Abstract
The gram-positive filamentous bacterium Streptomyces coelicolor has a complex developmental cycle with three distinct phases: growth of the substrate mycelium, development of reproductive structures called aerial hyphae, and differentiation of these aerial filaments into long chains of exospores. During a transposon mutagenesis screen, we identified a novel gene (devA) required for proper development. The devA mutant produced only rare aerial hyphae, and those that were produced developed aberrant spore chains that were much shorter than wild-type chains and had misplaced septa. devA encodes a member of the GntR superfamily, a class of transcriptional regulators that typically respond to metabolite effector molecules. devA forms an operon with the downstream gene devB, which encodes a putative hydrolase that is also required for aerial mycelium formation on R5 medium. S1 nuclease protection analysis showed that transcription from the single devA promoter was temporally associated with vegetative growth, and enhanced green fluorescent protein transcriptional fusions showed that transcription was spatially confined to the substrate hyphae in the wild type. In contrast, devAB transcript levels were dramatically upregulated in a devA mutant and the devA promoter was also active in aerial hyphae and spores in this background, suggesting that DevA might negatively regulate its own production. This suggestion was confirmed by gel mobility shift assays that showed that DevA binds its own promoter region in vitro.
The life cycle of the filamentous soil bacterium Streptomyces coelicolor begins when a germinating spore gives rise to a colony of substrate mycelium. Vegetative growth proceeds by hyphal tip extension and by branching until changes in nutritional status (29, 41) and the accumulation of extracellular signaling molecules (37, 38, 51) trigger formation of specialized reproductive structures called aerial hyphae. These multigenomic filaments grow from the colony surface into the air, subsequently subdividing through the synchronous deposition of multiple septa, with each resulting compartment maturing into a unigenomic spore (7, 8, 14, 30). Mutants involved in this complex developmental process can be classified into two broad groups: those blocked in formation of aerial hyphae (the bld mutants) and those able to form aerial hyphae but unable to complete their development into mature spores (the whi mutants).
Direct regulatory interactions between S. coelicolor developmental genes are beginning to be revealed. BldD is a key repressor, during vegetative growth, of several loci required for normal sporulation, including the sigma factor genes bldN and whiG (15), σBldN directs transcription of bldM (4), and σWhiG directs transcription of whiH and whiI (1, 46). Further, the response regulator RamR directly regulates the promoter of the RamCSAB operon, which specifies the synthesis of SapB, a modified peptide that functions as a surfactant, releasing surface tension at the air-water interface to allow nascent aerial hyphae to escape into the air (33, 39).
Although many bld and whi genes have been characterized, there is abundant evidence that there are many developmental loci still to be identified in S. coelicolor (19-21, 38, 47, 49). To date, most screens for developmental genes have been based on the isolation of spontaneous, UV-induced, or N-methyl-N′-nitro-N-nitrosoguanidine-induced mutants, followed by their complementation using shotgun libraries (or occasionally using map-based cloning). Although these methods are effective, they are very labor-intensive. More recently, successful screens for new developmental genes have been based on in vitro transposon mutagenesis of plasmid libraries of S. coelicolor DNA, followed by the delivery of these mutagenized libraries into S. coelicolor by transformation or conjugation (19-21, 49). However, a significant deficiency of the S. coelicolor“genetic tool kit” has been the absence of a robust in vivo transposon mutagenesis screening system; indeed, only one gene (bldK) has been identified by in vivo transposon tagging in S. coelicolor (37). Recently, a new in vivo transposon mutagenesis system has been developed for S. coelicolor, based on conjugation of a suicide delivery vector from Escherichia coli (16). Here we report the use of this system to identify devA, encoding a GntR-like transcriptional repressor required for normal development in S. coelicolor.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and conjugal transfer from E. coli to Streptomyces.
