Abstract
Interleukin-1 (IL-1) plays an important role in the inflammatory process. Some studies have demonstrated that IL-1 production was impaired in patients with chronic infections of hepatitis C virus (HCV), implying that IL-1 may play a role in viral clearance. Using an HCV subgenomic replicon cell line, we demonstrate that IL-1 can effectively inhibit HCV subgenomic RNA replication and viral protein expression, suggesting that IL-1 has direct antiviral activity. The inhibitory effect is associated with the extracellular regulatory kinase (ERK) activation. In addition, we also show that IL-1 can induce one of the interferon-stimulated genes (ISGs), 1-8U, which exhibits antiviral activity. However, it has no effect on the other ISG, 6-16, suggesting that IL-1 induces novel antiviral pathways within a cell.
The hepatitis C virus (HCV) is a plus-strand RNA virus that causes acute or chronic hepatitis, cirrhosis, and even hepatocellular carcinoma (20). The present treatment is alpha interferon (IFN-α)-based monotherapy or combination therapy with ribavirin. The overall response rate to the combination therapy is approximately 50 to 60%, with a lower response rate for HCV genotype 1 infection, the predominant genotype in Western countries (6, 22). One of the major clinical issues has been to manage patients who do not respond to IFN treatment. Although the viral molecular structure has been characterized (4, 16), the underlying pathogenesis of HCV infection and the detailed molecular mechanisms for IFN-α action are still not clear.
The histology of infected livers is characterized by inflammatory cell infiltration (predominantly lymphocytes), hepatocyte apoptosis, and various degrees of fibrosis (11). The inflammation caused by viral infection in the liver plays the central role in the pathogenesis, although the regulatory mechanisms are not fully defined. It is generally believed that the severity and extent of tissue inflammation are initiated and maintained by numerous cytokines. These cytokines are secreted by many types of inflammatory cells, and the cytokines themselves are also responsible for the migration and activation of inflammatory cells. Interleukin-1 (IL-1) is one of the most prominent proinflammatory cytokines involved in tissue inflammation (8). The key role of IL-1 appears to be related to its function concerning inflammatory cells, which is crucial for viral clearance and the host immune response. IL-1 has two isoforms, IL-1α and IL-1β, and both isoforms have similar biological functions. IL-1 can exert numerous biological effects on multiple cell types. The effect of IL-1 on noninflammatory cells, such as virus-infected cells, is still not clear.
Changes in IL-1 expression in HCV-infected patients have been well documented. Most studies showed that chronically infected patients had somewhat lower levels of spontaneous and stimulated IL-1 expression by the peripheral blood mononuclear cells (9, 23-25), although some other studies have suggested that patients with HCV had increased serum IL-1 levels (15). Moreover, Daniels et al. have shown that the resolution of the hepatitis B virus (HBV) infection during IFN-α treatment was accompanied by increasing production of IL-1, suggesting that IL-1 production is associated with the IFN therapy response rate (5). The precise biological roles of IL-1 in HCV infection are not defined. It was proposed that IL-1 could help viral clearance by regulating immune response, in particular, the antigen presentation process. In addition, molecular analysis has shown that IL-1 can enhance IFN-stimulated target gene expression, which could play a role in antiviral activity (14). Studies with HBV have shown that IL-1 affected HBV gene transcription in hepatocytes (19). However, the effect of IL-1 on HCV-infected hepatocytes has not been evaluated thoroughly.
IL-1 receptors are present on the membranes of human hepatocytes. They are capable of initiating the intracellular signaling cascade, which can potentially modulate the cellular physiological state (3, 30). We hypothesize that IL-1 may initiate a direct effect on viral RNA replication machinery in human hepatocytes. It has been difficult to ascertain the direct role of IL-1 in HCV replication due to the lack of an efficient cell culture system. The recent development of an HCV subgenomic viral RNA replicon provides a suitable model to test our hypothesis (2, 12, 21). The HCV replicon contains the HCV 5′ nontranslated region (5′NTR) fused to 12 amino acids of the capsid coding region, the neomycin phosphotransferase gene (neo) directed by the HCV internal ribosome entry site (IRES), HCV nonstructural proteins NS3 to NS5B directed by an IRES from encephalomyocarditis virus, and the HCV 3′NTR. Using this model, it has been demonstrated that IFN-α has direct antiviral activity in hepatoma cells (12). In this study, using this replicon cell culture system, we examined the effects of IL-1β on HCV RNA replication and the associated intracellular signaling pathways.
