., unpublished data). For the chloramphenicol acetyltransferase (CAT) assay, COS-1 cells were transfected with the pGEM plasmids encoding the polymerase subunits and NP and plasmid pT7NSCAT-RT or cat RNA, as described previously (53, 55). Cell extracts were used for the CAT assay with the phase separation method (10, 53). Competition of mutant PB2 proteins for CAT activity was performed as described previously (53).
RESULTS
Construction and expression of PB2 subunit point mutants.
In order to carry out a structure-function analysis of the PB2 subunit of influenza virus polymerase, we performed a multiple alignment of several PB2 protein sequences with proteins known to bind cap structures, eIF4E, and vaccinia virus VP39. The eIF4E subunit of eIF4F initiation factors from various origins could be aligned with PB2 proteins representative of the three influenza virus types, but the VP39 proteins could not. Out of the 38 positions strictly conserved among PB2 proteins in the region aligned (Fig. 1), 6 were also conserved with the eIF4E proteins, and many of these conserved positions corresponded to amino acids known to be involved in cap recognition, as determined from the structure of the cocrystal of eIF4E with a cap analog (40). This was the situation for positions W78, F130, R142, and K190 in type A PB2 protein. In addition, PB2 residue W49 was conserved in the alignment, and mutation of the corresponding amino acid in eIF4E protein abolishes cap binding (44).
FIG. 1.
Alignment of PB2 and eIF4E protein sequences. The figure shows the alignment of PB2 proteins from influenza A, B, and C type viruses with a series of eIF4E proteins from various origins (POMBE, Schizosaccharomyces pombe; YEAST, Saccharomyces cerevisiae). The positions of PB2 protein included in boxes were mutagenized as indicated in the text. Stars mark conserved residues of eIF4E involved in interaction with the cap structure (40). The solid arrowhead indicates a position conserved in eIF4E whose mutation altered cap recognition, although it does not contact the cap structure. The open arrowhead indicates a position conserved in eIF4E but not involved in cap recognition. The numbers at the top of the sequence indicate the positions in the A/Victoria/3/75 sequence. The consensus line indicates residues conserved among PB2 and eIF4E sequences.
Although eIF4E sequences could be aligned to other regions of PB2 with different alignment parameters, the amino acids relevant for cap binding did not align properly with homologous PB2 positions. Therefore, we decided to mutate the amino acids conserved in the PB2 gene indicated in Fig. 1 and analyzed the phenotypic consequences. As a control, we also mutated two amino acids that are conserved among PB2 proteins but are not conserved in eIF4E protein, T24 and D83, and one amino acid that is strictly conserved among PB2 and eIF4E proteins but not involved in cap recognition by eIF4E protein, W99 (Fig. 1).
The accumulation of the various mutant PB2 proteins was ascertained by Western blotting of extracts obtained from cells in which the viral polymerase, the NP, and a model vRNA template had been expressed. The results are presented in Fig. 2. Many of the mutant PB2 proteins were expressed at levels identical to wild-type PB2 (T24G, W49A, W49Y, F130A, F130Y, R142K, R142A K190R, and K190A). This result indicates that the stability of these mutant proteins is similar to that of wild-type PB2. Other mutants accumulated to a smaller extent (W78Y and W99A), and only a mutation at position W78 (W78A) abolished PB2 expression (data not shown). These mutants were not analyzed further.
FIG. 2.
Accumulation of wild-type (WT) and PB2 mutant proteins upon reconstitution of RNPs in vivo. Viral RNPs were reconstituted in vivo by transfection of plasmids encoding PB1, PB2, PA, and NP together with a ribozyme-containing plasmid that expresses a short vRNA template. Equal amounts of total cell extracts were assayed for accumulation of PB2 protein by Western blot with PB2-specific monoclonal antibodies. As a control (CTRL), RNP reconstitution was carried out in the absence of PB2-encoding plasmid.
Transcriptional and replicative activities of polymerase complexes containing PB2 subunit point mutants.
The functionality of mutant PB2 proteins in reconstituted viral RNPs was first tested by coexpression of the three subunits of the polymerase, the NP, and a model vRNA encoding the cat gene in negative polarity (43, 53). In this system, the level of CAT activity detected depends on the amplification of cat viral RNPs and their subsequent ability to transcribe cat pseudo-viral mRNA (28, 38). Nonconservative changes introduced at positions W49A, F130A, and R142A strongly reduced or abolished CAT expression, while conservative substitutions (W49Y and R142K) partially restored the wild-type phenotype or led to higher levels of CAT activity than the wild-type residue (F130Y) (data not shown).
Since alterations in CAT activity may reflect defects in either the replication or transcription step, the replication and transcription abilities of polymerase mutants with changes at positions 49, 130, 142, and 190 as well as at control positions 24 and 83 in PB2 protein were tested after RNP reconstitution in vivo as described previously (53, 55). Viral RNPs were generated by transfection of cDNAs encoding the polymerase subunits and the NP together with ribozyme constructs able to generate in vivo a short model RNA of 313 nucleotides resembling either a vRNA or a cRNA.
The accumulation of cRNA or vRNA in vivo was determined by an RNase protection assay with negative- and positive-polarity probes, respectively. In addition, the transcription capacity of the recombinant RNPs was determined in a similar way, by measuring the accumulation of viral mRNA in the polyadenylated fraction. The results of one representative experiment of several performed are presented in Fig. 3, and a summary of the results is presented in Table 1. The replication activity of RNPs reconstituted with control PB2 mutants T24G and D83T showed an imbalance in cRNA accumulation, which was higher than in the wild type, compared to vRNA accumulation, which was normal. RNPs with mutation K190A had lower cRNA but normal vRNA accumulations. However, the accumulation of vRNA and cRNA in cells expressing RNPs with PB2 mutants W49A and R142A was reduced, and mutation F130A abolished cRNA and vRNA accumulation.
FIG. 3.
Replication and transcription in vivo of viral RNPs containing wild-type or mutant PB2 subunits. The accumulation of mRNA and cRNA was determined by RNase protection assay of polyadenylated and nonpolyadenylated RNA, respectively, derived from reconstitution experiments in which vRNA-like template was provided in vivo. Likewise, vRNA accumulation was determined from reconstitution experiments in which cRNA-like template was provided in vivo. Control (CTRL) experiments lacked PB2-encoding plasmid in the reconstitution. P, probe used for RNase protection; MW, size markers (nucleotides).
