Abstract
Two-component regulatory systems are involved in processes important for bacterial pathogenesis. The proposed misR/misS (or phoP/phoQ) system is one of four two-component systems of the obligate human pathogen Neisseria meningitidis. Inactivation of this system results in loss of phosphorylation of the lipooligosaccharide inner core and causes attenuation in a mouse model of meningococcal infection. MisR and the cytoplasmic domain of MisS were purified as His6 and maltose binding protein fusion proteins, respectively. The MisS fusion was shown to be autophosphorylated in the presence of ATP, and the phosphoryl group was subsequently transferred to MisR. The phosphotransfer reaction was halted with a MisR/D52A mutation, while a MisS/H246A mutation prevented autophosphorylation. Specific interaction of phosphorylated MisR (MisR∼P) and MisR with the misR promoter was demonstrated by gel mobility shift assays, where MisR∼P exhibited higher affinity than did the nonphosphorylated protein. The transcriptional start site of the misRS operon was mapped, and DNase I protection assays revealed that MisR interacted with a 15-bp region upstream of the transcriptional start site that shared no similarity to binding motifs of other two-component systems. Transcriptional reporter studies suggested that MisR phosphorylation is critical for the autoinduction of the misRS operon. Limited Mg2+ concentration failed to induce expression of the misRS operon, which is the only operon now proven to be under the direct control of the MisRS two-component system. Thus, these results indicate that the meningococcal MisRS system constitutes a functional signal transduction circuit and that both components are critical in the autoregulation of their expression.
Two-component regulatory systems are one of the most common signal transduction mechanisms governing bacterial responses and adaptation to environmental changes (10). In addition, many such systems act as global regulators in coordinating the expression of virulence determinants. These signal transduction systems generally consist of a sensor histidine kinase and a response regulator protein (9). Upon sensing specific signals, the histidine kinase autophosphorylates the conserved histidine residue of the kinase domain, and the phosphoryl group is subsequently transferred to the invariant aspartate residue of the cognate response regulator. Regulation is through modulation of the phosphorylation status of the response regulator protein, thereby controlling its affinity for DNA motifs present in the promoter region of target genes.
Neisseria meningitidis is an obligate pathogen which inhabits the human nasopharynx but can rapidly disseminate to cause sepsis and meningitis during invasive infection (28, 32). In contrast to other gram-negative bacteria such as Escherichia coli, which encodes more than 30 two-component regulatory systems (20, 35), the meningococcus contains only four predicted pairs (1, 29), most likely due to the restricted human host environment encountered. The persistence of these few functional two-component regulatory systems implies an important role in regulating meningococcal colonization and virulence. Sequence comparison suggests that NMB0114/NMB0115 share amino acid sequence similarities to NtrY/NtrX (COG5000/COG2204 domain families), which are involved in the regulation of nitrogen metabolism in Azorhizobium caulinodans (21), while NMB1249/NMB1250 exhibit amino acid sequence similarities with NarQ/NarL, (34) and the equivalent genes in Neisseria gonorrhoeae have been characterized to regulate anaerobic growth (14). NMB1606/NMB1607 share sequence similarities with PilS/PilR that regulate piliation in Pseudomonas aeruginosa (8). We have reported that inactivation of the NMB0595/NMB0594 two-component regulatory system, designated MisR/MisS (or PhoP/PhoQ) (18), resulted in the loss of phosphoethanolamine substitutions on the lipooligosaccharide (LOS) inner core heptose II residue with an increased sensitivity to cationic antimicrobial peptides such as polymyxin B (31). Lipooligosaccharide, or endotoxin, is a major virulence factor of N. meningitidis, and structural changes in LOS are important in meningococcal pathogenesis.
Mutation of misR (NMB0595) encoding the response regulator protein in a serogroup C meningococcal strain resulted in a magnesium-dependent growth phenotype and sensitivity to antimicrobial peptides and other environmental stresses and is avirulent in a mouse model of infection (18). The serogroup C NMB0595 meningococcal mutant had increased membrane permeability and was unable to form colonies on solid media with low magnesium concentrations (19). Taken together with the observation that this mutant did not display magnesium-dependent changes in gene expression probed by microarray analysis (19), these data led the authors to conclude that this system functions analogously to the magnesium-sensing phoP/phoQ system in Salmonella (18) and is regulated by magnesium. However, no magnesium-dependent phenotypic differences have been observed in another NMB0595 mutant generated in a meningococcal serogroup B strain (31), suggesting possible strain-dependent variation in magnesium-related phenotypes. Further, magnesium, which was shown to interact with the periplasmic domain of PhoQ in Salmonella (3) and result in significant changes in PhoP-regulated gene expression, has not been demonstrated to cause changes in the expression of genes proven to be controlled by the meningococcal NMB0595/0594 two-component system.
Although sequence analysis suggests that MisR/MisS proteins are members of the two-component regulatory system family, no biochemical evidence is currently available to demonstrate their function. In this report, we demonstrate that MisS fusion protein underwent autophosphorylation in the presence of ATP and participated in phosphotransfer to MisR. In addition, MisR was shown to bind directly to the promoter region of the misRS operon, demonstrating an autoinduction mechanism. Further, no magnesium-dependent regulation was detected in the expression of the misR/S operon, the first promoter established to be directly regulated by the meningococcal MisRS two-component system, indicating that Mg2+ is not the inducing signal of the meningococcal MisRS two-component signal transduction system.
MATERIALS AND METHODS
Bacterial strains, medium, and reagents.
