Abstract
The proteins PomA, PomB, MotX, and MotY are essential for the motor function of Na+-driven flagella in Vibrio spp. Both MotY and MotX have the two cysteine residues (one of which is in a conserved tetrapeptide [CQLV]) that are inferred to form an intramolecular disulfide bond. The cysteine mutants of MotY prevented the formation of an intramolecular disulfide bond, which is presumably important for protein stability. Disruption of the disulfide bridge in MotX by site-directed mutagenesis resulted in increased instability, which did not, however, affect the motility of the cells. These lines of evidence suggest that the intramolecular disulfide bonds are involved in the stability of both proteins, but only MotY requires the intramolecular bridge for proper function.
Rotation of a helical flagellar filament is the most prevalent means of propulsion in bacteria (5, 6, 18). The driving force for this flagellar rotation is coupled to the flux of a specific ion across the cytoplasmic membrane (19). To date, proton and sodium ions have been shown to be used in this energy-coupling step (34). The motor, which is embedded in the cytoplasmic membrane at the base of a flagellum, is composed of stator and rotor components. In gram-negative bacteria, the basal-body structure consists of a rod and four surrounding ring structures called the L ring, the P ring, the MS ring, and the C ring (11). It is thought that multiple stator units surround the MS ring (7, 14, 24) and that the rotational force is generated by a stepping process (31).
Vibrio alginolyticus has two types of flagella in each bacterial cell: a lateral flagellum with a proton-driven motor and a polar flagellum with a sodium-driven motor (2, 13). The most extensively studied sodium-driven motors are the polar flagella of Vibrio spp. (22, 34). In these sodium-driven motors, PomA, PomB, MotX, and MotY have been identified as essential proteins for torque generation. PomA and PomB are membrane proteins that have four and one transmembrane segments, respectively. Furthermore, the C-terminal periplasmic domain of PomB contains a segment with high sequence similarity to the peptidoglycan-binding motif (1). Thus, PomB is believed to anchor and immobilize the PomA-PomB complex to the peptidoglycan layer in the inner membrane. A PomA-PomB complex contains four PomA subunits and two PomB subunits, and purified complexes have been shown to catalyze sodium influx when they were reconstituted into proteoliposomes (29, 30, 36). In a proton-driven motor of Escherichia coli, the torque-generating unit is composed of four MotA subunits and two MotB subunits, which are orthologues of PomA and PomB, respectively (9, 16). Unlike the sodium-driven polar-flagellar motor, however, the proton-driven motor of Escherichia coli does not require MotX and MotY for torque generation.
MotX and MotY were identified in Vibrio spp. as proteins specific for the sodium-driven motor (20, 21, 26, 28). Additionally, MotY homologues were recently reported to be part of the lateral proton-driven flagellar system of Vibrio spp. and the flagellum of Pseudomonas aeruginosa (12, 32). As with MotB and PomB, MotY contains a C-terminal peptidoglycan-binding motif (28). We have shown that the N-terminal segments of MotX and MotY contain secretion signals and that cleavage of these signals leaves the mature proteins outside the inner membrane (27). We also recently demonstrated direct interaction between MotX and MotY and that in the absence of MotY, the overproduction of MotX affected the membrane localization of PomB and the PomA-PomB complex, suggesting interaction between MotX and PomB (25).
Although there are several lines of evidence for the membrane localization and direct interaction of MotX and MotY, the precise roles of these proteins are still unclear. Since overproduction of MotX partially restored the motility of motY strains (26), we hypothesized that MotX is more directly involved than MotY in torque generation. In this report, to elucidate the functions of MotY, we randomly mutagenized a motY gene cloned on a plasmid and isolated three missense mutations that caused various swimming defects. Two of the mutations were found to change the two cysteine residues in MotY. Coincidently, MotX also has two cysteine residues, and MotX and MotY each contain a tetrapeptide sequence that begins with a cysteine (CQLV) (20). Both of the cysteine residues are highly conserved in either MotX or MotY of various species. To investigate the roles of the cysteine residues, we constructed mutants of MotX and MotY in which the cysteine residues were replaced by serine residues. We then characterized these mutants in terms of the swimming and swarming abilities of the bacteria, protein stability, and the MotX-MotY interaction.