TABLE 1.
Phenotypes of recombinant RNPs with point mutations in PB2a
| Virus | Protein expression | cRNA | vRNA | mRNA | In vitro transcription | Endonuclease activity |
|---|---|---|---|---|---|---|
| Wild type | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| T24G | ++++ | ++++++ | ++++ | ++++ | ND | ND |
| W49A | ++++ | + | + | + | ++++ | ++++ |
| W49Y | ++++ | +++ | +++ | +++ | ND | ND |
| W78A | +/− | ND | ND | ND | ND | ND |
| W78Y | +++ | ND | ND | ND | ND | ND |
| D83T | ++++ | ++++++ | ++++ | ++++ | ND | ND |
| W99A | ++++ | ND | ND | ND | ND | ND |
| W99Y | ++++ | ND | ND | ND | ND | ND |
| F130A | ++++ | − | − | − | NA | NA |
| F130Y | ++++ | ND | ND | ND | ND | ND |
| R142A | ++++ | + | + | + | ++++ | ++++ |
| R142K | ++++ | +++ | +++ | +++ | ND | ND |
| K190A | ++++ | ++++ | ++++ | ++++ | ND | ND |
| K190R | ++++ | ND | ND | ND | ND | ND |
ND, not determined; ++++, 100% of wild-type value; −, no activity. NA, not applicable.
With regard to transcription activity, the phenotypes of mutant RNPs exactly paralleled their replication phenotype, i.e., the accumulation of mRNA observed correlated with the corresponding accumulation of vRNA. These results suggest that a series of PB2 conserved amino acids, some of which could be aligned with positions in eIF4E proteins involved in cap recognition, are important for influenza virus RNA replication. However, the defect in replication activity in vivo did not allow us to determine directly the ability of these defective RNPs to transcribe.
To rule out the possibility that the phenotype of the mutant PB2 proteins would result from inability of mutant PB2 proteins to form polymerase complexes, the mutations W49A, F130A, R142K, and R142A were transferred to plasmid pGPB2His, in which a C-terminal His tag was incorporated into PB2. The tagged mutant PB2 proteins were coexpressed with wild-type PB1 and PA proteins as well as vRNA template by transfection of pGPB1, pGPA, mutant pGPB2His, and pT7NSΔCAT-RT plasmids. The formation of the polymerase complex was ascertained by pulldown experiments with Ni2+-nitrilotriacetic acid resin. As shown in Fig. 4A, the levels of expression of the various PB2 proteins were similar, including wild-type untagged PB2, used as a control. The formation of complex is indicated in Fig. 4B by Western blotting of the PB1 and PA proteins that were retained in the resin after pulldown. As expected, no retention was observed for untagged polymerase, while wild-type and mutant tagged polymerases were retained to similar extents. The quantitation of three independent transfections is shown in Fig. 4C. No differences were observed for complex formation of polymerase with PB2 mutants R142K and R142A, while a slight reduction was detected for mutant F130A.
FIG. 4.
Polymerase complex formation in vivo with wild-type and mutant PB2 proteins. The formation of polymerase complexes was determined by pulldown experiments from extracts of cells transfected with pGPB1, pGPA, pT7NSΔCAT-RT, and either wild-type (WT) or mutant pGPB2His plasmids, with Ni2+-nitrilotriacetic acid resin. As a control for binding specificity to the resin, extracts transfected with pT7NSΔCAT-RT, pGPB1, pGPB2, and pGPA plasmids were used. (A) Accumulation of PB2 proteins as revealed by Western blot with PB2-specific monoclonal antibodies. The star indicates the faster mobility of wild-type, untagged PB2 protein. (B) Complex formation as detected by Western blot of the proteins retained in the resin, with either PA-specific monoclonal antibodies (Mab) or PB1-specific serum (Pab). (C) Quantitation of three independent experiments performed as indicated in the panels above. The bars indicate the average and standard deviation of Western blot signals quantitated in a Bio-Rad Chemidoc, with the values obtained for wild-type PB2His complex used as a standard.
In vitro transcription and cap-snatching activities of purified RNPs containing PB2 subunit point mutants.
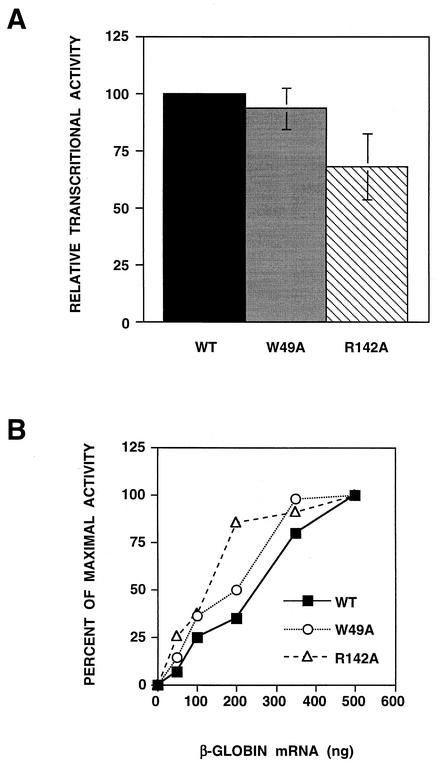
To obtain experimental data, we attempted the purification of wild-type and mutant recombinant RNPs by two cycles of sedimentation on glycerol gradients as described in Materials and Methods in order to determine their specific ability to transcribe in vitro when primed with β-globin mRNA (Fig. 5A). As expected from their replication-defective phenotype (Fig. 3), RNPs containing mutation W49A or R142A could only be obtained with a yield much lower than that of wild-type RNPs (see Fig. 6B), and RNPs containing mutation F130A could not be purified. The specific transcriptional activities, i.e., the cap-dependent synthesis of viral mRNA per unit mass of purified RNPs, of RNPs with mutations W49A and R142A were similar to that obtained for wild-type RNPs (Fig. 5A; Table 1). Furthermore, the dependence of mRNA synthesis on cap donor concentration was comparable for wild-type and mutant RNPs (Fig. 5B). These results suggest that mutant RNPs containing the PB2 W49A or R142A substitution are as efficient as wild-type RNPs in transcription in spite of being defective in RNA replication in vivo.
