Abstract
Inactivation or deletion of the RNase E-encoding rne gene of Escherichia coli results in the growth of bacterial cells as filamentous chains in liquid culture (K. Goldblum and D. Apirion, J. Bacteriol. 146:128-132, 1981) and the loss of colony-forming ability (CFA) on solid media. RNase E dysfunction is also associated with abnormal processing of ftsQAZ transcripts (K. Cam, G. Rome, H. M. Krisch, and J.-P. Bouché, Nucleic Acids Res. 24:3065-3070, 1996), which encode proteins having a central role in septum formation during cell division. We show here that RNase E regulates the relative abundances of FtsZ and FtsA proteins and that RNase E depletion results in decreased FtsZ, increased FtsA, and consequently an altered FtsZ/FtsA ratio. However, while restoration of the level of FtsZ to normal in rne null mutant bacteria reverses the filamentation phenotype, it does not restore CFA. Conversely, overexpression of a related RNase, RNase G, in rne-deleted bacteria restores CFA, as previously reported, without affecting FtsZ abundance. Our results demonstrate that RNase E activity is required to maintain a proper cellular ratio of the FtsZ and FtsA proteins in E. coli but that FtsZ deficiency does not account for the nonviability of cells lacking RNase E.
RNase E was initially discovered in Escherichia coli as a 9S RNA-processing enzyme (13) and later was shown to have a key role in the degradation of mRNA (32), the control of plasmid DNA replication (23), and the maturation/processing of a variety of small catalytic RNAs, including ssrA RNA (24), the M1 RNA subunit of RNase P (26), 5S rRNA (13), 16S rRNA (22, 43), and tRNA (33, 37). The catalytic activity of RNase E resides in its N-terminal half, while the C-terminal region serves as a scaffold for interaction with other proteins to form a multicomponent structure termed the “degradosome” (30, 35). The actions of RNase E are essential for E. coli cell viability, and inactivation of this enzyme results in the production of elongated filamentous structures consisting of multiple connected cell bodies (15) and the inability to form colonies on solid media (1). The loss of colony-forming ability (CFA) in RNase E deletion mutant strains can be reversed by overproduction of RNase G (9, 18), which was initially discovered because of its ability to induce cytoplasmic axial-filament formation (31). Subsequently, the RNase G protein was found to be a structural homologue of the amino-terminal segment of RNase E (28) and to have ribonucleolytic activity (22, 43).
Cell division in E. coli and other bacteria is affected by a complex network of regulatory mechanisms. The ftsZ (filamentation temperature-sensitive) gene, which is widely conserved among prokaryotes and the chloroplasts of plants, has a central and essential role in the cell division process (14, 27, 39). During cell division, the FtsZ protein, which is a GTPase and a structural homologue of tubulin, polymerizes and forms a scaffold for the assembly of proteins at the division site (3, 4, 10, 38). The shift of ftsZ mutant bacteria to a nonpermissive temperature results in perturbed cell division, the production of long filamentous chains of connected cells, and the loss of colony-forming ability (7). The E. coli FtsZ protein is encoded by polycistronic transcripts that also produce two other proteins, FtsA and FtsQ, which participate in the FtsZ-based scaffold at the division site (4, 8, 12). Earlier work has shown that RNase E cleaves the ftsQAZ transcripts at two sites between the contiguous ftsA and ftsZ genes and that RNase E dysfunction decreases the ratio of transcript segments encoded by the ftsZ and ftsA genes (5). As perturbation of the FtsZ/FtsA ratio can affect cell division (8), Cam et al. (5) speculated that the essentiality of RNase E for cell viability might result from its action on ftsQAZ transcripts. Recently, studies published while our investigations into the lethality of RNase E null mutations were in progress have concluded that a decreased level of FtsZ is responsible for the nonviability of RNase E-deficient cells (40).
Here, we show that cellular depletion of RNase E in E. coli does—as speculated by Cam et al. (5)—result in a decreased ratio of FtsZ to FtsA protein and results in the formation of long filamentous chains of cells but that neither filamentation nor FtsZ deficiency accounts for the lethality observed in RNase E-depleted cells. Reversal of filamentation by adventitious overproduction of FtsZ in rne mutant bacteria did not restore CFA, and conversely, restoration of CFA in rne-deleted cells by overexpression of RNase G did not eliminate the filamentation phenotype caused by decrease of FtsZ. Our results indicate that a decreased FtsZ level in cells that lack RNase E does not account for RNase E essentiality.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. The construction of E. coli strains with rne deleted that express full-length RNase E or its N-terminal fragment under the control of an arabinose-regulated promoter (KSL2000 and KSL2009, respectively) has been described previously (18, 21). pBAD-NRNE was constructed from pBAD-RNE and pNRNE4 as described previously (21). pBAD-RNE and pNRNE4 were constructed as follows (18).
TABLE 1.
