Abstract
Successful generation, extension, and removal of the plus-strand primer is integral to reverse transcription. For Moloney murine leukemia virus, primer removal at the RNA/DNA junction leaves the 3′ terminus of the plus-strand primer abutting the downstream plus-strand DNA, but this 3′ terminus is not efficiently reutilized for another round of extension. The RNase H cleavage to create the plus-strand primer might similarly result in the 3′ terminus of this primer abutting downstream RNA, yet efficient initiation must occur to synthesize the plus-strand DNA. We hypothesized that displacement synthesis, RNase H activity, or both must participate to initiate plus-strand DNA synthesis. Using model hybrid substrates and RNase H-deficient reverse transcriptases, we found that displacement synthesis alone did not efficiently extend the plus-strand primer at a nick with downstream RNA. However, specific cleavage sites for RNase H were identified in the sequence immediately following the 3′ end of the plus-strand primer. During generation of the plus-strand primer, cleavage at these sites generated a gap. When representative gaps separated the 3′ terminus of the plus-strand primer from downstream RNA, primer extension significantly improved. The contribution of RNase H to the initiation of plus-strand DNA synthesis was confirmed by comparing the effects of downstream RNA versus DNA on plus-strand primer extension by wild-type reverse transcriptase. These data suggest a model in which efficient initiation of plus-strand synthesis requires the generation of a gap immediately following the plus-strand primer 3′ terminus.
Shortly after entrance of the viral cores into the cytoplasm of a cell, the single-stranded plus-sense RNA genome of a retrovirus is converted into a double-stranded DNA molecule that subsequently integrates into the host cell genome (4). This process, termed reverse transcription, requires two distinct RNA primers to synthesize the double-stranded DNA. The first primer is a host cell-derived tRNA that is used for the initiation of minus-strand DNA synthesis. The second is a short RNA derived by RNase H cleavages within a purine-rich sequence in the viral genome called the polypurine tract (PPT). This primer is used to begin plus-strand synthesis and is referred to as the plus-strand primer or the PPT primer (reference 2 and references therein).
These RNA primers are extended by the viral-encoded reverse transcriptase, a multifunctional enzyme that carries out DNA polymerization, strand displacement synthesis, and the strand transfer reaction and that possesses an RNase H activity required at several steps during genome replication (4). The DNA polymerase activity of reverse transcriptase resides in the N-terminal two-thirds of the protein and utilizes either RNA or DNA as a template. Although displacement synthesis is not as efficient as nondisplacement synthesis (13, 15, 25, 43), reverse transcriptase can simultaneously extend the 3′ terminus of a DNA primer and displace a downstream nontemplate RNA or DNA strand, a function vital to complete reverse transcription. Previous studies have demonstrated that reverse transcriptase can access a 3′ primer terminus and initiate DNA and RNA displacement synthesis at a single-strand break or nick (21, 25, 43).
The RNase H activity of reverse transcriptase comprises the C-terminal one-third of the protein and cleaves RNA in an RNA-DNA hybrid (3). RNase H effectively degrades the RNA template both during and after minus-strand synthesis to facilitate strand transfer and plus-strand synthesis. Biochemical studies have shown that polymerization-dependent RNase H cleavages take place concomitant with DNA synthesis, are positioned by the polymerase domain, and occur in the RNA template strand 15 to 20 nucleotides (nt) from the DNA 3′ terminus (16, 17, 24). However, the polymerization-dependent form of RNase H activity is not sufficient to eliminate all RNA from the minus-strand DNA (6, 9, 10, 24).
The polymerization-independent form of RNase H activity cleaves RNA in the absence of any DNA synthesis. These RNase H cleavages can be positioned by the polymerase domain of reverse transcriptase binding to either a recessed DNA 3′ end on a longer RNA strand or a recessed RNA 5′ end on a longer DNA strand (5, 7, 8, 16, 24, 35). In the latter case, cleavages are referred to as 5′-end directed and generally occur about 15 to 20 nt from the RNA 5′ end. Recently, it has been proposed that primary 5′ end-directed cleavages are followed by two additional types of RNase H cleavages that appear to occur independent of other cleavages and with equal probability (46-48). These sequence-independent cleavages were classified either as secondary cuts that occur 7 to 10 nt from the RNA 5′ end or as 5-nt cuts that occur 4 to 7 nt from the RNA 3′ end.
The polymerization-independent form of RNase H activity can also cleave hybrids internally without positioning by an RNA 5′ end or a DNA 3′ end. An example is the generation of the PPT primer: RNase H efficiently generates the 3′ end of the PPT primer by a specific cleavage within the PPT at the plus-strand origin, herein referred to as the −1/+1 site. Importantly, this RNase H cleavage is sequence specific and can occur internally on an RNA-DNA hybrid without 5′ end-directed binding (14, 37). Sequence recognition directly contributes to RNase H specificity for at least two other sites as well. After plus-strand and minus-strand DNA syntheses have initiated, RNase H removes both the PPT primer and the tRNA primer at or near the RNA/DNA junctions (3). Other internal and potentially sequence-specific RNase H cleavage sites in the RNA genome have been observed but not characterized (26).
When RNase H removes the PPT primer from plus-strand DNA during reverse transcription, the 3′ end of the PPT primer is followed immediately by the 5′ end of the newly synthesized downstream DNA, which presents an interesting substrate for reverse transcriptase. Despite the ability of reverse transcriptase to carry out DNA displacement synthesis at a nick (21, 25, 43), the PPT primer is not efficiently reutilized for additional plus-strand synthesis (19, 37). A similar situation might arise when RNase H creates the PPT primer if RNase H were to make only a single cleavage at the −1/+1 site. This would offer reverse transcriptase a nick with the 3′ end of the PPT primer abutting downstream RNA as a substrate to initiate plus-strand DNA synthesis. Although numerous studies have addressed utilization of the PPT primer (3, 14, 18, 19, 22, 33, 34), none have considered the effect of extension at a nick with downstream RNA. In this report, we have used the reverse transcriptase of Moloney murine leukemia virus (M-MuLV) to ask whether displacement synthesis is sufficient to initiate plus-strand synthesis or whether further RNase H cleavages might facilitate extension of the PPT primer. The results of these experiments offer insights into how M-MuLV reverse transcriptase initiates plus-strand synthesis.
MATERIALS AND METHODS
Enzymes.
Recombinant wild-type M-MuLV reverse transcriptase, calf intestinal alkaline phosphatase, and T7 DNA polymerase (Sequenase version 2.0 [a modified form of phage T7 DNA polymerase lacking 3′ to 5′ exonuclease activity]) were purchased from Amersham Pharmacia Biotech. Superscript (RTΔH) and Superscript II (H− RT) were obtained from Invitrogen. T4 DNA polymerase, T4 polynucleotide kinase, T4 DNA ligase, and all restriction endonucleases were purchased from New England Biolabs.
Oligonucleotides.
RNA oligonucleotides were obtained from Oligos Etc., DNA oligonucleotides were purchased from Invitrogen, and all oligonucleotides were gel purified prior to use. The cleavage site that generates the 3′ terminus of the PPT primer is located on the plus-sense M-MuLV genome between positions 7815 and 7816 (39), which are designated as the −1 and +1 positions, respectively, in this study. The sequences, names, and positions of oligonucleotides and RNAs used are shown in Fig. 1. The names and sequences of DNA template strands are as follows: template 2, 5′-GTGGGGTCTTTCATTCCCCCCTTTTTCTGGAGACTAAATAAAATCTTTTATTTTATCTATGGCTCGTACGAGCTCCCGA-3′; template 2A, 5′-AGGTGGGGTCTTTCATTCCCCCCTTTTTCTGGAGACTAAATAAAATCTTTTATTTTATCTATGGCTCGTACGAGCTCCCGA-3′; template 2B, 5′-TACAGGTGGGGTCTTTCATTCCCCCCTTTTTCTGGAGACTAAATAAAATCTTTTATTTTATCTATGGCTCGTACGAGCTCCCGA-3′; template 2C, 5′-CCAAACCTACAGGTGGGGTCTTTCATTCCCCCCTTTTTCTGGAGACTAAATAAAATCTTTTATTTTATCTATGGCTCGTACGAGCTCCCGA-3′; template 5, 5′-ACAGGGACTTGAAAGCCCCCCTTTTTCTGGAGACTAAATAAAATCTTTTATTTTATCTATGGCTCGTACGAGCTCCCGA-3′; and template 5B, 5′-ACAGGGACTTGAAAGCTCATTCCCCCCTTTTTCTGGAGACTAAATAAAATCTTTTATTTTATCTATGGCTCGTACGAGCTCCCGA-3′.
