Abstract
Genetic evidence suggests that a family of bacterial and eukaryotic integral membrane proteins (referred to as Wzx and Rft1, respectively) mediates the transbilayer movement of isoprenoid lipid-linked glycans. Recent work in our laboratory has shown that Wzx proteins involved in O-antigen lipopolysaccharide (LPS) assembly have relaxed specificity for the carbohydrate structure of the O-antigen subunit. Furthermore, the proximal sugar bound to the isoprenoid lipid carrier, undecaprenyl-phosphate (Und-P), is the minimal structure required for translocation. In Escherichia coli K-12, N-acetylglucosamine (GlcNAc) is the proximal sugar of the O16 and enterobacterial common antigen (ECA) subunits. Both O16 and ECA systems have their respective translocases, WzxO16 and WzxE, and also corresponding polymerases (WzyO16 and WzyE) and O-antigen chain-length regulators (WzzO16 and WzzE), respectively. In this study, we show that the E. coli wzxE gene can fully complement a wzxO16 translocase deletion mutant only if the majority of the ECA gene cluster is deleted. In addition, we demonstrate that introduction of plasmids expressing either the WzyE polymerase or the WzzE chain-length regulator proteins drastically reduces the O16 LPS-complementing activity of WzxE. We also show that this property is not unique to WzxE, since WzxO16 and WzxO7 can cross-complement translocase defects in the O16 and O7 antigen clusters only in the absence of their corresponding Wzz and Wzy proteins. These genetic data are consistent with the notion that the translocation of O-antigen and ECA subunits across the plasma membrane and the subsequent assembly of periplasmic O-antigen and ECA Und-PP-linked polymers depend on interactions among Wzx, Wzz, and Wzy, which presumably form a multiprotein complex.
Transmembrane flipping of isoprenoid lipid-linked glycans is a common theme in the synthesis of glycoproteins and cell surface polysaccharides in prokaryotes, as well as in the synthesis of the glycan component of glycoproteins in eukaryotic cells (7, 8, 20, 21, 47). A family of bacterial and eukaryotic integral membrane proteins (referred to as Wzx and Rft1, respectively) is implicated in mediating the transbilayer movement of isoprenoid lipid-linked glycans (16, 21, 27, 41, 47). Rft1 is involved in the translocation of a dolichol-pyrophosphoryl (PP)-linked heptasaccharide across the membrane into the lumen of the endoplasmic reticulum (20). Wzx proteins, on the other hand, are found in bacterial systems that use a Wzy (polymerase)-dependent pathway for the assembly of O antigen and certain capsular polysaccharides (for recent reviews see references 39 and 47).
Lipopolysaccharide (LPS), a major component of the gram-negative bacterial outer membrane (38), consists of lipid A, core oligosaccharide, and, in some microorganisms, an O-specific polysaccharide (or O antigen) (39, 47). The core oligosaccharide is assembled on preformed lipid A while the O antigen is independently synthesized as a glycan-PP-undecaprenyl (Und)-linked intermediate. The O antigen is subsequently ligated onto the outer core domain of the lipid A-core oligosaccharide acceptor, with the concomitant release of Und-PP (47), a function attributed to the membrane protein WaaL (24, 39). Und-P is also the lipid carrier of cell surface glycan intermediates for the synthesis of enterobacterial common antigen (ECA) and cell wall peptidoglycan (42, 50).
The O-antigen assembly occurs by mechanisms referred to as Wzy (polymerase)-dependent and ATP-binding cassette (ABC)-dependent pathways, respectively. In the former, individual O-repeating subunits are synthesized on the cytosolic side of the plasma membrane. The repeating O units are subsequently translocated across the membrane by a mechanism that appears not to use ATP hydrolysis and is mediated by Wzx (formerly RfbX) (27, 39, 41, 47). On the periplasmic side of the plasma membrane, the translocated subunits polymerize to a certain length by the concerted functions of Wzy (O-antigen polymerase) and Wzz (O-antigen chain regulator). Finally, the polysaccharide is ligated “en bloc” to the lipid A-core oligosaccharide (30, 34, 37). The Wzy-dependent pathway coordinates the synthesis of many O antigens, especially those with repeating units made of different sugars (heteropolymeric O antigens) (25).
In the ABC-dependent pathways, the complete polymeric O antigen is formed on the cytoplasmic side of the inner membrane (for recent reviews, see references 39 and 47). Polymer export across the membrane needs an ABC transporter (5). The proteins Wzm and Wzt function as the permease and ATPase component of the ABC transporter, respectively (25, 39). A similar pathway is also found in group 2 and 3 exopolysaccharide capsules (53).
In contrast to the ABC transport pathway for O-antigen synthesis, no obvious ABC transporters have been identified in Wzy-dependent systems. Wzx-like proteins are also encoded in the biosynthetic gene clusters for some exopolysaccharides, such as colanic acid, and in the ECA biosynthesis cluster (3, 40). Apart from a loosely conserved region of approximately 208 amino acids (Polysacc_synt domain, Protein families database of alignments and HMMs; http://pfam.wustl.edu/cgi-bin/getdesc?acc=PF01943), protein alignments show relatively low conservation in the primary amino acid sequences of Wzx proteins. The low conservation among Wzx proteins and the absence of any characteristic motifs contrast with the general function they perform.
Unfortunately, rapid progress to uncover the mechanism of Wzx-mediated translocation of Und-PP-linked saccharides is hampered by a number of limitations. First, Wzx proteins are challenging to express in sufficient quantities for purification and structural analyses. Second, the functional analysis of the translocation process in vitro, which requires isoprenoid lipid phosphates representing a small fraction of the total membrane lipid content, has only been successful with soluble isoprenoid analogues (41, 43). From these studies, it has been proposed that Wzx facilitates the diffusion of undecaprenyl-bound O subunits across the plasma membrane (41). However, the isoprenoid analogues have different biophysical properties than the natural lipid carrier Und-P. An alternative model has recently been proposed by Zhou et al. (55). These authors have found that peptide-isoprenoid phosphate interactions can alter the membrane lipid bilayer by forming a tethered structure with the potential to create a membrane channel. Third, the in vitro reconstitution of the translocation using defined components suffers from several shortcomings and, in particular, the need for multiple enzymatic activities at both sides of the membrane, whose integrity cannot likely be preserved upon bacterial cell disruption.
In our laboratories, we have used genetic approaches to uncover the properties of the translocation process in an attempt to understand the basis for the relaxed specificity of the translocases and to determine whether they function in isolation or require additional components of the glycan assembly machinery. Previous work has elucidated common functional aspects of Wzx and other translocases, including the eukaryotic translocase Rft1 (21). We have also shown that Wzx proteins do not require a complete oligosaccharide unit to carry out their function, and, indeed, they can translocate a single sugar bound to Und-PP (16). Also, different Wzx proteins from various O-antigen systems that utilize N-acetylglucosamine (GlcNAc) or N-acetylgalactosamine (GalNAc) as the first sugar of the Und-PP-linked subunit could complement a wzx deletion in Escherichia coli K-12 (32). However, we could not establish whether this relaxed specificity was due to a possible interaction of these proteins with a common first enzyme in the biosynthesis pathway (WecA) or to their ability to recognize the structure of a common first sugar bound to Und-PP (GlcNAc or GalNAc) or both.