Bacterial strains and plasmidsa
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| N3433 | lacZ43(Fs) LAM−relA1 spoT1 thi-1 | 36 |
| MT2000 | Same as N3433 but rne::cat [pBAD-RNE] | 18 |
| KSL2000 | Same as N3433 but rne::cat recA::Tn10 [pBAD-RNE] | 18 |
| KSL2009 | Same as N3433 but rne::cat recA::Tn10 [pBAD-NRNE] | 21 |
| KSL2010 | Same as N3433 but rne::cat recA::Tn10 [pBAD-RNG] | This study |
| Plasmids | ||
| pKAN7 | pSC101 ori Kmr | This study |
| pBAD-RNE | pSC101 ori Kmr, rne under PBAD | 18 |
| pBAD-NRNE | pSC101 ori Kmr, N-rne under PBAD | 21 |
| pBAD-RNG | pSC101 ori Kmr, rng under PBAD | This study |
| pACYC177 | P15A ori Kmr Apr | 6 |
| pACYC177Δkan | P15A ori Apr | This study |
| pMT177Z | P15A ori Apr, ftsZ under natural promoter | This study |
See Materials and Methods for a detailed description of strains and plasmids.
pKAN7 was created by ligating the HincII fragment containing the replication origin from pSC101 and the larger Eco47III-FspI fragment from pKAN5 (J.-H. Yeom and K. Lee, in press). To construct pBAD-RNE, the EcoRI-XbaI fragment containing the coding region of rne plus 21 additional N-terminal amino acids, including the 10-His tag from pFUS1500 (29), was cloned into the same sites in pKAN7, which resulted in pBAD-N-His-RNE. Then, the original 5′ untranslated region of rne and the 5′ rne coding region encompassing the unique HindIII site were PCR amplified by using primers 5′ WT-RNE (5′-CAAGCTTGCGGCCGCAGGAGGTTACGATGAAAAGAATG) and RNE-1568R (5′-CGCTTACGTTCAGCGAACTC) with pFUS1500 as the DNA template. The PCR product was digested with HindIII and NotI and cloned into the same sites in pBAD-N-His-RNE, resulting in pBAD-RNE.
Plasmid pNRNE4 was constructed in three steps. (i) pNRNE1 was constructed by ligating EcoRI-StyI-digested pBR322 containing the replication origin and a β-lactamase gene with two DNA fragments: (a) a NotI-StyI-digested PCR DNA fragment containing the N-terminal 499 amino acids of the rne coding region (N-rne) plus a C-terminal hexahistidine tag using two primers, 5′ WT-RNE and C-RNE-HIS (5′-TCACCAAGGGCATGCTCTAGACTAGTGGTGGTGGTGGTGGTGGCTTAAGGTTGGGGGTTTC), and pFUS1500 as the template DNA and (b) an EagI-EcoRI-digested PCR DNA fragment containing lacIq and a lacUV5 promoter PCR amplified using two primers, lacI-5′ (5′-ATGAATTCCGACACCATCGAATGGTG) and lacI-3′ (5′-TAGCGGCCGCCAGGCATCCACACATTATACG), and plasmid p16ST (20) as the template DNA. (ii) To add the rrnB T1T2 transcriptional terminator (3a) at the 3′ end of the N-rne coding region, an SphI-XbaI fragment from p16ST was cloned into the same sites in pNRNE1 and resulted in pNRNE2. (iii) pNRNE4 was constructed by ligating two DNA fragments, a BamHI-NotI fragment containing lacIq and a lacUV5 promoter from p16ST and an EagI-MscI fragment containing sequences encoding C-terminally hexahistidine-tagged N-rne and an rrnB T1T2 transcriptional terminator from pNRNE2, with a BamHI- and StuI-digested pACYC177 (6) fragment containing the replication origin.
KSL2010 expresses RNase G from the arabinose-regulated BAD promoter (PBAD) on pBAD-RNG. Plasmid pBAD-RNG was constructed by replacing the NotI-NheI fragment of pBAD-NRNE containing the N-RNase E coding region plus a T1T2 transcriptional terminator with the NotI-NheI fragment containing the rng coding region plus a T1T2 transcriptional terminator from pRNG2. pRNG2 shares the same features as pRNG3 (18) but contains a P15A replication origin derived from pACYC177, whereas pRNG3 contains the replication origin of pSC101 (18). KSL2010 was obtained by a plasmid displacement procedure. Briefly, introduction of a kanamycin resistance plasmid (pBAD-RNG) expressing RNase G with a hexahistidine tag at the C terminus under the control of the BAD promoter into KSL2004 (18) and selection for the incoming plasmid by growing transformants containing both plasmids (pBAD-RNG and pRNG3) for 60 generations in the presence of kanamycin and 0.2% arabinose without IPTG (isopropyl-β-d-thiogalactopyranoside) resulted in displacement of the resident ampicillin resistance (Apr) plasmid by the kanamycin resistance (Kmr) RNase G-expressing plasmid.