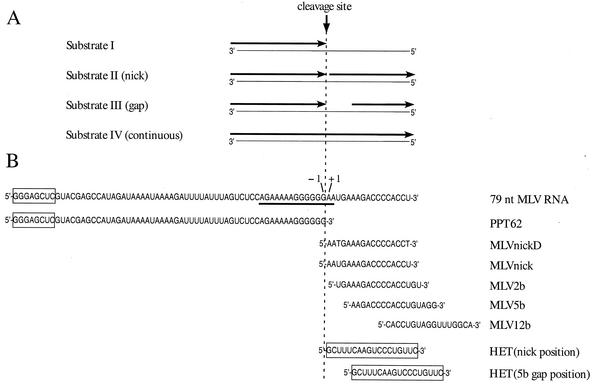
FIG. 1.
Model hybrid substrates with RNAs and oligonucleotides used in extension and cleavage analyses. (A) General structures of model hybrid substrates I to IV. Template strands are shown as thin lines with their 5′ and 3′ ends indicated. Oligonucleotides or RNAs annealed to template strands are shown as arrows originating at the 3′ ends. When applicable, positions of RNAs or oligonucleotides are described as upstream or downstream relative to the PPT primer cleavage site (indicated by vertical dashed line), which is based upon the plus-strand origin found within the PPT of the M-MuLV genome. Substrate I contains a 62-nt plus-strand primer (PPT62) with a 3′ end at position −1. Substrate II contains a downstream DNA or RNA oligonucleotide abutting the 3′ end of PPT62 and creating a nick. In substrate III, the 5′-end position of the downstream oligonucleotide creates a gap of 2, 5, or 12 bases from the 3′ end of PPT62. Substrate IV contains a 79-nt MLV RNA, which is a continuous RNA without a gap or nick. (B) The names and sequences of RNAs and oligonucleotides annealed to the template strands are shown relative to the −1 and +1 positions that border the plus-strand origin. The downstream oligonucleotides derived from the M-MuLV sequence are termed MLV, while the downstream oligonucleotide derived from an unrelated sequence is referred to as HET (for heterologous). The PPT sequence for the 79-nt MLV RNA is underlined. Sequences not derived from the M-MuLV genome are boxed. With the exception of the deoxyribonucleotide MLVnickD, all sequences are ribonucleotides.
Preparation of PPT62 and 79-nt MLV RNAs.
To prepare the transcription template for PPT62, the 62-nt RNA, a 70-mer DNA oligonucleotide(5′-AATTCGTCTCACCCCCCTTTTTCTGGAGACTAAATAAAATCTTTTATTTTATCTATGGCTCGTACGAGCT-3′) was annealed to a 62-mer DNA oligonucleotide (5′-CGTACGAGCCATAGATAAAATAAAAGATTTTATTTAGTCTCCAGAAAAAGGGGGGTGAGACG-3′) in 10 mM Tris-HCl (pH 7.5)-10 mM MgCl2-50 mM NaCl by heating to 90°C for 5 min and then slowly cooling to room temperature. The resulting duplex DNA was inserted into EcoRI- and SacI-digested pGEM9Zf(−) (Promega) to generate plasmid pGEM-BsmBI. Using the T7 MEGAshortscript kit (Ambion, Inc.) to generate the PPT62 RNA, which begins at the 5′ end with 8 nt (5′-GGGAGCUC-3′) of plasmid-derived sequence followed by 54 nt of M-MuLV sequence from positions −54 through −1 (39), the pGEM-BsmBI plasmid was linearized by BsmBI and transcribed in vitro by T7 RNA polymerase. Using the 19-mer DNA oligonucleotide (5′-TCGGGAGCTCGTACGAGCC-3′) for first-strand synthesis and the 32-mer DNA oligonucleotide (5′-GGAATTCGTCTCAAGGTGGGGTCTTTCATTCC-3′) for second-strand synthesis, the M-MuLV sequence from positions 7762 to 7831 (39) was amplified by PCR from template 2A to prepare the transcription template for the 79-nt MLV RNA. After digestion with EcoRI and SacI, the PCR product was introduced into EcoRI- and SacI-linearized pGEM9Zf(−). The resulting plasmid was linearized with BsmBI and transcribed in vitro as described above to generate the 79-nt MLV RNA, which begins at its 5′ end with the same 8 nt as PPT62 and ends at position +17.
In vitro-transcribed RNAs were purified by electrophoresis through 15% denaturing polyacrylamide gels, visualized by UV shadowing, and removed from gel slices by elution into TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The eluates were precipitated in 0.3 M sodium acetate (pH 5.2) with 2 volumes of 100% ethanol and resuspended in TE buffer. The 5′ triphosphate was removed by incubation with calf intestinal alkaline phosphatase in 50 mM Tris-HCl (pH 7.5)-10 mM MgCl2 for 60 min at 37°C, the RNA was recovered by extraction with phenol and chloroform and was ethanol precipitated, and concentrations were determined by UV spectroscopy.
5′- and 3′-end labeling of RNAs.
Using 5 to 10 pmol of [γ-32P]ATP (NEN Life Science Products) (3,000 Ci/mmol) as described previously (37, 38) for 5′-end labeling, 10 pmol of phosphatase-treated RNA was labeled in 20-μl reaction mixtures. After labeling, 5′ end-labeled 79-nt RNA was gel purified. For 3′-end labeling, 10 pmol of 79-nt MLV RNA was annealed to 20 pmol of template 2B in 25 mM Tris-HCl (pH 8.0)-200 mM NaCl by heating to 90°C for 3 min, slowly cooling to room temperature, and precipitating with ethanol and 2 μg of glycogen. The resulting hybrid contained a single-stranded extension with the sequence CAT-5′ and was incubated with T7 DNA polymerase and [α-32P]dGTP (NEN Life Science Products) (3,000 Ci/mmol) in 20 μl of extension buffer (50 mM Tris-HCl [pH 8.0], 50 mM KCl, 6 mM MgCl2, 5 mM dithiothreitol) at 37°C for 60 min and then chased with 200 μM dGTP at 37°C for 15 min. Reactions were stopped by the addition of 25 mM EDTA (pH 8.0), and the 3′ end-labeled RNA was gel purified from 10% denaturing polyacrylamide gels.
Preparation of hybrid substrates.
Hybrids containing primer PPT62 were annealed at a primer/template ratio of 1:1.5, and when required, downstream oligonucleotide generating a nick or 2-base gap was added at a 6-fold excess and downstream oligonucleotide generating a 5-base or 12-base gap was added at a 12-fold excess. Then, 79-nt MLV RNA was annealed at a primer/template ratio of 1:2. Hybrid substrates were made by heating in 10 mM Tris-HCl (pH 7.5)-200 mM KCl at 90°C for 3 min and slowly cooling to room temperature. The ends of all RNA-DNA hybrids had a 3′ overhang of 2 bases to preclude primer terminus positioning by reverse transcriptase.
Extension analysis of 5′ end-labeled PPT62.