The S. coelicolor strains used in this study are summarized in Table 1. All strains were cultivated on minimal medium containing mannitol (0.5% wt/vol), R5, or mannitol-soy flour agar (32). Conjugation from the E. coli strain ET12567 (dam dcm hsdS), containing the driver plasmid pUZ8002, was used to bypass the methyl-specific restriction system of S. coelicolor (40).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or comments | Source or reference |
|---|---|---|
| S. coelicolor strains | ||
| M600 | Prototrophic, SCP1− SCP2− | 32 |
| J3101 | M600 devA::Tn4560 | This work |
| J3102 | M600 devA::Tn5062 | This work |
| J3104 | M600 devB::Tn5062 | This work |
| Plasmids | ||
| pKay1 | Conjugative suicide vector for delivering Tn4560 from E. coli into Streptomyces | 16 |
| pSET152 | Plasmid integrating at phage φC31 attB site, carrying apramycin resistance (apr) | 5 |
| pMS82 | Plasmid integrating at phage φBT1 attB site, carrying hygromycin resistance (hyg) | 22 |
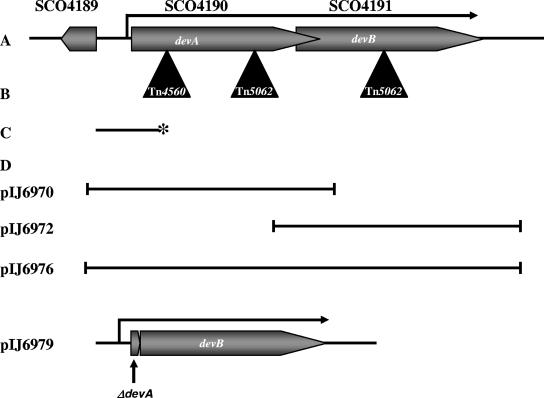
| pIJ6970 | pSET152 carrying 1.5-kb devA fragment (Fig.1) | This work |
| pIJ6972 | pMS82 carrying 1.3-kb devB fragment (Fig 1) | This work |
| pIJ6976 | pMS82 carrying 2-kb devAB fragment (Fig 1) | This work |
| pIJ6979 | pMS82 carrying devAB fragment with in-frame devA deletion (Fig 1) | This work |
| pIJ6973 | pET15b derivative expressing His6-DevA | This work |
| pIJ8660 | egfp-containing vector for transcriptional fusions | 50 |
| pIJ6974 | pIJ8660 derivative expressing a devAp-egfp transcriptional fusion | This work |
Random in vivo transposon mutagenesis.
Random in vivo transposition was performed using the Tn4556-derived, Tn3-like Streptomyces transposon Tn4560, carried on the conjugal suicide vector pKay1 (9, 16). pKay1 was conjugated from E. coli ET12567/pUZ8002 into S. coelicolor M600, and exconjugants were selected by overlaying with nalidixic acid (25 μg ml−1) and viomycin (30 μg ml−1). Mutants were checked for transposition (viomycin resistant) and loss of the delivery vector (apramycin and thiostrepton sensitive). In addition, mutants were checked for chloramphenicol resistance, and any chloramphenicol-sensitive strains were discarded (S. coelicolor is naturally chloramphenicol resistant, but sensitive derivatives arise through spontaneous deletion of segments near the “right-hand” end of the linear chromosome [32]).
Construction of devA and devB Tn5062 transposon mutants.
Derivatives of cosmid D66 carrying Tn5062 insertions in devA or devB (kind gifts of Paul Herron and Paul Dyson), generated using the in vitro transposition method of Bishop et al. (6), were introduced into S. coelicolor M600 by conjugation from E. coli ET12567/pUZ8002. Mutants exhibiting the double-crossover phenotype (apramycin resistant, kanamycin sensitive) were confirmed by Southern hybridization, and designated J3102 (devA::Tn4560) and J3104 (devB::Tn4560).
Plasmid construction.
Plasmids used in this work are described in Table 1. Plasmids were constructed as follows. To construct pIJ6970, a 1.5-kb fragment carrying devA (Fig. 1) was amplified from cosmid D66 by PCR using oligonucleotides 5′-AAGACCGCGGAGACCA-3′ and 5′-GGTCCCGGGTCTCCA-3′ and ligated into the EcoRV site of pSET152. For pIJ6972, a 1.3-kb fragment carrying devB and part of devA was amplified from cosmid D66 by using oligonucleotides 5′-GTACCTGCTCAACGGCGAGGA-3′ and 5′-ACGCAGATATCGGCAGACGACA-3′ and ligated into the EcoRV site of pSET152. To construct pIJ6976, a 2-kb fragment carrying devA and devB (Fig. 1) was amplified from cosmid D66 by using oligonucleotides 5′-AAGACCGCGGAGACCA-3′ and 5′-ACGCAGATATCGGCAGACGACA-3′ and ligated into the EcoRV site of pMS82. For pIJ6974, a 444-bp fragment carrying the SCO4189-devA intergenic region was amplified from cosmid D66 by using the oligonucleotides 5′-ACCGTACCTCCACTG-3′ and 5′-GGCTGCCCTTGCTGA-3′ and ligated into the EcoRV site of pIJ8660. The integrity of all subcloned fragments was confirmed by sequencing.
FIG. 1.