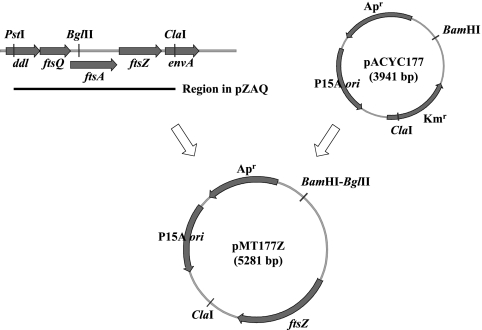
Plasmid pZAQ, which contains ftsQ, ftsA, and ftsZ, was kindly provided by Joe Lutkenhaus (45). To construct pACYC177Δkan, an NsiI fragment within the kanamycin resistance gene on pACYC177 was removed and the plasmid was self-ligated. To construct pMT177Z (Fig. 1), a pZAQ-derived BglII-ClaI fragment that contained the 3′ portion of ftsA and the full-length ftsZ region was introduced between the BamHI and ClaI sites of pACYC177.
FIG. 1.
Scheme for construction of the pMT177Z plasmid. A BglII-ClaI fragment from the pZAQ plasmid was ligated to the larger fragment of BamHI- and ClaI-cleaved pACYC177 DNA, as described in Materials and Methods.
Media and culture conditions.
Luria-Bertani (LB) medium was used for bacterial growth at 37°C. To express genes under the control of the BAD promoter, l-(+)-arabinose (Sigma) was added to the cultures to a final concentration of 0.1% to produce RNase E (strain KSL2000) or N-RNase E (strain KSL2009) as described previously (18, 21), and 0.2% to produce RNase G (strain KSL2010) at a cellular level that was approximately 5.3 times higher than that of the parental N3433 strain, as quantified by immunoblotting. In every experiment, appropriate antibiotics were used at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 10 μg/ml; kanamycin, 20 μg/ml; and tetracycline, 5 μg/ml.
Immunoblotting procedures.
For analysis of E. coli proteins (0.1 optical density at 600 nm [OD600] unit per lane), lysates containing total cellular proteins were electrophoresed on a 7% Tris-acetate sodium dodecyl sulfate-polyacrylamide gel (Bio-Rad) and electrophoretically transferred (0.5 A; 40 min) to nitrocellulose membranes (NitroPure, Supported, Pure Nitrocellulose 0.1 Micron; GE Osmonics) as described previously (41). FtsZ and FtsA were detected by using rabbit polyclonal antiserum against FtsZ (1:10,000 dilution), FtsA (1:10,000 dilution), or ribosomal protein S1 (1:20,000 dilution). Polyclonal anti-FtsZ antiserum and anti-FtsA antiserum were gifts from Cynthia A. Hale and Piet A. J. de Boer (16). Polyclonal anti-S1 was obtained as described previously (11). Anti-rabbit immunoglobulin G-horseradish peroxidase conjugate (1 mg/ml; Promega) was used as the secondary antibody (1:5,000 dilution in phosphate-buffered saline containing 0.1% bovine serum albumin and 0.1% Tween 20). Proteins were visualized by using Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer) and were exposed to 8- by 10-in. BioMax MR Film (Kodak) or a Versa Doc imaging system (Bio-Rad) and Quantity One software (version 4.5.1; Bio-Rad) to quantify the relative abundances of protein bands.
Microscopic observation of E. coli.
The procedures followed were as described previously (17) with slight modifications. A 30-μl sample from an E. coli culture was spread on a clean glass slide and dried at room temperature for 12 h. The sample was then fixed by immersion in methanol for 15 min and dried at room temperature for 2 h. The slides were washed several times with tap water (not distilled or deionized water) in a beaker and dried at room temperature for 2 h. One drop (approximately 20 μl) of poly-l-lysine (0.1% [wt/vol] in water; Sigma) was spread over the dried sample using a plastic tip, and the slide was dried again at room temperature for 1 h to fix all cells tightly to it. Sodium chloride (0.01 M in water) was dropped on the sample, and a glass coverslip was placed on the drop. After immersion oil was put between the coverslip and the objective lens, the cells were observed by light microscopy (Axiovert 100; Zeiss).
RESULTS
RNase E depletion alters the steady-state levels of FtsA and FtsZ proteins.