Hybrid substrate (0.1 pmol) was incubated with equal amounts of polymerase activity units (15 units) of wild-type reverse transcriptase, RTΔH, or H− RT or with 5 units of T7 DNA polymerase or with 0.5 units of T4 DNA polymerase in 20-μl reaction mixtures containing extension buffer and 200 μM dNTPs at 37°C for 15 min or for the indicated times. For each time point, 5-μl aliquots were added to 10 μl of formamide stop buffer (95% deionized formamide, 20 mM EDTA, 0.025% bromphenol blue, 0.025% xylene cyanol). Samples were analyzed in 15% denaturing polyacrylamide gels, visualized by PhosphorImager analysis, and quantified using ImageQuant software (Molecular Dynamics).
Extension analysis by incorporation of [α-32P]dGTP.
For analysis of DNA synthesis by wild-type reverse transcriptase, extension reactions identical to those for 5′ end-labeled PPT62 were carried out, except that reactions contained 313 nM [α-32P]dGTP (800 Ci/mmol) and 200 μM dATP, dCTP, and dTTP. At the indicated times, 5-μl aliquots were added to 5 μl of 40 mM EDTA, pH 8.0, to stop the reactions. Samples were treated with 0.3 N NaOH at 65°C for 30 min, neutralized with 0.3 M acetic acid, and mixed with 2 volumes of formamide stop buffer. Samples were analyzed in 20% denaturing polyacrylamide gels and visualized as described above.
Cleavage analysis of substrate containing the 79-nt MLV RNA.
Hybrid (0.2 pmol) containing either 5′ or 3′ end-labeled 79-nt MLV RNA was incubated with 2 pmol of wild-type reverse transcriptase in 20-μl reaction mixtures as previously described (37). Size ladders of labeled RNAs were prepared in 20-μl reactions containing 50 mM sodium acetate (pH 5.2), 200 ng of tRNA, and 5 ng of nuclease P1 (PL Biochemicals)/ml for 4 min at 37°C.
RESULTS
Downstream RNA impedes extension from the plus-strand primer.
The 3′ end of the PPT primer is generated by an RNase H cleavage at a specific site in the PPT between G and A residues, which are referred to herein as the −1 and +1 positions on the M-MuLV RNA genome (3). Accordingly, base positions upstream of the −1/+1 cleavage site are numbered as negative and base positions downstream are numbered as positive. To examine how downstream RNA might affect extension of the PPT primer, four different types of model hybrid substrates were created (substrates I to IV; Fig. 1A). The sequences of primer and nontemplate strand oligonucleotides and RNAs for these substrates are shown in Fig. 1B, while those of the template strands are given in Materials and Methods.
Initiation of plus-strand synthesis was assayed by extending a PPT-containing primer annealed to a template DNA strand. Initially we used a 13-mer RNA primer exclusively containing the PPT sequence with 5′ and 3′ ends at the −13 and −1 positions, respectively. However, when this 5′ end-labeled 13-mer was extended with wild-type reverse transcriptase, analysis of extension products was difficult due to poor utilization of this short primer and efficient primer removal by the RNase H activity (data not shown). Since reverse transcriptase utilizes a longer PPT primer more efficiently (37, 38), we improved the efficiency of extension by employing a 62-nt PPT primer with a 3′ end at position −1 (PPT62; Fig. 1) and eliminated the complications of primer removal by using reverse transcriptases deficient in RNase H. Importantly, it has been shown that RNase H-deficient reverse transcriptases have displacement synthesis activities comparable to those of the wild-type enzyme (13, 44).
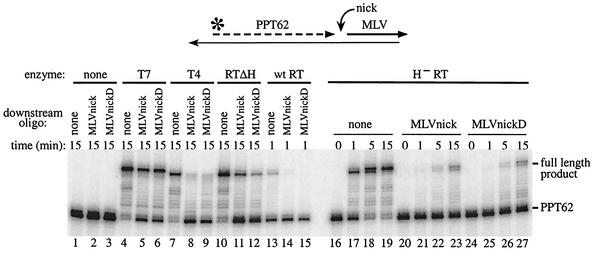
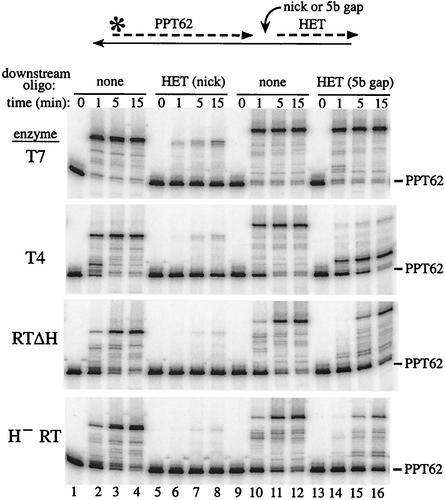
When RTΔH, a form of reverse transcriptase that contains a C-terminal deletion of the RNase H domain, was used to extend 5′ end-labeled PPT62 in the context of nondisplacement synthesis (substrate I), runoff extension products corresponding to primer length plus 15 to 16 nt were observed, representing full-length extension with some nontemplated addition of a single base (32) (Fig. 2, lane 10). When a 17-mer DNA corresponding to positions +1 to +17 of M-MuLV plus-strand DNA (MLVnickD) was placed downstream of PPT62 in substrate II, extension from PPT62 dropped sixfold (Fig. 2; compare lanes 10 and 12). When an equivalent 17-mer RNA (MLVnick) abutted PPT62 in substrate II, synthesis of full-length extension products decreased threefold (Fig. 2, lane 11). Similar results were observed in a time course assay with H− RT, an intact form of reverse transcriptase containing specific point mutations in the active site of the RNase H domain (Fig. 2, lanes 16 to 27). At the 1-min time points, 90-fold more full-length product was synthesized with substrate I than with both forms of substrate II (lanes 17, 21, and 25). By 16 min, the difference in synthesis of full-length extension products was five- to sevenfold (lanes 19, 23, and 27).
FIG. 2.
Extension from PPT62 at a nick followed by the M-MuLV RNA. 5′ end-labeled PPT62 without a downstream oligonucleotide (none) (lanes 1, 4, 7, 10, 13, and 16 to 19) or with downstream MLVnick (lanes 2, 5, 8, 11, 14, and 20 to 23) or MLVnickD (lanes 3, 6, 9, 12, 15, and 24 to 27) was annealed to template 2. Extensions were carried out with T7 DNA polymerase (T7; lanes 4 to 6), T4 DNA polymerase (T4; lanes 7 to 9), RTΔH (lanes 10 to 12), wild-type reverse transcriptase (wt RT; lanes 13 to 15) or H− RT (lanes 17 to 19, 21 to 23, and 25 to 27) for the indicated times. Substrates incubated without enzyme are shown in lanes 1 to 3, 16, 20, and 24. The products were analyzed in a 20% sequencing gel and visualized using a PhosphorImager. A schematic of the nicked substrate II tested is shown at top, and the positions of unextended PPT62 and the full-length extension product are indicated on the right.
In this assay, displacement synthesis through 4 nucleotides is required before the downstream oligonucleotide melts off and nondisplacement synthesis can proceed. To verify the structures of the model hybrid substrates, control extensions were performed with modified T7 DNA polymerase, which has strand displacement activity, and T4 DNA polymerase, which lacks significant strand displacement activity (43). T7 DNA polymerase efficiently extended PPT62 in substrate I but generated twofold less full-length product when MLVnick or MLVnickD was present in substrate II (Fig. 2, lanes 4 to 6). As expected, T4 DNA polymerase extended PPT62 in substrate I but poorly extended PPT62 in either form of substrate II (Fig. 2, lanes 7 to 9).