IL-1 inhibits HCV subgenomic RNA replication.
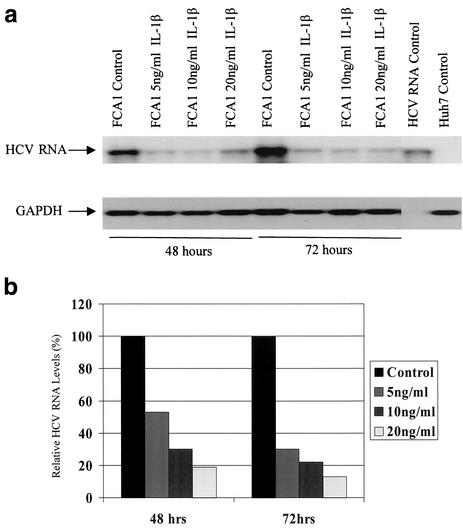
To investigate the direct antiviral activity of IL-1, we first incubated the replicon cell line FCA1 with different doses of IL-1β. The cells were harvested at different times for total RNA extraction. Total cellular RNA was extracted with TRIzol reagent, and 10 μg of RNA was used for Northern blot analysis. The 251-bp cDNA probe representing HCV 5′NTR generated by PCR was used to detect HCV subgenomic RNA. The 355-bp cDNA probe was used to detect glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which served as an internal control for total RNA loaded on the gel. We also examined the effect of IL-1 on viral protein production by Western blot analysis and immunohistochemistry using an NS5A-specific monoclonal antibody.
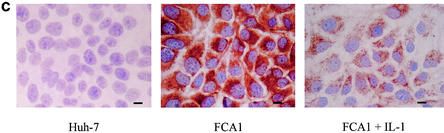
As shown in Fig. 1a, the replication level of the HCV subgenomic RNA replicon was significantly suppressed by treatment with IL-1β. The inhibitory effect was dose and time dependent, as shown in Fig. 1b. The inhibitory effect was more pronounced after 3 days of incubation. Consistent with the Northern blot result, HCV protein NS5A detected by an immunohistochemical stain using a monoclonal antibody against NS5A protein was also reduced by treatment with IL-1β (Fig. 1c).
FIG.1.
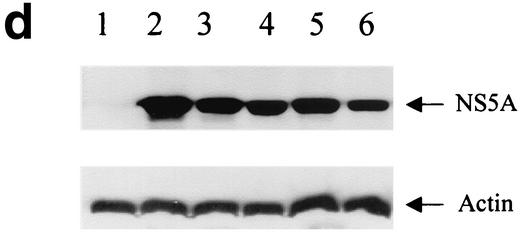
Effects of IL-1 on HCV RNA replication and viral protein expression. (a) Northern blot analysis of total cellular RNAs from FCA1 cells treated with IL-1β. GAPDH served as an internal control. RNA from parental Huh-7 cells served as a negative control. The positive control was subgenomic HCV replicon RNA generated by in vitro transcription. (b) Graphic representation of the Northern blot results. Cells were treated with 0, 5, 10, and 20 ng of IL-1β per ml (from left to right for each set of bars). The HCV RNA levels from the control cells were set at 100% at each time point. The data were normalized to the internal control (GAPDH) values. (c) Immunohistochemical staining of cells treated with IL-1β. Cells were cultured on coverslips in the presence or absence of 10 ng of IL-1β per ml. Cells were fixed with 95% ethanol-5% acetic acid and stained with a monoclonal antibody against NS5A. The parental Huh-7 cells are shown for a negative control. Bar, 20 μm. (d) Western blot analysis. Approximately 50 μg of total protein from FCA1 cells treated with IL-1β or IFN-α for 24 h was resolved on a gel by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a membrane. FCA1 cells were treated with 10 (lane 3) or 20 (lane 4) ng of IL-1β per ml or with 10 (lane 5) or 50 (lane 6) U of IFN-α per ml. Huh7 (lane 1) and FCA1 (lane 2) cells not treated with IL-1β or IFN-α were controls. An anti-HCV NS5A monoclonal antibody was used to detect NS5A protein. Actin served as an internal control.