Isolation of motY mutants and characterization of their motility.
Random mutagenesis of the motY gene was carried out by treating the plasmid pIO6 with hydroxylamine as described previously (17). The mutagenized plasmids were introduced into the motY mutant VIO542 (28), which possesses a nonmotile polar flagellum, though the mutation site has not been determined yet (15). We then isolated mutants that were completely or partially impaired in their polar-flagellar motility in a semisolid agar. Each of the nucleotide changes in the motY alleles was determined by DNA sequencing. In 10 isolates, no nucleotide change was found in the motY coding region of the plasmid, suggesting that the causative mutations likely affected the expression of motY. Introduced termination codons were found in an additional 16 isolates. We identified two isolates carrying single amino acid substitutions and one isolate with two amino acid substitutions: MotY-C147Y, MotY-R262Q, and MotY-C25Y(S−3N). It is noteworthy that two of the missense mutations affected one of the two cysteine residues found in MotY. The arginine residue at amino acid position 262 is located in a peptidoglycan-binding motif (28). The peptidoglycan-binding motif is conserved among the motor proteins MotB, PomB, and MotY, as well as several outer membrane proteins, such as PAL and OmpA (8, 10). Peptidoglycan binding by any of the flagellar-motor proteins, however, has not been demonstrated in vivo or in vitro; thus, the significance of these peptidoglycan-binding motifs for motor function remains unknown.
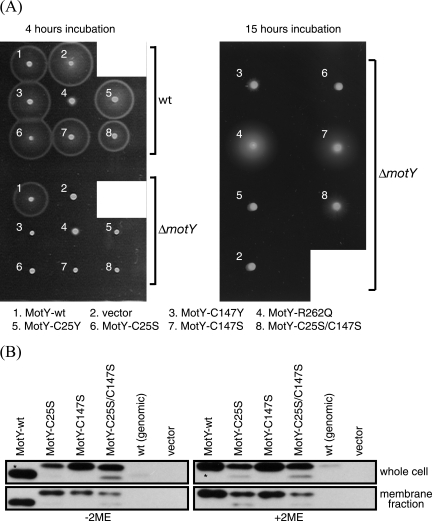
MotY-C147Y, MotY-R262Q, and MotY-C25Y(S−3N) were produced in VIO5 (a strain with wild-type polar flagella) (28) and GRF2 (a ΔmotY strain constructed from VIO5 in this study) (Table 1) cells, and the motility of the cells in 0.25% agar was analyzed (Fig. 1 and Table 2). Whereas wild-type MotY complemented the motility defect of the GRF2 cells, none of the three mutants was able to produce a wild-type level of motility in GRF2 cells. Among these MotY mutants, MotY-R262Q expression resulted in the most motile bacteria and also produced a negative dominance by multicopy effects (slower swarming speed) when it was coexpressed in VIO5 cells with the genomic copy of wild-type MotY.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or descriptiona | Source or reference |
|---|---|---|
| V. alginolyticus strains | ||
| VIO5 | VIK4 laf (Rifr Pof+ Laf−) | 28 |
| VIO542 | VIO5 motY (Rifr Laf− Mot−) | 28 |
| NMB94 | VIO5 motX (Rifr Laf− Mot−) | 26 |
| GRF2 | VIO5 ΔmotY (Rifr Laf− Mot−) | This study |
| E. coli strains | ||
| JM109 | recA1 endA1, gyrA96 thi hsdR17 relA1 supE44 λ− Δ(lac-proAB) F′ traD36 proAB lacIq ΔM15 | 33 |
| BL21(DE3) | Stratagene, San Diego, CA | |
| Plasmids | ||
| pSU38 | kan (Kmr) PlaclacZa | 4 |