E. coli and other enteric bacteria also produce ECA, a cell surface glycolipid that resembles a Wzy-dependent O antigen in its mechanism of synthesis (23). ECA is made of Und-PP-linked trisaccharide subunits composed of GlcNAc, N-acetyl-mannosaminuronic acid (ManNAcA), and N-acetylfucosamine (Fuc4NAc) (29). A dedicated wec (formerly rfe/rff) gene cluster (42) encodes the enzymes required for the ECA synthesis and assembly, including WzxE, WzyE, and WzzE. An O16 LPS-defective phenotype in strain CLM17 (ΔwzxO16), which lacks the WzxO16 translocase but carries a functional WzxE (32), suggests that the two proteins are not functionally interchangeable. In this study, we provide data demonstrating that WzxE can complement the phenotype of a ΔwzxO16 mutant if the majority of the wec gene cluster is deleted. We also show that reconstituting the expression of either the WzyE polymerase or the WzzE length regulator proteins dramatically reduces the complementing activity of WzxE. Furthermore, we demonstrate that this property also applies to other O-antigen translocases, since WzxO16 and WzxO7 can cross-complement translocase defects in the O16 and O7 antigen clusters as long as the corresponding Wzz and Wzy proteins are not coexpressed. We believe these experiments provide new genetic evidence to support the notion that Wzx, Wzz, and Wzy form a complex compartmentalizing the translocation of O antigen or ECA subunits across the plasma membrane.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
Strains and plasmids used in this study are described in Table 1. Bacteria were cultured in Luria broth (LB) supplemented with antibiotics at the following final concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 30 μg ml−1; kanamycin, 40 μg ml−1; spectinomycin, 80 μg ml−1; tetracycline, 20 μg ml−1; and trimethoprim, 50 μg ml−1. Chemicals and antibiotics were purchased from Sigma Aldrich, St. Louis, Mo., and Roche Diagnostics, Laval, Quebec. Oligonucleotide primers were purchased from Invitrogen Canada, Inc., and are listed in Table 2. Plasmids were introduced into electrocompetent cells by electroporation (13).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| E. coli Strain | ||
| DH5α | F− φ80lacZΔM15endA recA hsdR(rK−mK−) supE thi gyrArelA1Δ(lacZYA-argF)U169 | Laboratory stock |
| W3110 | rph-1 IN(rrnD-rrnE) | Laboratory stock |
| CLM17 | W3110, ΔwzxO16 | 32 |
| CLM24 | W3110, ΔwaaL | 17 |
| CLM35 | W3110, ΔwecAΔwzxO16 | This work |
| CLM37 | W3110, ΔwecA | 26 |
| CLM43 | W3110, ΔwzxE | This work |
| CLM45 | W3110, ΔwzxO16ΔwecAΔwzxE::Km(aph); Kmr | This work |
| CLM60 | W3110, ΔgmhA::Km(aph); Kmr | This work |
| CLM67 | W3110, ΔwzxO16 ΔECA | This work |
| EVV11 | W3110, ΔwzyO16 | 52 |
| EVV16 | W3110, ΔwzzO16::Km(aph), Kmr | 51 |
| EVV33 | EVV16, ΔwzzO16 | This work |
| Plasmids | ||
| PBAD24 | Cloning vector inducible with arabinose; Apr | 19 |
| pCE2 | wzxE cloned into pBADMycHis; Apr | 1 |
| pCM223 | wzxO16 cloned into pBAD24; Apr | 32 |
| pCM238 | wzxE cloned into pBAD24; Apr | This work |
| pCM241 | wzzE cloned into pSCRhaB3; Tpr | This work |
| pCM242 | wzyE cloned into pSCRhaB3; Tpr | This work |
| pCP20 | FLP+, λ cI857+, λ pR Repts, Apr Cmr | 12 |
| pEV6 | wzzO16 cloned into pBAD24; Apr | 51 |
| pEV7 | wzyO16 cloned into pBAD24; Apr | 51 |
| pEXT21 | Low-copy-number expression cloning vector; pSa replicon; Spr | 14 |
| pJHCV32 | Cosmid clone O7+; Tcr | 48 |
| pJHCV32::Tn3HoHo1-128 | wzx::Tn3HoHo1-128; Apr Tcr | 31 |
| pJV9 | wzxC cloned into pBADNTF; Apr | 32 |
| pKD4 | Template plasmid for mutagenesis; Apr Kmr | 12 |
| pKD46 | γ, β, and exo from λ phage; araC-ParaB; Apr | 12 |
| pKV1 | wecAFLAG/His cloned into pBAD24; Apr | K. Vigeant |
| pLDT36 | wecAFLAG/His from pKV1 cloned into pME6000; Tcr | This work |
| pMAV11 | wecA cloned into pMAV3; Cmr | 2 |
| pME6000 | Low-copy no. cloning vector; Tcr | S. Hebb |
| pMF19 | wbbLO16 cloned into pEXT21; Spr | 16 |
| pMF20 | wzxO16 cloned into pEXT21; Spr | 16 |
| pMF21 | wzxO7 cloned into pEXT21; Spr | 16 |
| pMF25 | wzyO7 cloned into pEXT21; Spr | 16 |
| pPR1474 | wbbLO16 cloned into pBR322; Apr | 28 |
| pSCRhaB3 | pSCRhaB2 containing a FLAG tag in the multiple cloning site; Tpr | 9 |
Ap, ampicillin; Sp, spectinomycin; Tc, tetracycline; Km, kanamycin; aph, aminoglycoside phosphotransferase; Tp, trimethoprim.
TABLE 2.
Primers used for mutagenesis experiments and for plasmid constructions
| Primer | Sequencea | Targeted geneb |
|---|---|---|
| Mutagenesis | ||
| 746 | 5′-AATTTACTGACAGTGAGTACTGATCTCATCAGTATTTTTGTGTAGGCTGGAGCTGCTTCG | wecA |
| 804 | 5′-CTGCATTTTGTCTATTACATTTATGCTGAAGGATATCCTCTGTGTAGGCTGGAGCTGCTT | gmhA |
| 805 | 5′-CCGGATGCGGCGTAAACGTCTTATCCGGCCTACGCCAGACCATATGAATATCCTCCTTAG | gmhA |
| 888 | 5′-GGTAATTGCGACTTTGTTGAACTACTTTTCCTGATATGTGTAGGCTGGAGCTGCTTCG | wzxE |
| 889 | 5′-GGGATATCCGATCCCAGTACGTGAATCAGTACAGTCATATGAATATCCTCCTTAG | wzxE |
| 1279 | 5′-GCGACGGAGTGACCACTCCGTCGCTTTACAAAGAGAGGACATATGAATATCCTCCTTAG | wecG |
| 1414 | 5′-CAAAATCATCGCTCCGGACGCAGGTTGAAGGATAACAATGTGTGTAGGCTGGAGCTGCTTC | wecG |
| Cloning | ||
| 890 | 5′-GATCTCTACAGTCATGCCCGCCTACGCC | wzxE |
| 966 | 5′-CTGATATGTCGTTGGCAAAAGCG | wzxE |
| 1732 | 5′-GACTCATATGACACAACCAATGCCT (NdeI) | wzzE |
| 1733 | 5′-CGCATCTAGATTTCGAGCAACGGCG (XbaI) | wzzE |
| 1734 | 5′-GACTCATATGAGTCTGCTGCAATTCAGT (NdeI) | wzyE |
| 1735 | 5′-GACTTCTAGATCCTTCAACCTGCGTCCG (XbaI) | wzyE |
Underlined nucleotides indicate the sequence homologous to pKD4; underlined and italicized nucleotides indicate the restriction endonuclease sites (shown in parenthesis) incorporated into the primer sequence.
The designations indicate the genes targeted for deletion or for cloning.
E. coli W3110 DNA was used as template to generate PCR products for plasmid constructions, and the sequences of the specific oligonucleotide primers used are indicated in Table 2. In all cases, PCR fragments were gel purified using a Gel Extraction kit from QIAGEN and subsequently ligated using a Rapid Ligation kit from Roche Diagnostics before the ligation mix was transformed into DH5α. Plasmid pCM238 was constructed by amplification of the wzxE gene with primers 890 and 966, followed by treatment of the amplicon with T4 kinase. This fragment was ligated into pBAD24, which was digested with SmaI. Plasmid pCM241 was constructed by amplification of the wzzE gene with primers 1732 and 1733, followed by digestion of the amplicon with NdeI and XbaI and ligation into pSCRhaB3, which was also digested with the same restriction enzymes. An identical approach was followed to construct pCM242, except that amplification of the wzyE fragment was done with primers 1734 and 1735. Plasmid pLDT36 was constructed by cloning a 2.6-kb fragment from pKV1, which encodes wecAFLAG-His under the control of the pBAD and the araC regulator. This fragment was digested with ClaI and Asp700 and ligated to the low-copy-number vector pME6000, which was first digested with XhoI, followed by treatment with the Klenow DNA polymerase to fill the ends and a subsequent digestion with ClaI. The details of the construction of pKV1 will be reported elsewhere.
Construction of deletion mutants.
Deletions in chromosomal genes were performed as described by Datsenko and Wanner (12). We generated primers of 40 to 45 nucleotides corresponding to regions adjacent to the gene targeted for deletion (Table 2). The primers also contained 20 additional nucleotides that annealed to the template DNA from plasmid pKD4, which carries a kanamycin-resistance gene (aph) flanked by FLP recognition target sites. Competent cells were prepared by growing E. coli strains carrying pKD46 in LB containing 0.5% (wt/vol) arabinose, and the PCR products were introduced by electroporation. The plasmid pKD46 encodes the red recombinase of the λ phage, which was placed under the control of the arabinose-inducible promoter pBAD. Kanamycin-resistant colonies were screened by PCR using primers annealing to regions outside of the mutated gene. The target gene in the deletion mutants, denoted as Δxxx::Km (where xxx refers to any given targeted gene), is replaced by the aph gene fragment encoding kanamycin resistance. To obtain an unmarked deletion of the target gene, the aph gene fragment was excised by introducing the plasmid pCP20 encoding the FLP recombinase. Plasmids pKD46 and pCP20 are both thermosensitive for replication and they were cured at 42°C.
LPS and ECA analysis.
LPS was prepared as previously described (33), and samples were separated on 14% Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. The gels were stained with silver nitrate as described previously (33) or transferred to nitrocellulose membranes for immunoblot analysis. The membranes were reacted with either O16- (The Gastroenteric Disease Center, Wiley Laboratory, University Park, Pennsylvania) or O7-specific (49) polyclonal rabbit antibodies, and the reacting bands were detected by fluorescence with an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, Neb.) using IRDye800CW affinity-purified anti-rabbit immunoglobulin G (IgG) antibodies (Rockland, Pa.). Densitometry analysis of the gels was performed using Odyssey software (Li-Cor Biosciences) as described before (51).
