Abstract
Bacillus subtilis produces and exports a peptide sporulation killing factor (SkfA) that induces lysis of sibling cells. skfA is part of the skf operon (skfA-H), which is responsible for immunity to SkfA, as well as for production and export of SkfA. Here we report that transcription of skfA is markedly induced when cells of B. subtilis are subjected to phosphate starvation. The role of PhoP in regulation of the skf operon was confirmed by in vitro gel shift assays, which showed that this operon is a new member of the PhoP regulon. A putative stem-loop structure in the skfA-skfB intergenic region is proposed to act as a stabilizer of an skfA-specific transcript.
Phosphate starvation leads to marked changes in the expression of genes in the PhoP and SigB (σB) regulons of Bacillus subtilis (2, 4, 8, 12, 16, 27). Genes in the PhoP regulon provide a specific response to phosphate starvation stress, while genes in the SigB regulon provide a general response to the resulting energy stress. Additionally, the PhoP and SigB regulons interact to modulate the level to which each regulon is activated (4, 27). The activity of the PhoP regulon is also influenced by Spo0A; activated Spo0A (Spo0A∼P) is responsible for the induction of sporulation and the repression of genes induced by the transition-phase regulator AbrB. The PhoP regulon is up-regulated in a spo0A null mutant that is unable to initiate sporulation (18, 25). Maximal induction of the PhoP regulon also requires an active ResD-ResE respiration signal transduction system (16). An spo0A abrB resD null mutant is not able to mount a specific response to phosphate starvation, showing that the induction of the PhoP regulon is dependent not only on the phosphate-specific PhoPR signal transduction system but also on this network of regulatory elements (30). If, despite these regulatory responses, phosphate starvation persists, Spo0A initiates sporulation and terminates the phosphate response by repressing the transcription of phoPR via AbrB and ResD-ResE (16, 17).
During the phosphate starvation-specific response, genes of the PhoP regulon are regulated by the PhoP-PhoR two-component signal transduction system (2, 25, 29). The PhoP response regulator is activated by its cognate sensor kinase, PhoR. Phosphorylated PhoP (PhoP∼P) induces the expression of the phoPR operon about threefold from a low constitutive level of expression (17, 26, 27) and is required for the induction or repression of other members of the PhoP regulon (16).
Fawcett et al. (9) and Molle and colleagues (21) have shown that Spo0A regulates the skf operon, which encodes the sporulation killing factor (SkfA). SkfA induces the lysis of sibling B. subtilis cells that have not entered the sporulation pathway (i.e., Spo0A inactive), providing a source of nutrients to support this key differentiation process.
Reporter gene analysis of the response of skfA to phosphate starvation.
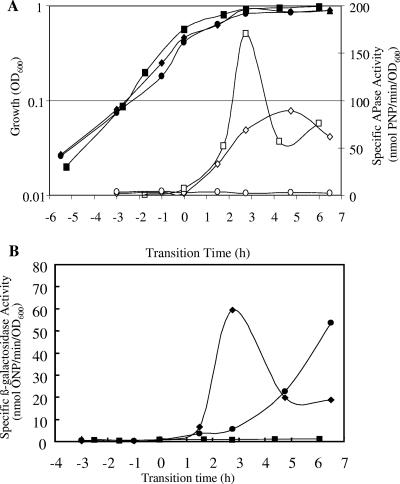
Previous DNA array analysis of the response of B. subtilis to phosphate starvation indicated that skfA was induced in response to phosphate starvation in a phoR-dependent manner (2). This induction was transient, and the level typically returned to a noninduced level within 3 h. The induction of skfA in response to phosphate starvation was confirmed with strain 168-SKFA (Table 1), in which skfA is transcriptionally fused to a lacZ reporter (skfA::lacZ). Cultures of 168-SKFA grown in LPM (0.42 mM Pi) were sampled to monitor the alkaline phosphatase (APase) and β-galactosidase activities (24, 27). Three independent experiments gave comparable results, and a representative data set is shown in Fig. 1. APase, an intrinsic reporter of the PhoP regulon, was induced during the transition from the exponential phase to stationary phase (T0), confirming that entry into stationary phase was due to phosphate starvation. skfA was induced (∼60 to 70 nmol of o-nitrophenol [ONP]/min/unit of optical density at 600 nm [OD600]) concomitantly with APase, as determined by the β-galactosidase activity of the reporter strain (Fig. 1).