Strains and plasmids used in this study are listed in Table 1. Primer sequences are listed in Table S1 in the supplemental material. Meningococcal strain NMB (CDC 8201085) is a serogroup B N. meningitidis strain originally isolated from the cerebrospinal fluid of a patient with meningococcal meningitis in Pennsylvania in 1982. Meningococcal strains were grown with 5% CO2 at 37°C unless specified otherwise. GC base agar (Difco), supplemented with 0.4% glucose and 0.68 mM Fe(NO3)3, or GC broth with the same supplements and 0.043% NaHCO3 was used. Brain heart infusion medium (37 g/liter brain heart infusion) with 1.25% fetal bovine serum was used when kanamycin selection was required. Antibiotic concentrations (μg/ml) used for E. coli strains were as follows: ampicillin, 100; kanamycin, 50; erythromycin, 300. Antibiotic concentrations (μg/ml) used for N. meningitidis were as follows: kanamycin, 80; erythromycin, 3. E. coli strain DH5α cultured on Luria Bertani (LB) medium was used for cloning and propagation of plasmids. Meningococci were transformed by the procedure of Janik et al. (12). E. coli strains were transformed by electroporation with a GenePulser (Bio-Rad) according to the manufacturer's protocol.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| N. meningitidis | ||
| NMB | B:2b:P1.2,5:L2 (CDC8201085) | 27 |
| NMBmisR | NMB with misR::erm mutation | 31 |
| YT0310 | NMB with ΔmisS::aphA-3 mutation | This study |
| YT0263 | NMB with promoterless lacZ fusion in the chromosomal 988 locus | This study |
| YT0321 | NMB with misR(−504/+104)::lacZ fusion in the chromosomal 988 locus | This study |
| YT0322 | NMB with misR(−348/+104)::lacZ fusion in the chromosomal 988 locus | This study |
| YT0323 | NMB with misR(−205/+104)::lacZ fusion in the chromosomal 988 locus | This study |
| YT0324 | NMB with misR(−89/+104 with internal deletion between −8 and +25)::lacZ fusion in the chromosomal 988 locus | This study |
| YT0326 | NMB with misR(−205/+3)::lacZ fusion in the chromosomal 988 locus | This study |
| YT0329 | NMB with misR(−46/+104)::lacZ fusion in the chromosomal 988 locus | This study |
| YT0330 | NMB with misR(−103/+104)::lacZ fusion in the chromosomal 988 locus | This study |
| YT0332 | NMB with misR(−89/+104)::lacZ fusion in the chromosomal 988 locus | This study |
| E. coli | ||
| DH5α | Cloning strain | New England Biolabs |
| BL21(DE3) | Host strain for PT7-controlled expression | Novagen |
| TB1 | Host strain for MBP fusion protein expression | New England Biolabs |
| Plasmids | ||
| pCR2.1 | TA cloning vector, Apr, Kmr | Invitrogen |
| pET28a(+) | PT7-controlled expression vector, Kmr | Novagen |
| pMal-c2 | N-terminal MBP fusion vector | New England Biolabs |
| pUC18k | Source of aphA-3 cassette | 17 |
| pYT298 | MisR in pET28a | This study |
| PYT311 | MisS (residue 202 to 469) fused to MalE at N terminus in pMal-c2 | This study |
| PYT310 | aphA-3 cassette (EcoRI-HincII blunted) replacing EcoRV-HincII within misS fragment of YT140-YT67 in pCR2.1 | This study |
| pXZ015 | MisR-(His)6 fusion with MisR52DA mutation in pET28a | This study |
| pXZ019 | MBP-MisS fusion with MisS246HA mutation in pMal-c2 | This study |
| pYT328 | Cloning vector for lacZ fusion in a chromosomal intergenic region | This study |
| pYT321 | misR::lacZ carrying 608-bp (−504/+104) misR promoter region | This study |
| pYT322 | misR::lacZ carrying 452-bp (−348/+104) misR promoter region | This study |
| pYT323 | misR::lacZ carrying 309-bp (−205/+104) misR promoter region | This study |
| pYT324 | misR::lacZ carrying 193-bp (−89/+104 with internal deletion between −8 and +25) misR promoter region | This study |
| pYT326 | misR::lacZ carrying 208-bp (−205/+3) misR promoter region | This study |
| pYT329 | misR::lacZ carrying 150-bp (−46/+104) misR promoter region | This study |
| pYT330 | misR::lacZ carrying 207-bp (−103/+104) misR promoter region | This study |
| pYT332 | misR::lacZ carrying 193-bp (−89/+104) misR promoter region | This study |
Construction of MisR and MisS overexpression strains.
The coding sequence of misR was amplified with primers YT124-NcoI and YT125-XhoI. The PCR product was digested with NcoI and XhoI and then cloned into pET28a(+), which had been digested with the same enzymes to yield pYT298. The plasmid was purified and transformed into E. coli expression strain BL21(DE3). The coding sequence of the soluble C-terminal domain from residues 202 to 469 of MisS was amplified by PCR using primers YT147-BamHI and YT148-HindIII. The PCR product was digested with BamHI and HindIII and then inserted in frame and downstream from the malE sequence in pMal-c2 that had been cut with the same enzymes to generate an N-terminal maltose-binding protein (MBP) fusion. The sequence of the resulting plasmid, pYT311, was verified by sequencing analysis with primer malE, and the plasmid was transformed into E. coli strain TB1 for expression.
Site-directed mutagenesis was performed to introduce single amino acid substitutions in the misR and misS coding sequence. To generate the MisR (D→A) mutation, the GAT→GCT codon change at amino acid residue 52 was incorporated into MisR by overlap PCR. Two internal fragments of MisR were amplified using the primer pairs T7p/YT165 and YT164/T7t. Equal amounts of the two purified PCR products were mixed and used as a template for the subsequent PCR amplification using T7p/T7t primers. The final PCR fragment was digested with NcoI and XhoI and then ligated with NcoI- and XhoI-digested pET-28a(+). Kanamycin-resistant colonies were examined by colony PCR, and the resulting plasmid, pXZ015, was verified to contain the desired mutation and correctly fused to a C-terminal His tag by sequencing analysis using primers T7p and T7t.
To make the MisS (H→A) mutation, the CAT→GCT mutation at amino acid residue 246 was introduced into MisS by overlap PCR. Two internal fragments of misS were amplified using the primer pairs YT147-BamHI/YT167 and YT166/YT148-HindIII. Equal amounts of these products were mixed together and used as the template for the subsequent PCR amplification with primers YT147-BamHI and YT148-HindIII. The resulting PCR fragment was digested with HindIII and BamHI and then ligated with pMal-c2 cut with the same enzymes. Ampicillin-resistant colonies were selected and analyzed by PCR. The resulting plasmid, named pXZ019, was verified to carry the desired mutation without additional PCR-introduced mutations by sequencing analysis.
Construction of mutants.
The construction of the misR::erm mutant has been previously described (31).
A 1,784-bp PCR product containing the entire misS coding sequence and flanking sequences was generated using primer pair YT140/YT67 and cloned into the pCR2.1 vector, yielding pTA140-67. An internal 825-bp fragment was released by EcoRV-HincII restriction and replaced by an aphA-3 cassette obtained from pUC18K with EcoRI-HincII digestion followed by Klenow treatment. Removal of the misS internal sequence and the presence of a correctly oriented aphA-3 cassette in the resulting pYT310 plasmid were confirmed by a panel of colony PCRs and sequencing analysis. This construct removed most of the periplasmic domain, the second transmembrane domain, and most of the cytoplasmic domain of MisS. To generate the meningococcal misS mutant, pYT310 was linearized with ScaI to disrupt the Ampr gene, and the digestion mixture was used to transform meningococcal strain NMB. Colonies were selected on brain heart infusion agar plates with kanamycin. Mutants were examined using colony PCR linking the aphA-3 cassette to a chromosome-specific primer and Southern blots to confirm correct allelic exchange at the chromosomal misRS locus.
Construction of reporter strains and β-galactosidase reporter assays.
Various regions of the misR promoter were obtained by PCR amplification using chromosomal DNA of the wild-type strain as a template and cloned into pCR2.1 by TOPO cloning (Invitrogen). Site-specific mutations of the MisR binding motif were constructed by megaprimer PCR mutagenesis (2) using the following mutagenic primers containing the desired base changes: YT183R-SmaI, YT183R-SnaBI, YT183R-SspI-1, YT183R-SspI-2, YT183R-SspI-3, YT183R-XhoI, YT183R-PstI. Each mutagenic primer was used in PCR amplification with YT66-ERI to generate the megaprimers. The PCR products were purified and used subsequently with YT118-ERI in the second run of the PCR to obtain the final PCR product containing the desired mutations. A 10-bp deletion was prepared using primer pairs YT184-d10-F and YT184-d10-R. The plasmids or mutagenized PCR products were digested with EcoRI and then subcloned into the pYT328 vector, which contains a unique EcoRI site upstream of the lacZ gene that is flanked by an ∼500-bp meningococcal sequence at either end. The ligation reaction products were transformed into E. coli strain DH5α, and erythromycin-resistant colonies were selected. Correct orientation of promoters relative to the lacZ gene was confirmed by colony PCR using an outward primer at the 5′ end of lacZ and a forward primer within the cloned promoter fragment. The resulting plasmids were linearized with NcoI digestion, the digestion mixtures were used directly to transform meningococcal strains, and erythromycin-resistant colonies were selected. The integration of the misR::lacZ fusion via homologous recombination into an irrelevant noncoding intergenic region was confirmed with colony PCR. β-Galactosidase assays were performed on mid-log-phase cultures in triplicate according to a previously published procedure (33). Peptone broth described by Johnson et al. (13) was used to grow the reporter strains when testing the effect of magnesium concentrations.