For the analysis of ECA, membrane fractions were prepared as previously described (3), and protein concentration was determined using the Bradford assay (Bio-Rad). Equal amounts of membranes were boiled for 10 min and then incubated overnight at 60°C with proteinase K (Roche Diagnostics, Laval, Quebec) to a final concentration of 1.6 μg/ml. Membranes were boiled again for 10 min, and sample buffer was added. ECA samples were separated on 14% Tricine-SDS-PAGE gels and transferred into nitrocellulose membranes. Blots were reacted with anti-O14 polyclonal antiserum (Staten's Serum Institut, Copenhagen, Denmark) and anti-LPS core monoclonal antibodies (HyCult Biotechnologies b.v., The Netherlands). Specific bands were detected by fluorescence using IRDye800CW affinity-purified anti-rabbit IgG antibodies (Rockland, Pennsylvania) and Alexa Fluor 680 anti-mouse IgG antibodies (Molecular Probes, Portland, Oregon) in the Odyssey infrared imaging system.
Growth curves.
The growth rate of wzxO16 and wzxE mutants was determined in the presence of arabinose and glucose by following the turbidity of the bacterial cultures in LB. Overnight cultures without sugars were diluted to an optical density at 600 nm of 0.1, the sugars were added, and growth was monitored every 30 min with a Klett-Summerson photoelectric colorimeter (Bel-Art, New Jersey).
RESULTS
WzxE cannot complement the synthesis of O16 LPS in a strain lacking the O16 antigen translocase.
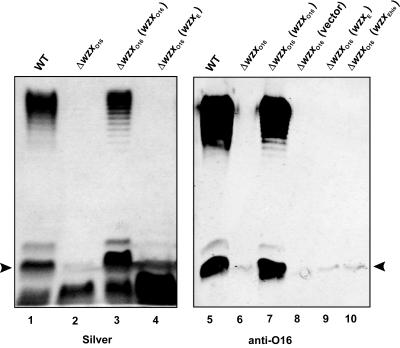
Previously (32), we showed that heterologous Wzx proteins can substitute for WzxO16 to mediate the translocation of O16 subunits. The complementation of O16 LPS production was particularly strong by Wzx proteins from strains with O antigens that utilize GlcNAc or GalNAc as the first sugar bound to the Und-PP-O subunit intermediate. Since WzxE is implicated in the translocation of a Fuc4NAc-ManNAc-GlcNAc-PP-Und intermediate for the assembly of ECA (23, 41), we investigated whether WzxE can also substitute for WzxO16. The O16 LPS synthesis in the E. coli K-12 strain W3110 and its derivatives was reconstituted by transformation with pMF19, a plasmid encoding the rhamnosyltransferase WbbL (16, 32). In contrast to the parental strain W3110(pMF19) (Fig. 1, lane 1), W3110ΔwzxO16 containing pMF19 did not form a full-length O side chain (Fig. 1, lane 2) but produced a small amount of a slow-migrating band in the region corresponding to one O16-specific subunit attached to the core lipid A (Fig. 1, arrowhead), as confirmed with anti-O16 rabbit antiserum (Fig. 1, lane 6). The partial restoration of O16 LPS production in the ΔwzxO16 background could be attributed to the chromosomal copy of wzxE. However, normal production of O16 LPS by W3110ΔwzxO16(pMF19) was not corrected with plasmids pCM238 and pCE2, both encoding wzxE under the control of an arabinose-inducible promoter (Fig. 1, lanes 4, 9, and 10), while O16 LPS production was fully restored by pCM223 encoding wzxO16 (Fig. 1, lanes 3 and 7). Lack of complementation was not due to the absence of WzxE protein expression since cells containing pCE2, which encodes a His6-tagged version of WzxE, produced a polypeptide of the expected mass that reacted with an anti-His6 monoclonal antibody (data not shown). Also, as expected, no O16 LPS was produced with the vector control (Fig. 1, lane 8).Together, these experiments demonstrated that WzxE could not fully restore the production of O16 LPS in a ΔwzxO16 background. Therefore, WzxE appears to be different from other Wzx proteins that function in systems where GlcNAc is the first sugar bound to the Und-PP-O subunit.
FIG. 1.
O16 LPS production in a ΔwzxO16 mutant of strain W3110. LPS was prepared from cultures induced with 0.2% arabinose. Samples were separated on a 14% Tricine-SDS-PAGE gel, and the gel was stained with silver nitrate or transferred into nitrocellulose membrane. Gel loading was normalized by densitometry. The blot was reacted with polyclonal anti-O16 antiserum, and O-antigen-specific bands were detected with fluorescence using IRDye800CW affinity-purified anti-rabbit IgG antibodies. Arrowheads indicate the region corresponding to the migration of one O-antigen unit attached to lipid A-core oligosaccharide. Lanes 1 and 5, W3110(pMF19); lanes 2 and 6, CLM17(pMF19); lanes 3 and 7, CLM17(pMF19, pCM223); lanes 4 and 9, CLM17(pMF19, pCM238); lane 8, CLM17(pMF19, pBAD24); lane 10, CLM17(pMF19, pCE2).
Deletion of the wzxE gene does not affect ECA production.
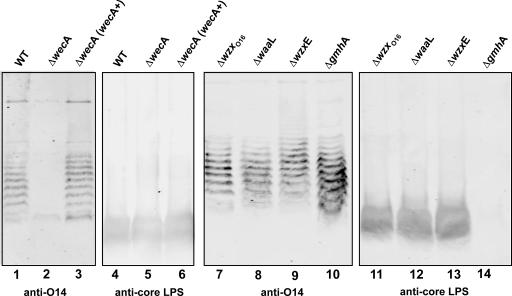
To assess the function of WzxE in more detail, we investigated ECA production by the parental strain W3110 and various isogenic mutant derivatives. Proteinase K-treated total membranes from W3110 produced a ladder-like banding material that reacted with the O14:K7-specific antiserum (Fig. 2, lane 1). E. coli O14:K7 is a strain that lacks a typical O antigen, in which the ECA is attached to the lipid A core (22, 44). Therefore, we concluded that the anti-O14 reactive bands corresponded to ECA polymers of various lengths, as described by Barr et al. (3). As a negative control, we used the strain W3110ΔwecA (CLM37) with a deletion of the wecA gene (Fig. 3). W3110ΔwecA did not produce ECA bands unless a functional wecA was provided in trans by the plasmid pMAV11 (Fig. 2, lanes 2 and 3, respectively). To confirm that the ECA is not associated to core lipid A, we conducted additional control experiments with strains W3110ΔwaaL (CLM24) and W3110ΔgmhA::Km (CLM60). W3110ΔwaaL carries a deletion of the waaL gene, which encodes the O-antigen ligase. No differences in the ECA banding profile were produced by W3110ΔwaaL (Fig. 2, lane 8), which is identical to the profile seen in the other strains that contain a functional waaL gene (Fig. 2, lanes 1, 3, 7, 9, and 10). In the strain W3110ΔgmhA::Km, a kanamycin resistance fragment replaces the gmhA gene, which encodes the first enzyme involved in the biosynthesis of ADP-heptose (6).W3110ΔgmhA::Km produces a truncated core oligosaccharide missing the acceptor site for the ligation of O polysaccharides (data not shown) but still produced ECA (Fig. 2, lane 10). Furthermore, only the bands corresponding to a complete lipid A-core region reacted with an anti-core LPS monoclonal antibody (Fig. 2, lanes 4 to 6 and lanes 11 to 13). As expected, the truncated lipid A-core oligosaccharide produced by the W3110ΔgmhA::Km strain did not react with the anti-core LPS monoclonal antibody (Fig. 2, lane 14), which does not recognize a core oligosaccharide lacking heptose. In conclusion, the combined results of these experiments support that the ECA polymers detected are not linked to lipid A-core and correspond to the phosphoglyceride-linked ECA (15). Strain W3110ΔwzxE also expressed ECA (Fig. 2, lane 9), suggesting that another cellular protein, possibly WzxO16, can compensate for the loss of WzxE.
FIG. 2.
Expression of ECA in W3110 wild-type and mutant strains. ECA extracts from total membranes were separated on a 14% Tricine-SDS-PAGE gel and transferred into nitrocellulose membrane. Gel loading was normalized by protein concentration. The blots were reacted simultaneously with polyclonal O14-specific antiserum and a monoclonal anti-LPS core antibody. The specific bands were detected with fluorescence using Alexa Fluor 680 anti-mouse IgG antibodies and IRDye800CW affinity-purified anti-rabbit IgG antibodies in the Odyssey infrared imaging system. Lanes 1 and 4, W3110; lanes 2 and 5, CLM37; lanes 3 and 6, CLM37(pMAV11); lanes 7 and 11, CLM17; lanes 8 and 12, CLM24; lanes 9 and 13, CLM43; lanes 10 and 14, CLM60.
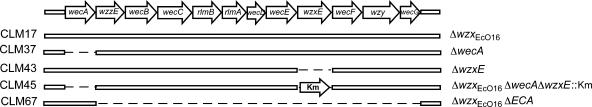
FIG. 3.