| pSU41 | kan (Kmr) PlaclacZa | 4 |
| pIO2 | pSU21; 20-kb SalI fragment (motY+) | 28 |
| pIO6 | pSU38; 1.0-kb HindIII-XbaI fragment (motY+) | 28 |
| pRK101 | pIO6; motY (C147Y) | This study |
| pRK102 | pIO6; motY (R262Q) | This study |
| pRK103 | pIO6; motY (C25Y; −3S of signal sequence to N) | This study |
| pRK201 | pSU38; 1.0-kb HindIII-XbaI fragment (motY C25S) | This study |
| pRK202 | pSU38; 1.0-kb HindIII-XbaI fragment (motY C147S) | This study |
| pRK203 | pSU38; 1.0-kb HindIII-XbaI fragment (motY C25S/C147S) | This study |
| pKS202 | pSU41; 1.0-kb EcoRI-XbaI fragment (6His-motY) | This study |
| pMO401 | pSU41; 700-bp ClaI-BamHI fragment (motX+) | 26 |
| pMO404 | pSU41; 700-bp ClaI-BamHI fragment (motX C35S) | This study |
| pMO405 | pSU41; 700-bp ClaI-BamHI fragment (motX C66S) | This study |
| pMO413 | pSU41; 700-bp ClaI-BamHI fragment (motX C35S/C66S) | This study |
Rifr, rifampin resistant; Kmr, kanamycin resistant; Cmr, chloramphenicol resistant; Plac, lac promoter; Pof+, normal polar-flagellar formation; Laf−, defective in lateral-flagellar formation.
FIG. 1.
Characterization of MotY mutants. (A) VPG (1% polypeptone, 0.4% K2HPO4, 3% NaCl, and 0.5% [wt/vol] glycerol) plates containing 0.25% agar and 100 μg/ml kanamycin were inoculated with fresh colonies of the wild-type strain (VIO5) or the ΔmotY mutant strain (GRF2) expressing the indicated proteins from plasmids. They were incubated at 30°C for 4 h or 15 h. The plasmids and strains used are listed in Table 1. (B) Whole-cell lysates and the membrane fractions of GRF2 cells (ΔmotY) expressing the indicated proteins from plasmids and the wild-type strain [VIO5; wt (genomic)] were prepared and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting as described previously (35) using anti-MotY antibodies (MotYB0079) in the absence (left) or the presence (right) of 2-mercaptoethanol (2ME). The asterisks (upper left and upper right) indicate that those samples were 1/10 the volume of the other samples. The anti-MotY antibody was raised against purified MotY produced from pKJ503 (25), which encoded C-terminally histidine-tagged MotY.
TABLE 2.
Swimming fractions
| Host/plasmid | Description | Swimming fraction (%)a |
|---|---|---|
| VIO5/pRK102 | Wild type/MotY-R262Q | 40 (n = 193) |
| GRF2/pIO6 | ΔmotY/wild-type MotY | 61 (n = 208) |
| GRF2/pRK102 | ΔmotY/MotY-R262Q | 29 (n = 192) |
| GRF2/pRK201 | ΔmotY/MotY-C25S | 0 |
| GRF2/pRK202 | ΔmotY/MotY-C147S | <1 |
| GRF2/pRK203 | ΔmotY/MotY-C25S-C147S | <1 |
Cells were grown at 30°C for 4 h in VPG broth medium. The swimming cells were counted under a dark-field microscope. The numbers in parentheses indicate the total cells counted. GRF2/pRK201 did not produce any motile cells, whereas GRF2/pRK202 and GRF2/pRK203 produced low but observable numbers of motile cells.
Conservation of cysteine residues in MotX and MotY.