Protein purification and antiserum production.
One liter of LB culture of the MisR overexpression strain was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h. The harvested cells were resuspended in 30 ml of lysis buffer (50 mM sodium phosphate, pH 8.0; 300 mM NaCl; 10 mM imidazole; 1 mM phenylmethylsulfonyl fluoride) and sonicated for 30 s 10 times with 30-s cooling intervals. The cell debris was removed by centrifugation at 14,000× g for 15 min. The crude extract was then incubated with 2 ml of Ni-nitrilotriacetic acid resin (QIAGEN) for 2 h before packing into a column. The column was washed with 10 ml (each) of 20 mM and 50 mM imidazole in lysis buffer and then eluted with 10 ml of 250 mM imidazole. The fractions were pooled after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, concentrated through a centricon-3 filter (Amicon), and dialyzed in storage buffer (50 mM HEPES, pH 7.5; 100 mM NaCl; 5 mM MgCl2; 1 mM EDTA) with two buffer changes. The protein concentration was determined by Bradford assay (Bio-Rad) with bovine serum albumin as a standard. Purified MisR protein was resolved by SDS-PAGE and visualized by brief Coomassie staining, and the gel slices containing MisR protein were used as an antigen for antiserum production in rabbits (Covance, Inc.).
For the purification of MBP-MisS fusion protein, a 1-liter culture of TB1/pYT311 grown in LB media containing 0.2% glucose was induced with 1 mM IPTG for 3 h and then collected by centrifugation. The cell pellet was suspended in 30 ml of column buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride) and lysed by sonication. After removing the cell debris by centrifugation at 9,000 × g for 30 min, the lysate was incubated with amylose resin (New England Biolabs) for 1 h and then the mixture was packed into a column. The column was washed with 12 volumes of column buffer and then eluted with 15 ml of column buffer containing 10 mM maltose. Fractions were combined after SDS-PAGE analysis, concentrated through a Centriprep 30 filter (Amicon), and then dialyzed with two changes of 1 liter of storage buffer, and saved at −80°C.
In vitro phosphorylation assays.
For autophosphorylation, MBP-MisS protein (2.5 or 5 μM) was incubated with [γ-32P]ATP (3,000 Ci mmol−1 [Amersham, New Jersey], 1.8 μCi per reaction, diluted with cold ATP, and used at a final concentration of 400 μM) in 5 μl phosphorylation buffer (50 mM Tris, pH 8.0; 20 mM MgCl2; 0.1 mM EDTA; 5% glycerol) at 37°C for 10 min. Ten microliters of MisR was added to a final concentration of 5 μM, and the reaction mixtures were incubated at 37°C for an additional 30 min. The reactions were quenched by adding 5 μl of 4× loading buffer (320 mM Tris-HCl, pH 6.8; 8% SDS; 100 mM EDTA; 100 mM DTT; 40% glycerol). SDS-PAGE analysis (12% gel) was carried out at 4°C, and the gels were dried under a vacuum followed by PhosphorImager analysis (Molecular Dynamics). Alternatively, MisS and MisR proteins were mixed together, and the reactions were initiated with the addition of ATP. Results were similar to those described above.
Primer extension.
Total RNA isolated with RNeasy midi kits (QIAGEN) was treated with DNase to remove contaminating genomic DNA. Primers misR-PE1 and misR-PE3 were designed and end-labeled using [γ-32P]ATP and T4 polynucleotide kinase, and excess free label was removed with a G-50 filtration spin column. RNA (40 μg) was suspended in 30 μl of hybridization buffer with 1 μl of labeled primer. Annealing of the primer with RNA was done by heating at 85°C for 10 min, slow cooling down to 30°C, and overnight incubation at 30°C. The mixture was precipitated and resuspended in 25 μl of elongation buffer containing deoxynucleoside triphosphate and RNasin (Promega), and 40 units of avian myeloblastosis virus reverse transcriptase was added to initiate the reaction. After a 90-min incubation at 42°C, the reaction was quenched with EDTA, cleaned up with a phenol-chloroform extraction, and precipitated by ethanol. The final product pellets were resuspended in loading buffer and analyzed on an 8% acrylamide sequencing gel along with a sequencing ladder obtained with the same primer.
Electrophoretic mobility shift assay (EMSA).
Promoter fragments of misR were obtained by PCR amplification and purified by gel electrophoresis and a QIAquick gel purification kit (QIAGEN), and the DNA concentration was quantified. Three picomoles of the promoter fragments was end-labeled for 30 min at 37°C with [γ-32P]ATP (3,000 Ci/mmol) using T4 polynucleotide kinase (New England Biolabs). The labeled DNA was purified through nucleotide removal kits (QIAGEN), and the specific activity of the probe was determined by scintillation counting. The binding reactions, consisting of 5 fmol of labeled DNA and various amounts of protein in a binding buffer (20 mM HEPES, pH 7.9, 60 mM KCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 0.3 mg/ml bovine serum albulin, 0.2 mg/ml salmon sperm DNA, and 10% glycerol), were carried out at 30°C for 20 min, and the reaction mixtures were directly loaded onto 6% nondenaturing polyacrylamide gels (0.5× TBE, 40:1 cross-linking), which had been prerun for 30 min. The electrophoresis was carried out for another 2.5 h at a constant 250 V. The gels were dried under a vacuum without prior fixing, and the resulting radioactive electrophoretic patterns were analyzed with a PhosphorImager. The binding reactions were examined with phosphorylated (50 mM acetyl phosphate for 30 min at 37°C) or nonphosphorylated MisR. Competition with excess specific competitors (unlabeled probes) and nonspecific competitors (593-bp internal coding sequences of misR, obtained by PCR amplification using primers YT45 and YT46) was performed to assess the specificity of the interaction.
DNase I protection.
Oligonucleotide primers corresponding to either the coding or the noncoding strand were end-labeled using T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol; Amersham) and purified with the QIAquick nucleotide removal kit (QIAGEN). PCR fragments corresponding to the promoter region were then amplified with the radiolabeled primer and the corresponding reverse primer, and the labeled DNA products were purified with a QIAquick PCR purification kit. Using the binding conditions defined in the EMSA studies, DNA-protein complex generated with labeled DNA and increasing amounts of purified MisR in the EMSA binding buffer without salmon sperm DNA was treated with diluted DNase for 1 min at room temperature before quenching with a phenol-chloroform extraction. Nucleic acids were recovered by ethanol precipitation, resuspended in sequencing loading solution, and analyzed on an 8 M urea-6% acrylamide sequencing gel along with a sequencing ladder obtained with the same labeled oligonucleotide. Both phosphorylated and nonphosphorylated MisR proteins were examined to determine whether the MisR protein exhibited a different binding specificity upon phosphorylation.
Immunoblots.
Bacteria (1 × 108 cells based on measurements of optical density at 550 nm) were collected from the wild-type strain and the misR and misS mutants, resuspended in loading buffer, and boiled for 4 min. Samples were resolved on a 12% SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Bio-Rad). Anti-MisR polyclonal sera were used at a dilution of 1:1,000, and anti-rabbit alkaline phosphatase-conjugated secondary antibody was used at a 1:5,000 dilution. The blots were developed according to the standard protocol.