Gene organization and structure of the wec cluster of E. coli K12/O16. The wec genes are as follows: wecA (UDP-N-acetylglucosamine transferase), wzzE (chain length regulator), wecB (UDP-N-acetylglucosamine epimerase), wecC (UDP-N-acetylmannosamine dehydrogenase), rlmB (TDP-glucose 4,6-dehydratase), rlmA (glucose-1-phosphate thymidylyltransferase), wecD (fucosamine acetyltransferase), wecE (TDP-4-oxo-6-deoxy-d-glucose transaminase), wzxE (ECA translocase), wecF (UDP-N-acetylfucosamine transferase), wzy (ECA polymerase), and wecG (UDP-N-acetylmannosaminuronic acid transferase). Km, kanamycin. Dashed lines indicate regions of the chromosome that have been deleted in the various deletion mutants of W3110. The strain designations and the relevant genotypes are indicated on the left and on the right of the figure, respectively.
WzxE can complement the synthesis of O16 LPS in the absence of the ECA cluster.
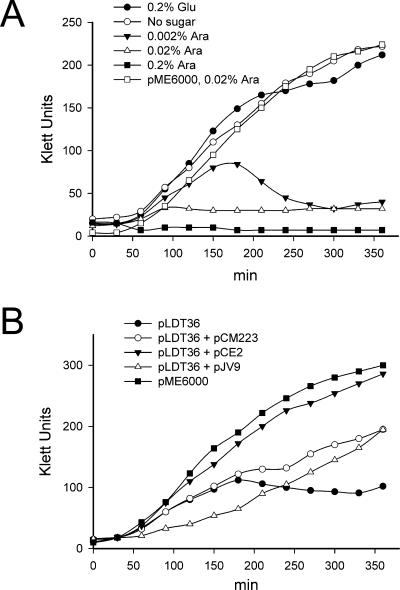
To understand better the different functional roles of WzxO16 and WzxE, a double wzxE and wzxO16 deletion was sought, but repeated attempts to obtain this mutant failed. In contrast, a double mutant in a background lacking the wecA gene (W3110ΔwecAΔwzx) was readily constructed, resulting in strain W3110ΔwecAΔwzxΔwzxE::Km (Fig. 3, CLM45). Previous work by Rick et al. (41) has demonstrated that the accumulation of ECA lipid-linked intermediates in an E. coli mutant with wecA::Tn10 and a large chromosomal deletion that eliminates wzxO16 and wzxC (encoding the translocase for the colanic acid capsule) was associated with a lethal phenotype in the presence of wecA gene expression. WecA is the UDP-GlcNAc:Und-P GlcNAc-1-P transferase responsible for the initiation of ECA (35) and O16 synthesis (46, 54). Therefore, since strain W3110 cannot produce O16 LPS but produces ECA, we concluded that, as previously shown by Rick et al. (41), a functional WecA protein without the WzxE and WzxO16 translocases causes a toxic accumulation of Fuc4NAc-ManNAc-GlcNAc-PP-Und intermediates on the cytosolic face of the plasma membrane. To validate this hypothesis we cloned a 2.6-kb fragment from pKV1 (Table 1), containing araC and the wecA gene under the control of the arabinose-inducible pBAD promoter, into the low-copy-number vector pME6000, generating plasmid pLDT36 (Table 1). The growth rate of the strain W3110ΔwecAΔwzxΔwzxE::Km containing pLDT36 with glucose or without any sugar in the medium was comparable to that of strain W3110ΔwecAΔwzxΔwzxE::Km containing the vector pME6000 (Fig. 4A). In contrast, W3110ΔwecAΔwzxΔwzxE::Km with pLDT36 did not grow with arabinose at concentrations of 0.2% and 0.02%. The culture grew slowly only at an arabinose concentration of 0.002%, although cell lysis occurred when cells reached the mid-logarithmic phase. Under phase-contrast microscopy, W3110ΔwecAΔwzxΔwzxE::Km(pLDT36) cells grown with arabinose looked longer and broader than those grown with glucose, and bacterial lysis was suggested by abundant cellular debris in the preparations (data not shown).
FIG. 4.
Growth rate curves for ΔwzxO16 and ΔwzxE mutants. Bacterial cultures were grown overnight in LB and then diluted to an optical density at 600 nm of 0.1. At this point, sugars (arabinose or glucose) were added, and the turbidity of each culture was determined with a Klett-Summerson photocolorimeter at 30-min intervals. (A) Growth rate of CLM45 (W3110ΔwzxO16ΔwecAΔwzxE::Km) containing pLDT36 at various concentrations of arabinose. (B) Growth rate of CLM45 (W3110ΔwzxO16ΔwecAΔwzxE::Km) complemented with pLDT36 (wecA+), pLDT36 and pCM223 (wzxO16+), pLDT36 and pCE2 (wzxE+), and pLDT36 and pJV9 (wzxC+) in the presence of 0.002% arabinose.
W3110ΔwecAΔwzxΔwzxE::Km containing pLDT36 provided us with a way to investigate whether the expression of WzxO16 or WzxE can rescue the growth-deficient phenotype of this strain. We transformed W3110ΔwecAΔwzxΔwzxE::Km(pLDT36)with pCM223 (encoding wzxO16) or pCE2 (encoding wzxE) and assessed the growth rate of the transformants in 0.002% arabinose. Full restoration of growth was observed with pCE2 (Fig. 4B), while W3110ΔwecAΔwzxΔwzxE::Km with both pLDT36 and CM223 grew at a slower rate (Fig. 4B). Since W3110ΔwecAΔwzxΔwzxE::Km containing only pCM223 grew normally (data not shown), we concluded that the inability to rescue the growth phenotype to the same rate as pCE2 was not due to an artifact caused by the overproduction of WzxO16 but, rather, by the accumulation in the plasma membrane of Fuc4NAc-ManNAc-GlcNAc-PP-Und that is not efficiently translocated by this protein. We also investigated whether WzxC, the translocase for the colanic acid capsule, can rescue the growth defect in W3110ΔwecAΔwzxΔwzxE::Km(pLDT36).The growth of this strain in the presence of pJV9 (encoding wzxC) was comparable to that found in the presence of wzxO16 (Fig. 4B). The combined results of the experiments presented in this part of our study demonstrated that translocases from the O16 antigen and colanic acid can substitute for WzxE to rescue the growth defect of a wzxE wzxO16 double deletion mutant with a functional WecA, although the growth rate of the mutant in these cases is somewhat slower than with WzxE. These results also explained why the ΔwzxE strain could still form ECA (Fig. 2, lane 9) and are in agreement with recent work by Kajimura et al. (23) showing that production of WzxO16 and WzxC can complement the synthesis of a cyclic form of ECA in the absence of wzxE expression.
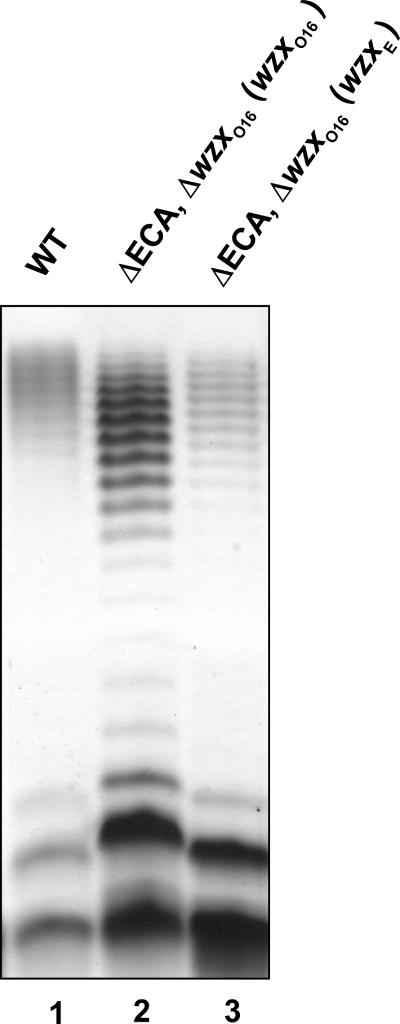
To reexamine the functionality of WzxE in the translocation of O16 antigen, we constructed the strain W3110ΔwzxO16ΔECA (CLM67), which lacks all the genes for ECA biosynthesis except wecA (Fig. 3) and does not produce ECA, as determined by anti-O14 immunoblotting (data not shown). As expected, transforming pCM223 (encoding wzxO16) and pMF19 into W3110ΔwzxO16ΔECA restored O16 production (Fig. 5, lane 2). However, transformation with pCE2 and pMF19 also restored O16 production, albeit at a lower level than with pCM223 (Fig. 5, lane 3). This result contrasted with the lack of complementation of O16 LPS production, previously found with pCE2 in W3100ΔwzxO16 (Fig. 1, lane 10). Since the only difference between these strains is the deletion of the majority of the wec cluster, we conclude that one or more wec gene products prevented WzxE from rescuing O16 LPS synthesis.
FIG. 5.
Expression of LPS in ΔwzxO16 and ΔECA mutants. LPS was prepared from cultures induced with 0.2% arabinose. Samples were separated on a 14% Tricine-SDS-PAGE gel, and the gel was stained with silver nitrate. Gel loading was normalized by densitometry. Lane 1, W3110(pMF19); lane 2, CLM67(pMF19, pCM223); lane 3, CLM67(pMF19, pCE2).
