Abstract
We report the results of cloning genes for two key biosynthetic enzymes of different 5-aminolevulinic acid (ALA) biosynthetic routes from Streptomyces. The genes encode the glutamyl-tRNAGlu reductase (GluTR) of the C5 pathway and the ALA synthase (ALAS) of the Shemin pathway. While Streptomyces coelicolor A3(2) synthesizes ALA via the C5 route, both pathways are operational in Streptomyces nodosus subsp. asukaensis, a producer of asukamycin. In this strain, the C5 route produces ALA for tetrapyrrole biosynthesis; the ALA formed by the Shemin pathway serves as a precursor of the 2-amino-3-hydroxycyclopent-2-enone moiety (C5N unit), an antibiotic component. The growth of S. nodosus and S. coelicolor strains deficient in the GluTR genes (gtr) is strictly dependent on ALA or heme supplementation, whereas the defect in the ALAS-encoding gene (hemA-asuA) abolishes the asukamycin production in S. nodosus. The recombinant hemA-asuA gene was expressed in Escherichia coli and in Streptomyces, and the encoded enzyme activity was demonstrated both in vivo and in vitro. The hemA-asuA gene is situated within a putative cluster of asukamycin biosynthetic genes. This is the first report about the cloning of genes for two different ALA biosynthetic routes from a single bacterium.
The 5-aminolevulinic acid (ALA) is the first committed precursor in the tetrapyrrole pathway that produces hemes, chlorophylls, billins, and corrinoids. It is synthesized by either of two major biosynthetic routes, the C5 pathway or the Shemin (C4) pathway. In the C5 pathway, the intact glutamate carbon backbone is transformed to ALA in three consecutive steps: (i) first, the charging of tRNAGlu with glutamate, (ii) its reduction to semialdehyde in a reaction catalyzed by glutamyl-tRNA reductase (GluTR), and (iii) final transamination. This C5 pathway operates in plants and in many bacteria. The Shemin pathway catalyzes the reaction between glycine and succinyl-coenzyme A (CoA) to create ALA in one step, catalyzed by the ALA synthase (ALAS). This route is present in animals, fungi, and in some, mostly photosynthetic, bacteria (4).
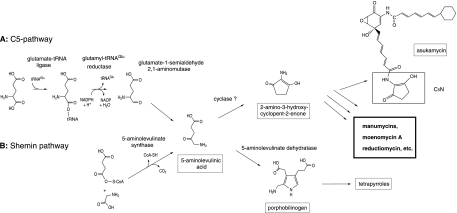
In addition to tetrapyrrole biosynthesis, ALA acts as a precursor in the formation of an unsaturated 2-amino-3-hydroxycyclopent-2-enone moiety (Fig. 1). This group, called a C5N unit, is a part of several secondary metabolites produced by certain Streptomyces strains. It has been shown that in the instance of asukamycin (34), reductiomycin (5), and moenomycin A (13), this moiety is formed by the cyclization of one ALA molecule formed by the Shemin pathway. Probably, the C5N units of other compounds are generated by the same mechanism. This group of metabolites involves almost all members of the manumycin antibiotic family (45, 53) and other compounds, such as moenomycin A (10), bafilomycins B1 and B2 (56), virustomycin (36), limocrocin (15), and enopeptins A and B (24).
FIG. 1.
C5 and Shemin pathways for ALA biosynthesis.
Although the first steps of the tetrapyrrole biosynthetic pathway were not studied in Streptomyces, the results from two recently finished genome-sequencing projects indicate that its ALA precursor is in the genus, as in many other eubacteria, synthesized via the C5 route (6, 21). Accordingly, the discovery of a second pathway in some strains raises a question about the presence of two ALA biosynthetic pathways in these bacteria or even about the heterogeneous distribution of these pathways in Streptomyces. It has already been shown that in rare cases both pathways can operate in one organism as they do in the green unicellular alga Euglena gracilis (27, 55). In this instance, however, the C5 route is confined to chloroplasts and supplies ALA for chlorophyll biosynthesis. The ALAS, representing the Shemin pathway, is encoded by the nuclear gene and is located in mitochondria, where it synthesizes the precursor for the heme biosynthesis. So far, the existence of two functional ALA pathways in one prokaryotic organism has been suggested only in Arthrobacter globiformis, where cells switch from the C5 pathway (predominant in early exponential phase cultures) to the Shemin pathway in stationary phase cultures (57). ALA is also synthesized by Rhodobacter sphaeroides in two ways; however, in these cases synthesis is performed by two ALAS isozymes, products of hemA and hemT genes, which are localized to different chromosomes (35, 52). Although the parallel occurrence of the two ALA biosynthetic pathways is rare in bacteria, the simultaneous presence of two different biosynthetic routes that form the same end product, which is then utilized in primary and secondary metabolism, has already been reported in Streptomyces (22).
In this study, we report the isolation and characterization of ALA biosynthetic genes for both pathways from Streptomyces nodosus subsp. asukaensis, the asukamycin producer. We examined whether the pathways are interchangeable with respect to the supplementation of precursors for the tetrapyrrole and asukamycin biosynthetic routes. Since secondary metabolite biosynthetic genes are often clustered in Streptomyces chromosomes, we also examined the neighborhood of the hemA-asuA gene and investigated the possible presence of such clusters in several strains.
MATERIALS AND METHODS
Bacteria, plasmids, media, and growth conditions.
Bacterial strains and plasmids used in the study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIqZΔM15 Tn10(Tetr)] | Stratagene |
| GM2929 | dam dcm recF hsdR | E. coli Genetic Stock Centera; 38 |
| K12 TB1 | F−ara Δ(lac-proAB) [φ80dlac Δ(lacZ)M15] rpsL(Strr) thi hsdR | New England Biolabs |
| HU227 | hemA41 relA1 spoT1 metB1 rrnB-2 mcrB1 creC510 Hem-p | E. coli Genetic Stock Center |
| Streptomyces strains | ||
| S. nodosus subsp. asukaensis ATCC 29757 | Wild-type strain | Heinz Flossb |
| S. xanthochromogenus AM 6201 | Wild-type strain | Heinz Floss |
| S. parvulus Tü64 | Wild-type strain | Heinz Floss |
| S. bambergiensis ATCC 13879 | Wild-type strain | DSMZ |
| S. ghanaensis ATCC 14672 | Wild-type strain | DSMZ |
| S. lividans TK24 | Wild-type strain | David Hopwoodc |
| S. coelicolor J1501 | hisA1 uraA1 strA1 pgl-1 SCP1- SCP2- | David Hopwood |
| S. nodosus MIP11 | gtr::aph1 | This work |
| S. nodosus MIP12 | hemA-asuA::apr | This work |
| S. nodosus MIP13 | hemA-asuA::apr gtr::aph1 | This work |
| S. coelicolor MIP14 | gtr::aph1 | This work |
| E. coli plasmids | ||
| pTZ19R | Cloning vector | Amersham Biosciences |
| pSL1180 | Cloning vector | Amersham Biosciences |
| pGEM-7Zf(+) | Cloning vector | Promega |
| pBluescript II SK(+) | Cloning vector | Stratagene |
| pQE31 | His-tagged expression vector | QIAGEN |
| pMAL-c2 | MBP-tagged expression vector | New England Biolabs |
| pNEO2 | aph1 from pIJ61 in pBluescript II SK(+) | This work |
| pAPR3 | aac(3)IV from pKC505 in pBluescript II SK(+) | This work |
| pIJ4070 | ermE* promoter in pTZ19R | David Hopwood |
| E68 | Cosmid clone with the gtr locus, S. coelicolor genomic library | 42 |
| pGF23 | Murine hemA gene-carrying plasmid | Gloria C. Ferreira; 14 |
| PMPH22 | gtr PCR fragment in pTZ19R | This work |
| pMPH25 | hemA-asuA PCR fragment in pTZ19R | This work |
| pMPA03 | hemA-asuA region; PstI fragment in pTZ19R | This work |
| pMPA01 | hemA-asuA region; BamHI fragment in pTZ19R | This work |
| pMPA10 | gtr region, BamHI fragment in pTZ19R | This work |
| PMPA11 | gtr gene; KpnI fragment in pTZ19R | This work |
| PMPH28 | BglII-BamHI fragment of E68 in pGEM-7Zf(+) | This work |
| pALS3 | hemA-asuA in pQE31 | This work |
| pALS5 | hemA-asuA in pMAL-c2 | This work |
| pGM160 | E. coli-Streptomyces shuttle; temperature-sensitive replicon; Tsrr | Günter Muth, University of Tübingen, Germany |
| Streptomyces plasmids | ||
| pIJ622 | pIJ702 derivative with an EcoRI linker inserted between mel and tsr genes | David Hopwood |
| pIJ61 | Streptomycete low-copy-number vector | David Hopwood; 17 |
| pIJ487 | Streptomycete high-copy-number vector | 54 |
| pALS4 | ermE*-(6xHis)hemA-asuA fusion in pIJ622 | This work |
| pALS1 | hemA-asuA in pIJ487 | This work |
| pALS2 | hemA-asuA ORFA in pIJ487 | This work |
| PMPA12 | gtr region; KpnI fragment in pIJ487 | This work |
New Haven, Conn.