TABLE 1.
Bacterial strains, plasmids, and primers
| Strain, plasmid, or primer | Relevant characteristic(s) or sequence (5′-3′)a | Positionb | Reference or source |
|---|---|---|---|
| B. subtilis strains | |||
| 168 | trpC2 | 3 | |
| 168-PR | trpC2 phoRΔBalI::Tcr | 26 | |
| 168-SKFA | trpC2 skfA::pYBCOdd Emr (previously YBCOdd) | T. Tanaka, Takai University | |
| 168-SKFA-PR | trpC2 phoRΔBalI::TcrskfA::pYBCOdd Emr | This study | |
| SWV215 | trpC2 pheA1 spo0A::Kmr | 31 | |
| 168-SKFA-OA | trpC2 spo0A::KmrskfA::pYBCOdd Emr | This study | |
| 168-SKFA-OA-PR | trpC2 spo0A::KmrphoRΔBalI::TcrskfA::pYBCOdd Emr | This study | |
| 168-SKFB | trpC2 skfB::pYBCPdd Emr (previously YBCPdd) | T. Tanaka, Takai University | |
| Plasmids | |||
| pET-PhoP | pET2816 containing a 722-bp insert of phoP Apr (6.386 kb) | 25 | |
| pET-PhoR231 | pET2816 containing a 1,049-bp insert of phoR Apr (6.713 kb) | 25 | |
| Primers | |||
| skfA-FOR | AAGAATGGGAATCTGTGA | 213942-213960 | |
| skfA-REV | CTAATACGACTCACTATAGGGAGATAGCAGGATGCGGAAG | 214081-214066 | |
| skfB-FOR | CCAGAGTAGCGGTTCAT | 214181-214166 | |
| skfB-REV | CTAATACGACTCACTATAGGGAGTTACCTGAGCTGGCATAG | 214550-214532 | |
| skfH-FOR | CGTGACAAGTTGTGACGA | 219909-219892 | |
| skfH-REV | CTAATACGACTCACTATAGGGAGGTTGACTGAATGGCCAAG | 219611-219628 | |
| YbcO-FOR | TATAACATAATGGACCGTCT | 213645-213664 | |
| YbcO-REV | CACGTGTCATAGCAATACTC | 214035-214055 | |
| YhaX-FOR | TAACGATATTAGGGAGAATGGC | 1056018-1056039 | |
| YhaX-REV | GAAGCAGCGCTCCATCTATATT | 1056207-1056228 |
Sequences in boldface type are specific for the T7 promoter used to initiate in vitro transcription.
Positions of the primers specific for the B. subtilis 168 chromosome as indicated in the SubtiList database (http://genolist.pasteur.fr/SubtiList) (22).
FIG. 1.
Growth and reporter gene activities of B. subtilis skfA-lacZ reporter gene fusion strains grown in LPM. (A) OD600 of mutants 168-SKFA-PR (phoPR null mutant) (•), 168-SKFA (⧫), and 168-SKFA-OA (spo0A null mutant) (▪) and APase activities of strains 168-SKFA (⋄), 168-SKFA-OA (□), and 168-SKFA-PR (○). (B) Transcriptional activities of skfA-lacZ: specific β-galactosidase activities in mutants 168-SKFA-PR (•), 168-SKFA-OA (▪), and 168-SKFA (⧫). PNP, p-nitrophenol.
To analyze skfA gene expression in a phoR null background, B. subtilis 168-SKFA was transformed with chromosomal DNA from 168-PR (a phoR null mutant) (26) to obtain strain 168-SKFA-PR. Strain 168-SKFA-PR was used to determine whether the response of skfA to phosphate starvation was dependent on the PhoPR regulatory system. Compared to the pattern of lacZ expression in 168-SKFA, the pattern of lacZ expression in 168-SKFA-PR was markedly different; low levels of β-galactosidase activity (>10 nmol ONP/min/OD600 unit) were observed during the transition phase, but the level increased to ∼50 nmol ONP/min/OD600 unit at 6 h after entry into stationary phase (T6) (Fig. 1).
Since skfA is a member of the Spo0A regulon (21), in order to determine whether Spo0A had a role in the induction of skfA during phosphate starvation, a spo0A null mutation was introduced into strain 168-SKFA (skfA::lacZ) (5). The spo0A null lesion eliminated the transcription of skfA but not the transcription of another member of the PhoP regulon, phoA, since alkaline phosphatase was induced normally during the transition and during early stationary phase (Fig. 1).