FIG. 5.
Transcriptional activity in vitro of purified RNPs containing wild-type (WT) or mutant PB2 subunits. Recombinant RNPs were generated by reconstitution in vivo as indicated above and purified by two cycles of glycerol gradient centrifugation. Purified RNPs were assayed for transcription in vitro with globin mRNA as the primer donor, and the results were corrected for the amount of RNPs used, as determined by Western blot with anti-PA monoclonal antibodies (see Fig. 6). (A) Maximal transcriptional activity was assayed with saturating amounts of primer donor. The results are presented as a percentage of the activity of wild-type RNPs and represent the average and range of two independent experiments. (B) Dose-response of primer donor. The transcriptional activity was measured with increasing amounts of primer donor. The results are presented as a percentage of maximal activity.
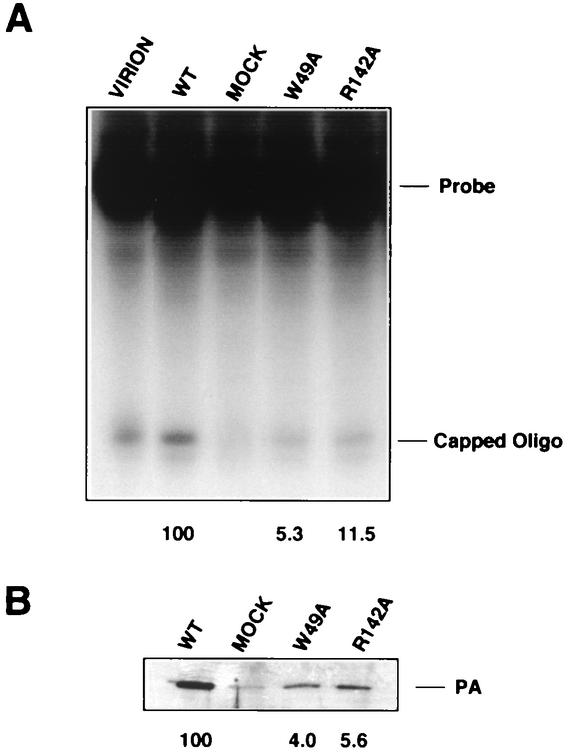
FIG. 6.
Cap-snatching activity in vitro of purified RNPs containing wild-type (WT) or mutant PB2 subunits. Recombinant RNPs, prepared and purified as indicated in the legend to Fig. 5, and virion-derived RNPs were tested for cap-snatching activity with a labeled probe capped and O-methylated in vitro. No cap snatching was observed in the absence of either capping or O-methylation of the probe. (A) The reaction products were separated in a 18% polyacrylamide-urea gel. The positions of the labeled probe and capped oligonucleotide product are indicated. (B) Western blot of parallel aliquots of the same RNPs used for cap snatching, with anti-PA monoclonal antibodies. MOCK, material obtained after purification of RNPs reconstituted in the absence of RNA template. The values are the percentage of capped oligonucleotide bands (A) or PA bands (B), with the wild-type value set at 100%.
These results were confirmed by assaying the cap-snatching activity of wild-type and mutant RNPs. Purified recombinant RNPs were used in a cap-dependent endonuclease assay with a 69-nucleotide synthetic cap donor (9). The results are summarized in Table 1, and a representative experiment is presented in Fig. 6A. When the lanes in Fig. 5A are compared to the corresponding ones in Fig. 6B, which show the amounts of purified RNPs used in the assays, it is clear that both W49A and R142A (PB2) mutant RNPs were active in this assay. Some residual PA protein was detected in the preparation from a negative control in which all subunits of the polymerase and NP were expressed but no template vRNA was provided (Fig. 6B, mock).
Phenotype of recombinant viruses containing point mutations in PB2 subunit.
To gain information about the consequences of the mutations in the PB2 subunit in the context of a virus infection, we attempted to rescue them into infectious virus. Since all experiments were carried out in the genetic background of the A/Victoria/3/75 strain, we used a collection of rescue plasmids derived from the pHH21 vector (46), which contained the cDNAs of the A/Victoria/3/75 strain as well as the corresponding polymerase and NP expression vectors (A. Falcón et al., unpublished data). Only the HA gene in the rescued viruses had a WSN origin, as a control to distinguish the recombinant viruses from other wild viruses used in the laboratory. Several of the mutations in PB2 analyzed above were transferred to the corresponding pHHVicPB2 plasmid, and virus was rescued as indicated in Materials and Methods.
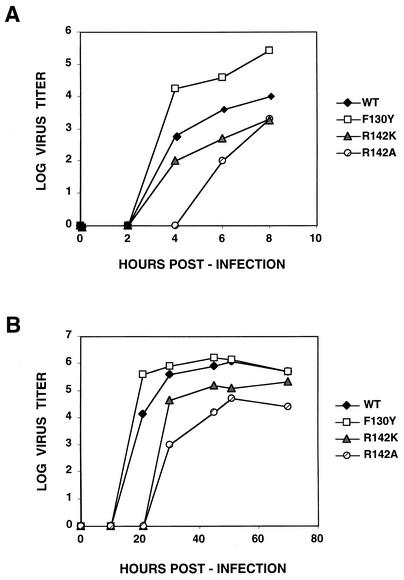
Viruses containing mutations F130Y, R142K, and R142A were viable, but rescue of mutations F130A, W49A, and W49Y failed repeatedly. Mutant F130Y virus grew faster and more efficiently than wild-type virus in a single-cycle infection, in agreement with its higher CAT activity, but the kinetics of virus production for the R142K and R142A mutants was somewhat delayed (Fig. 7A; Table 2). These phenotypes were more clearly observed in a multiple-cycle growth curve. The kinetics and final titer of the F130Y mutant were similar to those of the wild-type virus, whereas mutants R142K and especially R142A were delayed and reached a lower final titer (Fig. 7B; Table 2). In agreement with these results, the plaque size of mutants R142A and R142K was smaller than that of the wild-type virus, while that of mutant F130Y was larger (data not shown).