RESULTS
Autophosphorylation of MisS and phosphotransfer from MisS∼P to MisR.
To characterize the functions of MisR and MisS protein in vitro, both proteins were overexpressed and purified. To simplify purification of the MisS histidine kinase, which contains two transmembrane domains flanking a large periplasmic domain, the C-terminal soluble kinase domain downstream of the second transmembrane domain was fused at the N terminus with the maltose binding protein (MalE). Similarly, MisR was tagged at the C terminus with a (His)6 polypeptide to facilitate purification. Both fusion proteins, MisR-(His)6 and MBP-MisS202, were highly expressed in E. coli when induced and were purified to high degree (>95%) of homogeneity using the Ni-nitrilotriacetic acid and amylose affinity chromatography, respectively (see Fig. S1 in the supplemental material).
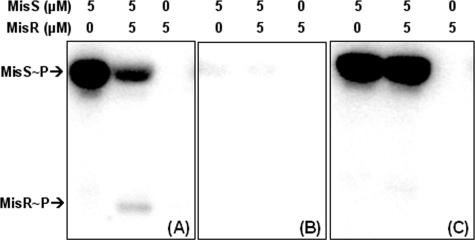
The MBP-MisS fusion protein was autophosphorylated in the presence of ATP (Fig. 1A), and upon the addition of MisR protein, MisS∼P transferred its phosphoryl group to MisR, confirming that these two proteins are paired partners in the phosphotransfer process. The phosphorylated MisR (MisR∼P) appeared to be unstable, as a net loss of protein-bound radiolabeled phosphate (MisS∼P and MisR∼P) was observed (Fig. 1A) and MisR∼P did not accumulate to a high level with prolonged incubation (data not shown). This apparent instability of MisR∼P might be due to the autophosphatase activity of MisR or a phosphatase activity mediated by the MisS kinase fusion protein, as many histidine kinases also have phosphatase activities (10). Thus, maintaining the level of MisR∼P may be an important control point in signal propagation.
FIG. 1.
In vitro phosphorylation of MisS and MisR. MisS was autophosphorylated with 0.4 mM ATP for 10 min prior to the addition of MisR, and the reaction was allowed to proceed for an additional 30 min before being quenched by SDS-PAGE loading buffer. The reaction mixtures were resolved on a 12% gel, and the radioactivity was detected by a phosphorimager. The final concentration of each protein is indicated above each panel. (A) Wild-type MisS and wild-type MisR; (B) MisS246HA and wild-type MisR; (C) wild-type MisS and MisR52DA.
Based on sequence homology with other two-component proteins, the conserved His residue at position 246 in MisS and the conserved Asp residue at position 52 in MisR were predicted to be the sites of phosphorylation. The involvement of these two residues in the phosphorylation reactions was examined by analysis of site-specific Ala mutant proteins. His246 and Asp52 in MisS and MisR, respectively, were altered by site-directed mutagenesis to create MisS246HA and MisR52DA. These mutant proteins were overexpressed and purified in parallel with the wild-type proteins to ensure that no activity difference was caused by the protein purification process. The MisS246HA mutant protein was defective in autophosphorylation (Fig. 1B), and similarly, mutation of Asp52 to Ala in MisR rendered the mutant protein unable to accept the phosphoryl group from MisS∼P (Fig. 1C). The results suggested that these two conserved residues are the likely phosphorylation sites in the autophosphorylation and phosphotransfer reactions of the MisS/MisR two-component system.
Characterization of the promoter region of the misRS operon.
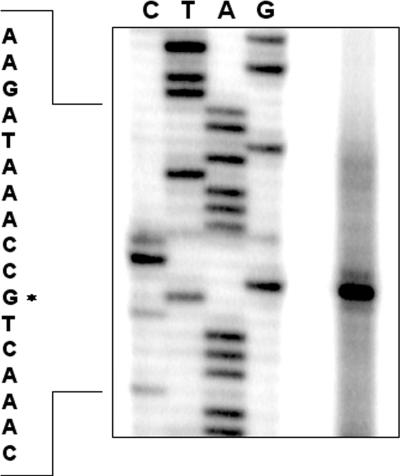
To determine the transcriptional start site of the misRS operon, primer extension experiments were conducted using total RNA isolated from the wild-type strain NMB and a reverse primer complementary to the coding strand of the misR gene. A single major product was observed corresponding to the −67 guanine nucleotide (Fig. 2), indicating a single promoter upstream of misR. The same start site was identified from a second complementary primer (data not shown). Examination of the sequence upstream of the transcriptional start site revealed appropriate promoter elements of a −10 (TAAGAT) and a −35 (TACGCT) sequence separated by 18-bp (see Fig. 7C).
FIG. 2.
Primer extension analysis using a primer complementary to misR and the total RNA isolated from the wild-type strain NMB. Lanes G, A, T, and C indicate the dideoxy sequencing reactions. The asterisk indicates the transcriptional start site.
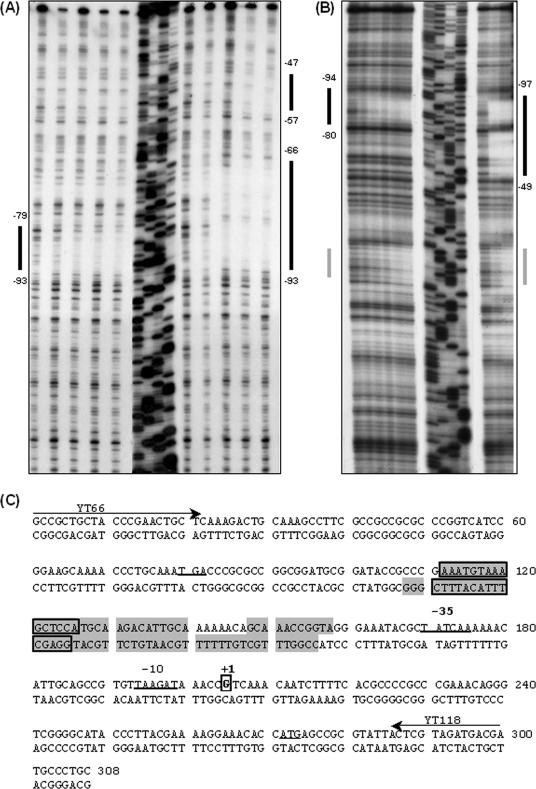
FIG. 7.
DNase I protection of the misR promoters by MisR∼P. The coding (A) or noncoding (B) strand of 32P-end-labeled misR promoter fragment between −205 to +104 with respect to the misR transcriptional start site was incubated with increasing amounts of MisR (left in panels A and B) or MisR∼P (right in panels A and B) for 20 min at 30°C and then subjected to DNase I digestion. The amounts of MisR used in panel A are 0, 85, 170, 255, and 340 pmol for both MisR and MisR∼P, while those used in panel B are 0, 170, 340, 680, 1,360, and 2,720 pmol for MisR and 0, 170, and 340 pmol for MisR∼P. Dideoxy chain termination sequences corresponding to the probes are shown in the order of G, A, T, and C. Black bars indicate regions protected, while the gray bar in panel B indicates a region of the noncoding strand that was not reproducibly protected in all footprinting experiments. (C) Sequence of the misR promoter region used in the DNase I footprinting experiments. The protected regions for MisR (boxed and shaded) and MisR∼P (shaded) are indicated for both strands. The transcriptional start site and −10 and −35 promoter elements are boxed, while the start codons are underlined. Arrows indicate primers used to generate the probes.