Transcription of skfA in response to phosphate starvation.
Northern blot analysis (14, 25) was used to monitor the expression of genes in the skf operon in response to phosphate starvation, using RNA extracted from the wild type and a phoR null mutant as described previously (7). The extracted RNA was probed with antisense RNA specific for sequences in the skfA, skfB, and skfH genes. Probes were prepared by PCR with primer pairs (Table 1) by incorporating a promoter sequence recognized by T7 polymerase into the reverse primer. The resulting amplicons were used as substrates for in vitro T7 RNA polymerase-directed synthesis of digoxigenin-labeled skf-specific RNA probes (1).
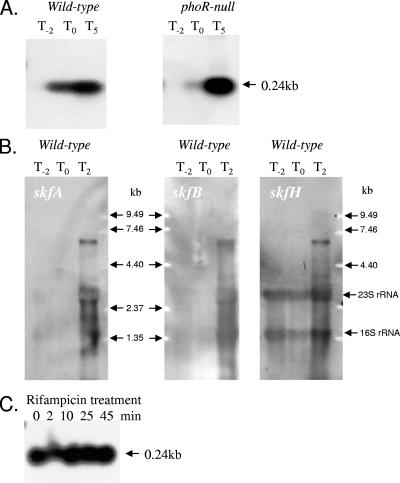
Irrespective of the strain, little or no skf-specific RNA was detected in samples taken 2 h before the onset of phosphate starvation (Fig. 2), confirming the low level of expression of this operon under phosphate-replete conditions (Pi concentration, >0.1 mM). When the skfA-specific probe was used, a very intense ∼0.25-kb transcript was detected during the transition to phosphate limitation (T0) and 5 h later (T5) in the wild-type strain (Fig. 2A). The size of this transcript is consistent with the predicted length of skfA. In the case of the phoR null mutant, an ∼0.25-kb skfA-specific transcript was detected at T5, but little or no transcript was detected at T0, confirming that the expression of this gene during phosphate starvation-induced entry into the transition phase is under the influence of the PhoPR two-component signal transduction system. When the gels were run long enough to remove the ∼0.25-kb transcript (the intensity of which obscured a significant portion of the gel), a larger less intense transcript was observed (Fig. 2B). The size of this transcript, ∼6.5 kb, was consistent with the hypothesis that it carried the entire skfA-H operon. When hybridization reactions were carried out with skfB- and skfH-specific probes, a similar ∼6.5-kb transcript was observed (Fig. 2B), but the ∼0.25-kb transcript was not observed. Additional bands at intermediate sizes could have been either processing products or mRNA entrapped in the rRNA bands (1).
FIG. 2.
(A) Northern blot analyses of the skf operon. Total RNA was isolated from wild-type strain B. subtilis 168 and a phoR null mutant. Bacteria were grown in LPM, and samples were taken 2 h before (T−2), at the time of (T0), and 5 h after (T5) entry into stationary growth phase provoked by phosphate starvation. Five micrograms of RNA was applied to each lane, and then after capillary blotting the filters were hybridized to skfA gene-specific riboprobes. (B) Northern blot analyses of the skf operon. RNA was isolated from wild-type strain B. subtilis 168. Bacteria were grown in LPM, and samples were taken 2 h before (T−2), at the time of (T0), and 2 h after (T2) entry into stationary growth phase provoked by phosphate starvation. Ten micrograms of RNA was applied per lane, and then after an alkaline-transblotting procedure the filters were hybridized to gene-specific riboprobes for skfA, skfB, and skfH. The 0.25-kb transcript was purposely run off the bottom of the gel to facilitate visualization of the weaker bands. (C) Transcriptional stability of skfA mRNA. RNA was isolated from wild-type strain B. subtilis 168, grown in LPM, and sampled 2 h after entry into stationary growth phase. RNA was isolated from samples harvested before and 2, 10, 25, 45 min after the addition of rifampin. Five micrograms of RNA was applied to each lane, and the membrane was hybridized to a skfA-specific riboprobe. Transcript sizes were determined by comparison with digoxigenin-labeled RNA size markers (Roche Diagnostics, Mannheim, Germany). An RNA molecular size ladder (0.24 to 9.5 kb) was purchased from Invitrogen.