These substrates were also extended with wild-type reverse transcriptase, but a shorter time period was required to visualize extension products due to RNase H cleavage between the primer RNA and the nascent DNA. When either form of substrate II containing PPT62 was extended with wild-type reverse transcriptase for 1 min, the amount of full-length product synthesized was reduced 20-fold or more compared to that resulting from extension with substrate I (Fig. 2, lanes 13 to 15).
Together, these data indicated that extension of a PPT primer by reverse transcriptase was impeded when either a DNA or an RNA nontemplate strand abutted the 3′ terminus of the RNA primer.
RNase H cleaves specifically downstream of the plus-strand primer.
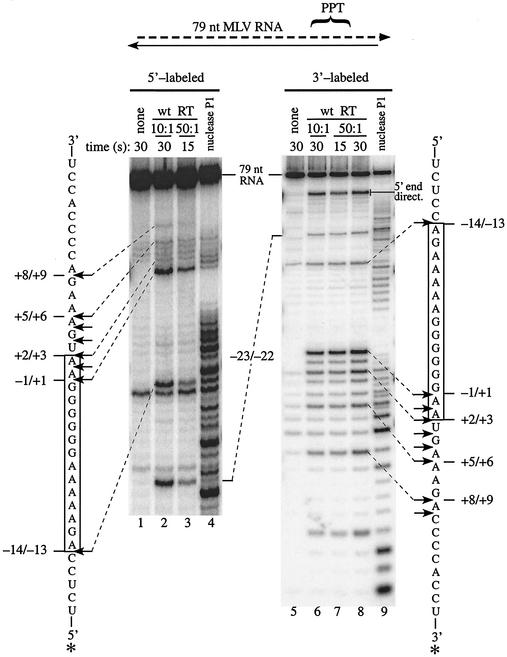
Since downstream RNA with a 5′ end at position +1 greatly diminished extension from a PPT primer, it seemed possible that RNase H cleavages in addition to the one at the −1/+1 site that generates the 3′ end of the PPT primer might facilitate the efficient initiation of plus-strand DNA synthesis. To ask whether RNase H exhibited specificity for any additional sites downstream of the −1 position, we employed an RNase H cleavage assay with substrate IV, which has a 79-nt RNA containing plus-sense M-MuLV sequences extending from positions −54 to +17 and therefore an internal PPT (Fig. 1A and B). When this 79-nt MLV RNA was 5′ end labeled in hybrids that were treated with wild-type reverse transcriptase, the most prominent bands resulted from internal cleavages at the −23/−22, −14/−13, and −1/+1 sites (Fig. 3, lanes 2 and 3). Significant cleavages sites were also apparent downstream of the −1/+1 site between positions +1 and +6 and at +8/+9. However, due to background bands in the untreated 5′ end-labeled 79-nt MLV RNA and strong cleavage at the −1/+1 site, it was necessary to confirm these observations by use of a 3′ end-labeled substrate.
FIG. 3.
RNase H cleavage sites in the vicinity of the −1/+1 cleavage site on the 79-nt MLV RNA containing an internal PPT. Either 5′ end-labeled (lanes 1 to 4) or 3′ end-labeled (lanes 5 to 9) 79-nt MLV RNA (containing M-MuLV sequence from −54 to +17) was annealed to template 2 in substrate IV (schematic at top), and these hybrids were incubated with a 10:1 (lanes 2 and 6) or 50:1 (lanes 3,7, and 8) molar ratio of wild-type reverse transcriptase/substrate for 15 or 30 s as indicated. Substrate incubated without enzyme is shown in lanes 1 and 5, and substrate digested with nuclease P1 to generate marker bands is shown in lanes 4 and 9. Samples were analyzed in a 20% sequencing gel and visualized using a PhosphorImager. Shown vertically at the left and right is the region of sequence containing the PPT (boxed) from the plus-sense M-MuLV genome (positions 7798 to 7832). In the sequence, the positions of observed cleavage sites are indicated with arrows. For prominent sites, the base positions bordering each site are given opposite the arrows and the arrows are connected to corresponding bands by dashed lines. For both 5′ and 3′ end-labeled substrates, positions of bands resulting from cleavage at the −23/−22 site are indicated in the center. In lanes 6 to 8, bands resulting from 5′ end-directed cleavages are indicated with a bar and line at right (5′ end direct.).
When the 79-nt MLV RNA was 3′ end labeled and used in an otherwise equivalent analysis, bands were evident that resulted from cleavages at the −1/+1, −14/−13, and −23/−22 sites and at every internucleotide linkage between positions +1 and +9 except at +6/+7 and +7/+8 (Fig. 3, lanes 6 to 8). Faint cleavage at +9/+10 was observed only with the 3′ end-labeled substrate (Fig. 3, lanes 6 to 8), presumably because the stronger upstream cleavage sites precluded visualization of this site with the 5′ end-labeled RNA. Importantly, each of these bands resulted from recognition and cleavage at a single internal site independent of positioning by the RNA 5′ end. In addition, 5′ end-directed cleavages were evident between positions −42 and −38 on the 79-nt MLV RNA (Fig. 3, lanes 6 to 8; data not shown for 5′ end-labeled substrate), and no significant internal cleavages were observed between positions −13 and −1 within the PPT when the 79-nt RNA was labeled at either end (Fig. 3, lanes 2, 3, and 6 to 8). Overall, these results indicated that the RNase H activity of reverse transcriptase specifically cleaves the M-MuLV RNA at multiple sites downstream of the PPT primer.
A gap downstream of the plus-strand primer improves extension efficiency.
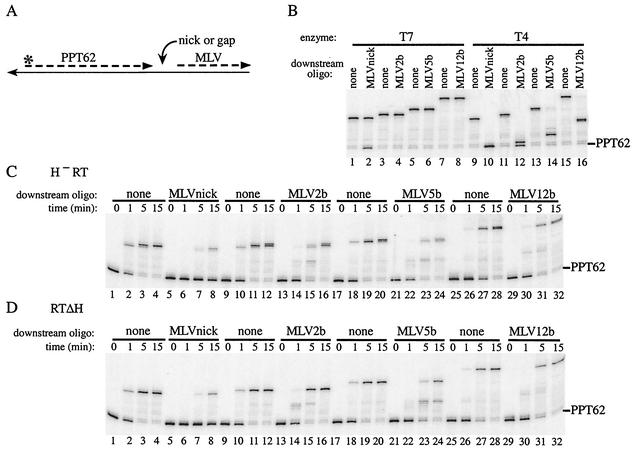
If multiple cleavages occurred on the same RNA immediately downstream of the −1/+1 site, these cleavages would create a gap beyond the 3′ terminus of the PPT primer. We addressed whether such a gap might facilitate extension from the PPT primer and thereby solve the impediment presented by downstream non-template-strand RNA. To test this possibility, substrates with three different gap sizes following the 3′ terminus of the PPT primer were constructed by using downstream 17-mer RNAs derived from the M-MuLV sequence. These 17-mers, termed MLV2b, MLV5b, and MLV12b, have 5′-end positions at +3, +6, or +13 to generate gaps of 2, 5, or 12 bases following the PPT primer 3′ end, respectively (Fig. 1A and B).
Extension of 5′ end-labeled PPT62 in the context of substrate III (containing a 2-, 5-, or 12-base gap) was compared to extension in both substrate II (containing a nick) and substrate I (which lacks downstream RNA) (Fig. 4A). In this experiment, the lengths of downstream duplex through which displacement synthesis had to occur were kept constant; therefore, extension products differed depending on the size of the gap. As described above, both forms of RNase H-deficient reverse transcriptase showed limited extension of PPT62 when downstream RNA was present (Fig. 4C and D; compare lanes 1 to 8 for H− RT and RTΔH). By contrast, introduction of a 2- or 5-base gap downstream of PPT62 significantly improved synthesis for RTΔH and H− RT (Fig. 4C and D; compare lanes 9 to 24). For both enzymes, a 12-base gap permitted synthesis levels approximately equivalent to those of nondisplacement synthesis with substrate I (Fig. 4C and D, lanes 25 to 32).