The above discussion mentioned the importance of two cysteine residues of MotY. The fact that MotX and MotY of V. alginolyticus share the same CQLV sequence and have two cysteine residues caused us to hypothesize the evolutionary importance of these cysteine residues for MotX and MotY. So far, MotX and/or MotY have been found or assigned only in some species of gammaproteobacteria, i.e., Vibrio species and Pseudomonas species. The CQLV sequence in MotY is conserved only in V. alginolyticus and Vibrio parahaemolyticus. According to the amino acid sequence alignment of these MotX or MotY proteins, the cysteine residues are conserved in all MotX and MotY proteins found (data not shown). These cysteine residues seem to be important for the proper function of MotX and MotY.
Intramolecular disulfide bond in MotY.
Since native MotY contains two cysteine residues, it is possible that these two cysteine residues are involved in intramolecular or intermolecular disulfide bond formation, which might be important for the function of the protein. To investigate this possibility, we constructed three motY mutants in which either or both cysteine residues were replaced by serine residues (pRK201 [C25S], pRK202 [C147S], and pRK203 [C25S/C147S]). The MotY-C25S mutant was not motile either on 0.25% agar plates or in broth medium, whereas the MotY-C147S and MotY-C25S/C147S mutants showed significantly reduced but observable spreads in the swarm assay after 15-h incubations (Fig. 1A). Since we did not find any revertant or pseudorevertant that would swarm at the rate of the wild type, it is not likely that those swimmers were the reverted wild-type cells. The swimming fraction of MotY-C147S and the double cysteine mutant in broth medium were below 1% (Table 2).
Next, we performed immunoblot assays to detect the mutant proteins. In the presence of the reducing agent 2-mercaptoethanol, all the MotY derivatives overexpressed with the lac promoter for enhanced visualization appeared as an approximately 30-kDa band, which was consistent with the expected size of the mature MotY protein. In the absence of 2-mercaptoethanol, however, there were slight differences in the mobilities of the wild-type and the MotY Cys mutant proteins (Fig. 1B). In the presence of a reducing agent, the native MotY band shifted upward to align with the other proteins. The faster mobility of the wild-type band in the absence of a reducing agent can be explained by a more compact conformation of the wild-type protein due to intramolecular disulfide bond formation. There were also significant differences in the band intensities. For the whole-cell preparations, the intensity of the overproduced wild-type MotY band was much stronger than the intensity of any of the Cys mutant protein bands. The differences in the band intensities from membrane fraction samples, however, were moderate. These results strongly suggest that MotY contains an intramolecular disulfide bridge. The reduced levels of the MotY Cys mutants suggest that the intramolecular disulfide bridge in MotY is required to maintain a stable conformation; without the disulfide bond, MotY is more susceptible to degradation and the amount of the protein is greatly reduced. However, the band intensities of the overproduced proteins with Cys replaced were still far greater than that of chromosomally expressed wild-type MotY. This might imply that the conformation achieved by the intramolecular cross-linking is necessary for the proper function of MotY, in addition to resistance to degradation. Whereas wild-type MotY did not show a sign of intermolecular cross-linking, MotY-C25S and MotY-C147S produced intermolecular-cross-link products in immunoblot assays; in particular, MotY-C25S showed numerous extra bands (data not shown). We speculate that the cysteine residue at 147 is more prone to a cross-link formation with other periplasmic proteins than C25 when an intramolecular cross-link is not possible and that the homo- or heterodimer formation interferes with its function.
Prevention of MotX degradation by MotY.