DNA microarray analysis (11) of the mRNA decay rates of ∼1,500 B. subtilis mRNA transcripts in early-stationary-phase cultures indicated that about 80% of them had a half-life of less than 7 min. However, the skfA transcript was among ∼30 mRNA species that were found to have a half-life of >15 min. We therefore attempted to determine the half-life of the phosphate starvation-induced skfA-specific transcript following treatment with rifampin, an inhibitor of transcription initiation. B. subtilis was grown in LPM, and rifampin was added to the culture 2 h after the transition to the phosphate starvation-induced transition phase (T2). Samples were harvested immediately before the addition of rifampin (zero time) and at 2, 10, 25, and 45 min after the addition of the antibiotic. The samples were hybridized with the skfA-specific probe. As shown in Fig. 2C, the skfA mRNA transcript was extremely stable, and no observable decrease in the amount of transcript was detected 45 min after the addition of rifampin. Analysis of the skfA-skfB intergenic region using the MFOLD program (32) predicted that the RNA in this region is able to form a stable stem-loop structure (ΔG = −23.3 kcal/mol) (data not shown).
Binding of PhoP and PhoP∼P to the region of the skfA promoter.
The B. subtilis genome sequence was interrogated using the pattern-matching function of SubtiList (http://genolist.pasteur.fr/SubtiList/) (22) to identify potential PhoP binding sequences. A 1-bp deviation from the PhoP consensus sequence (TTHACA3-7TTHACA, where H is A, C, or T) was allowed for each element of the search sequence, and only targets within 200 bp of a start codon were reported. The promoter region of skfA was found to contain at least one PhoP consensus sequence, and a possible second sequence was located immediately downstream (data not shown).
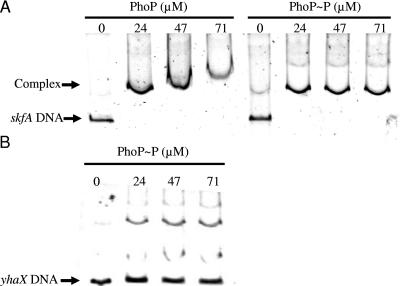
To determine whether the putative PhoP binding sequences identified were active in vitro, gel shift assays we used to analyze the binding of PhoP to the promoter region of skfA. Proteins PhoP-His6 and PhoR231-His6 were produced from Escherichia coli BL21(λD3) carrying pET-PhoP or pET-PhoR231 (25), as described previously (6). Fragments of the skfA and yhaX promoter regions were amplified with primers YbcO-FOR and YbcO-REV and with primers YhaX-FOR and YhaX-REV (Table 1). In the gel shift reactions, 4 μM PhoR231 and 0.1 μg of poly(dI-dC) (Sigma) per μl were incubated with 0, 24, 47, 71, and 95 μM PhoP in the presence or absence of 5 mM ATP for 15 min at room temperature (20). After addition of the DNA the mixture was incubated for a further 30 min. The samples were analyzed on a 6% native polyacrylamide gel by using Tris-glycine buffer (28) and were stained with Sybr GOLD (Molecular Probes).
Gel shift assays showed that both PhoP and PhoP∼P decreased the mobility of the DNA fragment encoding the skfA promoter region (Fig. 3A). As described previously (25), there was very little difference in the observed retardation of the probe when PhoP was phosphorylated, indicating either that PhoP and PhoP∼P bind to the skfA promoter region in vitro with similar efficiencies or that PhoP is spontaneously phosphorylated in E. coli (19). As a negative control, a gel shift assay was performed for the promoter region of yhaX (Fig. 3B), a gene that appears to be induced indirectly by PhoP. As shown previously (25), the mobility of yhaX was not influenced by PhoP∼P.
FIG. 3.
Gel shift assays of the skfA and yhaX promoter regions using PhoP and PhoP∼P. PhoR231 (4 μM) was incubated with PhoP (0, 24, 47, and 71 μM) in the absence or presence of ATP at room temperature for 15 min. skfA (A) and yhaX (B) promoter probes were added, and after binding (30 min), the samples were loaded onto a 6% native polyacrylamide gel to separate free DNA probe from DNA-protein complexes. The gels were then stained with Sybr GOLD used according to the manufacturer's instructions and were visualized by UV illumination. The amounts of PhoP and PhoP∼P added to the reaction mixtures are indicated above the lanes.
Conclusions.