To further confirm that expression of NS5A decreased in FCA1 cells treated with IL-1β, Western blot analysis was also performed. After FCA1 cells were treated with IL-1β for 2 days, total cellular protein was extracted and 50 μg of protein was used for electrophoresis. Western blotting was performed using chemiluminescent Luminol reagent (Santa Cruz Biotechnology, Santa Cruz, Calif.) according to the manufacturer's instructions. As shown in Fig. 1d, the levels of HCV NS5A protein expressed in FCA1 cells treated with IL-1β decreased significantly from the levels in the control FCA1 cells. The suppression level of 10 ng of IL-1β per ml is comparable to that of 10 ng of IFN-α per ml. The IL-1β concentration used in our experiments showed no apparent toxic effect to the cells. The overall cell numbers were similar for cells treated with IL-1β for 3 days and the corresponding controls, suggesting that the suppression of viral replication is the result of intracellular antiviral effect. The immunohistochemical staining in Fig. 1c clearly demonstrated the reduced viral protein expression in the cytoplasm. These results indicated that IL-1 directly inhibits HCV RNA replication machinery within human hepatoma cells.
IL-1 antiviral activity is associated with ERK MAP kinase activation.
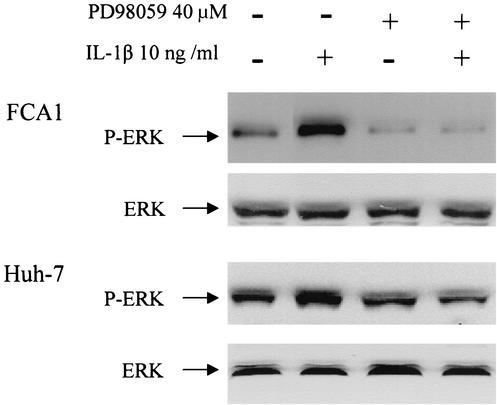
It is believed that binding of IL-1 with its receptor leads to activation of IL-1 receptor-associated kinases IRAK-1 and IRAK-2, followed by activation of mitogen-activated protein (MAP) kinase pathways, including extracellular regulatory kinase (ERK) and P38 pathways, and ultimately to the activation of NF-κB (1). To determine whether such signaling pathways are relevant in IL-1-induced antiviral activity in human hepatoma cells, we examined whether IL-1 antiviral activity is associated with the activation of ERK MAP kinase. We therefore analyzed the ERK protein phosphorylation status by Western blot analysis using a monoclonal antibody against phosphorylated ERK (p42/44) or an antibody against ERK. The parental Huh7 cells and the replicon FCA1 cells were treated with 10 ng of IL-1β per ml for 30 min, followed by protein extraction and Western blot analysis. As shown in Fig. 2, phosphorylated ERK was up-regulated at least three- to fourfold by IL-1β treatment. The basal levels of phosphorylated ERK in Huh-7 cells and FCA1 cells are comparable, suggesting that either viral replication itself or viral nonstructural proteins have no significant effect on ERK activation. These results suggested that activated ERK may be responsible for the antiviral activity. To further determine the causal relationship, an inhibitor of the ERK MAP kinase cascade was used to specifically block the activation of ERK. The flavone compound 2-(2′-amino-3′-methoxyphenyl)oxanaphthalen-4-one (PD98059) specifically inhibits mammalian MEK-1/2 by binding to a regulatory site on the enzyme and has been extensively used in investigations of the function of the ERK MAP kinase pathway (1, 18). As shown in Fig. 2, PD98059 could effectively inhibit IL-1-associated ERK phosphorylation.