University of Washington, Seattle, Wash.
John Innes Centre, Norwich, United Kindgom.
Streptomycete strains were grown in liquid GYM (48) or YEME (17) medium or on a solid GYM medium (2% agar) at 28°C. S. nodosus subsp. asukaensis cultures tested for asukamycin production were cultivated in the production medium for 72 h as described previously (18). When necessary, the medium was supplemented with thiostrepton (30 μg/ml), neomycin (10 μg/ml), or apramycin (25 μg/ml). For cultivation of mutants defective in GluTR, the medium was supplemented with ALA (10 μg/ml) or hemin (4 μg/ml). In the hemin-supplemented medium, bovine serum albumin (1 mg/ml) and cysteine (20 μg/ml) were added (30). Protoplasts were regenerated by plating on an R5 medium (17). Escherichia coli was grown in a liquid LB (Luria-Bertani) medium (37°C) or on a solid LB medium (2% agar) supplemented, when required, with ampicillin (100 μg/ml) and/or apramycin (100 μg/ml). The concentrations of antibiotic(s) were the same for both solid and liquid cultures.
DNA sequencing and analysis.
DNA manipulations followed standard procedures for both Streptomyces species (17) and E. coli (44).
Nucleotide sequencing reactions were performed with the ABI Prism BigDye terminator, version 3.1, cycle sequencing kit (Applied Biosystems, Warrington, United Kingdom) according to the manufacturer's instructions. Fragments were analyzed with an ABI Prism 3100 DNA sequencer (Applied Biosystems). The sequence edition, assembly, and translations were conducted with Lasergene, version 4 (DNASTAR, Inc., Madison, Wis.). The BLAST family of programs (1, 2) was used to compare nucleotide and deduced amino acid sequences against public sequence databases.
Pairwise and multiple alignments were carried out with BLAST2 (http://www.ncbi.nlm.nih.gov/BLAST/bl2seq/wblast2.cgi) and ClustalW (http://www.ebi.ac.uk/clustalw/).
DNA amplification and cloning.
S. nodosus subsp. asukaensis genomic DNA was amplified in reaction mixtures (total volume, 100 μl) containing 25 pmol of each primer, 1.5 mM MgCl2, 10% glycerol, a 0.2 mM concentration of each deoxynucleoside triphosphate, 2.5 U of Taq polymerase (Top-Bio, Prague, Czech Republic), PCR buffer, and 1 μg of genomic DNA. PCR was performed with a T Gradient Thermocycler 96 (Biometra GmbH, Göttingen, Germany). Oligonucleotides were purchased from VBC-Genomics (Vienna, Austria). Primers used to amplify the DNA fragment internal to a GluTR-encoding gene (gtr) were a 1:1 mixture of 5′-ACSTGYAACCGSACSGARMTSTAC-3′ and 5′-ACSTGYAACCGSCTSGARMTSTAC-3′ (rightward) and a 1:1 mixture of 5′-CCSCTYKGSGTCTAGGASCSSGTCCA-3′ and 5′-CCSCTYCCSGTCTAGGASCSSGTCCA-3′ (leftward). Primers used for amplification of DNA fragments internal to the gene for the ALAS (hemA-asuA) gene were a 1:1 mixture of 5′-GTSTGGTGYTCSAACGACTACCT-3′ and 5′-GTSTGGTGYRCSAACGACTACCT-3′ (rightward) and a 1:1 mixture of 5′-CCSAAGTAGAAGWGSTGSISSGAS-3′ and 5′-CCSAAGTAGAAGTSSTGSISSGAS-3′ (leftward). Redundancies were identified on the basis of the International Union of Biochemistry group codes, as follows: S = G + C, Y = C + T, and R = A + G; I stands for inosine. The 0.25-kb and 0.7-kb amplification products were size fractionated by agarose gel electrophoresis, recovered by elution by the glass milk method, blunt-ended with Klenow fragment, and cloned into SmaI-HincII-cleaved pTZ19R to produce pMPH22 (gtr fragment) and pMPH25 (hemA-asuA fragment).
Southern blotting and colony hybridization.
Southern blot analyses were performed with either [α-32P]dCTP- or digoxigenin-labeled DNA probes excised from pMPH22 as a SphI-SacI fragment (gtr probe) and from pMPH25 as a 0.3-kb SacII fragment (hemA-asuA probe), using kits obtained from Roche Diagnostics GmbH (Mannheim, Germany). Hybridizations were performed according to the supplier's instructions with the exception of when the probes (prepared from cloned fragments of S. nodosus subsp. asukaensis genomic DNA) were used for the detection of homologous genes in various streptomycete strains. In that case, the second posthybridization washing was performed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS) at 45°C. For cloning of the hemA-asuA and gtr genes, fragments between 6 and 8 kb in size from BamHI digestion and fragments approximately 7 kb in size from PstI digestion of chromosomal S. nodosus DNA were isolated from 0.8% agarose gel by glass milk elution and cloned into the BamHI site and PstI sites of pTZ19R. The resulting constructs were used to transform E. coli, and colonies positively hybridizing with the probes were subsequently isolated. Plasmids with 6.7-kb PstI and 7.7-kb BamHI inserts from the hemA-asuA region were designated pMPA03 and pMPA01, respectively; the plasmid with a cloned gtr region on a 6.4-kb BamHI fragment was labeled pMPA10.
Disruption of gtr and hemA-asuA genes in Streptomyces.
Gene replacements were achieved by following the procedures described by Muth et al. (33) by using vector pGM160 with a temperature-sensitive origin of replication. The principle has been described previously (23). The 3.4-kb BglII-BamHI DNA fragment bearing the gtr gene from Streptomyces coelicolor was first subcloned from the cosmid E68 in pGEM7-Zf(+), cleaved with BamHI to give plasmid pMPH28. In pMPH28, the internal 0.7-kb Asp718 (blunt-ended)-XhoI fragment was replaced with the XbaI (blunt-ended)-XhoI fragment of pNEO2 bearing the aphI gene. The DNA fragment with an aphI gene that is flanked by 5′ and 3′regions of the gtr gene was isolated after digestion with HindIII-XbaI and inserted into corresponding sites of the pGM160 vector with a temperature-sensitive origin of replication. The final construct was passaged in the E. coli GM2929 (lacking both dam and dcm) strain and thereafter used for the transformation of S. coelicolor J1501 protoplasts. Positive transformants were selected for resistance to 10 μg/ml neomycin and 30 μg/ml thiostrepton. Enrichment of mutants with gene replacement in the gtr region was achieved by growth at 39°C in YEME medium supplemented with 10 μg/ml neomycin, 20 μg/ml ALA, and 2.5 μg/ml hemin for 6 days. The screening for disruptants was performed after aliquots were plated on GYM plates with 10 μg/ml neomycin, 10 μg/ml ALA, and 2.5 μg/ml hemin. The positive disruptants, designated MIP14, were selected from the colonies that were unable to grow on the GYM plates with 30 μg/ml thiostrepton (Fig. 2C).