A. Genetic organization of the devA region. B. Position of Tn4560 in devA following random transposon mutagenesis and positions of the Tn5062 insertions inactivating devA and devB. C. S1 nuclease mapping probe for the devA promoter; 5′ 32P labeling of the probe is indicated by an asterisk. D. Fragments used in complementation experiments.
Construction of a devA in-frame deletion null mutant allele.
An in-frame deletion null mutant allele of devA was constructed by PCR targeting of linearized cosmid D66 in λ-RED-proficient E. coli, using the method of Gust et al. (23, 24). The genome of S. coelicolor contains only 27 AflII sites, which means that the single AflII site in Tn5062, used to make in vitro transposon mutants (6), can be exploited in PCR targeting experiments. A derivative of cosmid D66 carrying a Tn5062 insertion in devA (a kind gift of Paul Herron and Paul Dyson) was linearized within the transposon by digestion with AflII (the parent D66 cosmid itself contains no AflII sites). Uncut cosmid was eliminated by gel electrophoresis, and the linearized cosmid was coelectroporated into BW25113/pIJ790 along with a 100-mer oligonucleotide (5′-AAACAAGTTTCAAACAACTCCCTATAGGTAGGTCGAAGTTGTAGCGTTTGATCACAGAAGTGGTTCGACGCCCTCTGGGAAACCATCACCACGGACATGA-3′) consisting of two 50-nucleotide sequences corresponding to the upstream and downstream regions of the target gene (leaving the desired deletion junction 5′-TTGATCACAGAA-3′). Recircularization of the cosmid brought about by double crossing over between the 5′ and the 3′ ends of the oligonucleotide and the linearized cosmid resulted in colonies that were resistant to kanamycin (cosmid marker) and sensitive to apramycin (carried by Tn5062). Mutant cosmid D66 was confirmed by sequencing and was used to construct pIJ6979 as follows. A 1.2-kb fragment corresponding to the devA promoter region, four codons of devA (see above), and the complete devB coding sequence (Fig. 1) was amplified from the mutant cosmid D66 by PCR using oligonucleotides 5′-AAGACCGCGGAGACCA-3′ and 5′-ACGCAGATATCGGCAGACGACA-3′. The PCR product was ligated into the EcoRV site of pMS82. Sequencing confirmed the integrity of the fragment.
RNA isolation and S1 nuclease protection analysis.
RNA samples were isolated throughout the time course as described previously (31) and used to analyze expression of devAB. The devA transcription start point was mapped using a 444-bp probe that spanned the intergenic region between open reading frame 4189 and devA (Fig. 1), which was generated by PCR using the oligonucleotides 5′-ACCGTACCTCCACTGGA-3′ and 5′-GGCTGCCCTTGCTGA-3′. Oligonucleotide primer labeling and S1 nuclease protection assays were performed as described previously (32).
Overproduction and purification of DevA.
An 890-bp PCR-generated fragment with an engineered NdeI site overlapping the start codon and an XhoI site following the stop codon (generated using the primers 5′-CATATGGTCGTGACTCAGGA-3′ and 5′-ACTGTGAGAGGATCACTGAGCTC-3′) was blunt-end cloned into pGEM-T-Easy. The fragment was verified by sequencing, excised as an NdeI/XhoI fragment, and cloned into the expression vector pET15b (Novagen) cut with NdeI/XhoI, generating pIJ6973. pIJ6973 was introduced into E. coli Rosetta(DE3) (Novagen), and DevA expression was induced by addition of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) in exponentially growing cells (optical density at 600 nm of 0.5). The resulting N-terminally His-tagged DevA was purified by nickel affinity chromatography, and the molecular mass of the overproduced protein was confirmed by surface-enhanced laser desorption ionization mass spectrometry.
Gel mobility shift assays.
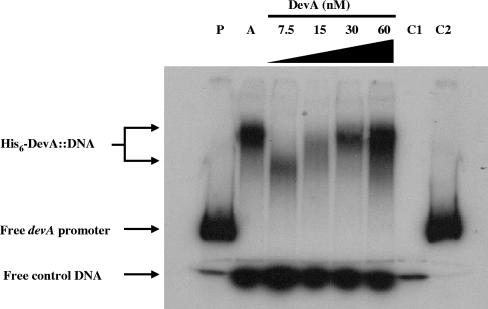
Protein-DNA gel retardation assays (gel shifts) were performed using the 444-bp PCR fragment used as the S1 nuclease protection assay probe (see above), containing the putative devA promoter region. A control fragment was amplified from the coding sequence of devA by using the oligonucleotides 5′-ACACCTCGGAGACGCTGA-3′ and 5′-CTCCTCGCCGTTGAGCA-3′. Fragments were labeled using [γ-32P]dATP and T4 polynucleotide kinase. Assays were performed with the following mixture in a total volume of 20 μl: 20 mM Tris-HCl (pH 7.5), 10 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 1 μg of bovine serum albumin, and 3.4 fmol 32P-labeled devA promoter DNA. The protein concentrations used are indicated in Fig. 6. The assays were initiated by addition of DevA diluted to the appropriate concentration, and incubation was at 30°C for 20 min. Samples was loaded onto a 6% polyacrylamide-Tris-borate-EDTA gel, and visualization was by autoradiography.