FIG. 2.
Effects of IL-1 on phosphorylated ERK in Huh-7 and FCA1 cells. Western blot analysis was performed on protein extracts from cells treated with 10 ng of IL-1 β per ml for 30 min (+). The positions of phosphorylated ERK (P-ERK) and total ERK are indicated by the arrows. The MEK-specific inhibitor PD98059 (40 μM) was added 15 min before the addition of IL-1β.
To determine whether inhibition of the ERK cascade would affect viral RNA replication and IL-1-induced antiviral activity, total RNA was extracted from cells treated with IL-1β in the presence of the PD98059 inhibitor and viral RNA replication levels were determined by Northern blotting. In addition, all cellular proteins were also extracted from these cells for Western blot analysis to determine viral NS5A protein expression.
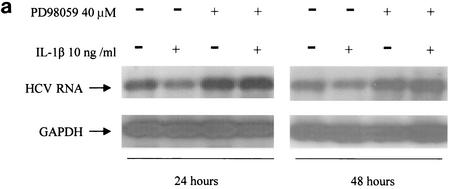
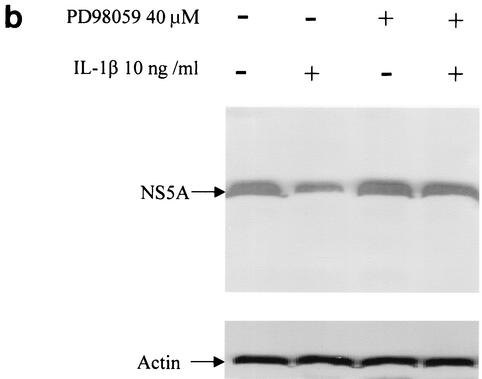
As demonstrated in Fig. 3, PD98059 inhibitor abolished IL-1-induced suppression of viral RNA replication and viral NS5A protein expression, suggesting that IL-1 antiviral activity is dependent on activation of ERK. Moreover, PD98059 treatment alone, which down-regulated the basal level of phosphorylated ERK, also enhanced viral replication efficiency, suggesting that activated ERK has an inhibitory effect on viral replication. The MAP kinase pathway has been implicated as a mechanism by which signals are transduced from the cell surface to the nucleus in response to a variety of different stimuli. The MAP kinase pathway participates in intracellular processes by further inducing phosphorylation of intracellular substrates, such as other protein kinases and transcription factors. The activation of this pathway may also affect the phosphorylation status of HCV proteins (27). Whether IL-1 functions by regulating HCV protein phosphorylation remains to be investigated. Our experiment clearly demonstrated the causal relationship between the antiviral effect of IL-1 and activation of the ERK pathway.
FIG. 3.
Effect of MAP kinase inhibitor PD98059 on IL-1β-induced antiviral activity. (a) Northern blot analysis of RNA extracted from FCA1 cells treated with IL-1β in the presence (+) or absence (−) of PD98059 (40 μM). (b) Western blot analysis of HCV NS5A protein expression in FCA1 cells after 48 h of IL-1β treatment.
IL-1 induces expression of IFN-responsive gene 1-8U but not of 6-16 gene.
Our results demonstrated that IL-1β decreased HCV replication in the HCV replicon cell line through activation of the ERK MAP kinase pathway, suggesting that target genes downstream of ERK may be responsible for the antiviral effect. ERK activates several genes in cells (13). The target genes responsible for the antiviral effect are not known. At this point in time, antiviral proteins have been characterized only in IFN-induced systems (17, 28). It is generally believed that IFN antiviral activity is performed by several IFN-stimulated genes (ISGs). The well-characterized proteins include double-stranded RNA-activated protein kinase (PKR), Mx, and RNase L (31). Mx protein has been shown to have no effect on HCV RNA replication (10). However, the functions of most of the ISGs are still not known (7). We have shown that several genes are activated by IFN-α in Huh7 and FCA1 cells (H. Z. Zhu and C. Liu, submitted for publication). Of these genes, 6-16 and 1-8U genes have higher expression levels in FCA1 cells treated with IFN-α. These two genes may play a role in the establishment of the intracellular antiviral state.