FIG. 7.
Kinetics of virus multiplication of wild-type (WT) and mutant viruses. Cultures of MDCK cells were infected at a multiplicity of 5 to 10 PFU/cell (A) or 10−2 PFU/cell (B). Aliquots of the culture supernatants were collected at the times indicated, and the virus titer was determined on MDCK cells.
TABLE 2.
Phenotypes of rescued viruses containing point mutations in PB2a
| Virus | Plaque size | Virus yield | Protein synthesis | cRNA | vRNA | Primary transcription |
|---|---|---|---|---|---|---|
| Wild type | ++++ | Normal | Normal | Normal | Normal | Normal |
| F130Y | +++++ | Increased | Normal | Increased | Increased | Normal |
| R142A | + | Delayed | Delayed | Delayed | Delayed | Normal |
| R142K | ++ | Delayed | Delayed | Delayed | Delayed | Normal |
See Table 1, footnote a.
As a first step in the phenotypic analysis of these mutant recombinant viruses, we studied the synthesis of viral proteins along a single-cycle infection. The results are presented in Fig. 8 and summarized in Table 2. They show that the kinetics of synthesis was slower for mutants R142K and R142A, although the final protein pattern was analogous to that of the wild type. Mutant F130Y showed a slightly faster kinetics of protein synthesis than wild-type virus. The delay in viral protein synthesis observed for mutants R142K and R142A could be due to a slower kinetics of viral RNA replication, viral transcription, or both. To check these alternatives, viral RNA replication was first analyzed. The concentrations of NP cRNA and vRNA were determined by real-time RT-PCR at various times after high-multiplicity infection with either wild-type or mutant viruses. The kinetics of NP cRNA accumulation was delayed in mutant R142K- and R142A-infected cells, whereas mutant F130Y accumulated slightly higher amounts of cRNA than wild-type virus (Fig. 9A; Table 2). The kinetics of NP vRNA accumulation showed a delay analogous to that obtained for cRNA accumulation (Fig. 9B; Table 2). These results are compatible with the data obtained by reconstitution in vivo of recombinant RNPs (Fig. 3) and again raise the question of whether the mutations analyzed affect not only the replication step but also the transcription step in the replication cycle, i.e., are mutations that diminish the general functionality of the polymerase or influence exclusively the replication step.
FIG. 8.
Kinetics of viral protein synthesis in wild-type (WT) and mutant virus-infected cells. Cultures of MDCK cells were infected at a multiplicity of 5 to 10 PFU/cell and labeled with [35S]methionine-cysteine. At the times indicated, total cell extracts were prepared in Laemmli sample buffer. The extracts were analyzed by polyacrylamide gel electrophoresis and autoradiography. The mobility of the polymerase subunits (POL) and the NP, NS1, and M1 proteins are indicated to the left. Numbers above each panel indicate the time (hours) after infection at which the extracts were prepared. C, control.
FIG. 9.
Kinetics of cRNA and viral vRNA in wild-type (wt) and mutant virus-infected cells. Cultures of MDCK cells were infected at a multiplicity of 5 to 10 PFU/cell. At the times indicated, total cell RNA was isolated. The accumulation of NP cRNA (A) and vRNA (B) was determined by real-time RT-PCR as indicated in Materials and Methods. The results are presented as a percentage of the maximal accumulation for wild-type virus-infected cells and are the averages and standard deviations of three to four determinations of the kinetics of infection.
To solve this problem, we studied primary transcription, i.e., transcription directed by the incoming parental RNPs in the absence of viral RNA amplification. To this aim, high-multiplicity infections with wild-type and mutant recombinant viruses were carried out in the presence of cycloheximide to avoid viral protein synthesis and hence repress viral RNA replication, and the accumulation of viral mRNA was determined at various times after infection. The results are presented in Fig. 10 and Table 2. The kinetics of NP mRNA accumulation was indistinguishable in cells infected with either wild-type or F130Y, R142K, or R142A mutant viruses (Fig. 10A). Under these experimental conditions, the top level of NP mRNA accumulation corresponded to an average of 23% of maximal mRNA accumulation in a normal infection. To control that no RNA amplification had occurred in the presence of cycloheximide, the accumulation of vRNA was determined in parallel. The results obtained are presented in Fig. 10B in comparison with a normal vRNA kinetics and clearly indicate that viral RNA replication was fully repressed. These results are in full agreement with the in vitro transcription data presented in Fig. 5 and 6 and demonstrate that mutations R142K and R142A do not alter the capacity of viral polymerase to transcribe the viral genome.
FIG. 10.
Kinetics of viral primary transcription in wild-type (wt) and mutant virus-infected cells. Cultures of MDCK cells were infected at a multiplicity of 5 to 10 PFU/cell. The cultures were maintained in the presence of cycloheximide (CHX, 100 μg/ml) from the time of virus adsorption. At the times indicated, total cell RNA was isolated. The accumulation of NP mRNA (A) and vRNA (B) was determined by real-time RT-PCR as indicated in Materials and Methods. The results are presented as a percentage of the maximal accumulation for wild-type virus-infected cells and are the averages and standard deviations of four determinations of the kinetics of infection. The hatched bars in B indicate the accumulation of vRNA in a parallel infection carried out in the absence of cycloheximide.
DISCUSSION
N-terminal region of PB2 is involved in RNA replication.
The PB2 subunit of influenza virus polymerase is a cap-binding protein involved in the initiation of viral transcription. Mapping the sites in PB2 that interact with capped RNA indicated that two regions of its primary sequence can be cross-linked to cap structures: an N-terminal-proximal site (25) and a C-terminal-proximal site close to position 550 (25, 36). In addition to this biochemical evidence, inhibition experiments indicated that cap snatching can be blocked with antibodies directed to the N terminus and C terminus of PB2 (7, 42), and monoclonal antibodies specific for the N-terminal 113 amino acids affect cap-dependent initiation of transcription in vitro (3, 48).