FIG. 4.
Extension from PPT62 at a nick or a gap followed by M-MuLV RNA. (A) Schematic of model substrates. (B, C, and D) 5′ end-labeled PPT62 was annealed with downstream oligonucleotide MLVnick to template 2 to create a nick, with downstream oligonucleotide MLV2b to template 2A to create a gap of 2 bases, with downstream oligonucleotide MLV5b to template 2B to create a gap of 5 bases, or with downstream oligonucleotide MLV12b to template 2C to create a gap of 12 bases following the 3′ end of PPT62, as indicated. Control extensions were carried out with 5′ end-labeled PPT62 without downstream oligonucleotide on each template strand (none), and all samples were analyzed as described in the legend to Fig. 2. Extensions were done with T7 DNA polymerase (T7; panel B, lanes 1 to 8) or T4 DNA polymerase (T4; panel B, lanes 9 to 16) for 15 min or with H− RT (C), or RTΔH (D) for the indicated times. The positions of unextended PPT62 are indicated on the right.
For T7 DNA polymerase, the levels of full-length product synthesized with all versions of substrate III matched those observed with substrate I, and as expected, control extensions with T4 DNA polymerase confirmed the structures of these substrates (Fig. 4B, lanes 1 to 16 for T7 and T4). These data indicated that a gap significantly improved the extension efficiency of the PPT primer.
Downstream sequence does not affect PPT primer extension at a nick or a gap.
To ask whether the downstream sequence specifically impaired PPT primer extension or whether the PPT primer was inherently recalcitrant to extension at a nick, extension of PPT62 was tested with a different downstream nontemplate strand. For the downstream RNA, an unrelated 18-mer RNA termed HET (Fig. 1B) was used in the context of the nicked substrate (substrate II). When compared to nondisplacement synthesis of PPT62 in substrate I, the efficiency of extension at a nick in substrate II was dramatically reduced for all of the enzymes tested, as shown in a time course assay (Fig. 5; compare lanes 1 to 8). Surprisingly, extension of PPT62 appeared even more inefficient with downstream HET than with downstream MLVnick (compare lanes 1 to 8 of Fig. 4C and D with lanes 1 to 8 of Fig. 5 for RTΔH and H− RT). This observation could not be attributed to a difference in the amount of downstream oligonucleotide annealed in these two substrates, as T4 DNA polymerase generated faint levels of full-length product with substrate II containing HET (Fig. 5, lanes 5 to 8) but synthesized no detectable full-length products with substrate II containing MLVnick (Fig. 4B, lane 10). The HET oligonucleotide itself was not intrinsically inhibitory to displacement synthesis, as T7 DNA polymerase extended a non-PPT RNA primer at a nick with downstream HET at a level comparable to that of nondisplacement synthesis (data not shown).
FIG. 5.
Extension from PPT62 at a nick or a gap followed by HET RNA. 5′ end-labeled PPT62 without a downstream oligonucleotide (none; lanes 1 to 4 and 9 to 12) or with downstream HET (lanes 5 to 8 and 13 to 16) was annealed to template 5 (lanes 1 to 8) or template 5B (lanes 9 to 16) to create a nick or a 5-base gap following the 3′ end of PPT62. The enzyme used for each set of extensions is indicated at the left. Experiments were performed and analyzed as described in the legend to Fig. 2. The positions of unextended PPT62 are indicated at right.
Since a gap preceding a nontemplate strand of viral RNA sequence improved the extension of PPT62, a gap of 5 bases was introduced between the 3′ end of PPT62 and the 5′ end of HET in substrate III. When this substrate was extended with T7 DNA polymerase, synthesis of full-length products equaled that carried out by nondisplacement synthesis (Fig. 5, lanes 9 to 16). For RTΔH and H− RT, the 5-base gap greatly improved extension, although the amounts of full-length extension products were still significantly less than those observed without downstream HET (Fig. 5, lanes 9 to 16).
These data indicated that the 3′ terminus of the PPT primer was poorly extended at a nick irrespective of the downstream sequence but that the presence of a gap beyond this primer consistently and significantly improved extension.
RNase H activity facilitates plus-strand primer extension by wild-type reverse transcriptase.
Since RNase H specifically recognized cleavage sites downstream of the PPT primer (Fig. 3) and gaps resulting from such RNase H activity facilitated plus-strand primer extension (Fig. 4 and 5), it was curious that either RNA or DNA positioned immediately downstream of the PPT primer appeared to equally impede extension by wild-type reverse transcriptase (Fig. 2, lanes 13 to 15). However, due to the highly efficient primer removal activity of RNase H, longer time periods to examine extension of 5′ end-labeled PPT62 were not feasible in this assay.
Using substrate I or substrate II containing MLVnick or MLVnickD in a time course analysis, plus-strand DNA synthesis was measured by the incorporation of [α-32P]dGTP into extension products to better assess extension of these substrates by the wild-type enzyme. For each time point, the extension products were treated with alkali to eliminate any RNA not removed by RNase H and samples were analyzed in 20% sequencing gels. Because primer removal by RNase H leaves a 5′ phosphate while RNA removal by alkali treatment leaves a 5′ hydroxyl, two species of bands were observed for each full-length and full-length-plus-one-nucleotide extension product. To simplify interpretation of these data, these bands were quantified together at each time point, and the accumulation of total full-length DNA extension products synthesized by wild-type reverse transcriptase is presented graphically in Fig. 6.
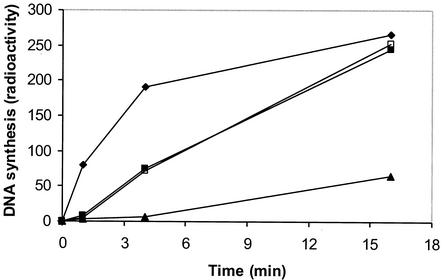
FIG. 6.
Plus-strand DNA synthesis by wild-type reverse transcriptase using model substrates. Extension assays were performed as described in the legend to Fig. 2, except that PPT62 was not 5′-end labeled and assays included [α-32P]dGTP. Samples were collected at 0, 1, 4, or 16 min, treated with alkali, and analyzed in a 20% sequencing gel. Extension products were visualized using a PhosphorImager, and bands representing full-length extension products were quantified with ImageQuant Software using volume integration. Total extension products are plotted as a function of time. Filled diamonds, PPT62 alone in substrate I; filled squares, PPT62 followed by downstream RNA MLVnick in substrate II; filled triangles, PPT62 followed by downstream DNA MLVnickD in substrate II; open squares, hybrid substrate containing continuous 79-nt MLV RNA in substrate IV.
Compared to nondisplacement synthesis using PPT62 in substrate I (Fig. 6), wild-type reverse transcriptase initially generated far fewer extension products with both forms of substrate II containing either downstream MLVnick or MLVnickD at the 1-min time point. This result was consistent with the data presented in Fig. 2, lanes 13 to 15. However, at later time points, a significant difference was observed between the extension reactions carried out with downstream RNA versus DNA. Compared to extension with substrate I at 4 min, extension with RNA downstream was only 2.5-fold lower but that with DNA downstream was approximately 20-fold lower. At 16 min, synthesis levels in the presence and absence of downstream RNA were almost equal but synthesis that required displacement of downstream MLVnickD was still fourfold lower.
Extension with substrate IV (containing the 79-nt MLV RNA with an internal PPT) was essentially identical to extension of PPT62 at a nick with downstream RNA (Fig. 6). In other experiments, synthesis with substrate III containing a 12-base gap was consistently higher than that seen when using a continuous RNA with an internal PPT (data not shown). This indicated that a preexisting nick at the −1/+1 position did not improve the efficiency of plus-strand primer extension and that additional RNase H cleavages facilitating this extension were required as well.