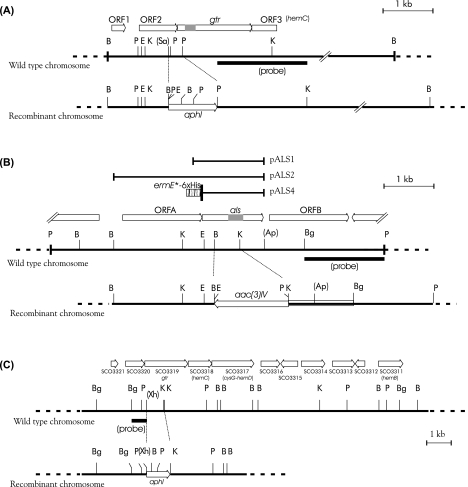
FIG. 2.
Restriction maps of the regions of S. nodosus genome containing the gtr and als genes (A and B) and the S. coelicolor region containing the gtr gene (C). Abbreviations for restriction sites: B, BamHI; Bg, BglII; E, EcoRI; K, KpnI; P, PstI; Sa, SalI; Xh, XhoI. Symbols in parentheses indicate that there are additional unmapped sites in the region. The original cloned PCR products containing portions of the gtr and als genes are shown as gray boxes within the arrows symbolizing the respective genes. Probes used for the detection of rearrangements in chromosomes upon recombination with gene replacement vectors are also shown below lines representing wild-type chromosomes. In panel A, the 6.4-kbp BamHI fragment cloned in pMPA10 is indicated in the wild-type chromosome as a thick line. Similarly, in panel B, the thick line shows the 6.7-kbp PstI fragment cloned in pMPA03. Other subclones are indicated above the map of the wild-type chromosome in panel B.
The same procedure was applied to disrupt the gtr and hemA-asuA regions of S. nodosus. In the case of gtr, the aphI gene was surrounded by a 1.2-kb BamHI-SalI fragment on one side and by a 1.8-kb PstI-KpnI fragment on the other side (Fig. 2A). In the matter of hemA-asuA gene disruption, the apramycin resistance gene aac(3)IV was used as a replacement; the XbaI-NheI fragment bearing the aac(3)IV gene was inserted between 2.4-kb BamHI and 2.8-kb KpnI-PstI fragments which had been subcloned in pTZ19R from the original plasmid obtained by colony hybridization (Fig. 2B). The cassettes with an antibiotic resistance gene flanked by S. nodosus chromosomal DNA fragments were subsequently inserted into pGM160 plasmid digested with HindIII-BamHI and, after amplification in the E. coli GM2929 strain, the resulting constructs were used to transform S. nodosus protoplasts. The transformants were selected after the plates were overlaid with appropriate antibiotics. The isolation of the disruptants (MIP11 and MIP12) was performed in the same way as in S. coelicolor. However, the GYM media employed for induction and selection of required chromosomal rearrangement in the hemA-asuA gene contained apramycin instead of neomycin.
The double mutant of S. nodosus MIP13 with the defect in both genes was prepared from a hemA-asuA::aac(3)IV mutant by replacing the gtr region with the aphI gene. The sources of the aphI and aac(3)IV genes were plasmids pNEO2 and pAPR3, which were constructed by subcloning the respective genes located on the blunt-ended ApaI-Asp718 fragment from pIJ61 and on the PstI-EcoRI fragment from pKC505 (41) into pBluescript SK(+) cleaved with HincII-EcoRV and with PstI-EcoRI, respectively. The original HindIII site in pAPR3 was converted to an NheI site by the religation of the plasmid first linearized with HindIII and subsequently blunt-ended with a Klenow fragment.
RNA isolation and analysis.
The total RNA from S. nodosus cells grown in the production medium (18) was isolated by a previously described method (40). The RiboPure-Bacteria RNA isolation kit from Ambion (Europe) Ltd. (Huntingdon, United Kingdom) was used for the total RNA isolation from the S. coelicolor J1501 cells grown in the liquid GYM medium. The isolation was performed according to the manufacturer's protocol. Dot blots were prepared according to the standard procedure (44) with 12 μg of each RNA sample applied to a nylon membrane in a Bio-Dot microfiltration apparatus (Bio-Rad, Hercules, CA). The membranes were scanned by a FUJI BAS5000 scanner.
Generation of constructs for ALAS expression.
PCR was used to modify the sequence upstream of the ATG site of the hemA-asuA gene. The rightward primer in the reaction with pMPA03 as a template contained a BglII restriction site (underlined) and the first five codons of the hemA-asuA gene including the start codon (in bold): 5′-ACAGATCTGATGAACAAGCACCTGG-3′. The leftward primer (5′-GAAGCCGGAGGGGAAGA-3′) was designed according to the sequence downstream of the BamHI site. The resulting 360-bp fragment was cleaved with BamHI-BglII and inserted into the BamHI site of vector pQE31. The recombinant plasmid with the PCR fragment inserted in the proper orientation was further cleaved with BamHI and ligated with a 1.1-kb BamHI-BclI fragment isolated from pMPA01 to give plasmid pALS3, later used to express the protein in E. coli. A similar procedure, except for the substitution of a different rightward primer (5′-TGACGATATCAACAAGCACCTGGACT-3′), was used to create an expression plasmid based on the pMAL-c2 vector. The fusion with the malE gene was achieved by ligation of the PCR fragment cleaved in the newly introduced EcoRV site with the XmnI site in the vector. The expression plasmid was designated as pALS5.
The fragment inserted in pALS3 along with the ribosome-binding site and the six-His tag-encoding sequence was removed by digestion with HindIII and by partial cleavage with EcoRI and then inserted into the corresponding sites of pGEM-7Zf(+). The resulting plasmid was cleaved with XbaI-HindIII, and the recombinant hemA-asuA gene was inserted downstream of the ermE* promoter (7) into plasmid pIJ4070 digested with the same enzymes. Finally, the fragment bearing the ermE*-(His6)hemA-asuA fusion was removed from the construct by digestion with SacI and ligated with a SacI-linearized pIJ622 vector to give pALS4, which was subsequently used to express the protein in Streptomyces.
The hemA-asuA gene alone (pALS1) or together with open reading frame A (ORFA) and the preceding putative promoter (pALS2) was inserted into a pIJ487 vector to examine the ALAS expression from its native promoter in Streptomyces (Fig. 2B). The murine hemA gene was removed from pGF23 by cleavage with SalI-BamHI and inserted into BglII-SalI-digested pSL1180 vector. The gene was next subcloned on an SalI-NsiI fragment in pMAL-c2 vector. To achieve an in-frame fusion with the malE gene, the construct was first partially cleaved with SalI, blunt-ended with Klenow fragment, cleaved with XmnI, and religated.
Other methods.
Thin-layer chromatography (TLC) analyses were performed on precoated silica gel plates (aluminum backing, 0.25-mm layer; UV-254 fluorescence) from Merck KGaA, Darmstadt, Germany. Compounds on plates were visualized under UV light at 254 and/or 310 nm. The fermentation broth (300 ml) from the S. nodosus production cultures was saturated with NaCl and extracted three times with ethyl acetate. The mycelium was extracted with acetone, the extract was concentrated, and the residue was extracted with ethyl acetate. The ethyl acetate extracts were combined, evaporated, and dissolved in 0.5 ml of chloroform. Ten microliters of each sample was applied on TLC plates and developed with benzene-acetone (3:2; asukamycin Rf, 0.56). ALAS assays were carried out by using modified colorimetric and continuous spectrophotometric methods (20, 26). To compare the rates of succinyl-CoA consumption and ALA formation, the activity of recombinant HemA-AsuA protein (0.25 to 2 mg) in a total volume of 1.0 ml was analyzed continuously for time periods of 20 and 40 min. Reactions were terminated by the addition of 125 μl of 50% trichloroacetic acid and analyzed for ALA content by colorimetric reaction. As a reference, a similar procedure was applied to 0.1 to 0.3 mg of recombinant murine ALAS enzyme; in this case, the reaction was terminated after 10 min.