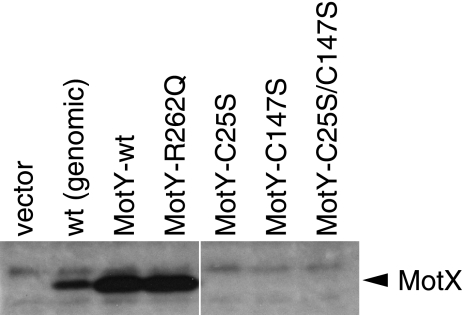
It has been shown that MotY directly interacts with MotX and that in the absence of MotY expression, MotX is not detectable by immunoblotting (25, 26). These lines of evidence led us to hypothesize that MotY acts to protect MotX against degradation. To evaluate our MotY mutants, we examined whether our MotY derivatives could prevent the degradation of MotX. As expected, increasing the amount of wild-type MotY elevated the level of detectable MotX without changing the motX expression on the chromosome (Fig. 2). Overexpression of MotY-R262Q also increased the detectable MotX level; it is likely that the R262 residue does not affect the interaction with MotX. On the other hand, although the levels of the Cys-substituted MotY mutants were greater than that of genomically expressed wild-type MotY, no MotX band was observed from cells expressing these mutant MotY proteins (Fig. 1B). This result implies that these Cys mutants are impaired in the ability to interact with MotX, and we postulate two distinct consequences: elevated susceptibility to periplasmic digestion and loss of a docking site for MotX. Since MotX appears to be more directly involved in flagellar-motor function than MotY (26), the direct reason for swimming deficiencies of these MotY Cys mutants seems to be the loss of MotX due to degradation.
FIG. 2.
Detection of MotX in strains coexpressing various MotY mutant proteins. Whole-cell lysates of the GRF2 strain (ΔmotY) expressing the indicated proteins from plasmids and the wild-type strain [VIO5; wt (genomic)] were prepared and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting as described previously (35) using anti-MotX antibodies (MotXB0080).
Characterization of the MotX Cys mutants.
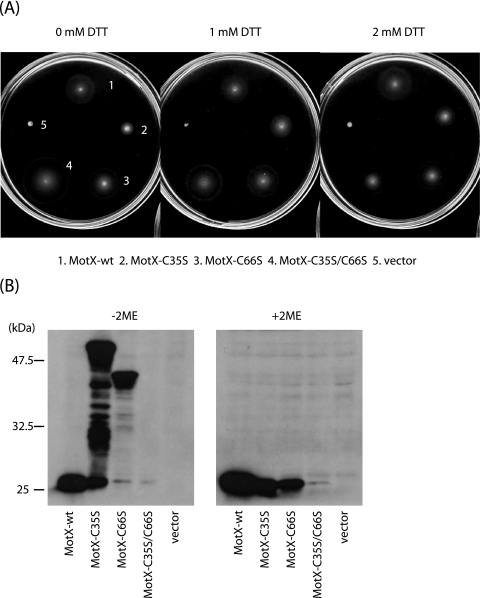
We constructed three motX mutants in which either or both of the cysteine residues were changed to serine residues by site-directed mutagenesis (pMO404 [C35S], pMO405 [C66S] and pMO413 [C35S/C66S]). All three MotX Cys mutant proteins conferred swarming ability on motX mutant cells (NMB94) (Fig. 3A). In the absence of the reducing agent dithiothreitol (DTT), the double mutant MotX-C35S/C66S produced a swarm ring that was unexpectedly larger than the one that resulted from overproduction of wild-type MotX. In the presence of DTT, MotX-C35S-expressing NMB94 cells formed a larger swarm ring than in the absence of DTT. For the other cells, the levels of the wild-type and C66S mutant proteins did not seem to be affected by the addition of DTT.
FIG. 3.
Characterization of the MotX mutants. (A) VPG plates containing 0.25% agar, kanamycin, and various concentrations of DTT were inoculated with fresh colonies of motX mutant cells (NMB94) expressing the indicated proteins from plasmids and incubated at 30°C for 4 h. (B) Whole-cell lysates of the NMB94 strain expressing the indicated proteins from plasmids were prepared and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting as described previously (35) using anti-MotX antibodies (MotXB0080) in the absence (−) and the presence (+) of 2-mercaptoethanol (2ME). The anti-MotX antibody was raised against purified MotX produced from pKJ402, which encoded C-terminally histidine-tagged MotX.