These data with wild-type reverse transcriptase are consistent with the results obtained using RNase H-deficient reverse transcriptases: the RNase H activity of reverse transcriptase can facilitate the extension of the PPT primer at a nick with downstream RNA but, as expected, does not enhance synthesis in the presence of downstream DNA.
DISCUSSION
Numerous studies have shown that specific cleavage at the −1/+1 site by RNase H creates the 3′ end of the PPT primer (reference 3 and references therein) and that the sequence of the PPT is sufficient to direct this cleavage (27, 33, 34). It is not known whether cleavage at the −1/+1 site occurs by the polymerization-dependent or polymerization-independent mode of RNase H activity during reverse transcription, but polymerization-dependent RNase H cleavages correlate with pause sites during RNA-dependent DNA synthesis (40, 41) and no strong pause sites corresponding to plus-strand primer generation are observed when the PPT region is copied (35, 51). The polymerization-independent activity of RNase H can cleave at the −1/+1 site in the PPT by an internal sequence-specific cleavage without 5′ end-directed positioning (12, 14, 37). Such a cleavage could generate a nick at this position, leaving the 3′ end of the PPT primer abutting downstream nontemplate RNA. To extend the 3′ terminus of this plus-strand primer, reverse transcriptase must either initiate displacement synthesis at a nick or alternatively generate a gap by additional RNase H cleavages prior to extension of the plus-strand primer.
Previous studies employing DNA primers to assess the effects of downstream RNA on plus-strand DNA synthesis at a nick have demonstrated that RNase H degradation of downstream RNA is not absolutely essential, but the presence of such RNA fragments can impede displacement synthesis by reverse transcriptase (13, 25). In our model hybrid substrates, we have examined how extension of the RNA plus-strand primer is affected by downstream nontemplate strand RNA and have characterized the specificity of RNase H for this downstream RNA sequence. We found that when the 3′ terminus of the PPT primer abuts nontemplate strand RNA of either an M-MuLV sequence or an unrelated sequence, initiation is remarkably inefficient for two RNase H-deficient forms of reverse transcriptase and, initially, even for the wild-type enzyme. In addition, T7 DNA polymerase, which possesses a robust displacement activity, is relatively inefficient at initiation from the plus-strand primer at a nick, irrespective of the downstream non-template-strand RNA. These data indicate that the PPT RNA primer itself is inherently difficult to extend in a configuration demanding immediate displacement synthesis of downstream RNA.
It is interesting to consider why initiation from the PPT primer at a nick with downstream RNA is intrinsically difficult. First, reverse transcriptase is much more facile at nondisplacement synthesis than displacement synthesis. Using DNA primers, nondisplacement synthesis is about 5- to 10-fold more efficient than displacement synthesis with a nontemplate DNA strand and at least 20-fold more efficient than displacement synthesis with a nontemplate RNA strand (1, 13, 15, 20, 21, 25, 43). In our experiments with RNase H-deficient reverse transcriptases, comparable differences were consistently observed between nondisplacement versus displacement synthesis in substrates containing the plus-strand RNA primer at a nick with downstream RNA. Second, reverse transcriptase extends DNA primers, including DNA versions of the PPT primer, much more efficiently than any RNA primers (14, 31, 35, 37). Third, the PPT assumes an unusual structure that might contribute to the difficulty in initiating plus-strand synthesis at a nick. This structural distortion promotes base unstacking and may render the majority of this sequence more resistant to RNase H degradation while facilitating RNase H cleavage at the −1/+1 site to generate the primer 3′ end (11, 28, 33, 36). The unusual structure of the PPT RNA-DNA hybrid does not require the binding of reverse transcriptase and is preserved whether the PPT sequence consists only of RNA or contains the RNA/DNA junction produced by plus-strand synthesis (28). It is conceivable that the viral nucleocapsid protein facilitates the initiation of RNA displacement synthesis at a nick without any additional RNase H cleavages. Although our results do not exclude this possibility, in a previous study only a modest effect of nucleocapsid on RNA displacement synthesis was observed (25).
Because the displacement synthesis activity of reverse transcriptase does not readily extend the PPT primer at a nick, it is likely that additional RNase H cleavages or a combination of RNase H cleavages and displacement synthesis must occur for efficient plus-strand synthesis. An investigation of the specificity of RNase H for the sequence immediately downstream of the −1/+1 site in the PPT revealed that multiple internal cleavage sites are recognized in this region. Most importantly, each of these sites is cleaved independently and without positioning by an RNA 5′ end in a hybrid containing the 79-nt MLV RNA. This reveals that like the −1/+1 site, sequence recognition is essential for cleavage of these downstream sites. These sites are clustered and occur from +1/+2 through +5/+6 and at +8/+9 and +9/+10. It is likely that a single cleavage at any one of these sites in combination with the −1/+1 cleavage would create a gap after the 3′ terminus of the PPT primer.
A gap downstream of the PPT primer might facilitate extension by allowing the polymerase domain of reverse transcriptase better access to the 3′ end of the PPT primer. Footprint analyses have shown that M-MuLV reverse transcriptase contacts the nontemplate strand from +1 through +7 or +8, extends up to 6 bases on the template strand ahead of the primer 3′ end, and melts the 2 bp immediately following the primer terminus (30, 45, 49, 50). In our substrates, the 2- and 5-base gaps fall within and the 12-base gap just outside the potential initial contact of reverse transcriptase on the nontemplate and template strands. Thus, these short gaps may promote easier recognition or binding of the enzyme to the 3′ terminus of the PPT primer, which might otherwise be difficult owing to the unusual configuration of the PPT.
Yet another possible benefit of a gap downstream of the PPT primer is that limited but sufficient extension from the plus-strand primer is permitted such that the nascent strand (instead of a less efficient RNA primer) is recognized as a preferred DNA 3′ terminus. The rate of polymerization for reverse transcriptase is affected by whether the primer strand is RNA or DNA (23, 29). Initiation of minus-strand synthesis has a distinct shift from a more distributive, slower mode of polymerization to a faster, more processive mode once the sixth nucleotide has been incorporated (29, 42). For plus-strand synthesis, significant pausing has been observed after a 1-nt extension and initiation becomes more efficient after the second nucleotide is incorporated (18, 19). Extension through a gap following the 3′ terminus of the PPT primer might allow reverse transcriptase sufficient distance to transition from the higher-fidelity mode of initiation to a more processive mode that extends the DNA 3′ terminus, which in turn promotes displacement of the downstream nontemplate strand. Consistent with this suggestion is the observation that DNA displacement synthesis with a DNA version of the PPT primer is much more efficient than that with an RNA PPT primer (37).
Our experiments comparing how wild-type reverse transcriptase carries out plus-strand DNA synthesis with different substrates directly test the hypothesis that RNase H activity facilitates the initiation of plus-strand DNA synthesis. First, plus-strand synthesis is initially delayed in the presence of downstream RNA but ultimately approximates synthesis carried out without downstream RNA. Second, displacement synthesis alone is not sufficient for efficient extension of the PPT primer, as plus-strand synthesis is impaired in the presence of downstream DNA, where the RNase H activity is useless. These data support the hypothesis that a combination of RNase H cleavages and displacement synthesis is required for efficient plus-strand synthesis.