B. subtilis has a range of adaptive responses which are induced under adverse conditions to maintain viability and, in some cases, to restore growth (1, 2, 4, 15). The role of SkfA is to bring about the lysis of cells that have not been induced to enter the sporulation pathway. This provides cells with activated Spo0A (Spo0A∼P) with a source of nutrients (including phosphorus from the cell wall, nucleic acids, membranes, and proteins) (9, 10, 21). The induction of skfA during the phosphate starvation-induced transition phase (T0 to T3) is dependent on PhoPR. This response is transient and is presumably terminated by Spo0A (18). A later induction event (T3 to T6) (Fig. 2) is PhoPR independent and likely to be associated with the onset of sporulation since skfA is a member of the Spo0A regulon (9, 21).
The binding of PhoP to the skfA promoter region was compared with the binding to the SigE-dependent yhaX promoter region (25). In contrast to the binding to yhaX, PhoP binds directly to the promoter region upstream of skfA, indicating that the skf operon is a member of the PhoP regulon. Northern blot hybridization (Fig. 2) detected a weak band corresponding to a full-length transcript of the skf operon (6.5 kb) and a much stronger band for the ∼0.25-kb transcript corresponding to skfA. Our results extend previous data (11) showing that the skfA transcript is extremely stable, with an estimated half-life of more than 45 min. Since the kinetics of induction and the relative amounts of β-galactosidase synthesized by strains 168-SKFA (skfA::lacZ) (Fig. 1) and 168-SKFB (skfB::lacZ) (not shown) in response to phosphate starvation were similar, it is likely that the genes are transcribed from the same promoter. By analogy with the pst operon (1), we propose that the stem-loop acts primarily as a barrier to 3′-5′ exoribonuclease activity with the skfA transcript and is therefore responsible for its extraordinary stability (13, 23). This provides a mechanism for coordinating the transcription of all the genes in the skf operon, while it allows preferential translation of skfA mRNA. Functionally, this should allow a large molar excess of the killing factor peptide compared with the other products of the skf operon.
Phosphate limitation is a common feature of life in the soil, the natural environment of B. subtilis. Induction of the sporulation killing factor by cells that have entered the sporulation pathway and the subsequent lysis of the noninduced portion of the population provide the former organisms with an important source of nutrients. The resulting influx of nutrients increases the survival potential of these cells and delays the onset of sporulation (9, 10, 21). We showed that the skf operon is induced in response to phosphate starvation in a PhoPR-dependent manner. This induction is distinct from that previously reported for Spo0A. An interesting aspect of this induction is its absolute dependence on Spo0A. This observation reinforces previous data which showed that the presence of fully functional PhoPR is not sufficient for maximal expression of the PhoP regulon, since an spo0A abrB resD null mutant does not show PhoP regulon activity (16). In the case of the skf operon, the requirement for Spo0A presumably arises from the need to coordinate its expression with sporulation since the sporulation killing factor's role is to provide nutrients for the subpopulation of cells that are committed to this differentiation process.
Acknowledgments
We thank T. Tanaka (Tokai University, Japan) for the gift of strain YBCOdd.
This work was funded by the European Commission (grant QLG2-CT-1999-01455) and the UK Biotechnology and Biological Sciences Research Council (grant 13/PRES/12179). N.E.E.A. was a recipient of a studentship from the UK Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Allenby, N. E., N. O'Connor, Z. Pragai, N. M. Carter, M. Miethke, S. Engelmann, M. Hecker, A. Wipat, A. C. Ward, and C. R. Harwood. 2004. Post-transcriptional regulation of the Bacillus subtilis pst operon encoding a phosphate-specific ABC transporter. Microbiology 150:2619-2628. [DOI] [PubMed] [Google Scholar]
- 2.Allenby, N. E., N. O'Connor, Z. Pragai, A. C. Ward, A. Wipat, and C. R. Harwood. 2005. Genome-wide transcriptional analysis of the phosphate starvation stimulon of Bacillus subtilis. J. Bacteriol. 187:8063-8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anagnastopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bron, S. 1990. Plasmids. John Wiley & Sons, Ltd, Chichester, United Kingdom.