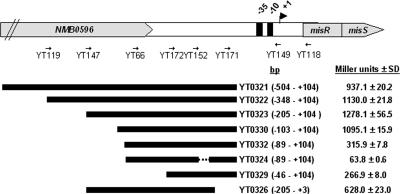
To delineate the required promoter length for transcriptional activation and further define the location of important motifs, a series of misR::lacZ transcriptional fusion constructs were made (Fig. 3). These fusions were inserted at a permissive heterologous chromosomal site as a single copy, and the expression of β-galactosidase was determined in culture grown to mid-log phase. As shown in Fig. 3, a misR promoter fragment that contains the nucleotide sequence between −504 and +104 bp (strain YT0321) generated similar activity to that of a strain carrying the −103 to +104 sequence (strain YT0330), indicating that the ∼100-bp sequence upstream of the transcriptional start site was sufficient for the misR promoter activation. However, removing the additional 14-bp upstream sequence (comparing strains YT0330 and YT0332) significantly reduced the promoter activity to a level similar to that of a strain with only the −35 and −10 promoter elements (strain YT0329), indicating a critical DNA motif was present in this upstream region. Interestingly, an ∼2-fold reduction of transcriptional activity was also observed when the downstream sequence within the untranslated region was deleted (comparing strains YT0323 and YT0326). The function of the −10 element was confirmed by the deletion of an internal 33-bp fragment (−8 to +25) in strain YT0324 that completely eliminated the promoter activity.
FIG. 3.
Transcriptional activities of various misR::lacZ promoter fusions. The schematic diagram of the misRS operon shows the locations of primers used to generate the promoter fragment. The cloned region with respect to the transcriptional start site (+1) is shown in parentheses. The dashed line indicates the 33-bp deleted region within the promoter in strain YT0324. β-Galactosidase activities of strains grown in GC broth to mid-log phase are expressed as Miller units ± standard deviation corresponding to mean values of triplicate measurements. The activity of a promoterless lacZ construct (strain 263) is 57.4 ± 1.7 Miller units.
MisR autogenously activates misRS expression.
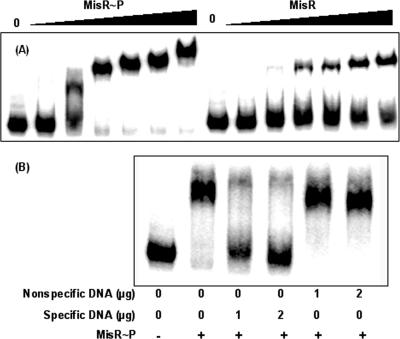
Many two-component regulatory systems are under the control of autoregulatory mechanism (4, 24, 26). To test whether MisR regulated its own expression, the direct involvement of MisR was examined using EMSA with a misR promoter fragment obtained by PCR amplification and end-labeled with 32P by T4 kinase. Similar to many two-component response regulators (16), the MisR protein can be phosphorylated with a small-molecule phosphoryl donor, acetyl phosphate, thus enabling the functional characterization of phosphorylated MisR. The phosphorylated and unphosphorylated MisR migrated differently during native gel electrophoresis. Estimation of the remaining unphosphorylated MisR after incubation with 50 mM acetyl phosphate followed by native gel electrophoresis and Coomassie blue staining revealed that ∼50% of the MisR protein was phosphorylated under these conditions (data not shown). This MisR∼P preparation was then used for the EMSAs. As shown in Fig. 4A, a dose-dependent shift of the misR probe was noted for both MisR and MisR∼P. A complete shift was observed with MisR∼P, while incomplete shifts were detected for unphosphorylated MisR at all protein concentrations tested. This result indicated that MisR∼P exhibited a significantly higher affinity for the misR promoter than MisR.
FIG. 4.
EMSA of MisR-misR promoter interaction. (A) A 608-bp PCR product (∼5 fmol) of a DNA fragment containing the misR promoter region (−504 to +104) was 32P-labeled by T4 kinase, mixed with increasing amounts of MisR∼P (left) and MisR (right) for 20 min at 30°C and then subjected to gel electrophoresis. MisR∼P was generated by incubation with 50 mM acetyl phosphate for 30 min at 37°C. The amounts of MisR protein are 0, 34, 68, 136, 204, 272, and 340 pmol. (B) Competition EMSA. 32P-labeled misR probe was mixed with the indicated amounts of either specific or nonspecific DNA, incubated with MisR∼P (102 pmol), and analyzed as described above.
To confirm that MisR binding to its promoter region was specific, unlabeled misR promoter DNA was used in a competition assay. As seen in Fig. 4B, an excess amount of specific misR promoter DNA eliminated the binding of MisR, while in the presence of an equal amount of a nonspecific DNA (an internal misR coding sequence), the specific binding activity of MisR to the misR promoter fragment was retained (Fig. 4B). These results thus confirmed that the binding of MisR to the misR promoter was sequence specific.
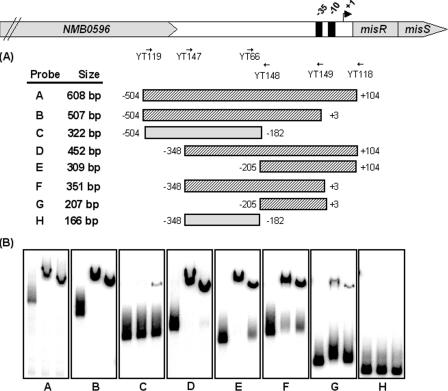
As a relatively large promoter fragment (608 bp) was used for the initial EMSA characterization, a more precise location of the misR promoter region important for binding was addressed. To delineate the minimal sequence essential for MisR binding, various overlapping misR promoter fragments were generated (Fig. 5A) and a series of electrophoretic mobility shift assays were performed utilizing these promoter fragments together with either nonphosphorylated MisR or phosphorylated MisR, and the results are shown in Fig. 5B. As expected, the original full-length misR promoter fragment (panel A) was shifted in the presence of MisR and MisR∼P. Truncation from the proximal region of the misR promoter to position −182 almost eliminated the binding of MisR protein, whereas removal of ∼300 bp from the distal region retained the MisR protein-promoter DNA interaction. Thus, approximately 200 to 300 bp of the promoter fragments was sufficient to give positive shifts in the EMSAs. Consistently, the MisR-DNA interaction region defined by EMSA correlated with the required promoter length in the in vivo transcriptional fusion studies (Fig. 3).
FIG. 5.
Mapping of the MisR binding site within the misR promoter region by EMSA using overlapping DNA fragments. (A) Schematic presentation of each probe used in this study with the primers used to generate these DNA fragments depicted above. Hatched boxes represent probes that are shifted by MisR, and gray boxes indicate PCR fragments that are not shifted. Numbers adjacent to the boxes indicate the endpoints of the fragment with respect to the start site of transcription. (B) In each panel, the left lane contains DNA probe only, the middle lane contains DNA probe and MisR∼P (340 pmol), and the right lane contains DNA probe and MisR (340 pmol). The binding conditions are as described in the legend to Fig. 4.
The MisS kinase is critical in autoregulation of the misRS operon.