The possible interference of 2-amino-3-hydroxycyclopent-2-enone or its potential degradation products with the colorimetric assay was analyzed by adding excessive amounts of the chemically synthesized compound (up to 3.5 mM) to the reaction mixture, while omitting succinyl-CoA. Since the compound was quite unstable, it was freshly prepared prior to usage by a procedure described previously (12), neutralized with NaOH, and immediately used in the reaction.
Blue-native polyacrylamide gel electrophoresis (PAGE) was prepared and performed as described by Schägger et al. (46). Affinity purifications of N-terminally His6- and maltose-binding protein (MBP)-tagged proteins were performed according to the manufacturer's instructions on Talon (Clontech, Heidelberg, Germany) and on amylose resins (New England Biolabs, Frankfurt a. M., Germany).
The protein concentrations were determined according to Bradford (9).
Nucleotide sequence accession numbers.
The nucleotide sequences of the S. nodosus subsp. asukaensis hemA-asuA and gtr genes have been deposited in the GenBank database under accession numbers AY240962 and AY953282, respectively.
RESULTS
Detection and cloning of gtr and hemA-asuA genes from S. nodosus subsp. asukaensis.
With the help of primers based on the amino acid sequences TCNR(T/L)E(I/L)Y and GE(T/G/P)QIL(G/A)QV of two conservative regions on the polypeptide chain of GluTR and S. nodosus chromosomal DNA as a template, a 220-bp fragment was amplified by PCR and cloned in E. coli. The comparison of the corresponding DNA sequence with nonredundant GenBank cDNA sequence translations by the BLASTX program confirmed that the cloned fragment was most likely a part of the gtr gene [98% similarity to the relevant part of a putative GluTR from S. coelicolor A3(2)]. Subsequently, a 6.4-kb BamHI DNA fragment of genomic S. nodosus DNA was identified by Southern hybridization using the above-described 220-bp PCR product as a probe. The 6.4-kb BamHI fragment was cloned in E. coli and identified by screening a partial genomic library made of size-fractioned BamHI-digested S. nodosus DNA inserted into the pTZ19R vector with the help of the same probe.
A similar procedure was applied to the hemA-asuA gene. We used primers derived from two ALAS regions with two short conservative sequences, VWC(S/G)NDYL and GFIF(T/S)T(T/S/A)LPP. These regions were determined after a comparison of the amino acid sequences of seven enzymes from the following organisms: Bradyrhizobium japonicum (GenBank accession no. M16751), Rhodobacter capsulatus (X53309), Saccharomyces cerevisiae (J03556), rat (J03190), human (housekeeping 5-aminolevulinate synthase; X56351), chicken (SwissProt accession no. Q9XT75), and Agrobacterium tume-Faciens (PIR accession no. S15996). A 700-bp fragment was amplified by PCR using S. nodosus chromosomal DNA as a template. No amplification was observed with the control genomic DNA from S. coelicolor A3(2). The fragment was inserted into the pTZ19R vector to give plasmid pMPH25 and cloned in E. coli XL1-Blue cells. The DNA fragment sequence comparison with the translated GenBank database revealed a strong similarity with the ALAS-encoding genes. The 6.7-kb PstI and the 7.7-kb BamHI fragments were identified by a Southern hybridization of digested genomic S. nodosus DNA with the labeled probe generated from pMPH25. The same probe was used to screen a partial genomic library generated by inserting an appropriate fraction of PstI fragments into pTZ19R and by cloning in E. coli XL1-Blue cells. The restriction maps of the genomic regions surrounding the sequences that hybridized with the gtr-specific and hemA-asuA-specific probes are presented in Fig. 2A and B.
Sequence analysis of cloned gtr and hemA-asuA genes and adjacent regions.
The nucleotide sequence of more than 3,300 bp out of the 6.4-kb BamHI fragment that hybridized with the gtr probe revealed three complete and one truncated ORF. The deduced amino acid sequences of their products and also the organization of these ORFs were highly similar to those found in the putative gtr loci of completely sequenced chromosomes of S. coelicolor A3(2) (Fig. 2) and Streptomyces avermitilis. In all three cases, the putative gtr gene is preceded by an ORF encoding a potential DNA-binding protein and more distantly by a putative redoxin-encoding gene. The ORF located immediately downstream is similar to the hemC genes encoding porphobilinogen deaminases. The gtr genes are over 80% identical in the first half of the sequence and also in the last third. The regions encoding the central part of the protein are variable in the sequence and in their length. The deduced amino acid sequence of the gtr gene from S. nodosus (499 residues) is the shortest among the three streptomycete enzymes determined thus far; the other two from S. coelicolor and S. avermitilis have chain lengths of 569 and 581 residues, respectively (data not shown). The conservative N-terminal region of the protein sequence constitutes a domain with catalytic and NAD(P)H-binding sites; the invariant residues of the C-terminal domain are putatively implicated in GluTR dimerization (32).
The sequence information obtained from the analysis of the 6.7-kb PstI fragment hybridizing with the hemA-asuA probe revealed the presence of three complete ORFs showing the common patterns of GC bias and preferred codon usage for ORFs in Streptomyces (Fig. 2B). The central ORF (referred to here as hemA-asuA) putatively encodes a polypeptide of 409 amino acids with a molecular mass of 43.7 kDa. The translation start of this ORF overlaps with the termination codon of the preceding ORF and is located 10 bp downstream of an imperfect putative ribosome binding site GAGtTGA (51).
By using the BLAST program, we compared the derived amino acid sequence of the hemA-asuA-encoded protein with the nonredundant database of protein sequences. The strongest observed similarities were with four previously characterized enzymes from R. sphaeroides (50% and 45% identities over 382 and 387 amino acids with hemT and hemA products, respectively) (35), Agrobacterium radiobacter (49% identity over 399 amino acids) (11), and Bradyrhizobium japonicum (47% identity over 388 amino acids) (29). Despite the high degree of overall sequence identities, in the S. nodosus enzyme, Ser83 and Ser352 replace Thr residues strictly conserved in all other ALA synthases. The adjacent ORFs temporarily designated ORFA and ORFB appear to encode amide synthase and a protein similar to long-chain fatty acid-CoA ligases, respectively (data not shown).
Disruption of gtr and hemA-asuA genes in S. nodosus and gtr gene in S. coelicolor.
In order to confirm the gtr gene function in both streptomycetes and also to further define the roles of both gene products in S. nodosus, the internal regions of the respective genes were deleted from the chromosomes. Mutants with deletions in the gtr regions were constructed from the parental strains S. coelicolor J1501 or S. nodosus subsp. asukaensis by homologous recombination as described in Materials and Methods. The expected deletion in the S. coelicolor J1501 host genome, formed upon double crossing-over, spans 0.7 kb. The aphI marker with the transcriptional terminator removed was oriented in the same direction as the original gtr gene to promote a potential transcription of downstream genes that are likely a part of the same operon. The gene replacement was confirmed by a Southern blot analysis of chromosomal DNA. The 0.7-kb EcoRI-XhoI-labeled fragment hybridized with a 0.7-kb PstI chromosomal DNA fragment from the MIP14 mutant strain in place of a 2.6-kb fragment detected in the wild-type strain. Also, in the case of MIP14, a 9-kb BamHI fragment was detected instead of a 12-kb fragment.
The mutants deficient in heme biosynthesis usually fall into two phenotypic classes. Those blocked in ALA synthesis can grow when fed ALA, whereas the mutants blocked in late steps of heme synthesis have an absolute requirement for heme. The isolated mutant of S. coelicolor grew on a medium supplemented with ALA, suggesting that the expression of downstream biosynthetic genes was not substantially affected by the deletion-substitution. The mutant also grew on the complex GYM medium supplemented with hemin but only for a few generations, probably due to the deficiency of other tetrapyrrole cofactors, such as siroheme, which cannot be directly synthesized from the hemin precursor.