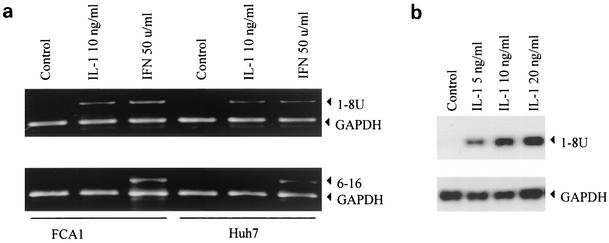
To determine whether IL-1β affects the 6-16 and 1-8U genes, reverse transcription-PCR was performed using specific primers for each gene. As shown in Fig. 4a, IFN induced expression of both 6-16 and 1-8U genes, while IL-1 induced only 1-8U expression, not 6-16 expression. That 1-8U expression was induced by IL-1β was further confirmed by a Northern blot analysis. As shown in Fig. 4b, the level of 1-8U expressed after treatment with 20 ng of IL-1β per ml was at least 30-fold higher than that for the control. The results indicated that IL-1 can actually activate genes that are considered ISGs, suggesting that some overlap signaling pathways exist. The biological functions of both 6-16 and 1-8U are not known (26).
FIG. 4.
Effects of IL-1 and IFN on 1-8U and 6-16 gene expression in FCA1 and Huh7 cells. (a) The cells were treated with 10 ng of IL-1β per ml or with 50 U of IFN-α per ml for 24 h. Total RNA was extracted and used for semiquantitative reverse transcription-PCR analysis. Primers specific for the 6-16 and 1-8U genes and the GAPDH primer were used. The PCR products were resolved on 1.5% agarose gels. The positions of the gene-specific bands are indicated by arrowheads. (b) Northern blot analysis of 1-8U mRNA induction by IL-1β. FCA1 cells were treated with various doses of IL-1β for 24 h. The total RNAs were extracted for Northern blot analysis. GAPDH served as an internal control.
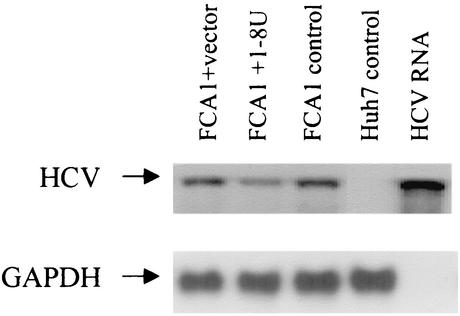
To determine whether 1-8U has antiviral activity, we cloned the 1-8U gene in the expression vector pEF6TOPO (Invitrogen, Carlsbad, Calif.) and transfected it into FCA1 cells. As shown in Fig. 5, 1-8U reduced HCV RNA by approximately twofold compared to the cells transfected with vector controls. This result suggested that 1-8U has antiviral activity. The mechanisms underlying the interactions of the 1-8U protein with viral replication machinery remain to be determined. It would also be interesting to see how the regulatory elements of the 1-8U gene respond to IL-1 and IFN stimulation. Interaction between IL-1 and IFN signaling pathways has been reported recently. One report suggested that IL-1 inhibited Mx protein expression in mouse liver (29). Another report indicated that IL-1 potentiated IFN-induced 2′5′-oligoadenylate synthetase and PKR gene expression (14). Our results suggested that IL-1 alone could activate IFN-responsive gene 1-8U. All these observations support the notion that IL-1 and IFN may share some common pathways in initiating an antiviral state within a cell. To fight viral infection, the cells more likely develop multiple, unique, and redundant pathways to establish an antiviral state. Whether and how IL-1 can induce such intracellular pathways to inhibit HCV replication remain to be investigated.