Earlier work showed that RNase E cleaves ftsQAZ transcripts and that such cleavage affects the absolute and relative abundances of mRNA segments encoding the FtsA and FtsZ open reading frames (ORFs) (5). DNA microarray analysis from our laboratory also demonstrated that the abundances of mRNA segments encoding the FtsA and FtsZ ORFs were increased in the absence of RNase E and were restored to normal by expression of full-length RNase E or N-RNase E, but not by RNase G (http://sncohenlab.stanford.edu/ecoli/info.html; 18). To learn the consequences of these alterations for fts gene-encoded protein levels and for cellular physiology, we used a strain with rne deleted complemented by a plasmid expressing rne under the control of an arabinose-inducible promoter (18). Transfer of these cells to media lacking arabinose resulted in the loss of CFA but a continued increase in the OD600 of liquid cultures, as previously described (18). Consistent with the effects of RNase E depletion on the decay of ftsQAZ transcripts reported by Cam et al. (5), we found that the cellular abundances of the FtsA and FtsZ proteins were altered when the PBAD-controlled plasmid-borne rne gene was turned off in rne null mutant bacteria by transfer to media lacking arabinose. However, the alterations were in opposite directions for the two Fts proteins: the steady-state level of FtsA was increased approximately 1.5-fold (1.51 in KSL2000, 1.45 in KSL2009, and 1.51 in KSL2010) (Fig. 2, IC, IIC, and IIIC) relative to both a parental rne+ strain (N3433) and a strain (KSL2000) complemented by arabinose-induced expression of RNase E. Additionally, FtsZ was decreased to 0.65 of normal in KSL2000, 0.66 of normal in KSL2009, and 0.48 of normal in KSL2010. Consequently, the FtsZ/FtsA protein ratio was reduced in these strains to less than half of what was observed in the parental rne+ strain, N3433.
FIG. 2.
Immunoblot comparison of FtsA and FtsZ protein abundances in complemented and uncomplemented rne null mutant bacteria. Bacteria were grown overnight on LB plates containing arabinose (0.1% for KSL2000 and KSL2009; 0.2% for KSL2010) and the following day were inoculated into LB media in the presence or absence of arabinose, cultured at 37°C, and harvested in mid-log phase after 10 to 12 h; during this period, the cultures were diluted several times to maintain them in mid-log phase (OD600 = 0.5 to 1.0). Immunoblotting was performed as described in Materials and Methods. The same membranes probed with anti-FtsA antibody were stripped and reprobed with anti-FtsZ antibody. Analysis of bands was carried out as described in Materials and Methods. The ratios of protein abundance relative to the N3433 strain are shown below the lanes; standard deviations (in parentheses) were calculated from two bands derived from different cultures and normalized according to the abundance of ribosomal protein S1. Note that under the experimental conditions employed, RNase E was not detected by immunoblotting 2 to 3 h after the transfer of cells to medium lacking arabinose (data not shown), whereas increase in OD600 continued for 13 to 14 h after the transfer of cells to arabinose-deficient liquid LB media.
CFA was restored (Table 2) and the observed alteration in the abundance of the FtsZ protein relative to that of the FtsA protein under RNase E depletion conditions was reversed by expression of a plasmid-borne gene encoding either the full-length RNase E protein (Fig. 2, IB) or a truncated N-terminal RNase E fragment (Fig. 2, IIB) that carried the catalytic site of the enzyme (N-RNase E) (28) but lacked the necessary scaffold domain for degradosome formation (25, 34, 42). These observations indicate that degradosome formation is not required for restoration of either CFA or normal levels of the FtsZ and FtsA proteins. In contrast, RNase G overexpression in RNase E-depleted cells restored CFA without restoring a normal growth rate, as previously reported (19). While RNase G overproduction had no detectable effect on FtsZ abundance, it decreased FtsA abundance to a normal level (from 1.51 to 0.95) (Fig. 2, IIIB) and resulted in an increase in the FtsZ/FtsA ratio (from 0.32 to 0.58) (Fig. 2, IIIB).
TABLE 2.
Cell lengths and CFU numbers of different strains with rne deleted
| Strain | Cell length (μm)a | No. of CFU/ml/OD600 unitb |
|---|---|---|
| N3433 + pKAN7 + pACYC177Δkan | 1.80 ± 0.47 | (610 ± 33) × 106 |
| N3433 + pKAN7 + pMT177Z | 1.65 ± 0.56 | (794 ± 23) × 106 |
| KSL2000 + pACYC177 | 1.61 ± 0.42 | (379 ± 28) × 106 |
| KSL2000 + pMT177Z | 1.35 ± 0.22 | (655 ± 40) × 106 |
| KSL2009 + pACYC177 | 1.86 ± 0.42 | (52 ± 3) × 106 |
| KSL2009 + pMT177Z | 1.50 ± 0.44 | (318 ± 43) × 106 |
| KSL2010 + pACYC177 | 4.09 ± 2.34 | (33 ± 15) × 106 |
| KSL2010 + pMT177Z | 1.80 ± 0.46 | (131 ± 21) × 106 |
Cells were grown in LB medium at 37°C in the presence of arabinose (0.1% for N3433, KSL2000, and KSL2009; 0.2% for KSL2010) and harvested in mid-log phase. Slides were prepared as described in Materials and Methods. After photographs were taken under a microscope, the average (± standard deviation) of a random 50 cells chosen was calculated.
Cells were grown to mid-log phase in LB medium at 37°C in the presence of arabinose (0.1% for N3433, KSL2000, and KSL2009; 0.2% for KSL2010). The cells were diluted 10−4 to 10−6 and plated on LB plates containing arabinose. The average of colonies from three LB plates was calculated. The values shown indicate CFU per ml of culture (± standard deviation) adjusted for OD600 units.