In S. nodosus, gtr and hemA-asuA gene disruptions were also achieved by transforming the wild-type strain with neomycin and apramycin resistance markers, respectively, surrounded by DNA sequences flanking the region to be deleted. The deletion in MIP11 was expected to remove the beginning of the gtr gene along with the terminal part of the preceding ORF; in MIP12, a 0.65-kb fragment internal to the structural part of the hemA-asuA gene was removed. Gene deletion-substitutions were confirmed by Southern analyses using a 130-bp KpnI-BglII labeled fragment for the gtr detection and a 1.7-kb SacI-KpnI DNA probe for the hemA-asuA detection (Fig. 2). The gtr probe hybridized with 5.5-kb BamHI and with 3.8-kb MluI fragments instead of the 6.4-kb and 2.9-kb fragments from the wild-type strain DNA digested with the same enzymes. The hemA-asuA probe hybridized with 9.0-kb KpnI and with 7.1-kb BamHI fragments instead of the 7.0-kb and 7.7-kb fragments from the wild-type DNA.
Phenotypes of S. nodosus disruptants.
Growth of the S. nodosus MIP11 strain was absolutely dependent on supplementation with ALA or heme. In the presence of ALA, the parental strain and the gtr mutant grow at the same rate (data not shown). Upon prolonged cultivation (14 days) on unsupplemented GYM plates densely inoculated with spores of the mutant strain, sparsely growing colonies of prototrophic revertants appeared that were no longer dependent on ALA or heme supplementation. No such revertants emerged on plates inoculated with the S. coelicolor MIP14 mutant.
The S. nodosus MIP12 strain displayed essentially the same growth capabilities of the parental strain in both complex and chemically defined media. The MIP12 strain, however, did not produce any detectable levels of asukamycin (Fig. 3B). Surprisingly, the addition of ALA to the culture medium did not restore asukamycin production. To exclude the possibility that the gene disruption-replacement affected the expression of downstream genes involved in antibiotic biosynthesis, the mutant strain was transformed with plasmid pALS4, where the hemA-asuA expression was directed by an ermE* promoter. The transformant regained the ability of asukamycin production and synthesized the antibiotic in larger amounts than the parental strain (Fig. 3).
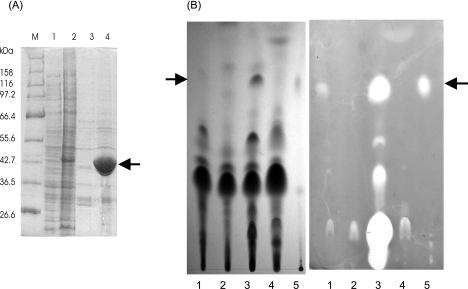
FIG. 3.
Overexpression of His6-tagged hemA-asuA in S. nodosus and its effect on asukamycin production. (A) The SDS-PAGE gel analysis (10% gel; 20 μg of the protein per lane was applied) shows cell extracts of S. nodosus carrying pIJ622 vector as a negative control (lane 1) or pALS4 plasmid (lane 2). The extracts from 1.5 ml of culture were subjected to affinity purification using TALON resin according to the manufacturer's instructions for native purification. The results are shown in lanes 3 (control) and 4 (His6-ALAS). The position of the ALAS band is indicated by an arrow. The molecular sizes of the marker (M) proteins are indicated. (B) The presence of the antibiotic in the cultivation broth was detected by TLC and bioassay against Bacillus subtilis 0453-52-9 (Difco) grown on a nutrient agar plate (CM0003; Oxoid). The left part shows a chromatogram visualized under UV light (254 nm); the right part represents the bioassay (chromatogram was laid on the surface of the agar plate for 15 min, and the plate was incubated overnight at 37°C). Into each lane, 10 μl of the extract was applied. The extracts were prepared from the following cultures: lane 1, parental S. nodosus strain; lane 2, MIP12 mutant; lane 3, MIP12 × pALS4; lane 4, MIP12 mutant supplemented with 20 μg/ml ALA. Asukamycin (10 μg) was applied as a control to lane 5. The position of asukamycin is indicated by an arrow. (The inhibition zone in the start of lane 3 is caused by thiostrepton, which was coextracted with asukamycin from the supplemented medium.)
The double mutant S. nodosus MIP13 depended on ALA or heme supplementation and, like the S. coelicolor MIP14 mutant, did not revert to the prototrophic phenotype.
Expression of the S. nodosus hemA-asuA gene in E. coli and in Streptomyces.
Features of the isolated hemA-asuA gene were investigated after recombinant ALAS expression in E. coli and in Streptomyces. Expression plasmids were constructed as described in Materials and Methods. SDS-PAGE analysis of a cell extract generated from E. coli TG1/pALS3 taken 5 h after induction with IPTG (isopropyl-β-d-thiogalactopyranoside) revealed an intense band of a 45-kDa protein, which was mainly located in an insoluble fraction. The amount of soluble protein was not increased by either reducing the growth temperature or lowering the IPTG concentration. Therefore, another system based on the pMAL-c2 expression vector was applied. The majority of the MBP-tagged ALAS (87 kDa) isolated from the E. coli TB1/pALS5 cells cultivated for 18 h at 14°C was in the soluble fraction. The fusion protein forms homodimers migrating as a single band of approximately 175 kDa on blue-native PAGE (data not shown).
In contrast to E. coli, the soluble and active His-tagged ALAS was obtained when the hemA-asuA gene was expressed under the ermE* promoter in Streptomyces lividans TK24 and in S. nodosus using the plasmid construct pALS4 (Fig. 3A). Specific activity of the purified enzyme isolated from the recombinant strains as measured in an in vitro assay was low. According to the colorimetric assay, the enzyme purified from the recombinant E. coli TB1/pALS5 strain had a final specific activity of 25 U/mg, with 1 unit defined as 1 nmol of 5-aminolevulinic acid formed per hour at 30°C (26). No enzyme activity was detected when glycine was replaced with alanine or threonine in the reaction mixture. The murine ALAS expressed by the same E. coli system was used to compare the specific enzymatic activities. The enzyme activity of this fusion protein was estimated to be 1,460 U/mg at 37°C. The data shown in the figures represent mean values for specific activities estimated in triplicate, and the standard deviation did not exceed 6%. Similar values of specific activities were obtained by using the continuous spectrophotometric assay in direct combination with the colorimetric assay (20). This allowed us to compare the rates of CoA formation from succinyl-CoA with ALA synthesis rates. The specific activity of the streptomycete enzyme was higher when estimated using the continuous assay: 66 ± 6% compared to 25 ± 6% U/mg obtained from the subsequent colorimetric activity determination. In the case of the murine ALAS, the estimated activity based on the continuous measurement was 1,395 ± 2% U/mg compared to 1,320 ± 3% U/mg obtained from the subsequent colorimetric determination. Addition of the chemically synthesized C5N unit up to a final concentration of 3.5 mM into the reaction mixture did not result in any color reaction in the colorimetric ALAS assay.
Complementation of Streptomyces and E. coli mutants with gtr and hemA-asuA genes.
Plasmid constructs containing the genes under the control of their native or suitable heterologous promoters were introduced into E. coli HU227 and S. coelicolor MIP14 mutant strains defective in the gene for glutamyl-tRNA transferase. The growth of both strains is fully dependent on ALA supplementation. Multicopy plasmid constructs pMPA11 and pMPA12, carrying the KpnI fragment from S. nodosus with the entire gtr gene controlled by its own promoter, complemented well the growth defects of E. coli and S. coelicolor, respectively. No complementation of the defect in ALA synthesis was observed when the S. coelicolor MIP14 mutant was transformed with either of two plasmid constructs based on the pIJ487 vector in which the hemA-asuA gene alone (pALS1) or together with ORFA and the preceding putative native promoter (pALS2) was inserted. However, when the hemA-asuA expression was controlled by a strong promoter (in pALS4 and pALS5), it was able to restore the growth of S. coelicolor and E. coli mutants.