FIG. 5.
Effect of 1-8U on HCV RNA replication in FCA1 cells. FCA1 cells were transfected by the pEF6TOPO vector containing the 1-8U gene or by pEF6TOPO vector alone. Total RNA was extracted after 48 h, and Northern blot analysis was performed. Huh7 cells served as a negative control. The FCA1 cells and in vitro-transcribed HCV subgenomic replicon RNA served as positive controls.
In conclusion, our study provides the first evidence of direct antiviral IL-1 activity in human hepatoma cells. The activity seems to be associated with the activation of the ERK MAP kinase pathway. We demonstrated that the IFN-α-stimulated gene 1-8U was induced by IL-1β, while the ISG 6-16 was not, suggesting that IL-1 and IFN may have common and diverse antiviral pathways in hepatocytes. Moreover, we also showed that 1-8U protein had antiviral activity. Although IL-1 is a cytokine that causes tissue inflammation, its potent antiviral activity could be employed for antiviral treatment in patients with chronic HCV infections, particularly in the setting of IFN resistance. Further work should be aimed at enhancing IL-1 antiviral activity while limiting its proinflammatory activity.
Acknowledgments
We thank Christopher Seeger and Ju-Tao Guo for the cell line, Charles Rice for the HCV replicon plasmid, Johnson Lau for the anti-NS5A antibody, and James Crawford, Nao Terada, and David Nelson for helpful discussions.
C.L. was partially supported by a grant from the National Institutes of Health (K08 DK02958) and received The Charles Trey MD Memorial Liver Scholar Award from the American Liver Foundation.
REFERENCES
- 1.Baud, V., Z. G. Liu, B. Bennett, N. Suzuki, Y. Xia, and M. Karin. 1999. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 13:1297-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 3.Cao, Z., W. J. Henzel, and X. Gao. 1996. IRAK: a kinase associated with the interleukin-1 receptor. Science 271:1128-1131. [DOI] [PubMed] [Google Scholar]
- 4.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 5.Daniels, H. M., A. Meager, A. L. Eddleston, G. J. Alexander, and R. Williams. 1990. Spontaneous production of tumour necrosis factor alpha and interleukin-1 beta during interferon-alpha treatment of chronic HBV infection. Lancet 335:875-877. [DOI] [PubMed] [Google Scholar]
- 6.Davis, G. L. 1999. Combination treatment with interferon and ribavirin for chronic hepatitis C. Clin. Liver Dis. 3:811-826. [DOI] [PubMed] [Google Scholar]
- 7.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinarello, C. A. 2000. Proinflammatory cytokines. Chest 118:503-508. [DOI] [PubMed] [Google Scholar]
- 9.Dumoulin, F. L., L. Leifeld, U. Honecker, T. Sauerbruch, and U. Spengler. 1999. Intrahepatic expression of interleukin-1beta and tumor necrosis factor-alpha in chronic hepatitis C. J. Infect. Dis. 180:1704-1708. [DOI] [PubMed] [Google Scholar]
- 10.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 11.Goodman, Z. D., and K. G. Ishak. 1995. Histopathology of hepatitis C virus infection. Semin. Liver Dis. 15:70-81. [DOI] [PubMed] [Google Scholar]
- 12.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howe, A. K., A. E. Aplin, and R. L. Juliano. 2002. Anchorage-dependent ERK signaling-mechanisms and consequences. Curr. Opin. Genet. Dev. 12:30-35. [DOI] [PubMed] [Google Scholar]
- 14.Ichikawa, T., K. Nakao, K. Nakata, M. Yamashita, K. Hamasaki, M. Shigeno, S. Abiru, H. Ishikawa, N. Ishii, and K. Eguchi. 2002. Involvement of IL-1beta and IL-10 in IFN-alpha-mediated antiviral gene induction in human hepatoma cells. Biochem. Biophys. Res. Commun. 294:414-422. [DOI] [PubMed] [Google Scholar]