We performed immunoblot assays to detect the MotX proteins in the presence or absence of the reducing agent 2-mercaptoethanol. In all the mutant and wild-type MotX samples, we detected 24-kDa bands, which corresponded to the mature MotX protein (Fig. 3B). In the absence of the reducing agent, MotX-C35S and MotX-C66S produced extra bands of various sizes, probably due to the intermolecular-cross-link formation. As in the case of MotY Cys mutants, we can see that C66 is more likely than C35 to form intermolecular cross-links, which might explain the slower swarming of MotX-C35S than the other two mutants and its greater recovery by the addition of the reductant. Although the double Cys mutant (MotX-C35S/C66S) conferred better swarming ability than the wild-type protein when overproduced, the level of the double mutant was significantly reduced. It has been reported that an excessive amount of MotX forms aggregates in V. alginolyticus (25). We speculate that aggregation and/or intermolecular cross-linking by single Cys mutants of MotX can raise its resistance to degradation. Therefore, among these MotX derivatives, only the double Cys mutant showed significant reduction in the amount detected. It has also been reported that the overexpression of MotX in E. coli kills the cells (20). In addition, Vibrio cells overexpressing MotX exhibit aberrant morphologies (data not shown). These types of evidence suggest that the aggregation of MotX affects the shape of the cells, possibly by disrupting the peptidoglycan layer or outer membrane integrity. We have also observed that MotX overproduction affected the motility—both the swimming and swarming abilities—of the cells (data not shown). The better swarming ability of the MotX-C35S/C66S-expressing cells than the cells overproducing wild-type MotX, therefore, might be explained by the increased susceptibility of this mutant protein to degradation, which reduced the damaging effects of the aggregation.
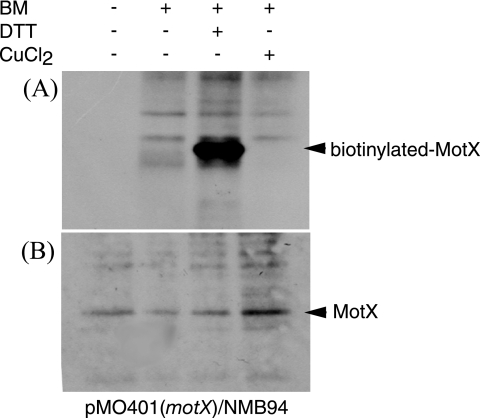
Unlike MotY, wild-type MotX exhibited the same gel mobilities in the presence and absence of the reductant. We then treated MotX with biotin maleimide, which binds to free thiol groups, and the labeled biotin was detected using horseradish peroxidase-conjugated streptavidin. The labeled biotin was detected only when DTT was added to the sample (Fig. 4). Since the wild-type protein seemed to be a monomer in the absence of the reductant (Fig. 3B), biotin labeling only in the presence of DTT suggests that MotX contains an intramolecular disulfide bond. In conclusion, the presence of the intramolecular disulfide bond is not essential for the function of MotX but instead affects the susceptibility of the protein to degradation.
FIG. 4.
Biotin maleimide labeling of MotX cysteine residues. NMB94 harboring pMO401 (MotX wild type) was treated with biotin maleimide (BM) and DTT or CuCl2. The immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. Biotinylated MotX was detected with streptavidin-conjugated horseradish peroxidase and chemiluminescence (A) or immunoblotting using anti-MotX antibodies (B).
Concluding remarks.
MotX and MotY each contain two cysteine residues (Fig. 5), which are highly conserved among several species of gammaproteobacteria. We speculated that these cysteine residues are critical for the functions of these proteins and determined that both MotX and MotY form intramolecular disulfide bonds. In vivo, disulfide bond formation in the prokaryotic periplasm is a catalyzed process, which is crucial for the folding and stability of many proteins and protein complexes (23). Failure to form proper disulfide bonds is likely to lead to protein aggregation and degradation by proteases. Disulfide bonds are donated directly to unfolded polypeptides by the DsbA protein, whereas DsbA is reoxidized by DsbB. For example, dsbA mutant strains exhibit reduced levels of alkaline phosphatase, β-lactamase, or the outer membrane protein OmpA (3). It seems that both MotX and MotY acquired the cysteine residues for the same reason the other secreted proteins did. However, we have also shown that disulfide bond formation is not sufficient for MotX to be fully stabilized. The interaction with MotY is the key for MotX to fold correctly and to function. For example, when MotX is far in excess of MotY, MotX aggregates, which in turn affects swimming ability, cell shape, and cell growth, as stated above. Therefore, we speculate that the MotX-MotY interaction is another level of control on the amount of MotX in the periplasm. As MotX is essential for motor rotation in Vibrio and is likely to interact with PomB (25), we believe that MotX modulates the activity of the sodium channel, a stator complex composed of PomA and PomB. On the other hand, the role of MotY may be limited to the anchoring and/or stabilization of MotX.