In view of this experimental evidence, it was highly suggestive to find a positive, albeit subtle, alignment of the eIF4E sequence to positions conserved in PB2 among type A, B, and C viruses (Fig. 1). In this alignment, some amino acid residues of eIF4E known to be involved in cap recognition are conserved in the PB2 subunit from all influenza viruses. To test whether such alignment is meaningful, drastic and conservative mutations were introduced at these sites in the PB2 gene as well as in control sites that are conserved in PB2 genes but do not align with eIF4E proteins. Many of these mutant PB2 proteins accumulated to levels similar to that of wild-type PB2 in RNP reconstitution experiments in vivo (Fig. 2). Some of the mutant recombinant RNPs generated were active in transcription-replication in vivo, as determined by the expression of CAT activity from a pseudo-viral cat mRNA. Furthermore, many of the CAT-negative mutations behaved as dominant-negative in competition experiments, suggesting that they were able to form RNPs but were biologically inactive (data not shown).
Interestingly, nonconservative mutations at positions W49, F130, and R142 in PB2, which aligned with amino acids important for cap recognition in eIF4E, reduced the CAT activity of the mutant RNP reconstituted in vivo, whereas conservative substitutions or mutations in control positions (T24 and D83) did not (data not shown). These results were suggestive for a role of the N-terminal region of PB2 in cap RNA recognition and anticipated a defect in these mutants for viral RNA transcription. However, the analysis of vRNA, cRNA, and mRNA accumulation in vivo after reconstitution of wild-type and mutant RNPs indicated that RNA replication was affected (Fig. 3; Table 1). The results obtained in vivo suggest that transcription is not affected, since the accumulation of mRNA paralleled that of vRNA, but the reduced level of replication shown by mutant RNPs made it difficult to reach any firm conclusion. In agreement with their reduced capacity to replicate, some of the mutant RNPs could only be purified with low yields after reconstitution in vivo. This was not the consequence of a defect in the interaction among the subunits, since polymerase complex formation was not affected by mutations at position 142 and only partially reduced for mutant F130A, which was completely inactive (Fig. 4). When the specific transcriptional activity of purified mutant RNPs was measured in vitro, no obvious alteration was observed (Fig. 5; Table 1). Furthermore, cap-snatching activity was indistinguishable for wild-type and mutant RNPs (Fig. 5; Table 1).
Although the experiments discussed above clearly suggest a replication-defective phenotype for the mutations introduced in PB2, the interpretation suffers from the drawback that they were not carried out in the context of a normal virus infection. Therefore, we rescued these mutations in infectious virus to analyze their behavior in vivo. Rescue was carried out in the genetic background of A/Victoria/3/75 virus to be consistent with the experiments carried out by transfection. In agreement with the complete lack of replication activity of the F130A mutant in transfection experiments (Fig. 3), the corresponding virus could not be rescued. Surprisingly, we could not rescue mutations W49Y or W49A in spite of the partial phenotype they showed by transfection (Fig. 3; Table 1). This negative result may suggest that in virus-infected cells, this mutation shows other defects not related to RNA replication. The phenotypes of mutants F130Y, R142K, and R142A essentially paralleled those obtained by transfection and in vitro experiments: the kinetics of cRNA and vRNA accumulation for the F130Y mutant were slightly faster than that of the wild-type virus, whereas they were delayed for mutants R142K and R142A (Fig. 9; Table 2). However, primary transcription was normal for all mutant viruses (Fig. 10; Table 2). As a consequence of the alterations in viral RNA replication, the kinetics of virus gene expression and virus production were delayed in mutants R142K and R142A and accelerated in mutant F130Y (Fig. 7 and 8; Table 2).
These are the first point mutations of PB2 that affect RNA replication and do not alter RNA transcription. Their locations in the gene and their phenotypes indicate that PB2 protein plays a role in the RNA replication process, in agreement with previous results that established a need for PB2 in replication (54), and mapped this replicative function of PB2 to its N-terminal region. These results contrast with those recently reported by Honda et al. (26) on the in vitro activity of PB1-PA or PB1-PB2 binary polymerase complexes. The replicative synthesis of RNA obtained with short templates indicates that PB2 is not absolutely required for the initiation of replication. However, since no NP was present in the system and no direct comparison with the activity of normal RNPs was provided, the significance of these results in vivo is unknown at present. On the other hand, the reported capacity of the PB1-PB2 binary complex to perform cap-dependent transcription is in disagreement with the phenotype of a point mutation in the PA subunit of the polymerase that is specifically affected in cap snatching (14). This phenotype, together with the phenotype of PB2 mutants presented in this report and the structural information of the polymerase (E. Area et al., unpublished data), indicates that the complex is very compact and residues from various subunits may contribute to specific functions of the polymerase in the transcription and replication processes.
Possible roles for PB2 in RNA replication.
The results presented in this report indicate that mutations designed to alter a potential cap-binding domain at the N-terminal region of PB2 affect viral RNA replication but do not modify the capacity of mutant RNPs to initiate transcription. It is conceivable that the mutations introduced alter the recognition of the RNA template by the polymerase and therefore affect its capacity for replication. It has been shown that the PB2 subunit can be cross-linked to the vRNA template (15). Although the interacting region has not been mapped, it is reasonable to assume that the N-terminal region is involved, since it interacts with the C-terminal region of PB1 (20, 73) and this segment of PB1 is capable of interacting with template vRNA (18, 35). Our data indicate that the recognition of both termini of the vRNA template by mutants W49 and R142 is correct as far as reconstituting an RNP functional for transcription. Furthermore, virus mutants affected in position R142 show normal primary transcription but delayed RNA replication. However, we cannot exclude that template-polymerase interaction for these mutants is not appropriate for RNA replication.
Since it has been shown that PB2 protein interacts in vitro with several sites in the NP molecule (4), an alternative possibility to explain the replication-defective phenotype of these mutants is that the PB2-NP interaction is affected by the mutations and the resulting RNPs have a reduced capacity to replicate. Alternatively, interaction of the polymerase with host cell factors, for instance hCLE (29), might be affected by the mutations analyzed.