Analysis of gtr and hemA-asuA transcription in Streptomyces.
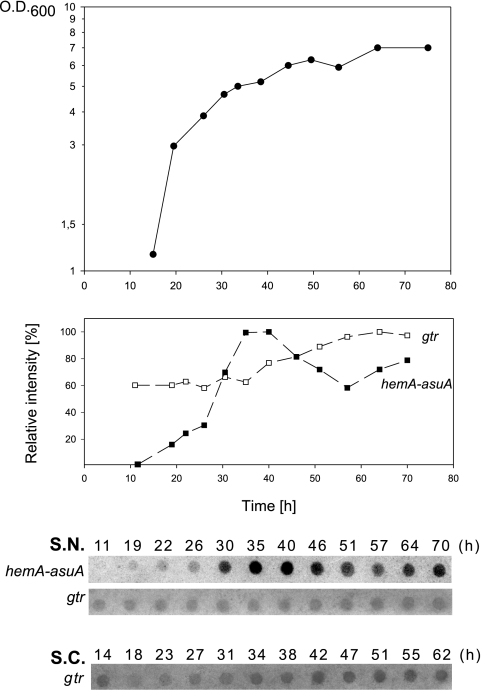
An effect of the growth stage on the steady state of hemA-asuA and gtr transcripts was observed in S. coelicolor and S. nodosus strains by using RNA dot blots (Fig. 4). The relative concentration of the transcripts was estimated in the RNA extracted from the cells at different growth stages. The results of these experiments show that the relative concentrations of gtr transcripts remain fairly constant throughout the growth curve. In contrast, a steady-state level of S. nodosus hemA-asuA transcripts steadily increased from the initial undetectable level and peaked as the cells entered stationary phase.
FIG. 4.
Analysis of the amount of steady-state gtr and hemA-asuA transcripts in S. nodosus. The bottom of the figure shows RNA dot blots of total RNA isolated from cultures at different time points on the growth curve. Twelve micrograms of total RNA was applied in each dot, and the blots were hybridized with appropriate probes. The gene probes are indicated in parentheses. The growth curve of S. nodosus (S.N.) and relative intensities of corresponding RNA dots are plotted on the top. The intensities of the dot signals were quantified using the AIDA Bio Package (Raytest, Straubenhardt, Germany). The relative intensities were calculated using the maximum intensity value of each row as 100%. For comparison, an RNA dot blot with gtr-specific probe obtained by the same procedure from S. coelicolor (S.C.) is shown. The signal intensities in individual blot strips do not reflect the concentration ratios between particular gene transcripts.
Inspection of Streptomyces genomic DNA for the presence of hemA-asuA gene.
The genomic DNA of other streptomycetes was inspected for the presence of hemA-asuA homologues. The same labeled probe that was used to detect the hemA-asuA gene in S. nodosus was further employed to identify homologous genes in other streptomycete producers of secondary metabolites with an attached C5N unit and also in 20 randomly chosen strains from the laboratory stock. So far, only genomic DNA from strains producing compounds with a specific moiety hybridized positively on Southern blots. These strains included Streptomyces parvulus Tü64, a producer of manumycin A; Streptomyces xanthochromogenus, a producer of reductiomycin; and Streptomyces bambergiensis, a producer of moenomycin A. The regions homologous to hemA-asuA were partially sequenced in these strains, and an identical ORFA-hemA-asuA-ORFB gene arrangement was found in all cases (data not shown).
DISCUSSION
5-Aminolevulinic acid, normally a biosynthetic precursor to tetrapyrroles, is used in some Streptomyces to form a distinct C5N moiety (Fig. 1), a component of several types of secondary metabolites. It has been shown that for asukamycin, reductiomycin, and moenomycin, ALA, which next undergoes an intramolecular cyclization, originates from glycine and succinyl-CoA. The utilization of such substrates indicates that the functional Shemin pathway is present in strains producing these secondary metabolites, and it is therefore likely that the two most common ALA biosynthetic pathways are simultaneously present in certain Streptomyces strains.
As an initial step for proving this assumption, we cloned genes for glutamyl-tRNAGlu reductase (gtr) and ALAS (hemA-asuA) from the asukamycin producer S. nodosus subsp. asukaensis. Since both the above genes encode the first step in heme biosynthesis, they are often designated hemA in other organisms. To discriminate them in Streptomyces, we have used an alternative designation. Gene organization in the gtr chromosomal locus of S. nodosus is similar to that of S. coelicolor and S. avermitilis, where the putative hemC, hemD, and closely spaced hemB are immediately downstream of the gtr gene. Consequently, the locus may then encode the first half of the tetrapyrrole biosynthetic pathway, with the exception of the hemL gene coding for glutamate-1-semialdehyde 2,1-aminomutase located in a different place. A similar organization of the genes encoding the biosynthesis of uroporphyrinogen III is found also in other phylogenetically diverse bacteria (16, 43).
The role that gtr plays in S. nodosus and in S. coelicolor was confirmed by its deletion in these strains. Both gtr mutants could not grow without feeding the exogenous ALA or heme, which demonstrates that the gtr gene is required for tetrapyrrole biosynthesis in both Streptomyces species. These results, supported by a high degree of gtr similarity with already characterized homologous genes (25, 31, 39, 47, 50) and the fact that it complements the hemA mutation in E. coli, suggest that the gene indeed encodes a functional GluTR. The results also indicate that Streptomyces requires the functional C5 pathway for tetrapyrrole synthesis.
In contrast to S. coelicolor, an ALAS-encoding gene representing the Shemin pathway was isolated from S. nodosus. The plasmid encoding an N-terminally His-tagged ALAS transcribed from a strong constitutive ermE* promoter complemented the growth defect of an S. coelicolor gtr mutant. Also, a plasmid construct directing the expression of the MBP-ALAS complemented the defect in the E. coli HU227 strain. This demonstrates that the gene does in fact encode a functional enzyme. The estimated specific activity of the enzyme was about 60 times lower than that of the murine recombinant ALAS prepared under the same conditions and about 500 times lower than was reported for the HemA enzyme from R. sphaeroides prepared from a recombinant E. coli strain (8). Whether the replacements of two otherwise strictly invariant Thr residues (83 and 352) by Ser could account for the low specific activity remains to be proved experimentally. A recently published three-dimensional model of ALAS from Rhodobacter capsulatus illustrates that side chains of both corresponding Thr residues contribute to the active site formation: the first one is crucial for the glycine binding specificity, and the latter appears essential both for active-site closure and for succinyl-CoA stabilization (3, 49).
The S. nodosus MIP12 mutant did not produce any asukamycin. However, a small amount of the asukamycin biosynthetic pathway intermediate with a carboxyl group in place of the C5N unit could be detected in the cultivation medium (T.-W. Yu, personal communication). This observation fits well with the asukamycin biosynthetic model, where the attachment of the C5N unit is the step preceding the final oxidation of the aromatic ring to the epoxyquinol moiety (18, 19). Surprisingly, no antibiotic was formed upon adding the exogenous ALA to the liquid culture of the S. nodosus MIP12. However, when the mutant strain was transformed with the plasmid construct, where the hemA-asuA gene expression was directed by the ermE* promoter, it produced an increased amount of asukamycin into the medium (Fig. 3). ALA uptake problems should be ruled out, since the compound is taken up well by Streptomyces gtr mutants. These results, thus, could indicate that the presence of functional ALAS is also necessary for ALA cyclization, which the enzyme could perform either by itself or in conjunction with other enzyme(s). As the C5N unit is quite unstable, it is difficult to prove this hypothesis directly even in the in vitro reaction mixture. In order to investigate the possibility that the C5N unit is in fact the major product of the enzymatic reaction, the rates of substrate consumption were compared with those of ALA formation in the ALAS reaction mixtures. In the case of the streptomycete enzyme, succinyl-CoA utilization was about 2.5 times higher than ALA formation. This result argues against the cyclization hypothesis that presumes a substantial overproduction of the free C5N unit. Moreover, the addition of the chemically prepared compound to the reaction, even in a molar excess of 100 times the amount of formed ALA, did not result in the positive color reaction. This also means that the ALA formed in the reaction does not originate from the C5N decomposition. We cannot exclude the possibility that small amounts of the C5N unit may be formed by the HemA-AsuA enzyme. However, the difference between the HemA-AsuA activities estimated by the two methods can also be caused by the interference of high protein concentrations used in the continuous assay, as discussed previously (20). The possible participation of another enzyme, such as fatty acid-CoA ligase, on the ALA cyclization should be taken into account, as has already been suggested (28). The genes putatively encoding these enzymes are always closely linked with ALAS genes in the strains producing C5N-containing compounds (M. Petříček, unpublished data). In connection with that, substrate channeling should be considered as a possible mechanism for how the flow of ALA into the asukamycin biosynthetic route is controlled.