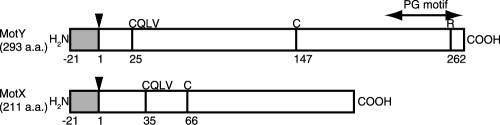
FIG. 5.
Schematic diagram of MotX and MotY. MotX and MotY contain two cysteine residues and the conserved tetrapeptide CQLV. The arrowheads indicate the cleavage sites of the signal sequences (gray sections). The double-headed arrow demarcates the region containing the putative motif for peptidoglycan binding (PG motif). a.a., amino acids.
Acknowledgments
We are grateful to Seiji Kojima for critically reading the manuscript.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan; from the Japan Science and Technology Corporation (to M.H. and T.Y.); and from the Soft Nano-Machine Project of the Japan Science and Technology Agency (to T.Y. and M.H.).
REFERENCES
- 1.Asai, Y., S. Kojima, H. Kato, N. Nishioka, I. Kawagishi, and M. Homma. 1997. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J. Bacteriol. 179:5104-5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atsumi, T., Y. Maekawa, H. Tokuda, and Y. Imae. 1992. Amiloride at pH 7.0 inhibits the Na+-driven flagellar motors of Vibrio alginolyticus but allows the cell growth. FEBS Lett. 314:114-116. [DOI] [PubMed] [Google Scholar]
- 3.Bardwell, J. C., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581-589. [DOI] [PubMed] [Google Scholar]
- 4.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACY184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 5.Berg, H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19-54. [DOI] [PubMed] [Google Scholar]
- 6.Berg, H. C. 1995. Torque generation by the flagellar rotary motor. Biophys. J. 68:S163-S167. [PMC free article] [PubMed] [Google Scholar]
- 7.Blair, D. F., and H. C. Berg. 1988. Restoration of torque in defective flagellar motors. Science 242:1678-1681. [DOI] [PubMed] [Google Scholar]
- 8.Bouveret, E., H. Benedetti, A. Rigal, E. Loret, and C. Lazdunski. 1999. In vitro characterization of peptidoglycan-associated lipoprotein (PAL)-peptidoglycan and PAL-TolB interactions. J. Bacteriol. 181:6306-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, T. F., L. Q. Al-Mawsawi, S. Kojima, and D. F. Blair. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 43:35-45. [DOI] [PubMed] [Google Scholar]
- 10.De Mot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 11.Derosier, D. J. 1998. The turn of the screw: the bacterial flagellar motor. Cell 93:17-20. [DOI] [PubMed] [Google Scholar]
- 12.Doyle, T. B., A. C. Hawkins, and L. L. McCarter. 2004. The complex flagellar torque generator of Pseudomonas aeruginosa. J. Bacteriol. 186:6341-6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawagishi, I., Y. Maekawa, T. Atsumi, M. Homma, and Y. Imae. 1995. Isolation of the polar and lateral flagellum-defective mutants in Vibrio alginolyticus and identification of their flagellar driving energy sources. J. Bacteriol. 177:5158-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan, S., M. Dapice, and T. S. Reese. 1988. Effects of mot gene expression on the structure of the flagellar motor. J. Mol. Biol. 202:575-584. [DOI] [PubMed] [Google Scholar]
- 15.Kojima, S., Y. Asai, T. Atsumi, I. Kawagishi, and M. Homma. 1999. Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by mutations of putative channel components. J. Mol. Biol. 285:1537-1547. [DOI] [PubMed] [Google Scholar]
- 16.Kojima, S., and D. F. Blair. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26-34. [DOI] [PubMed] [Google Scholar]