The decrease in the FtsZ/FtsA ratio observed during RNase E depletion (Fig. 2, IC, IIC, and IIIC) was reversed by adventitious expression of FtsZ from the ftsZ gene carried by plasmid pMT177Z (Fig. 1), which contains the 3′ half of the ftsA encoding region plus the full-length FtsZ ORF and produces only the intact FtsZ protein from transcripts initiated at the promoter located in its ftsA region (12). pMT177Z not only restored FtsZ abundance to the normal level (from 0.65 to 1.08 in KSL2000, from 0.66 to 0.92 in KSL2009, and from 0.48 to 0.94 in KSL2010) (Fig. 2, IE, IIE, and IIIE), but also partially decreased FtsA abundance (from 1.51 to 1.26 in KSL2000, from 1.45 to 1.14 in KSL2009, and from 1.51 to 1.24 in KSL2010) (Fig. 2, IE, IIE, and IIIE). Reduced FtsZ abundance in the KSL2010 strain (0.55) (Fig. 2, IIIB), even in the presence of 0.2% arabinose, was also restored to a normal level by the introduction of pMT177Z (0.98) (Fig. 2, IIID).
Effects of RNase E, N-RNase E, RNase G, and FtsZ on the filamentation phenotype in cells with rne deleted.
Consistent with previously reported results (15, 44), long filamentous chains of bacterial cells (>50 μm) were observed during culture in liquid media of bacteria with rne deleted (Fig. 3A, E, and I). As expected, this long-filamentation phenotype was completely reversed by complementation with full-length RNase E or N-RNase E (Fig. 3B and F and Table 2). Elevated RNase G expression, which mitigated the increase in FtsA protein observed in rne null mutant bacteria but did not affect FtsZ protein abundance, reduced filamentation (to 4.09 μm on average) (Fig. 3J and Table 2). However, elevated RNase G expression caused reversion of only about 20% of the cell population to normal size, and in the RNase G-complemented strain, approximately 1% of the population formed very long filaments (longer than 40 μm) (Fig. 3J). The persistence of filamentation in RNase G-complemented cells that had a reduced FtsZ protein is consistent with earlier evidence that a subnormal level of FtsZ in E. coli leads to the filamentous morphology (7).
FIG. 3.
Effects of adventitious expression of ftsZ on cellular morphologies of rne deletion mutant strains. Each strain was transformed with pACYC177 or pMT177Z DNA. Liquid cultures were started from single transformant colonies, grown in the presence (0.1% for KSL2000 and KSL2009; 0.2% for KSL2010) or absence of arabinose, and harvested in the mid-log phase after 10 to 12 h; during this period, the cultures were diluted several times to maintain the cells in mid-log-phase growth (OD600 = 0.5 to 1.0). Slides were prepared as described in Materials and Methods. (A to D) KSL2000 (full-length RNase E complementation); (E to H) KSL2009 (N-RNase E complementation); (I to L) KSL2010 (RNase G complementation); (M to P) MT2000 (recA+ strain) (full-length RNase E complementation).
Earlier work showed that adventitious expression of FtsZ in cells having a decreased FtsZ level reverses the cell filamentation phenotype and restores a normal rate of growth (7, 8). However, while introduction of the FtsZ-expressing plasmid pMT177Z into otherwise-uncomplemented cells with rne deleted resulted in replacement of long filaments (>50 μm) (Fig. 3A, E, and I) by short filaments (<10 μm) (Fig. 3C, G, and K), normal cell morphology was not restored. Further experiments indicated that the observed residual filamentation was attributable to the recA deletion we had introduced into our test strains to avoid homologous recombination between the chromosome and the plasmid carrying the complementing genes (18). In a corresponding recA+ RNase E-depleted strain, filamentation was much reduced (to approximately 5 to 10 μm [Fig. 3M] versus >50 μm [Fig. 3A, E, and I]) and was fully reversed by adventitious FtsZ expression (Fig. 3O). Additionally, the short filaments observed in cultures of the RNase G-complemented strain (KSL2010) were absent after the introduction of pMT177Z (Fig. 3L and Table 2), resulting in a phenotype that was indistinguishable from the wild type. However, notwithstanding the absence of filamentation in the rne deletion strain complemented by adventitious expression of FtsZ from pMT177Z, these cells did not acquire CFA in either recA mutant or recA+ bacteria in the absence of further complementation by plasmid-encoded RNase E, N-RNase E, or RNase G (Fig. 4A).
FIG. 4.