Analysis of steady-state hemA-asuA mRNA transcripts of the S. nodosus wild-type strain revealed that the gene is transcriptionally regulated; its transcription reaches the maximum in the beginning of the stationary phase. This corresponds well with the timing of the major phase of antibiotic production (37). The steady state of the gtr transcript remained fairly constant throughout the whole growth period of S. nodosus and differed somewhat from that of S. coelicolor, which showed a slight increase in specific mRNA concentration in the stationary phase. Accordingly, the possibility cannot be excluded that in the later growth stages of S. nodosus, a part of ALA from the Shemin pathway is utilized in the porphyrin biosynthetic route, as in the case of A. globiformis (57). Consistently, the hemA-asuA gene complemented the gtr mutation in the S. coelicolor MIP14 strain only when its native promoter was replaced with ermE*. This observation is in accordance with data obtained from the mRNA analysis and indicates that its transcription may be positively regulated in S. nodosus.
In this work, we showed that streptomycete strains producing unsaturated 2-amino-3-hydroxycyclopent-2-enone-derived amides possess both major routes of ALA biosynthesis. While the C5 pathway is strictly used for tetrapyrrole biosynthesis in S. nodosus, the Shemin route is indispensable for the formation of a C5N unit. Therefore, the presence of the hemA-asuA gene in Streptomyces is specifically linked with the production of this group of secondary metabolites. Furthermore, since the gene is obviously located within the asukamycin biosynthetic cluster, it can serve as an extremely simple clue both to detect new producers of C5N-containing compounds and to localize corresponding biosynthetic clusters on their chromosomes. This hypothesis is also supported by a recently described biosynthetic gene cluster of the antibiotic ECO-02301 from Streptomyces aizunensis, also containing a C5N unit, where the hemA-asuA gene homologue was identified as well (28).
Acknowledgments
This work was supported in part by the Grant Agency of the Czech Republic (research grant 310/03/0285), the Czech Ministry of Education (grant ME707), and by the Grant Agency of the Czech Academy of Sciences (postdoctoral research grant B5020306 for K.P.). The support by the Institutional Scientific Program AV0Z50200510 is also acknowledged.
We are grateful to Heinz Floss and Tin-Wei Yu (University of Washington) for the gift of asukamycin, for providing streptomycete strains, and for sharing their unpublished data with us. We also thank Gloria Ferreira for providing the plasmid pGF23 and Kateøina Hejduková for technical assistance.
REFERENCES
- 1.Altschul, S. F., M. S. Boguski, W. Gish, and J. C. Wootton. 1994. Issues in searching molecular sequence databases. Nat. Genet. 6:119-129. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Astner, I., J. O. Schulze, J. van den Heuvel, D. Jahn, W. D. Schubert, and D. W. Heinz. 2005. Crystal structure of 5-aminolevulinate synthase, the first enzyme of heme biosynthesis, and its link to XLSA in humans. EMBO J. 24:3166-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avissar, Y. J., J. G. Ormerod, and S. I. Beale. 1989. Distribution of δ-aminolevulinic acid biosynthetic pathways among phototrophic bacterial groups. Arch. Microbiol. 151:513-519. [DOI] [PubMed] [Google Scholar]
- 5.Beale, J. M., J. P. Lee, A. Nakagawa, S. Omura, and H. G. Floss. 1986. Biosynthesis of the antibiotic reductiomycin. J. Am. Chem. Soc. 108:331-332. [Google Scholar]
- 6.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 7.Bibb, M. J., G. R. Janssen, and J. M. Ward. 1985. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene 38:215-226. [DOI] [PubMed] [Google Scholar]
- 8.Bolt, E. L., L. Kryszak, J. Zeilstra-Ryalls, P. M. Shoolingin-Jordan, and M. J. Warren. 1999. Characterization of the Rhodobacter sphaeroides 5-aminolaevulinic acid synthase isoenzymes, HemA and HemT, isolated from recombinant Escherichia coli. Eur. J. Biochem. 265:290-299. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Donnerstag, A., S. Marzian, D. Müller, P. Welzel, D. Bottger, A. Stark, H. W. Fehlhaber, A. Markus, Y. van Heijenoort, and J. van Heijenoort. 1995. A structurally and biogenetically interesting moenomycin antibiotic. Tetrahedron 51:1931-1940. [Google Scholar]
- 11.Drolet, M., and A. Sasarman. 1991. Cloning and nucleotide sequence of the hemA gene of Agrobacterium radiobacter. Mol. Gen. Genet. 226:250-256. [DOI] [PubMed] [Google Scholar]
- 12.Ebenezer, W. J. 1991. Colabomycin co-metabolites: synthesis of 2880-II, a metabolite related to ferulic acid. Synth. Commun. 21:351-358. [Google Scholar]
- 13.Endler, K., U. Schuricht, L. Hennig, and P. Welzel. 1998. Exploratory investigations into the biosynthesis of the antibiotic moenomycin A. Tetrahedron Lett. 39:13-16. [Google Scholar]
- 14.Ferreira, G. C., and H. A. Dailey. 1993. Expression of mammalian 5-aminolevulinate synthase in Escherichia coli: overproduction, purification, and characterization. J. Biol. Chem. 268:584-590. [PubMed] [Google Scholar]
- 15.Hanajima, S., K. Ishimaru, K. Sakano, S. K. Roy, Y. Inouye, and S. Nakamura. 1985. Inhibition of reverse transcriptase by limocrocin. J. Antibiot. (Tokyo) 38:803-805. [DOI] [PubMed] [Google Scholar]
- 16.Hansson, M., L. Rutberg, I. Schröder, and L. Hederstedt. 1991. The Bacillus subtilis hemAXCDBL gene cluster, which encodes enzymes of the biosynthetic pathway from glutamate to uroporphyrinogen III. J. Bacteriol. 173:2590-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces. A laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 18.Hu, Y. 2000. The biosynthesis of manumycin type metabolites. Ph.D. thesis. Department of Chemistry, University of Washington, Seattle.