- 6.Chastanet, A., J. Fert, and T. Msadek. 2003. Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol. Microbiol. 47:1061-1073. [DOI] [PubMed] [Google Scholar]
- 7.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eymann, C., H. Mach, C. R. Harwood, and M. Hecker. 1996. Phosphate-starvation-inducible proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study. Microbiology 142:3163-3170. [DOI] [PubMed] [Google Scholar]
- 9.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Pastor, J. E., E. C. Hobbs, and R. Losick. 2003. Cannibalism by sporulating bacteria. Science 301:510-513. [DOI] [PubMed] [Google Scholar]
- 11.Hambraeus, G., C. von Wachenfeldt, and L. Hederstedt. 2003. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics 269:706-714. [DOI] [PubMed] [Google Scholar]
- 12.Hecker, M., and U. Volker. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigma B regulon. Mol. Microbiol. 29:1129-1136. [DOI] [PubMed] [Google Scholar]
- 13.Higgins, C. F., S. W. Peltz, and A. Jacobson. 1992. Turnover of mRNA in prokaryotes and lower eukaryotes. Curr. Opin. Genet. Dev. 2:739-747. [DOI] [PubMed] [Google Scholar]
- 14.Homuth, G., S. Masuda, A. Mogk, Y. Kobayashi, and W. Schumann. 1997. The dnaK operon of Bacillus subtilis is heptacistronic. J. Bacteriol. 179:1153-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hulett, F. M. 2002. The Pho regulon, p. 193-203. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 16.Hulett, F. M. 1996. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:933-939. [DOI] [PubMed] [Google Scholar]
- 17.Hulett, F. M., J. Lee, L. Shi, G. Sun, R. Chesnut, E. Sharkova, M. F. Duggan, and N. Kapp. 1994. Sequential action of two-component genetic switches regulates the PHO regulon in Bacillus subtilis. J. Bacteriol. 176:1348-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen, K. K., E. Sharkova, M. F. Duggan, Y. Qi, A. Koide, J. A. Hoch, and F. M. Hulett. 1993. Bacillus subtilis transcription regulator, Spo0A, decreases alkaline phosphatase levels induced by phosphate starvation. J. Bacteriol. 175:3749-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ladds, J. C., K. Muchova, D. Blaskovic, R. J. Lewis, J. A. Brannigan, A. J. Wilkinson, and I. Barak. 2003. The response regulator Spo0A from Bacillus subtilis is efficiently phosphorylated in Escherichia coli. FEMS Microbiol. Lett. 223:153-157. [DOI] [PubMed] [Google Scholar]
- 20.Liu, W., and F. M. Hulett. 1997. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J. Bacteriol. 179:6302-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molle, V., M. Fujita, S. T. Jensen, P. Eichenberger, J. E. Gonzalez-Pastor, J. S. Liu, and R. Losick. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683-1701. [DOI] [PubMed] [Google Scholar]
- 22.Moszer, I. 1998. The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett. 430:28-36. [DOI] [PubMed] [Google Scholar]
- 23.Newbury, S. F., N. H. Smith, and C. F. Higgins. 1987. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell 51:1131-1143. [DOI] [PubMed] [Google Scholar]
- 24.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. H. S. M. Cutting (ed.), Molecular biological methods for Bacillus. Wiley, Chichester, United Kingdom.
- 25.Prágai, Z., N. E. Allenby, N. O'Connor, S. Dubrac, G. Rapoport, T. Msadek, and C. R. Harwood. 2004. Transcriptional regulation of the phoPR operon in Bacillus subtilis. J. Bacteriol. 186:1182-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prágai, Z., C. Eschevins, S. Bron, and C. R. Harwood. 2001. Bacillus subtilis NhaC, an Na+/H+ antiporter, influences expression of the phoPR operon and production of alkaline phosphatases. J. Bacteriol. 183:2505-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prágai, Z., and C. R. Harwood. 2002. Regulatory interactions between the Pho and sigma(B)-dependent general stress regulons of Bacillus subtilis. Microbiology 148:1593-1602. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1987. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Seki, T., H. Yoshikawa, H. Takahashi, and H. Saito. 1988. Nucleotide sequence of the Bacillus subtilis phoR gene. J. Bacteriol. 170:5935-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun, G., S. M. Birkey, and F. M. Hulett. 1996. Three two-component signal-transduction systems interact for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:941-948. [DOI] [PubMed] [Google Scholar]
- 31.Xu, K., and M. A. Strauch. 1996. In vitro selection of optimal AbrB-binding sites: comparison to known in vivo sites indicates flexibility in AbrB binding and recognition of three-dimensional DNA structures. Mol. Microbiol. 19:145-158. [DOI] [PubMed] [Google Scholar]
- 32.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]