Effects of adventitious expression of ftsZ carried by the pMT177Z plasmid on the bacterial growth rate in liquid culture and on the CFA of rne deletion mutant strains. (A) Effects of ftsZ complementation on colony formation. KSL2000(pBAD-RNE) or MT2000 (recA+ strain; pBAD-RNE) containing the pMT177Z plasmid were grown to mid-log phase at 37°C in LB medium supplemented with 0.1% arabinose and antibiotics. After being washed with LB, the cells were diluted (approximately 10−5) and the same volume of culture was spread on LB plates containing or lacking 0.1% arabinose. The plates were incubated at 37°C for 72 h. Colonies were seen on the plate containing 0.1% arabinose after 12 h. (B) Effects of pMT177Z on cell growth in strains also complemented by expression of full-length RNase E, N-RNase E, or RNase G. N3433(pKAN7), KSL2000(pBAD-RNE), KSL2009(pBAD-NRNE), and KSL2010(pBAD-RNG) containing either the pACYC177 (pACYC177Δkan for N3433) plasmid or the pMT177Z plasmid were grown at 37°C in LB medium supplemented with arabinose and antibiotics as indicated in Materials and Methods. Increases in cell density were monitored by measuring the OD600. At time zero, the OD600 values of all cultures were adjusted to 0.1.
Effects of RNase E, N-RNase E, RNase G, and FtsZ expression on cell growth rate and colony formation.
The effects of FtsZ expression on growth of cells with rne deleted complemented by full-length RNase E, N-RNase E, or RNase G, on the growth rate in LB liquid cultures, and on the ability to form colonies on LB plates were examined (Fig. 4 and Table 2). Expression of full-length RNase E (strain KSL2000) restored growth to the rate seen for the parental strain, N3433. Although N-RNase E and RNase G complementation also restored CFA to rne null mutant bacteria (Table 2), the rate of growth was reduced (Fig. 4B). Expression of FtsZ from pMT177Z had no significant effect on the rates of growth of these complemented strains. Occasional colonies were observed when strain KSL2000 was plated at high concentration (>104 CFU/plate) even in the absence of arabinose, whether the FtsZ-expressing plasmid was present or absent. Further analysis of several randomly selected colonies indicated that all of them had acquired arabinose-independent induction of the BAD promoter and were expressing RNase E in the absence of arabinose (data not shown).
DISCUSSION
RNase E has long been known to be essential for the propagation of E. coli cells in liquid cultures and for colony formation on solid media (1, 15). Because RNase E carries out the processing of 9S rRNA (13), tRNA (33, 37), and the precursor of M1 RNA (26), defective processing and decay of these various RNAs have been investigated as potential causes of the loss of CFA in cells lacking RNase E activity. The evidence accumulated from such studies argues that RNase E actions on these RNAs are not the basis for the essentiality of the RNase (9, 18, 34). As RNase E also affects the decay of bulk mRNA (32) and the degradation of a large number of individual RNAs (2, 18), it has been speculated that RNase E essentiality may be the consequence of its activity on one or more mRNA species.
Alteration of the ratio of the FtsZ and FtsA proteins has been known for more than a decade to lead to the growth of E. coli cells in long filamentous chains (8). The filamentation phenotype observed in cells lacking RNase E function (15, 44) and the observation that RNase E is involved in the processing and degradation of transcripts of the polycistronic ftsQAZ operon (5, 12), which includes genes required for the physical separation of cells at the time of division, together have raised the possibility that abnormal cell division resulting from fts operon dysfunction may account for the essentiality of RNase E in E. coli. These considerations, plus earlier results from our laboratory indicating that complementation of an rne deletion mutant by overproduced RNase G restores CFA (18) but not a normal growth rate (19), prompted the investigations reported here.
Our findings indicate that RNase E depletion results in an increase of FtsA protein and a decrease of FtsZ protein. As earlier work has shown that alteration in the FtsZ/FtsA ratio is associated with cell filamentation (8), we suggest that the filamentation phenotype observed during RNase E deficiency may be mediated through this changed ratio. However, while the imbalance of FtsZ/FtsA and the filamentation phenotype were reversed by adventitious FtsZ expression, as expected, CFA was not restored, indicating that the actions of RNase E in maintaining a proper relative abundance of the FtsZ and FtsA proteins is not the basis of RNase E essentiality. Additionally, the observed ability of RNase G to confer CFA on an rne deletion mutant without altering the cellular level of FtsZ or eliminating FtsZ-induced filamentation further supports the conclusion that FtsZ deficiency and the filamentation associated with it do not account for the nonviability of RNase E-depleted E. coli. Immunoblot analysis showed that RNase E-depleted bacteria carrying pMT177Z had a FtsZ/FtsA ratio that resembled that of the parental rne+ strain and that was higher than the ratio observed in the RNase G-complemented KSL2010 strain.
Takada et al. recently reported that artificial expression of the FtsZ protein from a plasmid-borne ftsZ gene partially suppressed the temperature sensitivity of an rne(Ts) mutant strain and consequently concluded that a decreased cellular level of FtsZ is responsible for the nonviability of RNase E-deficient cells (40). Our results, which were obtained using an rne deletion mutant, do not support this conclusion.