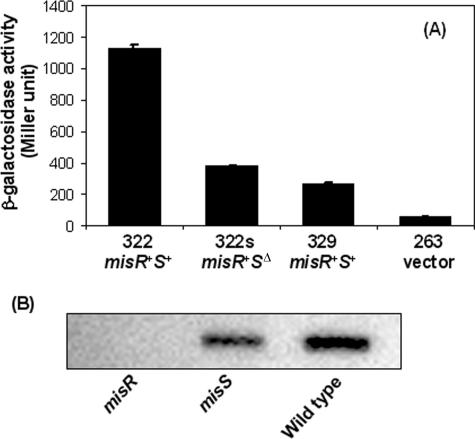
To examine the plausibility of an autoactivation mechanism, transcriptional misR::lacZ fusion constructs were made in the wild-type strain and the misS mutant. As shown in Fig. 6A, inactivating misS (strain YT322s) diminished the promoter activity to a level similar to that of a construct containing only the −35 and −10 promoter elements (strain YT329). These data indicated that phosphorylation of MisR by MisS was critical in the autoregulation of the misRS operon and that the MisRS system was the major activating mechanism of its expression. These data also suggested that there was minimal, if any, cross-phosphorylation of MisR by other two-component kinases present in meningococci or by the intracellular pool of acetyl phosphates. The importance of MisR and MisS in the transcription of its own operon was also confirmed by quantitative real-time PCR experiments using the SYBR green detection method. The misR probe was designed to locate upstream from the erm cassette insertion site, thus avoiding any transcriptional effect caused by the antibiotic cassette. The expression of misR decreased significantly in both the misR null mutant and the misS mutant (data not shown).
FIG. 6.
Transcription of misR (A) and MisR protein production (B) are reduced in the misS mutant. (A) β-Galactosidase activities of misR::lacZ fusions in various genetic backgrounds. The misRS genotypes are indicated below the strains. Strain YT263 is the promoterless negative control. (B) Western blotting and probing with polyclonal anti-MisR antisera were performed on whole-cell lysates of the wild-type strain, the misR mutant, and the misS mutant. Equal cell numbers (1 × 108 cells) were loaded in each sample.
In correlation with the in vivo transcriptional reporter studies, the reduced MisR expression in the misS mutant was also demonstrated at the protein level. Western blots were performed on whole-cell lysates derived from the wild type, misR mutant, and misS mutant utilizing polyclonal antisera raised against the MisR-His6 recombinant protein (Fig. 6B). As expected, MisR was absent in the misR null mutant, and a reduced expression was seen in the misS sample compared to that of the wild-type lysate. Taken together, these data demonstrated that autoregulation of the misRS operon requires both MisS and MisR.
Identification of the MisR-binding sequence within the misR promoter region.
To identify the precise location of MisR binding site(s), DNase I protection assays were performed on both strands of a 309-bp 32P-end-labeled misR promoter probe (−205 to +104 with respect to the misR transcriptional start site, probe E) (Fig. 5). In addition, the effect of phosphorylation on DNA-binding activity of MisR was analyzed with a MisR∼P preparation generated by incubation with 150 mM acetyl phosphate. In the presence of increasing amounts of MisR, a 15-bp region (AAATGTAAAGCTCCA, −93 to −79) of the coding strand and the noncoding strand (−94 to −80) was protected from DNase I digestion (Fig. 7A and B). Upon interaction with MisR∼P, a larger region (−93 to −66) and an additional 11-bp (−57 to −47) downstream sequence of the coding strand was protected (Fig. 7A). Similar patterns of protection by MisR∼P were observed on the noncoding strand (Fig. 7B). A region of DNase I hypersensitivity was also observed on the coding strand upon MisR∼P binding. These data suggested that a higher degree of MisR oligomerization likely occurred upon phosphorylation and that the interaction of the MisR∼P oligomers with its target DNA resulted in more extensive protection from DNase I digestion. In addition, the results also indicated that the 15-bp region protected by nonphosphorylated MisR was potentially a high-affinity binding site for MisR.
The protected region of the misR promoter in the DNase I footprinting experiments fell within the sequence deemed important in the EMSAs and the transcriptional reporter assays. However, no clear secondary structure of the MisR-binding motif, such as direct repeats or inverted repeats, was identified from these protected sequences (Fig. 7C). Additionally, the defined MisR-binding sequence did not share any sequence similarity to binding motifs of other two-component regulatory systems.
When correlated with the in vivo transcriptional fusion studies, the importance of the sequence identified by DNase I footprinting was confirmed. While the misR promoter within strain YT0332 was only 14 bp shorter at the 5′ end than that of strain YT0330, the transcriptional activation by MisR was completely eliminated (Fig. 3). The omitted region in strain YT0332 overlapped (4 bp) with the 5′ end of the 15-bp sequence identified above (Fig. 7C) and thus demonstrated the importance of this sequence in MisR-DNA interaction.
To characterize the importance of the MisR-binding sequences identified through DNase I protection assays, individual nucleotides were mutated by site-directed mutagenesis and introduced into the reporter strain YT0323, and the resulting promoter activities were compared to that of the wild-type reporter strain (see Fig. S2 in the supplemental material). All nucleotide changes introduced within the 15-bp high-affinity site resulted in an approximately 50% reduction in promoter activities. While nucleotide changes only had modest effects, deletion of four bases from the 5′ end of the 15-bp motif (strain YT0332) eliminated the activation by MisR∼P. Removal of the proximal 10-bp sequence protected only by MisR∼P also caused ∼50% reduction in promoter activity. As the mutation did not completely eliminate MisR-DNA interaction, this analysis suggested that the studied nucleotides are not essential in MisR binding and that there are likely multiple contacts between MisR protein and its target sequence.
Different magnesium concentrations did not affect expression of the misRS operon.
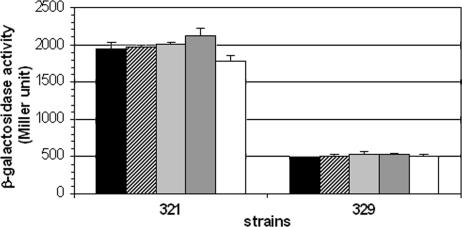
The misR/S system has been proposed by Newcombe and coworkers to be the magnesium-sensing meningococcal phoP/Q system (13, 18, 19). However, magnesium-mediated regulation of genes under the control of the MisR/S system was not clearly demonstrated. As we have shown, expression of the misRS operon is directly controlled by MisR/S through an autoinduction mechanism. Thus, if magnesium were an environmental signal detected by the histidine kinase, varying magnesium concentrations in the growth media would cause changes in misR transcription. Using the YT321 reporter strain, which contains the longest misR promoter sequence, and the YT329 strain, which carries the minimal misR promoter element, the effect of various magnesium concentrations in the peptone broth described by Newcombe et al. (18, 19) was examined. As shown in Fig. 8, magnesium concentrations ranging from 20 mM to trace amounts (no exogenous addition of magnesium) has no effect on the expression of misR promoter fusions. These data demonstrated that magnesium is not the signal sensed by the MisR/S two-component regulatory system, thus MisR/S is not a functional homologue of the PhoP/Q system in Salmonella.
FIG. 8.
The misR promoter activity is not affected by different magnesium concentrations. β-Galactosidase activities of reporter strains grown in peptone broth supplemented with various amounts of magnesium were determined. Black bar, 0 mM; dotted bar, 1 mM; hatched bar, 2 mM; gray bar, 10 mM; white bar, 20 mM. A representative result from two independent experiments is shown where each condition was assayed in four replicates. Error bars indicate standard deviations.