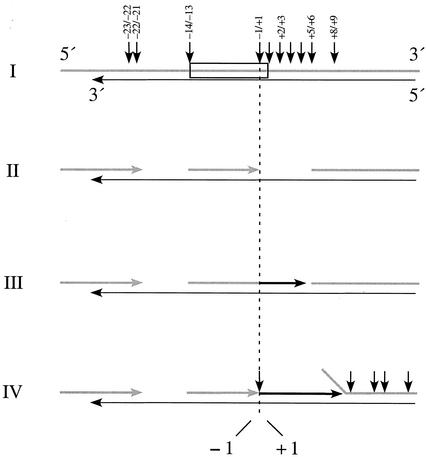
Together, these results contribute to a better understanding of how the M-MuLV reverse transcriptase efficiently initiates plus-strand DNA synthesis (Fig. 7). After minus-strand DNA synthesis has copied the RNA genome through the PPT, reverse transcriptase must bind the RNA strand of the hybrid near the PPT to generate the plus-strand primer. Instead of a single cleavage at the −1/+1 site to generate the 3′ end of the PPT primer, we propose that several RNase H cleavages occur at specific sites in the PPT region both upstream and downstream of the plus-strand origin (step I). Downstream cleavages occur at multiple sites between positions +1 and +9, likely progress in the 3′ to 5′ direction, and ultimately cleave at the −1/+1 site. These cleavages result in a gap following the 3′ terminus of the plus-strand primer. If cleavages also were to occur at the −23/−22, −22/−21, and −14/−13 sites as previously reported (37), this might result in a gap upstream of the PPT primer as well (step II). After RNase H has created a gap in the RNA sequence downstream of the plus-strand primer 3′ terminus, reverse transcriptase rebinds the hybrid with the polymerase domain positioned to initiate plus-strand synthesis from the PPT primer by efficient nondisplacement synthesis (step III). After the incorporation of 5 to 8 nucleotides (depending upon the gap size), displacement synthesis appears sufficient to continue DNA synthesis but additional RNase H cleavages further downstream likely would provide additional assistance (step IV) (13, 25). After limited extension, the PPT primer is removed from the nascent DNA by cleavage at the RNA/DNA junction.
FIG. 7.
Model for initiation of M-MuLV plus-strand RNA synthesis. In the diagram, plus-strand RNA is represented by thick gray lines, minus-strand DNA is represented by thin black arrows pointing to the left, nascent plus-strand DNA is represented by thick black arrows, and the PPT is boxed. RNase H cleavage sites are denoted by vertical arrows, and several specific sites are indicated. The model is described in the Discussion.
Acknowledgments
We thank all members of the Champoux laboratory for helpful discussions and especially thank Christian Lanciault for critically reading the manuscript.
This work was supported by National Institutes of Health grant CA51605.
REFERENCES
- 1.Amacker, M., M. Hottiger, and U. Hubscher. 1995. Feline immunodeficiency virus reverse transcriptase: expression, functional characterization, and reconstitution of the 66- and 51-kilodalton subunits. J. Virol. 69:6273-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arts, E. J., and S. F. J. LeGrice. 1998. Interaction of retroviral reverse transcriptase with template-primer duplexes during replication. Prog. Nucleic Acid Res. Mol. Biol. 58:339-393. [DOI] [PubMed] [Google Scholar]
- 3.Champoux, J. J. 1993. Roles of ribonuclease H in reverse transcription, p. 103-118. In A. M. Skalka and S. P. Goff (ed.), Reverse transcriptase. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 4.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 5.DeStefano, J. J. 1995. The orientation of binding of human immunodeficiency virus reverse transcriptase on nucleic acid hybrids. Nucleic Acids Res. 23:3901-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeStefano, J. J., R. G. Buiser, L. M. Mallaber, T. W. Myers, R. A. Bambara, and P. J. Fay. 1991. Polymerization and RNase H activities of the reverse transcriptases from avian myeloblastosis, human immunodeficiency, and Moloney murine leukemia viruses are functionally uncoupled. J. Biol. Chem. 266:7423-7431. [PubMed] [Google Scholar]
- 7.DeStefano, J. J., J. V. Cristofaro, S. Derebail, W. P. Bohlayer, and M. J. Fitzgerald-Heath. 2001. Physical mapping of HIV reverse transcriptase to the 5′ end of RNA primers. J. Biol. Chem. 276:32515-32521. [DOI] [PubMed] [Google Scholar]
- 8.DeStefano, J. J., L. M. Mallaber, P. J. Fay, and R. A. Bambara. 1993. Determinants of the RNase H cleavage specificity of human immunodeficiency virus reverse transcriptase. Nucleic Acids Res. 21:4330-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeStefano, J. J., L. M. Mallaber, P. J. Fay, and R. A. Bambara. 1994. Quantitative analysis of RNA cleavage during RNA-directed DNA synthesis by human immunodeficiency and avian myeloblastosis virus reverse transcriptases. Nucleic Acids Res. 22:3793-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudding, L. R., N. C. Nkabinde, and V. Mizrahi. 1991. Analysis of the RNA- and DNA-dependent DNA polymerase activities of point mutants of HIV-1 reverse transcriptase lacking ribonuclease H activity. Biochemistry 30:10498-10506. [DOI] [PubMed] [Google Scholar]
- 11.Fedoroff, O. Y., Y. Ge, and B. R. Reid. 1997. Solution structure of r(gaggacug):d(CAGTCCTC) hybrid: implications for the initiation of HIV-1 (+)-strand synthesis. J. Mol. Biol. 269:225-239. [DOI] [PubMed] [Google Scholar]
- 12.Finston, W. I., and J. J. Champoux. 1984. RNA-primed initiation of Moloney murine leukemia virus plus strands by reverse transcriptase in vitro. J. Virol. 51:26-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuentes, G. M., P. J. Fay, and R. A. Bambara. 1996. Relationship between plus strand DNA synthesis and removal of downstream segments of RNA by human immunodeficiency virus, murine leukemia virus and avian myeloblastoma virus reverse transcriptases. Nucleic Acids Res. 24:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuentes, G. M., L. Rodriguez-Rodriguez, P. J. Fay, and R. A. Bambara. 1995. Use of an oligoribonucleotide containing the polypurine tract sequence as a primer by HIV reverse transcriptase. J. Biol. Chem. 270:28169-28176. [DOI] [PubMed] [Google Scholar]
- 15.Fuentes, G. M., L. Rodriguez-Rodriguez, C. Palaniappan, P. J. Fay, and R. A. Bambara. 1996. Strand displacement synthesis of the long terminal repeats by HIV reverse transcriptase. J. Biol. Chem. 271:1966-1971. [DOI] [PubMed] [Google Scholar]
- 16.Furfine, E. S., and J. E. Reardon. 1991. Reverse transcriptase.RNase H from the human immunodeficiency virus. Relationship of the DNA polymerase and RNA hydrolysis activities. J. Biol. Chem. 266:406-412. [PubMed] [Google Scholar]
- 17.Gopalakrishnan, V., J. A. Peliska, and S. J. Benkovic. 1992. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc. Natl. Acad. Sci. USA 89:10763-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gotte, M., M. Kameoka, N. McLellan, L. Cellai, and M. A. Wainberg. 2001. Analysis of efficiency and fidelity of HIV-1 (+)-strand DNA synthesis reveals a novel rate-limiting step during retroviral reverse transcription. J. Biol. Chem. 276:6711-6719. [DOI] [PubMed] [Google Scholar]
- 19.Gotte, M., G. Maier, A. M. Onori, L. Cellai, M. A. Wainberg, and H. Heumann. 1999. Temporal coordination between initiation of HIV (+)-strand DNA synthesis and primer removal. J. Biol. Chem. 274:11159-11169. [DOI] [PubMed] [Google Scholar]
- 20.Hottiger, M., V. N. Podust, R. L. Thimmig, C. McHenry, and U. Hubscher. 1994. Strand displacement activity of the human immunodeficiency virus type 1 reverse transcriptase heterodimer and its individual subunits. J. Biol. Chem. 269:986-991. [PubMed] [Google Scholar]