Whereas the growth rate of KSL2009 (N-RNase E) was much higher than that of KSL2010 (RNase G), the CFU per ml per OD600 unit of KSL2009 interestingly was similar to that of KSL2010, indicating that the growth rate in liquid culture does not necessarily quantitatively parallel the CFA. Additionally, whereas concurrent overproduction of FtsZ and RNase G in rne null mutant bacteria (KSL2010) reversed filamentation, the lowered growth rate was unaffected and the observed CFU per ml per OD600 was only 20% of the level seen for the parental N3433 strain. Together, these data suggest that the loss of CFA associated with rne deletion is not per se a consequence of altered cell morphology.
Our results indicate that RNase E activity, as was speculated by Cam et al. (5), is required to maintain a proper cellular ratio of the FtsZ and FtsA proteins and further suggest that perturbation of this ratio is a cause of the filamentation observed in rne null mutant bacteria. While we additionally found that adventitious production of FtsZ in RNase E-depleted cells reverses the filamentation phenotype, restoration of a normal cellular level of FtsZ and a proper FtsZ/FtsA ratio were not sufficient to confer CFA on cells lacking RNase E.
Acknowledgments
We thank Björn Sohlberg and Johan Kers for helpful discussion and suggestions. We also thank Roberta Peterson for her assistance with the manuscript and Munetaka Kawamoto for his secretarial support.
This study was supported by NIH-NIGMS grant GM54158 to S.N.C. and in part by a research fellowship from the Japan Society for the Promotion of Science for Young Scientists and the Yoshida Foundation for Science and Technology to M.T.
REFERENCES
- 1.Apirion, D. 1978. Isolation, genetic mapping and some characterization of a mutation in Escherichia coli that affects the processing of ribonuleic acid. Genetics 90:659-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein, J. A., A. B. Khodursky, P. H. Lin, S. Lin-Chao, and S. N. Cohen. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99:9697-9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bramhill, D., and C. M. Thompson. 1994. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc. Natl. Acad. Sci. USA 91:5813-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 4.Buddelmeijer, N., and J. Beckwith. 2002. Assembly of cell division proteins at the E. coli cell center. Curr. Opin. Microbiol. 5:553-557. [DOI] [PubMed] [Google Scholar]
- 5.Cam, K., G. Rome, H. M. Krisch, and J. P. Bouche. 1996. RNase E processing of essential cell division genes mRNA in Escherichia coli. Nucleic Acids Res. 24:3065-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai, K., and J. Lutkenhaus. 1991. ftsZ is an essential cell division gene in Escherichia coli. J. Bacteriol. 173:3500-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai, K., and J. Lutkenhaus. 1992. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J. Bacteriol. 174:6145-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deana, A., and J. G. Belasco. 2004. The function of RNase G in Escherichia coli is constrained by its amino and carboxyl termini. Mol. Microbiol. 51:1205-1217. [DOI] [PubMed] [Google Scholar]
- 10.de Boer, P., R. Crossley, and L. Rothfield. 1992. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359:254-256. [DOI] [PubMed] [Google Scholar]
- 11.Feng, Y., H. Huang, J. Liao, and S. N. Cohen. 2001. Escherichia coli poly(A)-binding proteins that interact with components of degradosomes or impede RNA decay mediated by polynucleotide phosphorylase and RNase E. J. Biol. Chem. 276:31651-31656. [DOI] [PubMed] [Google Scholar]
- 12.Flärdh, K., P. Palacios, and M. Vicente. 1998. Cell division genes ftsQAZ in Escherichia coli require distant cis-acting signals upstream of ddlB for full expression. Mol. Microbiol. 30:305-315. [DOI] [PubMed] [Google Scholar]
- 13.Ghora, B. K., and D. Apirion. 1978. Structural analysis and in vitro processing to p5 rRNA of a 9S RNA molecule isolated from an rne mutant of E. coli. Cell 15:1055-1066. [DOI] [PubMed] [Google Scholar]
- 14.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15:R514-R526. [DOI] [PubMed] [Google Scholar]
- 15.Goldblum, K., and D. Apirion. 1981. Inactivation of the ribonucleic acid-processing enzyme ribonuclease E blocks cell division. J. Bacteriol. 146:128-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hale, C. A., and P. A. J. de Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiraga, S., H. Niki, T. Ogura, C. Ichinose, H. Mori, B. Ezaki, and A. Jaffe. 1989. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J. Bacteriol. 171:1496-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, K., J. A. Bernstein, and S. N. Cohen. 2002. RNase G complementation of rne null mutation identifies functional interrelationships with RNase E in Escherichia coli. Mol. Microbiol. 43:1445-1456. [DOI] [PubMed] [Google Scholar]