DISCUSSION
Neisseria meningitidis, an obligate human pathogen, remains a leading cause of meningitis and rapidly fatal sepsis, usually in otherwise healthy individuals. To successfully colonize and survive within the human host, N. meningitidis responds to signals emanating from the host environment, a task often carried out in bacteria by environmental sensing two-component regulatory systems. The meningococcus contains only four pairs of predicted two-component regulatory systems, which presumably sense the restricted human host environment encountered. It has previously been shown that the N. meningitidis MisR/MisS two-component system is involved in phosphorylation of the LOS inner core (31) and, when mutated, resulted in sensitivity to antimicrobial peptides (13, 31) and attenuation of virulence in a mouse model of infection (18). However, the genes regulated by the MisR/S proteins have yet to be characterized. In this report, using purified proteins, we confirmed that MisR/MisS communicates via paired phosphoryl transfer reactions between the conserved His246 residue of the kinase and the invariable Asp52 residue of the response regulator. This is the first detailed biochemical characterization of a meningococcal two-component signal transduction system.
The regulation of the misRS operon, which encodes the transcriptional response regulator MisR and the histidine sensor kinase MisS, was investigated. We established that the meningococcal misRS is transcribed from a single promoter, which is positively autoregulated by both MisR and MisS through a direct interaction between the phosphorylated MisR response regulator and a 15-bp sequence motif upstream of the promoter. The footprint of MisR extended just upstream of the −35 region of the misR promoter, in keeping with its role as a transcriptional activator of the misRS operon. Thus, the misRS operon is the first promoter shown to be under the direct control of this two-component system, and an autoregulatory mechanism was demonstrated. Autoregulation is an important feature of many two-component regulatory systems and has been characterized in detail in several systems, such as phoPQ of Salmonella enterica serovar Typhimurium (26), cpxRA of E. coli (4) and bvgAS of Bordetella pertussis (24). Autoregulation allows positive feedback amplification of the signals leading to increased levels of both MisR and MisS and concomitant modulation of MisR-controlled gene expression in response to the detection of environmental changes.
Several two-component response regulators have also been shown to bind DNA in their nonphosphorylated states (6, 11, 15, 23); however, very few were reported to activate gene transcription without phosphorylation (6, 7). In the presence of acetyl phosphate, MisR clearly showed increased affinity to the misR promoter region in both EMSA and footprinting experiments, although MisR phosphorylation is incomplete under the experimental conditions and MisR∼P appears to be labile. The in vivo transcriptional fusion data suggest that nonphosphorylated MisR may not play a critical role in the activation of its own expression, since no substantial activation occurred without MisS. The role of MisS in autoregulation was also revealed at the protein level, since a misS null mutant expressed a lower basal level of MisR protein, probably due to the absence of MisS to activate the system. Thus, our results are consistent with a model where the phosphorylated form of MisR is the major transcriptional activator of the misRS operon and unphosphorylated MisR is not likely to compete with MisR∼P for the MisR-binding site upstream of the misRS promoter in vivo. Phosphorylation of response regulators often leads to an oligomeric status that provided cooperative binding to their target DNA sequences, thus resulting in an enhanced affinity. Interestingly, slower-mobility bands of the misR probes in EMSA were observed at higher concentrations of MisR∼P (Fig. 4). Similarly, increased shift positions were seen with higher concentrations of MisR, albeit to a lesser extent. This phenomenon may be caused by further oligomerization of MisR∼P on DNA or by aggregation of the protein-DNA complex, and the difference in shifts of MisR∼P and MisR may indicate different levels of oligomerization. As shown in the DNase I footprinting analysis, MisR∼P exhibits increased binding affinity and protects a larger region, consistent with the oligomerization of MisR∼P upon DNA binding.
The majority of PhoP-activated genes are transcriptionally regulated by the concentration of Mg2+ in the growth medium. It has been shown (25) that the ratio of β-galactosidase activity for cells grown in low (μM range) versus high (mM range) Mg2+ concentrations ranges from 4 to 287. The laboratory of Newcombe and coworkers reported (13, 19) that neither the wild type nor the response regulator mutant of two different serogroup C meningococcal strains grew well at micromolar concentrations (<0.1 mM) (13) of magnesium; thus, comparison of magnesium regulation was performed either at 0.16 mM versus 10 mM (13) or 2 mM versus 20 mM (19). Through microarray analyses, the authors observed a total of 281 genes that were deregulated in the NMB0595 (misR) mutant compared to the wild type when grown on blood agar, and a total of 106 genes were identified as genes significantly regulated by magnesium when comparing 20 mM versus 2 mM concentrations. Only ∼12% of the genes presumably controlled by NMB0595 were regulated by magnesium (19). NMB0595/NMB0594 (misR/misS) encoding the two-component system were shown to be upregulated ∼2.0 and 1.5-fold, respectively, at 2 mM magnesium (19). However, the microarray observation has not been further confirmed by other assays of transcriptional activity, such as reporter fusion or real-time quantitative PCR. Comparing the upstream promoter and the coding nucleotide sequence of the serogroup B misR/S locus to that of the assembled genome sequence of a serogroup C strain FAM18 (www.sanger.ac.uk) revealed nearly 100% identity, suggesting that the misRS operon in the serogroup C strain is likely controlled by the same autoactivation mechanism. Without the knowledge of which genes among the 281 genes reported to be controlled by the MisRS (PhoPQ) system are under direct control, attempts to examine the upstream sequences and identify additional MisR binding motifs were of limited success. As the MisR binding motif is A/T rich, a better understanding of the important nucleotides that interact directly with MisR, using assays such as the uracil interference assay, would facilitate this bioinformatics approach to understanding the scope of regulation of the MisRS two-component system.
In contrast, no magnesium-dependent growth differences were observed in the serogroup B meningococcal strain NMB used in our studies, and the reporter strains derived from NMB grew well in peptone broth without magnesium supplementation. In addition, we showed that the transcription of the misR/S operon is under the autoregulatory control of the MisR response regulator, yet magnesium did not cause a change in the expression of misR when examined by transcriptional lacZ fusion assays. If magnesium is indeed the inducing signal and the system is under positive feedback regulation, one would expect a significant upregulation of the misRS expression upon signal detection. In silico structural homology analysis of the NMB0595 (MisR) protein indicated that it belongs to the OmpR subfamily of response regulators, which includes E. coli PhoP, and this analysis was used (19) to support the conclusion that NMB0595/NMB0594 is a functional homologue of the Salmonella phoP/Q system. However, the members of the response regulator family are highly homologous due to the conserved phosphotransfer reaction mechanism. Only the identification of the inducing signal, the characterization of its regulon, or the functional complementation of its true ortholog in other bacterial systems will define the real signaling function of the two-component regulatory systems. Genes perturbed in the NMB0595 mutant, as described by Newcombe et al. (19), overlap those identified as being regulated by several two-component systems in E. coli, including ArcAB, PhoPQ, CpxRA, and BasRS (35). If magnesium were not the inducing signal, it would be misleading to name the NMB0595/NMB0594 system the PhoPQ system.
Many response regulators bind to DNA consensus sequences that were determined by compiling multiple binding sites (11, 23, 26, 30). The consensus DNA binding sequences are often composed of direct repeats, inverted repeats, or multiple highly homologous sites spaced throughout the promoter region. For example, a consensus sequence, 5′-GTAAA(N5)GTAA-3′, has been defined for CpxR (22), while a direct repeat consensus sequence (T/G)GTTTA-(N5)-(T/G)GTTTA was defined as the PhoP box (5). Thus far, only one DNA binding site defined in the misR promoter region is known. This site does not show homology with previously identified consensus DNA binding sequences of other two-component systems. Efforts are under way to identify additional MisR-regulated genes and characterize MisR binding sites. The misR promoter represents the first MisR-activated promoter of this two-component system. Understanding the regulatory circuitry of the MisR/MisS meningococcal two-component system provides a foundation for further characterization of regulatory networks in this human pathogen.