- 21.Huber, H. E., J. M. McCoy, J. S. Seehra, and C. C. Richardson. 1989. Human immunodeficiency virus 1 reverse transcriptase. Template binding, processivity, strand displacement synthesis, and template switching. J. Biol. Chem. 264:4669-4678. [PubMed] [Google Scholar]
- 22.Huber, H. E., and C. C. Richardson. 1990. Processing of the primer for plus strand DNA synthesis by human immunodeficiency virus 1 reverse transcriptase. J. Biol. Chem. 265:10565-10573. [PubMed] [Google Scholar]
- 23.Isel, C., J. M. Lanchy, S. F. Le Grice, C. Ehresmann, B. Ehresmann, and R. Marquet. 1996. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J. 15:917-924. [PMC free article] [PubMed] [Google Scholar]
- 24.Kati, W. M., K. A. Johnson, L. F. Jerva, and K. S. Anderson. 1992. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 267:25988-25997. [PubMed] [Google Scholar]
- 25.Kelleher, C. D., and J. J. Champoux. 1998. Characterization of RNA strand displacement synthesis by Moloney murine leukemia virus reverse transcriptase. J. Biol. Chem. 273:9976-9986. [DOI] [PubMed] [Google Scholar]
- 26.Kelleher, C. D., and J. J. Champoux. 2000. RNA degradation and primer selection by Moloney murine leukemia virus reverse transcriptase contribute to the accuracy of plus strand initiation. J. Biol. Chem. 275:13061-13070. [DOI] [PubMed] [Google Scholar]
- 27.Klarmann, G. J., H. Yu, X. Chen, J. P. Dougherty, and B. D. Preston. 1997. Discontinuous plus-strand DNA synthesis in human immunodeficiency virus type 1-infected cells and in a partially reconstituted cell-free system. J. Virol. 71:9259-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kvaratskhelia, M., S. R. Budihas, and S. F. Le Grice. 2002. Pre-existing distortions in nucleic acid structure aid polypurine tract selection by HIV-1 reverse transcriptase. J. Biol. Chem. 277:16689-16696. [DOI] [PubMed] [Google Scholar]
- 29.Lanchy, J. M., G. Keith, S. F. Le Grice, B. Ehresmann, C. Ehresmann, and R. Marquet. 1998. Contacts between reverse transcriptase and the primer strand govern the transition from initiation to elongation of HIV-1 reverse transcription. J. Biol. Chem. 273:24425-24432. [DOI] [PubMed] [Google Scholar]
- 30.Metzger, W., T. Hermann, O. Schatz, S. F. Le Grice, and H. Heumann. 1993. Hydroxyl radical footprint analysis of human immunodeficiency virus reverse transcriptase-template primer complexes. Proc. Natl. Acad. Sci. USA 90:5909-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palaniappan, C., J. K. Kim, M. Wisniewski, P. J. Fay, and R. A. Bambara. 1998. Control of initiation of viral plus strand DNA synthesis by HIV reverse transcriptase. J. Biol. Chem. 273:3808-3816. [DOI] [PubMed] [Google Scholar]
- 32.Peliska, J. A., and S. J. Benkovic. 1992. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science 258:1112-1118. [DOI] [PubMed] [Google Scholar]
- 33.Powell, M. D., and J. G. Levin. 1996. Sequence and structural determinants required for priming of plus-strand DNA synthesis by the human immunodeficiency virus type 1 polypurine tract. J. Virol. 70:5288-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pullen, K. A., A. J. Rattray, and J. J. Champoux. 1993. The sequence features important for plus strand priming by human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 268:6221-6227. [PubMed] [Google Scholar]
- 35.Randolph, C. A., and J. J. Champoux. 1994. The use of DNA and RNA oligonucleotides in hybrid structures with longer polynucleotide chains to probe the structural requirements for Moloney murine leukemia virus plus strand priming. J. Biol. Chem. 269:19207-19215. [PubMed] [Google Scholar]
- 36.Sarafianos, S. G., K. Das, C. Tantillo, A. D. Clark, Jr., J. Ding, J. M. Whitcomb, P. L. Boyer, S. H. Hughes, and E. Arnold. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20:1449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz, S. J., M. Zhang, C. D. Kelleher, and J. J. Champoux. 2000. Analysis of plus-strand primer selection, removal, and reutilization by retroviral reverse transcriptases. J. Biol. Chem. 275:32299-32309. [DOI] [PubMed] [Google Scholar]
- 38.Schultz, S. J., M. Zhang, C. D. Kelleher, and J. J. Champoux. 1999. Polypurine tract primer generation and utilization by Moloney murine leukemia virus reverse transcriptase. J. Biol. Chem. 274:34547-34555. [DOI] [PubMed] [Google Scholar]
- 39.Shinnick, T. M., R. A. Lerner, and J. G. Sutcliffe. 1981. Nucleotide sequence of Moloney murine leukaemia virus. Nature 293:543-548. [DOI] [PubMed] [Google Scholar]
- 40.Suo, Z., and K. A. Johnson. 1997. Effect of RNA secondary structure on RNA cleavage catalyzed by HIV-1 reverse transcriptase. Biochemistry 36:12468-12476. [DOI] [PubMed] [Google Scholar]
- 41.Suo, Z., and K. A. Johnson. 1997. Effect of RNA secondary structure on the kinetics of DNA synthesis catalyzed by HIV-1 reverse transcriptase. Biochemistry 36:12459-12467. [DOI] [PubMed] [Google Scholar]
- 42.Thrall, S. H., R. Krebs, B. M. Wohrl, L. Cellai, R. S. Goody, and T. Restle. 1998. Pre-steady-state kinetic characterization of RNA-primed initiation of transcription by HIV-1 reverse transcriptase and analysis of the transition to a processive DNA-primed polymerization mode. Biochemistry 37:13349-13358. [DOI] [PubMed] [Google Scholar]
- 43.Whiting, S. H., and J. J. Champoux. 1998. Properties of strand displacement synthesis by Moloney murine leukemia virus reverse transcriptase: mechanistic implications. J. Mol. Biol. 278:559-577. [DOI] [PubMed] [Google Scholar]
- 44.Whiting, S. H., and J. J. Champoux. 1994. Strand displacement synthesis capability of Moloney murine leukemia virus reverse transcriptase. J. Virol. 68:4747-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winshell, J., and J. J. Champoux. 2001. Structural alterations in the DNA ahead of the primer terminus during displacement synthesis by reverse transcriptases. J. Mol. Biol. 306:931-943. [DOI] [PubMed] [Google Scholar]
- 46.Wisniewski, M., M. Balakrishnan, C. Palaniappan, P. J. Fay, and R. A. Bambara. 2000. The sequential mechanism of HIV reverse transcriptase RNase H. J. Biol. Chem. 275:37664-37671. [DOI] [PubMed] [Google Scholar]
- 47.Wisniewski, M., M. Balakrishnan, C. Palaniappan, P. J. Fay, and R. A. Bambara. 1983. 2000. Unique progressive cleavage mechanism of HIV reverse transcriptase RNase H. Proc. Natl. Acad. Sci. USA 97:11978-11983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wisniewski, M., Y. Chen, M. Balakrishnan, C. Palaniappan, B. P. Roques, P. J. Fay, and R. A. Bambara. 2002. Substrate requirements for secondary cleavage by HIV-1 reverse transcriptase RNase H. J. Biol. Chem. 277:28400-28410. [DOI] [PubMed] [Google Scholar]
- 49.Wohrl, B. M., M. M. Georgiadis, A. Telesnitsky, W. A. Hendrickson, and S. F. Le Grice. 1995. Footprint analysis of replicating murine leukemia virus reverse transcriptase. Science 267:96-99. [DOI] [PubMed] [Google Scholar]
- 50.Wohrl, B. M., C. Tantillo, E. Arnold, and S. F. Le Grice. 1995. An expanded model of replicating human immunodeficiency virus reverse transcriptase. Biochemistry 34:5343-5356. [DOI] [PubMed] [Google Scholar]
- 51.Wu, W., L. E. Henderson, T. D. Copeland, R. J. Gorelick, W. J. Bosche, A. Rein, and J. G. Levin. 1996. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J. Virol. 70:7132-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]