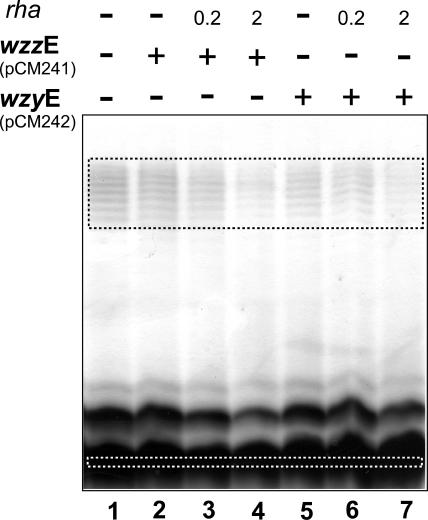
Both O16 and ECA need Wzx, Wzy, and Wzz proteins for their assembly (39, 47). In the strain W3110, the O16 antigen and ECA systems have their corresponding sets of wzy and wzz genes. Thus, we investigated the possibility that WzzE and WzyE could influence the functionality of WzxE. We first constructed the plasmids pCM241 and pCM242, which encode wzzE and wzyE, respectively, under the control of a rhamnose-inducible promoter in a low-copy-number vector (Table 1, pSCRhaB3,). Each of these plasmids was individually transformed into the W3100ΔwzxO16ΔECA mutant also containing pMF19 and pCE2. Cultures were grown with 0.005% arabinose (for WzxE expression) and various rhamnose concentrations to induce the expression of WzzE or WzyE. The amount of O16 LPS was reduced when either WzzE (lane 4) or WzyE (lane 7) was expressed with 2% rhamnose (Fig. 6). Densitometric comparisons of the amount of O antigen relative to the amount of lipid A-core indicated a threefold reduction in O16 antigen levels (Fig. 6, lanes 4 and 7), compared to the parental strain (Fig. 6, lane 1). The reduced O16 LPS production was not due to the overexpression of WzzE or WzyE alone, since no changes were observed in the O16 LPS made by W3110 with pMF19 and either pCM241 or pCM242 (data not shown). Also, expression of Wzy and Wzz did not alter the banding pattern of O16 LPS, which displayed the same modality as seen in the parental strain. Therefore, we concluded from these experiments that the reconstitution of WzzE or WzyE protein expression in strain W3110ΔwzxO16ΔECA containing pMF19 and pCE2 is associated with a reduced O16 LPS-complementing activity of WzxE. Furthermore, these experiments suggested an explanation to our original observation that WzxE cannot complement O16 LPS production in a ΔwzxO16 genetic background, even though the O16 LPS and ECA systems share a GlcNAc residue in the first position of the unit bound to Und-PP.
FIG. 6.
Expression of LPS in ΔwzxEcO16 and ΔECA mutants transformed with plasmids encoding wzzE and wzyE. LPS was prepared from strain CLM67 (ΔECAΔwzxO16) containing pMF19 and pCE2 (wzxE+) alone and also with either pCM241 (wzzE+) or pCM242 (wzyE+). Gene expression was induced with 0.005% arabinose and 0.2% or 2% rhamnose (rha). Samples were separated on a 14% Tricine-SDS-PAGE gel, and the gel was stained with silver nitrate. Gel loading was normalized by densitometry. In addition, the relative amounts of polymerized O antigen and core-lipid A in each of the lanes were calculated by densitometric analysis of the regions indicated by the dotted squares. The pixels of the core-lipid A band was divided by the total pixels of the O ladder region, and the results were compared to the values obtained for the parental strain in lane 1.
Wzx translocases function in the context of their corresponding Wzy and Wzz proteins.
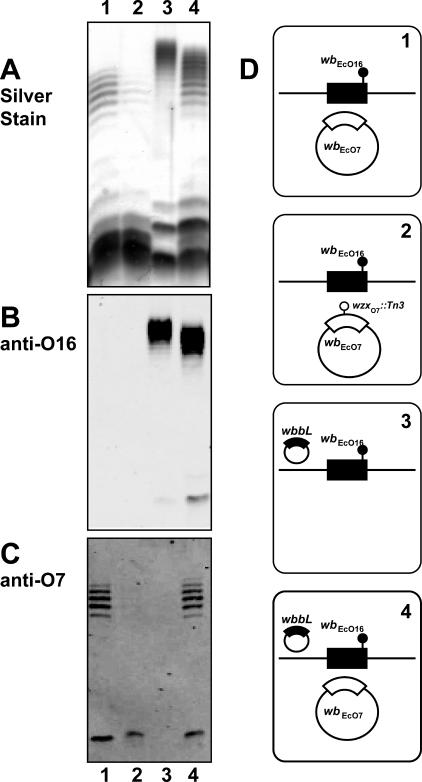
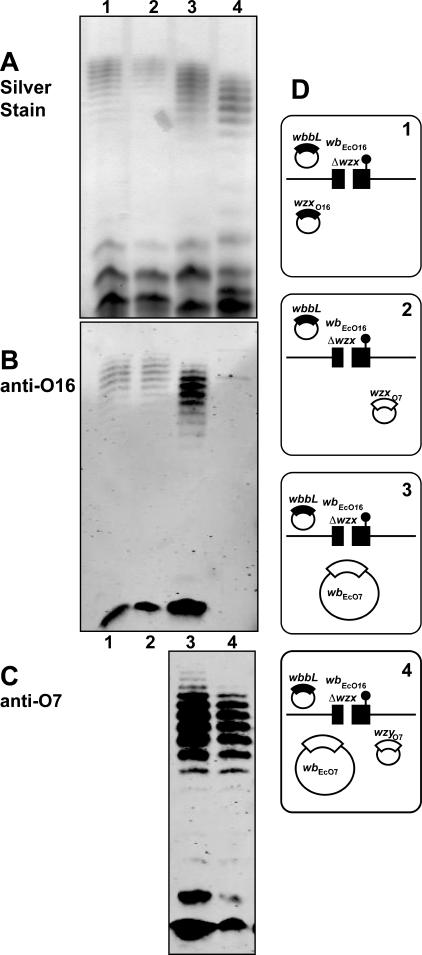
The ability of WzxE to substitute for WzxO16 only without WzyE and WzzE expression suggests a more general notion that the Wzx translocases may interact preferentially with their corresponding Wzy and Wzz proteins. To investigate this hypothesis we coexpressed the O16 and O7 antigen clusters in strain W3110. These two O antigens have different O units with only the first GlcNAc residue in common (16, 32). Production of the O7 antigen was achieved with plasmid pJHCV32 (48). Figure 7, lane 4 (panel A) shows that the simultaneous coproduction of O16 and O7 LPS in W3110 with pJHCV32 and pMF19 causes a combined banding pattern by silver staining. However, immunodetection of each individual O antigen demonstrated their characteristic banding patterns (Fig. 7, lane 4, panels B and C). As controls, Fig. 7 also shows the O7-specific (lane 1, panels A and C) and the O16-specific (lane 3, panels A and B) banding patterns when each of the O antigens is independently produced. The reduced average length of the O16 polysaccharide in W3110 containing pJHCV32 and pMF19 (Fig. 7, lane 4) relative to that of O16 in W3110 containing only pMF19 (Fig. 7, lane 3) was reproducible and is probably due to a competition for a stable pool of nucleotide sugar precursors in the strain producing two O antigens.
FIG. 7.
Coproduction of O16- and O7-specific LPS in strain W3110 transformed with various combinations of pJHCV32 (wbEcO7), pJHCV32::Tn3HoHo1-128 (wzxO7::Tn3), and pMF19 (wbbL). LPS fractions were separated on a 14% Tricine-SDS-PAGE gel, and the gel was stained with silver nitrate or transferred into nitrocellulose membranes. Gel loading was normalized by densitometry. Specific bands were immunodetected with fluorescence using IRDye800CW affinity-purified anti-rabbit IgG antibodies in the Odyssey infrared imaging system. (A) Silver stain. (B) Blot reacted with polyclonal anti-O16 rabbit antiserum. (C) Blot reacted with polyclonal anti-O7 rabbit antiserum. (D) Cartoons corresponding to each lane indicate the relevant genetic background of each strain used for LPS analysis. E. coli K-12/O16 chromosomal genes (wbEcO16) are indicated in black. Plasmid-encoded O7 genes are indicated in white. Chromosomal wbbL::IS5 and plasmid wzxO7::Tn3 insertions are also indicated.
Using the O16/O7 LPS coproduction experimental system, we first determined the ability of WzxO16 to complement a wzxO7 defect with WzyO16 and WzzO16 overexpression. Strain W3110(pMF19) was transformed with pJHCV32::Tn3HoHo1-128 (wzxO7::Tn3). The latter plasmid carries a transposon insertion inactivating wzxO7 (31). Therefore, W3110 carrying pJHCV32::Tn3HoHo1-128 (wzxO7::Tn3) produces only a small amount of O7 LPS (Fig. 7, lane 2) detectable by silver staining (panel A) and immunoblotting (panel C), which is due to the complementing activity of WzxO16 (32).