- 19.Lee, K., and S. N. Cohen. 2003. A Streptomyces coelicolor functional orthologue of Escherichia coli RNase E shows shuffling of catalytic and PNPase-binding domains. Mol. Microbiol. 48:349-360. [DOI] [PubMed] [Google Scholar]
- 20.Lee, K., C. A. Holland-Staley, and P. R. Cunningham. 2001. Genetic approaches to studying protein synthesis: effects of mutations at Psi516 and A535 in Escherichia coli 16S rRNA. J. Nutr. 131:2994S-3004S. [DOI] [PubMed] [Google Scholar]
- 21.Lee, K., X. Zhan, J. Gao, J. Qiu, Y. Feng, R. Meganathan, S. N. Cohen, and G. Georgiou. 2003. RraA, a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell 114:623-634. [PubMed] [Google Scholar]
- 22.Li, Z., S. Pandit, and M. P. Deutscher. 1999. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 18:2878-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin-Chao, S., and S. N. Cohen. 1991. The rate of processing and degradation of antisense RNAI regulates the replication of ColE1-type plasmids in vivo. Cell 65:1233-1242. [DOI] [PubMed] [Google Scholar]
- 24.Lin-Chao, S., C. L. Wei, and Y. T. Lin. 1999. RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc. Natl. Acad. Sci. USA 96:12406-12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez, P. J., I. Marchand, S. A. Joyce, and M. Dreyfus. 1999. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 33:188-199. [DOI] [PubMed] [Google Scholar]
- 26.Lundberg, U., and S. Altman. 1995. Processing of the precursor to the catalytic RNA subunit of RNase P from Escherichia coli. RNA 1:327-334. [PMC free article] [PubMed] [Google Scholar]
- 27.Margolin, W. 2005. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell. Biol. 6:862-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDowall, K. J., and S. N. Cohen. 1996. The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA-binding site. J. Mol. Biol. 255:349-355. [DOI] [PubMed] [Google Scholar]
- 29.McDowall, K. J., V. R. Kaberdin, S. W. Wu, S. N. Cohen, and S. Lin-Chao. 1995. Site-specific RNase E cleavage of oligonucleotides and inhibition by stem-loops. Nature 374:287-290. [DOI] [PubMed] [Google Scholar]
- 30.Miczak, A., V. R. Kaberdin, C. L. Wei, and S. Lin-Chao. 1996. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. USA 93:3865-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okada, Y., M. Wachi, A. Hirata, K. Suzuki, K. Nagai, and M. Matsuhashi. 1994. Cytoplasmic axial filaments in Escherichia coli cells: possible function in the mechanism of chromosome segregation and cell division. J. Bacteriol. 176:917-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono, M., and M. Kuwano. 1979. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J. Mol. Biol. 129:343-357. [DOI] [PubMed] [Google Scholar]
- 33.Ow, M. C., and S. R. Kushner. 2002. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev. 16:1102-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ow, M. C., Q. Liu, and S. R. Kushner. 2000. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol. Microbiol. 38:854-866. [DOI] [PubMed] [Google Scholar]
- 35.Py, B., C. F. Higgins, H. M. Krisch, and A. J. Carpousis. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169-172. [DOI] [PubMed] [Google Scholar]
- 36.Ray, A., and D. Apirion. 1980. Cloning the gene for ribonuclease E, an RNA processing enzyme. Gene 12:87-94. [DOI] [PubMed] [Google Scholar]
- 37.Ray, B. K., and D. Apirion. 1981. Transfer RNA precursors are accumulated in Escherichia coli in the absence of RNase E. Eur. J. Biochem. 114:517-524. [DOI] [PubMed] [Google Scholar]
- 38.RayChaudhuri, D., and J. T. Park. 1992. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359:251-254. [DOI] [PubMed] [Google Scholar]
- 39.Romberg, L., and P. A. Levin. 2003. Assembly dynamics of the bacterial cell division protein FTSZ: poised at the edge of stability. Annu. Rev. Microbiol. 57:125-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takada, A., K. Nagai, and M. Wachi. 2005. A decreased level of FtsZ is responsible for inviability of RNase E-deficient cells. Genes Cells 10:733-741. [DOI] [PubMed] [Google Scholar]
- 41.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanzo, N. F., Y. S. Li, B. Py, E. Blum, C. F. Higgins, L. C. Raynal, H. M. Krisch, and A. J. Carpousis. 1998. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12:2770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wachi, M., G. Umitsuki, M. Shimizu, A. Takada, and K. Nagai. 1999. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5′ end of 16S rRNA. Biochem. Biophys. Res. Commun. 259:483-488. [DOI] [PubMed] [Google Scholar]
- 44.Wang, M., and S. N. Cohen. 1994. ard-1: a human gene that reverses the effects of temperature-sensitive and deletion mutations in the Escherichia coli rne gene and encodes an activity producing RNase E-like cleavages. Proc. Natl. Acad. Sci. USA 91:10591-10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward, J. E., Jr., and J. Lutkenhaus. 1985. Overproduction of FtsZ induces minicell formation in E. coli. Cell 42:941-949. [DOI] [PubMed] [Google Scholar]