Supplementary Material
Acknowledgments
This work was supported by grants R01 AI061031 (Y.T.), R01 AI033517, and R01 AI40247 (D.S.S.) from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Achtman, M., K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, B. G. Barrell, and J. Parkhill. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 2.Barik, S. 1995. Site-directed mutagenesis by double polymerase chain reaction. Mol. Biotechnol. 3:1-7. [DOI] [PubMed] [Google Scholar]
- 3.Chamnongpol, S., M. Cromie, and E. A. Groisman. 2003. Mg2+ sensing by the Mg2+ sensor PhoQ of Salmonella enterica. J. Mol. Biol. 325:795-807. [DOI] [PubMed] [Google Scholar]
- 4.De Wulf, P., O. Kwon, and E. C. Lin. 1999. The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J. Bacteriol. 181:6772-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haydel, S. E., W. H. Benjamin, Jr., N. E. Dunlap, and J. E. Clark-Curtiss. 2002. Expression, autoregulation, and DNA binding properties of the Mycobacterium tuberculosis TrcR response regulator. J. Bacteriol. 184:2192-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Himpens, S., C. Locht, and P. Supply. 2000. Molecular characterization of the mycobacterial SenX3-RegX3 two-component system: evidence for autoregulation. Microbiology 146(Pt. 12):3091-3098. [DOI] [PubMed] [Google Scholar]
- 8.Hobbs, M., E. S. Collie, P. D. Free, S. P. Livingston, and J. S. Mattick. 1993. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 7:669-682. [DOI] [PubMed] [Google Scholar]
- 9.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165-170. [DOI] [PubMed] [Google Scholar]
- 10.Hoch, J. A., and T. J. Silhavy. 1995. Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 11.Holman, T. R., Z. Wu, B. L. Wanner, and C. T. Walsh. 1994. Identification of the DNA-binding site for the phosphorylated VanR protein required for vancomycin resistance in Enterococcus faecium. Biochemistry 33:4625-4631. [DOI] [PubMed] [Google Scholar]
- 12.Janik, A., E. Juni, and G. A. Heym. 1976. Genetic transformation as a tool for detection of Neisseria gonorrhoeae. J. Clin. Microbiol. 4:71-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, C. R., J. Newcombe, S. Thorne, H. A. Borde, L. J. Eales-Reynolds, A. R. Gorringe, S. G. Funnell, and J. J. McFadden. 2001. Generation and characterization of a PhoP homologue mutant of Neisseria meningitidis. Mol. Microbiol. 39:1345-1355. [DOI] [PubMed] [Google Scholar]
- 14.Lissenden, S., S. Mohan, T. Overton, T. Regan, H. Crooke, J. A. Cardinale, T. C. Householder, P. Adams, C. D. O'Conner, V. L. Clark, H. Smith, and J. A. Cole. 2000. Identification of transcription activators that regulate gonococcal adaptation from aerobic to anaerobic or oxygen-limited growth. Mol. Microbiol. 37:839-855. [DOI] [PubMed] [Google Scholar]
- 15.Liu, W., and F. M. Hulett. 1997. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J. Bacteriol. 179:6302-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCleary, W. R., and J. B. Stock. 1994. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 269:31567-31572. [PubMed] [Google Scholar]
- 17.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newcombe, J., L. J. Eales-Reynolds, L. Wootton, A. R. Gorringe, S. G. Funnell, S. C. Taylor, and J. J. McFadden. 2004. Infection with an avirulent phoP mutant of Neisseria meningitidis confers broad cross-reactive immunity. Infect. Immun. 72:338-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newcombe, J., J. C. Jeynes, E. Mendoza, J. Hinds, G. L. Marsden, R. A. Stabler, M. Marti, and J. J. McFadden. 2005. Phenotypic and transcriptional characterization of the meningococcal PhoPQ system, a magnesium-sensing two-component regulatory system that controls genes involved in remodeling the meningococcal cell surface. J. Bacteriol. 187:4967-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oshima, T., H. Aiba, Y. Masuda, S. Kanaya, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46:281-291. [DOI] [PubMed] [Google Scholar]
- 21.Pawlowski, K., U. Klosse, and F. J. de Bruijn. 1991. Characterization of a novel Azorhizobium caulinodans ORS571 two-component regulatory system, NtrY/NtrX, involved in nitrogen fixation and metabolism. Mol. Gen. Genet. 231:124-138. [DOI] [PubMed] [Google Scholar]
- 22.Pogliano, J., A. S. Lynch, D. Belin, E. C. Lin, and J. Beckwith. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11:1169-1182. [DOI] [PubMed] [Google Scholar]
- 23.Roy, C. R., and S. Falkow. 1991. Identification of Bordetella pertussis regulatory sequences required for transcriptional activation of the fhaB gene and autoregulation of the bvgAS operon. J. Bacteriol. 173:2385-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scarlato, V., A. Prugnola, B. Arico, and R. Rappuoli. 1990. Positive transcriptional feedback at the bvg locus controls expression of virulence factors in Bordetella pertussis. Proc. Natl. Acad. Sci. USA 87:10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soncini, F. C., E. Garcia Vescovi, F. Solomon, and E. A. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soncini, F. C., E. G. Vescovi, and E. A. Groisman. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177:4364-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens, D. S., J. S. Swartley, S. Kathariou, and S. A. Morse. 1991. Insertion of Tn916 in Neisseria meningitidis resulting in loss of group B capsular polysaccharide. Infect. Immun. 59:4097-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephens, D. S., and S. M. Zimmer. 2002. Pathogenesis, therapy, and prevention of meningococcal sepsis. Curr. Infect. Dis. Rep. 4:377-386. [DOI] [PubMed] [Google Scholar]
- 29.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 30.Tsung, K., R. E. Brissette, and M. Inouye. 1989. Identification of the DNA-binding domain of the OmpR protein required for transcriptional activation of the ompF and ompC genes of Escherichia coli by in vivo DNA footprinting. J. Biol. Chem. 264:10104-10109. [PubMed] [Google Scholar]
- 31.Tzeng, Y. L., A. Datta, K. D. Ambrose, J. K. Davies, R. W. Carlson, D. S. Stephens, and C. M. Kahler. 2004. The MisR/MisS two-component regulatory system influences inner core structure and immunotype of lipooligosaccharide in Neisseria meningitidis. J. Biol. Chem. 279:35053-35062. [DOI] [PubMed] [Google Scholar]
- 32.Tzeng, Y.-L., and D. S. Stephens. 2000. Epidemiology and pathogenesis of Neisseria meningitidis. Microbes Infect. 6:687-700. [DOI] [PubMed] [Google Scholar]
- 33.Tzeng, Y. L., J. S. Swartley, Y. K. Miller, R. E. Nisbet, L. J. Liu, J. H. Ahn, and D. S. Stephens. 2001. Transcriptional regulation of divergent capsule biosynthesis and transport operon promoters in serogroup B Neisseria meningitidis. Infect. Immun. 69:2502-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unden, G., and J. Bongaerts. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217-234. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto, K., K. Hirao, T. Oshima, H. Aiba, R. Utsumi, and A. Ishihama. 2004. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 280:1448-1456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.