- 17.Kojima, S., M. Kuroda, I. Kawagishi, and M. Homma. 1999. Random mutagenesis of the pomA gene encoding the putative channel component of the Na+-driven polar flagellar motor of Vibrio alginolyticus. Microbiology 145:1759-1767. [DOI] [PubMed] [Google Scholar]
- 18.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 19.Manson, M., P. Tedesco, H. Berg, F. Harold, and C. Van der Drift. 1977. A protonmotive force drives bacterial flagella. Proc. Natl. Acad. Sci. USA 74:3060-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarter, L. L. 1994. MotX, the channel component of the sodium-type flagellar motor. J. Bacteriol. 176:5988-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarter, L. L. 1994. MotY, a component of the sodium-type flagellar motor. J. Bacteriol. 176:4219-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messens, J., and J. F. Collet. 2006. Pathway of disulfide bond formation in Escherichia coli. Int. J. Biochem. Cell. Biol. 38:1050-1062. [DOI] [PubMed] [Google Scholar]
- 24.Muramoto, K., S. Sugiyama, E. J. Cragoe, and Y. Imae. 1994. Successive inactivation of the force-generating units of sodium-driven bacterial flagellar motors by a photoreactive amiloride analog. J. Biol. Chem. 269:3374-3380. [PubMed] [Google Scholar]
- 25.Okabe, M., T. Yakushi, and M. Homma. 2005. Interactions of MotX with MotY and with the PomA/PomB sodium ion channel complex of the Vibrio alginolyticus polar flagellum. J. Biol. Chem. 280:25659-25664. [DOI] [PubMed] [Google Scholar]
- 26.Okabe, M., T. Yakushi, Y. Asai, and M. Homma. 2001. Cloning and characterization of motX, a Vibrio alginolyticus sodium-driven flagellar motor gene. J. Biochem. (Tokyo) 130:879-884. [DOI] [PubMed] [Google Scholar]
- 27.Okabe, M., T. Yakushi, M. Kojima, and M. Homma. 2002. MotX and MotY, specific components of the sodium-driven flagellar motor, colocalize to the outer membrane in Vibrio alginolyticus. Mol. Microbiol. 46:125-134. [DOI] [PubMed] [Google Scholar]
- 28.Okunishi, I., I. Kawagishi, and M. Homma. 1996. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 178:2409-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato, K., and M. Homma. 2000. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:5718-5722. [DOI] [PubMed] [Google Scholar]
- 30.Sato, K., and M. Homma. 2000. Multimeric structure of PomA, the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:20223-20228. [DOI] [PubMed] [Google Scholar]
- 31.Sowa, Y., A. D. Rowe, M. C. Leake, T. Yakushi, M. Homma, A. Ishijima, and R. M. Berry. 2005. Direct observation of steps in rotation of the bacterial flagellar motor. Nature 437:916-919. [DOI] [PubMed] [Google Scholar]
- 32.Stewart, B. J., and L. L. McCarter. 2003. Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 185:4508-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 34.Yorimitsu, T., and M. Homma. 2001. Na+-driven flagellar motor of Vibrio. Biochim. Biophys. Acta 1505:82-93. [DOI] [PubMed] [Google Scholar]
- 35.Yorimitsu, T., K. Sato, Y. Asai, I. Kawagishi, and M. Homma. 1999. Functional interaction between PomA and PomB, the Na+-driven flagellar motor components of Vibrio alginolyticus. J. Bacteriol. 181:5103-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yorimitsu, T., T. Yakushi, and M. Homma. 2004. Multimeric structure of the PomA/PomB complex, a channel component of the Na+-driven flagellar motor of Vibrio alginolyticus. J. Biochem. (Tokyo) 135:43-51. [DOI] [PubMed] [Google Scholar]