A speculative and intriguing alternative would be that the viral polymerase contains two independent cap-binding sites, one of them involved in recognition of the capped RNA primer and required for cap snatching, and the other involved in RNA replication. Cross-linking experiments with capped RNA probes have indicated that the recognition of the transcription primer is mediated by sequences located close to position 550 in PB2 (25, 36), as predicted on the basis of sequence comparisons (11), but an additional site that is able to cross-link capped RNA has been found closer to the N terminus of PB2 (25). It is possible that the latter cross-linking site represents an independent cap-binding place that is not related to transcription initiation but is involved in RNA replication. In support of this possibility, it is worth mentioning that antibodies specific for the C terminus of PB2 can block cap snatching but not cap binding (7) and that the activity of viral polymerase in vitro can be stimulated by free cap1 structures (32, 52). This stimulation is mechanistically different from priming by ApG or capped RNA, since the free cap1 structure is not incorporated into the RNA product and stimulation is additive to priming (32, 52). Furthermore, a virus mutant with lesions in the NS and PB2 RNA segments could not be stimulated by cap1 structures in vitro, although it was capable of using capped RNAs as primer donors for transcription (52).
In conclusion, our results indicate that the PB2 subunit of influenza virus polymerase plays a role in viral RNA replication and are compatible with the N-terminal region of the protein's being one of the domains responsible for this activity.
Acknowledgments
We are indebted to J. A. Melero, A. Nieto, and A. Portela for critical comments on the manuscript. We thank P. Gershon, B. Moss, and P. Palese for providing biological materials. The technical assistance of Y. Fernández and J. Fernández is gratefully acknowledged.
P.G. was a fellow from Gobierno Vasco. A.M.F. was a fellow from Programa Nacional de Formación de Personal Investigador, MCYT. This work was supported by Programa Sectorial de Promoción General del Conocimiento (grants PB97-1160 and BMC01-1223) and grant PTR1995-0564-OP.
REFERENCES
- 1.Asano, Y., and A. Ishihama. 1997. Identification of two nucleotide binding domains on the PB1 subunit of influenza virus RNA polymerase. J. Biochem. 122:627-634. [DOI] [PubMed] [Google Scholar]
- 2.Asano, Y., K. Mizumoto, T. Maruyama, and A. Ishihama. 1995. Photoaffinity labeling of influenza virus RNA polymerase PB1 subunit with 8-azido GTP. J. Biochem. Tokyo 117:677-682. [DOI] [PubMed] [Google Scholar]
- 3.Bárcena, J., M. Ochoa, S. de la Luna, J. A. Melero, A. Nieto, J. Ortín, and A. Portela. 1994. Monoclonal antibodies against influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J. Virol. 68:6900-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas, S. K., P. L. Boutz, and D. P. Nayak. 1998. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol. 72:5493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas, S. K., and D. P. Nayak. 1994. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 68:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaas, D., E. Patzelt, and E. Keuchler. 1982. Identification of the cap binding protein of influenza virus. Nucleic Acids Res. 10:4803-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blok, V., C. Cianci, K. W. Tibbles, S. C. Inglis, M. Krystal, and P. Digard. 1996. Inhibition of the influenza virus RNA-dependent RNA polymerase by antisera directed against the carboxy-terminal region of the PB2 subunit. J. Gen. Virol. 77:1025-1033. [DOI] [PubMed] [Google Scholar]
- 8.Braam, J., I. Ulmanen, and R. M. Krug. 1983. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell 34:609-618. [DOI] [PubMed] [Google Scholar]
- 9.Cianci, C., L. Tiley, and M. Krystal. 1995. Differential activation of the influenza virus polymerase via template RNA binding. J. Virol. 69:3995-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Luna, S., J. Martín, A. Portela, and J. Ortín. 1993. Influenza virus naked RNA can be expressed upon transfection into cells coexpressing the three subunits of the polymerase and the nucleoprotein from SV40 recombinant viruses. J. Gen. Virol. 74:535-539. [DOI] [PubMed] [Google Scholar]
- 11.de la Luna, S., C. Martinez, and J. Ortin. 1989. Molecular cloning and sequencing of influenza virus A/Victoria/3/75 polymerase genes: sequence evolution and prediction of possible functional domains. Virus Res. 13:143-155. [DOI] [PubMed] [Google Scholar]
- 12.Detjen, B. M., C. St. Angelo, M. G. Katze, and R. M. Krug. 1987. The three influenza virus polymerase (P) proteins not associated with viral nucleocapsids in the infected cell are in the form of a complex. J. Virol. 61:16-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Digard, P., V. C. Blok, and S. C. Inglis. 1989. Complex formation between influenza virus polymerase proteins expressed in Xenopus oocytes. Virology 171:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fodor, E., M. Crow, L. J. Mingay, T. Deng, J. Sharps, P. Fechter, and G. G. Brownlee. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fodor, E., B. L. Seong, and G. G. Brownlee. 1993. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J. Gen. Virol. 74:1327-1333. [DOI] [PubMed] [Google Scholar]
- 16.Fuerst, T. R., P. L. Earl, and B. Moss. 1987. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol. Cell. Biol. 7:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gluzman, Y. 1981. SV40 transformed simian cells support the replication or early SV40 mutants. Cell 23:175-182. [DOI] [PubMed] [Google Scholar]