- 19.Hu, Y. D., and H. G. Floss. 2004. Further studies on the biosynthesis of the manumycin-type antibiotic, asukamycin, and the chemical synthesis of protoasukamycin. J. Am. Chem. Soc. 126:3837-3844. [DOI] [PubMed] [Google Scholar]
- 20.Hunter, G. A., and G. C. Ferreira. 1995. A continuous spectrophotometric assay for 5-aminolevulinate synthase that utilizes substrate cycling. Anal. Biochem. 226:221-224. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki, T., T. Kuzuyama, K. Furihata, N. Itoh, H. Seto, and T. Dairi. 2003. A relationship between the mevalonate pathway and isoprenoid production in actinomycetes. J. Antibiot. (Tokyo) 56:957-966. [DOI] [PubMed] [Google Scholar]
- 23.Kieser, T., and D. A. Hopwood. 1991. Genetic manipulation of Streptomyces: integrating vectors and gene replacement. Methods Enzymol. 204:430-458. [DOI] [PubMed] [Google Scholar]
- 24.Koshino, H., H. Osada, T. Yano, J. Uzawa, and K. Isono. 1991. The structure of enopeptins A and B, novel depsipeptide antibiotics. Tetrahedron Lett. 32:7707-7710. [Google Scholar]
- 25.Li, J. M., C. S. Russell, and S. D. Cosloy. 1989. Cloning and structure of the hemA gene of Escherichia coli K-12. Gene 82:209-217. [DOI] [PubMed] [Google Scholar]
- 26.Lien, L. F., and D. S. Beattie. 1982. Comparisons and modifications of the colorimetric assay for δ-aminolevulinic acid synthase. Enzyme 28:120-132. [DOI] [PubMed] [Google Scholar]
- 27.Mayer, S. M., and S. I. Beale. 1992. Succinyl-coenzyme A synthetase and its role in δ-aminolevulinic acid biosynthesis in Euglena gracilis. Plant Physiol. 99:482-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McAlpine, J. B., B. O. Bachmann, M. Piraee, S. Tremblay, A. M. Alarco, E. Zazopoulos, and C. M. Farnet. 2005. Microbial genomics as a guide to drug discovery and structural elucidation: ECO-02301, a novel antifungal agent, as an example. J. Nat. Prod. 68:493-496. [DOI] [PubMed] [Google Scholar]
- 29.McClung, C. R., J. E. Somerville, M. L. Guerinot, and B. K. Chelm. 1987. Structure of the Bradyrhizobium japonicum gene hemA encoding 5-aminolevulinic acid synthase. Gene 54:133-139. [DOI] [PubMed] [Google Scholar]
- 30.McConville, M. L., and H. P. Charles. 1979. Isolation of haemin-requiring mutants of Escherichia coli K12. J. Gen. Microbiol. 113:155-164. [DOI] [PubMed] [Google Scholar]
- 31.Moberg, P. A., and Y. J. Avissar. 1994. A gene cluster in Chlorobium vibrioforme encoding the first enzymes of chlorophyll biosynthesis. Photosynth. Res. 41:253-259. [DOI] [PubMed] [Google Scholar]
- 32.Moser, J., W. D. Schubert, D. W. Heinz, and D. Jahn. 2002. Structure and function of glutamyl-tRNA reductase involved in 5-aminolaevulinic acid formation. Biochem. Soc. Trans. 30:579-584. [DOI] [PubMed] [Google Scholar]
- 33.Muth, G., B. Nußbaumer, W. Wohlleben, and A. Pühler. 1989. A vector system with temperature-sensitive replication for gene disruption and mutational cloning in streptomycetes. Mol. Gen. Genet. 219:341-348. [Google Scholar]
- 34.Nakagawa, A., T.-S. Wu, P. J. Keller, J. P. Lee, S. Omura, and H. G. Floss. 1985. Biosynthesis of asukamycin. Formation of the 2-amino-3-hydroxycyclopent-2-enone moiety. J. Chem. Soc. Chem. Commun. p. 519-521.
- 35.Neidle, E. L., and S. Kaplan. 1993. Expression of the Rhodobacter sphaeroides hemA and hemT genes, encoding 2 5-aminolevulinic acid synthase isozymes. J. Bacteriol. 175:2292-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Omura, S., N. Imamura, K. Hinotozawa, K. Otoguro, G. Lukacs, R. Faghih, R. Tolmann, B. H. Arison, and J. L. Smith. 1983. The structure of virustomycin A. J. Antibiot. (Tokyo) 36:1783-1786. [DOI] [PubMed] [Google Scholar]
- 37.Omura, S., C. Kitao, H. Tanaka, R. Oiwa, and Y. Takahashi. 1976. A new antibiotic, asukamycin, produced by Streptomyces. J. Antibiot. (Tokyo) 29:876-881. [DOI] [PubMed] [Google Scholar]
- 38.Palmer, B. R., and M. G. Marinus. 1994. The dam and dcm strains of Escherichia coli—a review. Gene 143:1-12. [DOI] [PubMed] [Google Scholar]
- 39.Petricek, M., L. Rutberg, I. Schröder, and L. Hederstedt. 1990. Cloning and characterization of the hemA region of the Bacillus subtilis chromosome. J. Bacteriol. 172:2250-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petricek, M., P. Tichy, and M. Kuncova. 1992. Characterization of the α-amylase-encoding gene from Thermomonospora curvata. Gene 112:77-83. [DOI] [PubMed] [Google Scholar]
- 41.Rao, R. N., M. A. Richardson, and S. Kuhstoss. 1987. Cosmid shuttle vectors for cloning and analysis of Streptomyces DNA. Methods Enzymol. 153:166-198. [DOI] [PubMed] [Google Scholar]
- 42.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 43.Rhie, G. E., Y. J. Avissar, and S. I. Beale. 1996. Structure and expression of the Chlorobium vibrioforme hemB gene and characterization of its encoded enzyme, porphobilinogen synthase. J. Biol. Chem. 271:8176-8182. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Sattler, I., R. Thiericke, and A. Zeeck. 1998. The manumycin-group metabolites. Nat. Prod. Rep. 15:221-240. [DOI] [PubMed] [Google Scholar]
- 46.Schägger, H., W. A. Cramer, and G. VonJagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 47.Schröder, I., L. Hederstedt, G. Kannangara, and S. Gough. 1992. Glutamyl-tRNA reductase activity in Bacillus subtilis is dependent on the hemA gene product. Biochem. J. 281:843-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shima, J., A. Hesketh, S. Okamoto, S. Kawamoto, and K. Ochi. 1996. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 178:7276-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shoolingin-Jordan, P. M., S. Al Daihan, D. Alexeev, R. L. Baxter, S. S. Bottomley, I. D. Kahari, I. Roy, M. Sarwar, L. Sawyer, and S. F. Wang. 2003. 5-Aminolevulinic acid synthase: mechanism, mutations and medicine. Biochim. Biophys. Acta 1647:361-366. [DOI] [PubMed] [Google Scholar]
- 50.Srivastava, A., V. Lake, L. A. Nogaj, S. M. Mayer, R. D. Willows, and S. I. Beale. 2005. The Chlamydomonas reinhardtii gtr gene encoding the tetrapyrrole biosynthetic enzyme glutamyl-tRNA reductase: structure of the gene and properties of the expressed enzyme. Plant Mol. Biol. 58:643-658. [DOI] [PubMed] [Google Scholar]
- 51.Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tai, T. N., M. D. Moore, and S. Kaplan. 1988. Cloning and characterization of the 5-aminolevulinate synthase gene(s) from Rhodobacter sphaeroides. Gene 70:139-151. [DOI] [PubMed] [Google Scholar]
- 53.Thiericke, R., A. Zeeck, A. Nakagawa, S. Omura, R. E. Herrold, S. T. S. Wu, J. M. Beale, and H. G. Floss. 1990. Biosynthesis of the manumycin group antibiotics. J. Am. Chem. Soc. 112:3979-3987. [Google Scholar]
- 54.Ward, J. M., G. R. Janssen, T. Kieser, M. J. Bibb, and M. J. Buttner. 1986. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol. Gen. Genet. 203:468-478. [DOI] [PubMed] [Google Scholar]
- 55.Weinstein, J. D., and S. I. Beale. 1983. Separate physiological roles and subcellular compartments for two tetrapyrrole biosynthetic pathways in Euglena gracilis. J. Biol. Chem. 258:6799-6807. [PubMed] [Google Scholar]
- 56.Werner, G., H. Hagenmaier, H. Drautz, A. Baumgartner, and H. Zähner. 1984. Metabolic products of microorganisms. 224. Bafilomycins, a new group of macrolide antibiotics. Production, isolation, chemical structure and biological activity. J. Antibiot. (Tokyo) 37:110-117. [DOI] [PubMed] [Google Scholar]
- 57.Yang, H. S., and J. K. Hoober. 1995. Divergent pathways for δ-aminolevulinic acid synthesis in two species of Arthrobacter. FEMS Microbiol. Lett. 134:259-263. [Google Scholar]