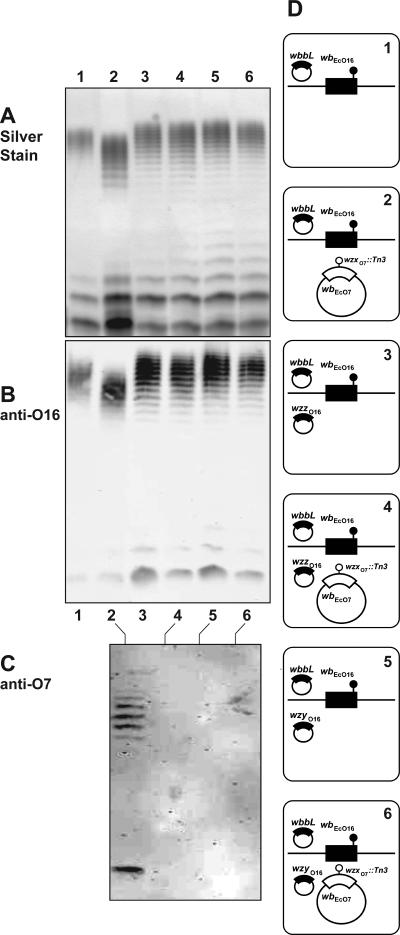
A similar finding was observed in Fig. 8, lane 2 (panels A, B, and C), which shows that this strain produced O16 LPS and also a reduced amount of O7 LPS. However, introduction of either pEV6 (expressing WzzO16) or pEV7 (expressing WzyO16) abolished the production of O7 LPS (Fig. 8, lanes 4 and 6, respectively). The amounts and migration pattern of the O16 LPS were not affected in W3110(pMF19) containing pEV6 (Fig. 8, panels A and B, lanes 3) and pEV7 (Fig. 8, panels A and B, lanes 5), indicating that the overexpression of these proteins did not alter O16 LPS synthesis. Therefore, as shown before with WzxE and its corresponding WzzE and WzyE proteins, WzzO16 and WzyO16 proteins appear to modulate the functionality of WzxO16.
FIG. 8.
Suppression of O7 LPS production by WzxO16 in the presence of WzzO16 and WzyO16 overexpression. W3110 containing pMF19 was transformed with various combinations of pJHCV32::Tn3HoHo1-128 (wzxO7::Tn3), pEV6 (wzzO16), or pEV7 (wzyO16). LPS was prepared from cultures induced with 0.2% arabinose. Samples were separated on a 14%-Tricine SDS-PAGE gel, and the gel was stained with silver nitrate or transferred into a nitrocellulose membrane. Gel loading was normalized by densitometry. Specific bands were immunodetected with fluorescence using IRDye800CW affinity-purified anti-rabbit IgG antibodies in the Odyssey infrared imaging system. (A) Silver stain. (B) Blot reacted with polyclonal anti-O16 rabbit antiserum. (C) Blot reacted with polyclonal anti-O7 rabbit antiserum. (D) Cartoons corresponding to each lane indicate the relevant genetic background of each strain used for LPS analysis. E. coli K-12/O16 chromosomal genes (wbEcO16) and the cloned wzzO16 and wzyO16 genes are indicated in black. Plasmid-encoded O7 genes are indicated in white. Chromosomal wbbL::IS5 and plasmid wzxO7::Tn3 insertions are also indicated.
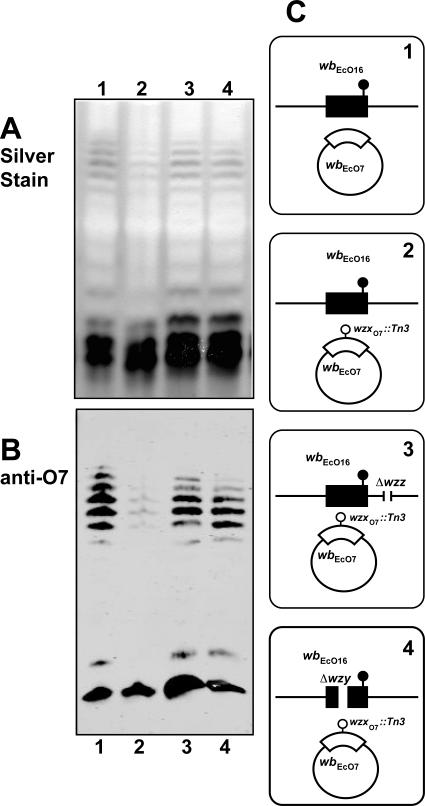
We also examined whether WzyO7 could modulate the ability of WzxO7 to complement a wzxO16 defect. For these experiments, we used the strain W3110ΔwzxO16 (CLM17). As a control, we demonstrated first, as we have previously reported (32), that O16 LPS synthesis in W3110ΔwzxO16 containing pPR1474 was complemented with either WzxO16 (encoded by pMF20) (Fig. 9, lane 1) or WzxO7 (encoded by pMF21) (Fig. 9, lane 2). Also both O16 and O7 LPS were coproduced in W3110ΔwzxO16 carrying pPR1474 and pJHCV32 (Fig. 9, lane 3). In contrast, O16 LPS production was virtually suppressed with pMF25 (Table 1), which encodes wzyO7 (Fig. 9, lane 4). The amounts and migration pattern of the O7 LPS were not affected in W3110ΔwzxO16 containing pJHCV32 and pMF25 (Fig. 9, lane 4, panels A and C), indicating that an excess of WzyO7 had no effect on O7 LPS production. Therefore, we concluded that WzyO7 could also modulate the functionality of WzxO7, as in the case of the O16 and ECA systems.
FIG. 9.
Suppression of O16 LPS production by WzxO7 in the presence of WzyO7 over expression. Strain W3110ΔwzxO16 (CLM17) containing pPR1474 (wbbL) was transformed with various combinations of pJHCV32 (wbEcO7), pMF20 (wzxO16), pMF21 (wzxO7), and pMF25 (wzyO7). LPS was prepared from cultures induced with 0.2% arabinose. Samples were separated on a 14% Tricine-SDS-PAGE gel, and the gel was stained with silver nitrate. LPS fractions were separated on a 14% Tricine-SDS-PAGE gel and transferred into nitrocellulose membrane. Gel loading was normalized by densitometry. Specific bands were immunodetected with fluorescence using IRDye800CW affinity-purified anti-rabbit IgG antibodies in the Odyssey infrared imaging system. (A) Silver stain. (B) Blot reacted with polyclonal anti-O16 rabbit antiserum. (C) Blot reacted with polyclonal anti-O7 rabbit antiserum. (D) Cartoons corresponding to each lane indicate the relevant genetic background of each strain used for LPS analysis. E. coli K12/O16 of each strain used for LPS analysis. E. coli K12/O16 background of each strain used for LPS analysis. E. coli K12/O16 chromosomal genes (wbEcO16) and the cloned wzxO16 are indicated in black. The chromosomal wbbL::IS5 insertion and the deletion of the wzx gene in the E. coli K12/O16 cluster are also indicated. Plasmid-encoded O7 genes (wbEcO7, wzxO7, and wzyO7) are indicated in white.
The combined results of the previous experiments demonstrate interactions of Wzx translocases with their corresponding Wzy and Wzz proteins, suggesting the possibility that these proteins may form a complex. To provide additional support to this idea, we transformed pJHCV32::Tn3HoHo1-128(wzxO7::Tn3) into the parental strain W3110, as well as in its ΔwzzO16 (EVV33) and ΔwzyO16 (EVV11) isogenic derivatives. A small amount of O7-specific LPS was produced in W3110 containing pJHCV32::Tn3HoHo1-128 (Fig. 10, lane 2), compared to W3110 carrying the parental plasmid pJHCV32 (Fig. 10, lane 1). As we have demonstrated previously, the expression of O7 LPS in W3110(pJHCV32::Tn3HoHo1-128) is due to the complementing activity of WzxO16. In contrast, the amount of O7 LPS produced by the ΔwzzO16 and ΔwzyO16 mutants, both carrying pJHCV32::Tn3HoHo1-128 (Fig. 10, lanes 3 and 4, respectively), was comparable to that of W3110 with pJHCV32 (lane 1). Since wzxO16, wzzO16, and wzyO16 are all expressed in W3110, we interpreted these experiments as an indication that in the absence of either WzzO16 or WzyO16 there is more WzxO16 that can presumably interact with WzyO7, WzzO7, or both proteins, which are encoded by pJHCV32::Tn3HoHo1-128, thus effectively increasing the production of O7 LPS.
FIG. 10.
Increased O7 LPS production mediated by WzxO16 in the absence of WzzO16 and WzyO16. Strains W3110 and the deletion mutants ΔwzzO16 (EVV33) and ΔwzyO16 (EVV11) were transformed with pJHCV32 (wbO7) or pJHCV32::Tn3HoHo1-128 (wzxO7::Tn3) as indicated by the plus and minus signs in each specific lane. LPS samples were separated on a 14% Tricine-SDS-PAGE gel, and the gel was stained with silver nitrate or into nitrocellulose membrane. Gel loading was normalized by densitometry. Specific bands were immunodetected with fluorescence using IRDye800CW affinity-purified anti-rabbit IgG antibodies in the Odyssey infrared imaging system. (A) Silver stain. (B) Blot reacted with polyclonal anti-O7 rabbit antiserum. (C) Cartoons corresponding to each lane indicate the relevant genetic background of each strain used for LPS analysis. E. coli K-12/O16 chromosomal genes (wbEcO16) are indicated in black. The chromosomal wbbL::IS5 insertion and the deletion of the wzy gene in the E. coli K-12/O16 cluster are also indicated. Plasmid-encoded O7 genes with or without the wzxO7::Tn3 insertion are indicated in white (wbEcO7).