- 18.González, S., and J. Ortín. 1999. Characterization of influenza virus PB1 protein binding to viral RNA: two separate regions of the protein contribute to the interaction domain. J. Virol. 73:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González, S., and J. Ortín. 1999. Distinct regions of influenza virus PB1 polymerase subunit recognize vRNA and cRNA templates. EMBO J. 18:3767-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González, S., T. Zürcher, and J. Ortín. 1996. Identification of two separate domains in the influenza virus PB1 protein responsible for interaction with the PB2 and PA subunits: a model for the viral RNA polymerase structure. Nucleic Acids Res. 24:4456-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagen, M., T. D. Chung, J. A. Butcher, and M. Krystal. 1994. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J. Virol. 68:1509-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara, K., M. Shiota, H. Kido, Y. Ohtsu, T. Kashiwagi, J. Iwahashi, N. Hamada, K. Mizoue, N. Tsumura, H. Kato, and T. Toyoda. 2001. Influenza virus RNA polymerase PA subunit is a novel serine protease with Ser624 at the active site. Genes Cells 6:87-97. [DOI] [PubMed] [Google Scholar]
- 23.Hay, A. J., J. J. Skehel, and J. McCauley. 1982. Characterization of influenza virus RNA complete transcripts. Virology 116:517-522. [DOI] [PubMed] [Google Scholar]
- 24.Herz, C., E. Stavnezer, R. M. Krug, and T. Gurney. 1981. Influenza virus, an RNA virus, synthesizes its messenger RNA in the nucleus of infected cells. Cell 26:391-400. [DOI] [PubMed] [Google Scholar]
- 25.Honda, A., K. Mizumoto, and A. Ishihama. 1999. Two separate sequences of PB2 subunit constitute the RNA cap-binding site of influenza virus RNA polymerase. Genes Cells 4:475-485. [DOI] [PubMed] [Google Scholar]
- 26.Honda, A., K. Mizumoto, and A. Ishihama. 2002. Minimum molecular architectures for transcription and replication of the influenza virus. Proc. Natl. Acad. Sci. USA 99:13166-13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda, A., J. Mukaigawa, A. Yokoiyama, A. Kato, S. Ueda, K. Nagata, M. Krystal, D. P. Nayak, and A. Ishihama. 1990. Purification and molecular structure of RNA polymerase from influenza virus A/PR8. J. Biochem. Tokyo 107:624-628. [DOI] [PubMed] [Google Scholar]
- 28.Huang, T. S., P. Palese, and M. Krystal. 1990. Determination of influenza virus proteins required for genome replication. J. Virol. 64:5669-5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huarte, M., J. J. Sanz-Ezquerro, F. Roncal, J. Ortin, and A. Nieto. 2001. PA subunit from influenza virus polymerase complex interacts with a cellular protein with homology to a family of transcriptional activators. J. Virol. 75:8597-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson, D. A., A. J. Caton, S. J. McCready, and P. R. Cook. 1982. Influenza virus RNA is synthesized at fixed sites in the nucleus. Nature 296:366-368. [DOI] [PubMed] [Google Scholar]
- 31.Kato, A., K. Mizumoto, and A. Ishihama. 1985. Purification and enzymatic properties of an RNA polymerase-RNA complex from influenza virus. Virus Res. 3:115-127. [DOI] [PubMed] [Google Scholar]
- 32.Kawakami, K., K. Mizumoto, A. Ishihama, Y. K. Shinozaki, and K. Miura. 1985. Activation of influenza virus-associated RNA polymerase by cap-1 structure (m7GpppNm). J. Biochem. 97:655-661. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi, M., T. Toyoda, and A. Ishihama. 1996. Influenza virus PB1 protein is the minimal and essential subunit of RNA polymerase. Arch. Virol. 141:525-539. [DOI] [PubMed] [Google Scholar]
- 34.Krug, R. M., B. A. Broni, and M. Bouloy. 1979. Are the 5′-ends of influenza viral mRNAs synthesized in vivo donated by host mRNAs? Cell 18:329-334. [DOI] [PubMed] [Google Scholar]
- 35.Li, M. L., B. C. Ramírez, and R. M. Krug. 1998. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 17:5844-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, M. L., P. Rao, and R. M. Krug. 2001. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 20:2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo, G. X., W. Luytjes, M. Enami, and P. Palese. 1991. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J. Virol. 65:2861-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luytjes, W., M. Krystal, M. Enami, J. D. Parvin, and P. Palese. 1989. Amplification, expression and packaging of a foreign gene by influenza virus. Cell 59:1107-1113. [DOI] [PubMed] [Google Scholar]
- 39.Mahy, B. W. J. 1983. Mutants of influenza virus, p. 192-253. In P. Palese and D. W. Kingsbury (ed.), Genetics of influenza viruses. Springer Verlag, Vienna, Austria.
- 40.Marcotrigiano, J., A. C. Gingras, N. Sonenberg, and S. K. Burley. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89:951-961. [DOI] [PubMed] [Google Scholar]
- 41.Marión, R. M., T. Zürcher, S. de la Luna, and J. Ortín. 1997. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J. Gen. Virol. 78:2447-2451. [DOI] [PubMed] [Google Scholar]
- 42.Masunaga, K., K. Mizumoto, H. Kato, A. Ishihama, and T. Toyoda. 1999. Molecular mapping of influenza virus RNA polymerase by site-specific antibodies. Virology 256:130-141. [DOI] [PubMed] [Google Scholar]
- 43.Mena, I., S. de la Luna, C. Albo, J. Martín, A. Nieto, J. Ortín, and A. Portela. 1994. Synthesis of biologically active influenza virus core proteins with a vaccinia-T7 RNA polymerase expression system. J. Gen. Virol. 75:2109-2114. [DOI] [PubMed] [Google Scholar]
- 44.Morino, S., H. Hazama, M. Ozaki, Y. Teraoka, S. Shibata, M. Doi, H. Ueda, T. Ishida, and S. Uesugi. 1996. Analysis of cap-binding ability of human eukaryotic initiation factor-4E by use of recombinant wild-type and mutant forms. Eur. J. Biochem. 239:597-601. [DOI] [PubMed] [Google Scholar]
- 45.Naffakh, N., P. Massin, and S. van der Werf. 2001. The transcription/replication activity of the polymerase of influenza A viruses is not correlated with the level of proteolysis induced by the PA subunit. Virology 285:244-252. [DOI] [PubMed] [Google Scholar]
- 46.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nieto, A., S. de la Luna, J. Bárcena, A. Portela, and J. Ortín. 1994. Complex structure of the nuclear translocation signal of the influenza virus polymerase PA subunit. J. Gen. Virol. 75:29-36. [DOI] [PubMed] [Google Scholar]
- 48.Ochoa, M., J. Bárcena, S. de la Luna, J. A. Melero, A. R. Douglas, A. Nieto, J. Ortín, J. J. Skehel, and A. Portela. 1995. Epitope mapping of cross-reactive monoclonal antibodies specific for the influenza A virus PA and PB2 polypeptides. Virus Res. 37:305-315. [DOI] [PubMed] [Google Scholar]