- 15.Kaplanski, G., C. Farnarier, M. J. Payan, P. Bongrand, and J. M. Durand. 1997. Increased levels of soluble adhesion molecules in the serum of patients with hepatitis C. Correlation with cytokine concentrations and liver inflammation and fibrosis. Dig. Dis. Sci. 42:2277-2284. [DOI] [PubMed] [Google Scholar]
- 16.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, A., A. Middleton, T. C. Chambers, and K. D. Mehta. 1998. Differential roles of extracellular signal-regulated kinase-1/2 and p38MAPK in interleukin-1beta- and tumor necrosis factor-alpha-induced low density lipoprotein receptor expression in HepG2 cells. J. Biol. Chem. 273:15742-15748. [DOI] [PubMed] [Google Scholar]
- 19.Lara-Pezzi, E., P. L. Majano, M. Gomez-Gonzalo, C. Garcia-Monzon, R. Moreno-Otero, M. Levrero, and M. Lopez-Cabrera. 1998. The hepatitis B virus X protein up-regulates tumor necrosis factor alpha gene expression in hepatocytes. Hepatology 28:1013-1021. [DOI] [PubMed] [Google Scholar]
- 20.Liang, T. J., B. Rehermann, L. B. Seeff, and J. H. Hoofnagle. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann. Intern. Med. 132:296-305. [DOI] [PubMed] [Google Scholar]
- 21.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 22.McHutchison, J. G., M. Manns, K. Patel, T. Poynard, K. L. Lindsay, C. Trepo, J. Dienstag, W. M. Lee, C. Mak, J. J. Garaud, and J. K. Albrecht. 2002. Adherence to combination therapy enhances sustained response in genotype-1-infected patients with chronic hepatitis C. Gastroenterology 123:1061-1069. [DOI] [PubMed] [Google Scholar]
- 23.Mendoza, E. C., T. G. Paglieroni, and J. B. Zeldis. 1996. Decreased phorbol myristate acetate-induced release of tumor necrosis factor-alpha and interleukin-1 beta from peripheral blood monocytes of patients chronically infected with hepatitis C virus. J. Infect. Dis. 174:842-844. [DOI] [PubMed] [Google Scholar]
- 24.Muller, C., P. Knoflach, and C. C. Zielinski. 1993. Reduced production of immunoreactive interleukin-1 by peripheral blood monocytes of patients with acute and chronic viral hepatitis. Dig. Dis. Sci. 38:477-481. [DOI] [PubMed] [Google Scholar]
- 25.Ozyilkan, E., G. Tatar, A. Hacibektasoglu, B. Kayhan, and H. Telatar. 1994. Impaired lipopolysaccharide-induced interleukin-1-beta production in patients with anti-hepatitis C virus antibody-positive chronic liver disease. Scand. J. Gastroenterol. 29:280-283. [DOI] [PubMed] [Google Scholar]
- 26.Porter, A. C., and J. E. Itzhaki. 1993. Gene targeting in human somatic cells. Complete inactivation of an interferon-inducible gene. Eur. J. Biochem. 218:273-281. [DOI] [PubMed] [Google Scholar]
- 27.Reed, K. E., A. E. Gorbalenya, and C. M. Rice. 1998. The NS5A/NS5 proteins of viruses from three genera of the family Flaviviridae are phosphorylated by associated serine/threonine kinases. J. Virol. 72:6199-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 29.Tian, Z., X. Shen, H. Feng, and B. Gao. 2000. IL-1 beta attenuates IFN-alpha beta-induced antiviral activity and STAT1 activation in the liver: involvement of proteasome-dependent pathway. J. Immunol. 165:3959-3965. [DOI] [PubMed] [Google Scholar]
- 30.Yamin, T. T., J. M. Ayala, and D. K. Miller. 1996. Activation of the native 45-kDa precursor form of interleukin-1-converting enzyme. J. Biol. Chem. 271:13273-13282. [DOI] [PubMed] [Google Scholar]
- 31.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]