DISCUSSION
Recently, we discovered that Wzx proteins from systems where N-acetylhexosamines, especially GlcNAc and GalNAc, are in the first position of the lipid-linked intermediate could complement a deletion mutant of wzxO16 and restore O16 LPS synthesis to wild-type levels (32). However, it was puzzling that WzxE, which has similar characteristics to other Wzx proteins, did not complement O16 LPS synthesis. WzxE functions in ECA synthesis, which also employs GlcNAc-PP-isoprenoid intermediates. We undertook the present study to investigate this phenomenon in more detail. Once again, we took advantage of our ability to reconstitute O-antigen synthesis in E. coli K-12/O16 (16, 32), which allowed us to work in an isogenic background under carefully controlled conditions of plasmid expression. We also paid special attention to the quantitative loading of LPS gels, and for this purpose all lanes were loaded with samples that had been previously normalized by densitometry (51). Our results clearly demonstrate that WzxE could not restore the production of full-size O16 LPS in the ΔwzxO16 background and argue against the generality of our previous conclusion that Wzx proteins that can recognize GlcNAc-PP-Und have the ability to complement O16 LPS production in the ΔwzxO16 mutant (32). However, in these experiments the complementing Wzx proteins were expressed in isolation from their corresponding components of each O-antigen system, while the ECA system is intact in the ΔwzxO16 mutant. Therefore, we hypothesized that although Wzx proteins have relaxed specificity for GlcNAc, specific components of each system have an influence in the assembly of LPS O antigen. Given that the initiating enzyme WecA is common to both O16 LPS and ECA systems, we suspected that other membrane proteins from the Wzy-dependent pathway, such as the WzyE polymerase and/or the WzzE polysaccharide chain regulator, could be involved in modulating the ability of WzxE to effectively complement O16 LPS production in the ΔwzxO16 mutant. Two key predictions arising from our hypothesis were that WzxE could complement O16 LPS production in the absence of WzyE and WzzE, and, conversely, reintroduction of any of these components would reduce or eliminate the WzxE complementing activity. Our results showing that WzxE complemented O16 LPS production in a mutant with a deletion of the majority of the ECA gene cluster but that the complementation was reduced in the presence of plasmids reconstituting the expression of either WzyE or WzzE demonstrate that both predictions are correct. Both O16 and ECA use GlcNAc-Und-PP as the initial sugar acceptor for biosynthesis of the respective subunits. Thus, it is conceivable that the inability of WzxE to complement O16 LPS production in the ΔwzxO16 mutant is due to an interaction with the corresponding WzyE, WzzE, or both that ultimately favors the synthesis of ECA. Presumably, this interaction may involve the formation of a membrane protein complex sequestering WzxE.
We also examined the generality of this model using two O-antigen systems, O16 and O7, which were coexpressed in the same bacterial cell. Our results show that WzxO16 and WzxO7 can cross-complement O-antigen synthesis in the presence of gene defects of wzxO16 or wzxO7, respectively. However, the complementing activity of each translocase is lost when their corresponding Wzy and Wzz proteins are overexpressed. Wzy and Wzz proteins could presumably form a complex that recruits the corresponding Wzx protein. Such an interaction would ensure that the glycan intermediates are exported at the site where further processing will occur. This hypothesis is further supported by two additional observations. First, WzxO16, WzxE, and WzxO7 can mediate the translocation of GlcNAc-PP-Und and the subsequent transfer of the GlcNAc residue onto core-lipid A (16) (C. L. Marolda and M. Valvano, unpublished data), indicating that their translocating activity is not dramatically different. Second, deletions eliminating WzyO16 and WzzO16 result in enhanced O7 LPS production in a strain containing a defective wzxO7 but intact wzxO16, wzyO7, and wzzO7 genes. Therefore, it would appear that Wzx proteins in isolation are competent for translocation and have a relaxed specificity for the O unit. Conversely, in the presence of the corresponding Wzz and Wzy proteins, Wzx translocases may be unable to mediate the translocation of additional lipid-linked glycans except for those from their own system.
The notion that the assembly proteins of the Wzy-dependent pathway function as multiprotein complexes has been proposed previously for O antigens of Yersinia enterocolitica (4), Shigella flexneri (11, 36), E. coli O7 (18), and Pseudomonas aeruginosa (10). Also, it is possible that WzzE, WzyE, and a putative cyclase involved in the assembly of a cyclic form of ECA exist together in the plasma membrane as a complex (23). Furthermore, direct evidence exists for oligomerization in vivo of at least one of these proteins, Wzz, in S. flexneri (11), E. coli K-12/O16 (45), and P. aeruginosa (10). We believe the present study provides strong genetic evidence suggesting that Wzx proteins not only appear to recognize the sugar-PP-Und intermediate in the first position of the glycan moiety (32) but also may interact with additional components of their corresponding systems. It is possible that multiprotein complexes at the plasma membrane exist for the translocation and assembly of the O antigen, presumably organized in membrane microdomains. Experiments to obtain direct biochemical evidence for the existence of such complexes are currently under way in our laboratories.
Acknowledgments
The authors thank D. Stoykova for technical assistance; S. Saldías, R. Flannagan, M. Feldman, and C. Creuzenet for critical reading of the manuscript; and the colleagues referenced or mentioned in Table 1 for the gift of strains and plasmids.
This work was supported by grant MOP-10206 from the Canadian Institutes of Health Research (to M.A.V.) and by grants 3100170-105541 and 3100-057082 from the Swiss National Science Foundation (to M.A.). The Infectious Diseases Research Group Microscopic Facility was supported by grants from the Academic Development Fund of the University of Western Ontario and the Canadian Institutes of Health Research. M.A.V. holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis.
REFERENCES
- 1.Alaimo, C., I. Catrein, L. Morf, C. L. Marolda, N. Callewaert, M. A. Valvano, M. F. Feldman, and M. Aebi. 2006. Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J. 25:967-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, D. C., and M. A. Valvano. 1994. Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J. Bacteriol. 176:7079-7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr, K., J. Klena, and P. D. Rick. 1999. The modality of enterobacterial common antigen polysaccharide chain lengths is regulated by o349 of the wec gene cluster of Escherichia coli K-12. J. Bacteriol. 181:6564-6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bengoechea, J. A., E. Pinta, T. Salminen, C. Oertelt, O. Holst, J. Radziejewska-Lebrecht, Z. Piotrowska-Seget, R. Venho, and M. Skurnik. 2002. Functional characterization of Gne (UDP-N-acetylglucosamine-4-epimerase), Wzz (chain length determinant), and Wzy (O-antigen polymerase) of Yersinia enterocolitica serotype O:8. J. Bacteriol. 184:4277-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronner, D., B. R. Clarke, and C. Whitfield. 1994. Identification of an ATP-binding cassette transport system required for translocation of lipopolysaccharide O-antigen side-chains across the cytoplasmic membrane of Klebsiella pneumoniae serotype O1. Mol. Microbiol. 14:505-519. [DOI] [PubMed] [Google Scholar]
- 6.Brooke, J. S., and M. A. Valvano. 1996. Biosynthesis of inner core lipopolysaccharide in enteric bacteria identification and characterization of a conserved phosphoheptose isomerase. J. Biol. Chem. 271:3608-3614. [DOI] [PubMed] [Google Scholar]
- 7.Bugg, T. D., and P. E. Brandish. 1994. From peptidoglycan to glycoproteins: common features of lipid-linked oligosaccharide biosynthesis. FEMS Microbiol. Lett. 119:255-262. [DOI] [PubMed] [Google Scholar]
- 8.Burda, P., and M. Aebi. 1999. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426:239-257. [DOI] [PubMed] [Google Scholar]
- 9.Cardona, S. T., and M. A. Valvano. 2005. An expression vector containing a rhamnose-inducible promoter provides tightly regulated gene expression in Burkholderia cenocepacia. Plasmid 54:219-228. [DOI] [PubMed] [Google Scholar]
- 10.Daniels, C., C. Griffiths, B. Cowles, and J. S. Lam. 2002. Pseudomonas aeruginosa O-antigen chain length is determined before ligation to lipid A core. Environ. Microbiol. 4:883-897. [DOI] [PubMed] [Google Scholar]
- 11.Daniels, C., and R. Morona. 1999. Analysis of Shigella flexneri wzz (Rol) function by mutagenesis and cross-linking: Wzz is able to oligomerize. Mol. Microbiol. 34:181-194. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dykxhoorn, D. M., R. St. Pierre, and T. Linn. 1996. A set of compatible tac promoter expression vectors. Gene 177:133-136. [DOI] [PubMed] [Google Scholar]
- 15.Erbel, P. J., K. Barr, N. Gao, G. J. Gerwig, P. D. Rick, and K. H. Gardner. 2003. Identification and biosynthesis of cyclic enterobacterial common antigen in Escherichia coli. J. Bacteriol. 185:1995-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman, M. F., C. L. Marolda, M. A. Monteiro, M. B. Perry, A. J. Parodi, and M. A. Valvano. 1999. The activity of a putative polyisoprenol-linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J. Biol. Chem. 274:35129-35138. [DOI] [PubMed] [Google Scholar]
- 17.Feldman, M. F., M. Wacker, M. Hernandez, P. G. Hitchen, C. L. Marolda, M. Kowarik, H. R. Morris, A. Dell, M. A. Valvano, and M. Aebi. 2005. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. USA 102:3016-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaspar, J. A., J. A. Thomas, C. L. Marolda, and M. A. Valvano. 2000. Surface expression of O-specific lipopolysaccharide in Escherichia coli requires the function of the TolA protein. Mol. Microbiol. 38:262-275. [DOI] [PubMed] [Google Scholar]
- 19.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helenius, J., and M. Aebi. 2002. Transmembrane movement of dolichol linked carbohydrates during N-glycoprotein biosynthesis in the endoplasmic reticulum. Semin. Cell. Dev. Biol. 13:171-178. [DOI] [PubMed] [Google Scholar]
- 21.Helenius, J., D. T. Ng, C. L. Marolda, P. Walter, M. A. Valvano, and M. Aebi. 2002. Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature 415:447-450. [DOI] [PubMed] [Google Scholar]
- 22.Jensen, S. O., and P. R. Reeves. 2004. Deletion of the Escherichia coli O14:K7 O antigen gene cluster. Can. J. Microbiol. 50:299-302. [DOI] [PubMed] [Google Scholar]
- 23.Kajimura, J., A. Rahman, and P. D. Rick. 2005. Assembly of cyclic enterobacterial common antigen in Escherichia coli K-12. J. Bacteriol. 187:6917-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaniuk, N. A., E. Vinogradov, and C. Whitfield. 2004. Investigation of the structural requirements in the lipopolysaccharide core acceptor for ligation of O antigens in the genus Salmonella: WaaL “ligase” is not the sole determinant of acceptor specificity. J. Biol. Chem. 279:36470-36480. [DOI] [PubMed] [Google Scholar]
- 25.Keenleyside, W. J., and C. Whitfield. 1999. Genetics and biosynthesis of lipopolysaccharide O-antigens, p. 331-358. In H. Brade, D. C. Morrison, S. Vogel, and S. Opal (ed.), Endotoxin in health and disease. Marcel Dekker, Inc., New York, N.Y.