- 49.Ortega, J., J. Martín-Benito, T. Zürcher, J. M. Valpuesta, J. L. Carrascosa, and J. Ortín. 2000. Ultrastructural and functional analyses of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J. Virol. 74:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ortín, J., R. Nájera, C. López, M. Dávila, and E. Domingo. 1980. Genetic variability of Hong Kong (H3N2) influenza viruses: spontaneous mutations and their location in the viral genome. Gene 11:319-331. [DOI] [PubMed] [Google Scholar]
- 51.Parvin, J. D., P. Palese, A. Honda, A. Ishihama, and M. Krystal. 1989. Promoter analysis of influenza virus RNA polymerase. J. Virol. 63:5142-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Penn, C. R., and B. W. Mahy. 1984. Capped mRNAs may stimulate the influenza virion polymerase by allosteric modulation. Virus Res. 1:1-13. [DOI] [PubMed] [Google Scholar]
- 53.Perales, B., S. de la Luna, I. Palacios, and J. Ortín. 1996. Mutational analysis identifies functional domains in the influenza A virus PB2 polymerase subunit. J. Virol. 70:1678-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perales, B., and J. Ortín. 1997. The influenza A virus PB2 polymerase subunit is required for the replication of viral RNA. J. Virol. 71:1381-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perales, B., J. J. Sanz-Ezquerro, P. Gastaminza, J. Ortega, J. Fernández-Santarén, J. Ortín, and A. Nieto. 2000. The replication activity of influenza virus polymerase is linked to the capacity of the PA subunit to induce proteolysis. J. Virol. 74:1307-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pérez, D. R., and R. O. Donis. 1995. A 48-amino-acid region of influenza A virus PB1 protein is sufficient for complex formation with PA. J. Virol. 69:6932-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piccone, M. E., S. A. Fernandez, and P. Palese. 1993. Mutational analysis of the influenza virus vRNA promoter. Virus Res. 28:99-112. [DOI] [PubMed] [Google Scholar]
- 58.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1990. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poon, L. L., E. Fodor, and G. G. Brownlee. 2000. Polyuridylated mRNA synthesized by a recombinant influenza virus is defective in nuclear export. J. Virol. 74:418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poon, L. L., D. C. Pritlove, J. Sharps, and G. G. Brownlee. 1998. The RNA polymerase of influenza virus, bound to the 5′ end of virion RNA, acts in cis to polyadenylate mRNA. J. Virol. 72:8214-8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poon, L. L. M., D. C. Pritlove, E. Fodor, and G. G. Brownlee. 1999. Direct evidence that the polyadenylated tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 73:3473-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pritlove, D. C., L. L. Poon, E. Fodor, J. Sharps, and G. G. Brownlee. 1998. Polyadenylation of influenza virus mRNA transcribed in vitro from model virion RNA templates: requirement for 5′ conserved sequences. J. Virol. 72:1280-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pritlove, D. C., L. L. M. Poon, L. J. Devenish, M. B. Leahy, and G. G. Brownlee. 1999. A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of polyadenylated mRNA in vitro. J. Virol. 73:2109-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robertson, J. S., M. Schubert, and R. A. Lazzarini. 1981. Polyadenylation sites for influenza mRNA. J. Virol. 38:157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Romanos, M. A., and A. J. Hay. 1984. Identification of the influenza virus transcriptase by affinity-labeling with pyridoxal 5′-phosphate. Virology 132:110-117. [DOI] [PubMed] [Google Scholar]
- 66.Rose, J. K., L. Buonocore, and M. A. Whitt. 1991. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques 10:520-525. [PubMed] [Google Scholar]
- 67.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 68.Sanz-Ezquerro, J. J., S. de la Luna, J. Ortín, and A. Nieto. 1995. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J. Virol. 69:2420-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanz-Ezquerro, J. J., J. Fernández Santarén, T. Sierra, T. Aragón, J. Ortega, J. Ortín, G. L. Smith, and A. Nieto. 1998. The PA influenza polymerase subunit is a phosphorylated protein. J. Gen. Virol. 79:471-478. [DOI] [PubMed] [Google Scholar]
- 70.Sanz-Ezquerro, J. J., T. Zürcher, S. de la Luna, J. Ortin, and A. Nieto. 1996. The amino-terminal one-third of the influenza virus PA protein is responsible for the induction of proteolysis. J. Virol. 70:1905-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schnierle, B. S., P. D. Gershon, and B. Moss. 1994. Mutational analysis of a multifunctional protein, with mRNA 5′ cap-specific (nucleoside-2′-O-)-methyltransferase and 3′-adenylyltransferase stimulatory activities, encoded by vaccinia virus. J. Biol. Chem. 269:20700-20706. [PubMed] [Google Scholar]
- 72.Shi, L., J. M. Galarza, and D. F. Summers. 1996. Recombinant-baculovirus-expressed PB2 subunit of the influenza A virus RNA polymerase binds cap groups as an isolated subunit. Virus Res. 42:1-9. [DOI] [PubMed] [Google Scholar]
- 73.Toyoda, T., D. M. Adyshev, M. Kobayashi, A. Iwata, and A. Ishihama. 1996. Molecular assembly of the influenza virus RNA polymerase: determination of the subunit-subunit contact sites. J. Gen. Virol. 77:2149-2157. [DOI] [PubMed] [Google Scholar]
- 74.Ulmanen, I., B. A. Broni, and R. M. Krug. 1981. The role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc. Natl. Acad. Sci. USA 78:7355-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zürcher, T., S. de la Luna, J. J. Sanz-Ezquerro, A. Nieto, and J. Ortín. 1996. Mutational analysis of the influenza virus A/Victoria/3/75 PA protein: studies of interaction with PB1 protein and identification of a dominant negative mutant. J. Gen. Virol. 77:1745-1749. [DOI] [PubMed] [Google Scholar]
- 76.Zürcher, T., R. M. Marión, and J. Ortín. 2000. The protein synthesis shut-off induced by influenza virus infection is independent of PKR activity. J. Virol. 74:8781-8784. [DOI] [PMC free article] [PubMed] [Google Scholar]