- 26.Linton, D., N. Dorrell, P. G. Hitchen, S. Amber, A. V. Karlyshev, H. R. Morris, A. Dell, M. A. Valvano, M. Aebi, and B. W. Wren. 2005. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 55:1695-1703. [DOI] [PubMed] [Google Scholar]
- 27.Liu, D., R. A. Cole, and P. R. Reeves. 1996. An O-antigen processing function for Wzx (RfbX): A promising candidate for O-unit flippase. J. Bacteriol. 178:2102-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, D., and P. R. Reeves. 1994. Escherichia coli K12 regains its O antigen. Microbiology 140:49-57. [DOI] [PubMed] [Google Scholar]
- 29.Lugowski, C., E. Romanowska, L. Kenne, and B. Lindberg. 1983. Identification of a tri-saccharide repeating unit in the enterobacterial common antigen. Carbohydr. Res. 118:171-181. [DOI] [PubMed] [Google Scholar]
- 30.Marino, P. A., B. C. McGrath, and M. J. Osborn. 1991. Energy dependence of O-antigen synthesis in Salmonella typhimurium. J. Bacteriol. 173:3128-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marolda, C. L., M. F. Feldman, and M. A. Valvano. 1999. Genetic organization of the O7-specific lipopolysaccharide biosynthesis cluster of Escherichia coli VW187 (O7:K1). Microbiology 145:2485-2496. [DOI] [PubMed] [Google Scholar]
- 32.Marolda, C. L., J. Vicarioli, and M. A. Valvano. 2004. Wzx proteins involved in O antigen biosynthesis function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology 150:4095-4105. [DOI] [PubMed] [Google Scholar]
- 33.Marolda, C. L., J. Welsh, L. Dafoe, and M. A. Valvano. 1990. Genetic analysis of the O7-polysaccharide biosynthesis region from the Escherichia coli O7:K1 strain VW187. J. Bacteriol. 172:3590-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGrath, B. C., and M. J. Osborn. 1991. Localization of the terminal steps of O-antigen synthesis in Salmonella typhimurium. J. Bacteriol. 173:649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier-Dieter, U., K. Barr, R. Starman, L. Hatch, and P. D. Rick. 1992. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. Molecular cloning of the rfe-rff gene cluster. J. Biol. Chem. 267:746-753. [PubMed] [Google Scholar]
- 36.Morona, R., L. van den Bosch, and P. A. Manning. 1995. Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J. Bacteriol. 177:1059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulford, C. A., and M. J. Osborn. 1983. An intermediate step in translocation of lipopolysaccharide to the outer membrane of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 80:1159-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikaido, H. 1996. Outer membrane, p. 29-47. In F. C. Neidhardt, R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 39.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rahman, A., K. Barr, and P. D. Rick. 2001. Identification of the structural gene for the TDP-Fuc4NAc:lipid II Fuc4NAc transferase involved in synthesis of enterobacterial common antigen in Escherichia coli K-12. J. Bacteriol. 183:6509-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rick, P. D., K. Barr, K. Sankaran, J. Kajimura, J. S. Rush, and C. J. Waechter. 2003. Evidence that the wzxE gene of Escherichia coli K-12 encodes a protein involved in the transbilayer movement of a trisaccharide-lipid intermediate in the assembly of enterobacterial common antigen. J. Biol. Chem. 278:16534-16542. [DOI] [PubMed] [Google Scholar]
- 42.Rick, P. D., and R. P. Silver. 1996. Enterobacterial common antigen and capsular polysaccharides, p. 104-122. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 43.Rush, J. S., K. van Leyen, O. Ouerfelli, B. Wolucka, and C. J. Waechter. 1998. Transbilayer movement of Glc-P-dolichol and its function as a glucosyl donor: protein-mediated transport of a water-soluble analog into sealed ER vesicles from pig brain. Glycobiology 8:1195-1205. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt, G., B. Jann, and K. Jann. 1974. Genetic and immunochemical studies on Escherichia coli O14:K7:H. Eur. J. Biochem. 42:303-309. [DOI] [PubMed] [Google Scholar]
- 45.Stenberg, F., P. Chovanec, S. L. Maslen, C. V. Robinson, L. Ilag, G. von Heijne, and D. O. Daley. 2005. Protein complexes of the Escherichia coli cell envelope. J. Biol. Chem. 280:34409-34419. [DOI] [PubMed] [Google Scholar]
- 46.Stevenson, G., B. Neal, D. Liu, M. Hobbs, N. H. Packer, M. Batley, J. W. Redmond, L. Lindquist, and P. Reeves. 1994. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb cluster. J. Bacteriol. 176:4144-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valvano, M. A. 2003. Export of O-specific lipopolysaccharide. Front. Biosci. 8:s452-471. [DOI] [PubMed] [Google Scholar]
- 48.Valvano, M. A., and J. H. Crosa. 1989. Molecular cloning and expression in Escherichia coli K-12 of chromosomal genes determining the O7 lipopolysaccharide antigen of a human invasive strain of E. coli O7:K1. Infect. Immun. 57:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valvano, M. A., and C. L. Marolda. 1991. Relatedness of O-specific lipopolysaccharide side chain genes from strains of Shigella boydii type 12 belonging to two clonal groups and from Escherichia coli O7:K1. Infect. Immun. 59:3917-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Heijenoort, J. 2001. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 18:503-519. [DOI] [PubMed] [Google Scholar]
- 51.Vinés, E., C. L. Marolda, A. Balachandran, and M. A. Valvano. 2005. Defective O-antigen polymerization in tolA and pal mutants of Escherichia coli in response to extracytoplasmic stress. J. Bacteriol. 187:3359-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wacker, M., M. F. Feldman, N. Callewaert, B. R. Clarke, M. Hernandez, E. D. Vines, M. A. Valvano, C. Whitfield, and M. Aebi. 2006. Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proc. Natl. Acad. Sci. USA 103:7088-7093. [Online.] doi: 10.1073/pnas.0509207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 54.Yao, Z., and M. A. Valvano. 1994. Genetic analysis of the O-specific lipopolysaccharide biosynthesis region (rfb) of Escherichia coli K-12 W3110: identification of genes that confer group 6 specificity to Shigella flexneri serotypes Y and 4a. J. Bacteriol. 176:4133-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, G. P., and F. A. Troy. 2005. NMR study of the preferred membrane orientation of polyisoprenols (dolichol) and the impact of their complex with polyisoprenyl recognition sequence peptides on membrane structure. Glycobiology 15:347-359. [DOI] [PubMed] [Google Scholar]