FIG. 6.
Gel retardation of the devA promoter by DevA. Results of mobility shift assays in which 3.4 fmol 32P-labeled probe was incubated with 20 ng crude extract from E. coli Rosetta carrying pIJ6973 after induction with IPTG (A) or with 7.5 to 60 nM purified DevA are shown. P, probes alone; C1, control probe (internal to the devA coding sequence) alone; C2, devA promoter probe alone.
DevA in silico analysis.
Secondary-structure predictions, multiple alignment, and phylogenetic studies of DevA were done as described previously (44). The GntR proteins used in phylogenetic analysis are listed in Table 2.
TABLE 2.
GntR proteins used in phylogenetic analysis
| Protein | Function | Organism | Length (amino acids) | Accession no. | Subfamily |
|---|---|---|---|---|---|
| FadR | Fatty acid metabolism regulator | Escherichia coli | 238 | P09371 | FadR |
| PdhR | Pyruvate dehydrogenase complex repressor | Escherichia coli | 254 | P06957 | |
| FarR | Histidine utilization repressor | Pseudomonas putida | 248 | P22773 | HutC |
| XlnR | Unknown | Streptomyces lividans | 252 | Q9ACN8 | |
| YvoA | Unknown | Bacillus subtilis | 243 | O34817 | |
| SAV4023 | Unknown | Streptomyces avermitilis | 297 | Q99SM3 | DevA |
| SAV4021 | Unknown | Streptomyces avermitilis | 292 | Q82G77 | |
| SCO4188 | Unknown | Streptomyces coelicolor | 303 | Q9FCI0 | |
| DevA | Developmental regulator | Streptomyces coelicolor | 291 | Q9FCH9 | |
| MocR | Probable rhizopine catabolism regulator | Rhizobium meliloti | 493 | P49309 | MocR |
| YtrA | Acetoine utilization gene cluster repressor | Bacillus subtilis | 130 | O34712 | YtrA |
| PlmA | Plasmid maintenance regulator | Anabaena sp. strain PCC 7120 | 328 | Q8YXY0 | PlmA |
| AraR | Transcriptional repressor of the l-arabinose operon | Bacillus subtilis | 362 | P96711 | AraR |
Microscopy.
Light microscopy, scanning electron microscopy (SEM), and transmission electron microscopy (TEM) were performed as described previously (35).
RESULTS
Disruption of SCO4190 (devA) causes a developmental defect.
A screen for developmentally impaired mutants of S. coelicolor was undertaken using the in vivo transposon mutagenesis system developed by Fowler (16). The Tn3-like Streptomyces transposon Tn4560 was introduced by conjugation from E. coli into S. coelicolor M600 on a suicide vector (pKay1), as described in Materials and Methods, and the resulting exconjugants were screened for developmental defects, which were determined by the lack of fuzzy white aerial mycelium and/or the inability to develop the gray polyketide spore pigment characteristic of wild-type spores. One such mutant, J3101, produced a very sparse aerial mycelium, and the developmental defect of this strain was confirmed by light microscopy observation, showing rare, short, aberrantly formed aerial hyphae. The Tn4560 insertion site in J3101 was identified by ligation-mediated PCR, using one primer internal to Tn4560 and sequencing of the resulting PCR product, followed by BLAST searching against the S. coelicolor genome (www.sanger.ac.uk/projects/s_coelicolor/). The transposon was found near the middle of the chromosome at position 4599109, within the gene SCO4190, which was designated devA (Fig. 1).
devA encodes a member of the GntR family of transcriptional regulators.
devA encodes a 291-amino-acid protein with a predicted N-terminal helix-turn-helix motif at residues 43 to 66 (Fig. 2A) (Score 7.17) (10). Global similarity searches of the EMBL and Pfam databases showed that this protein belongs to the GntR family of transcriptional regulators (Pfam entry PF00392 gntR) (2, 25). Where they have been investigated, GntR regulators have been found to act as dimeric repressors and/or activators that respond to effector molecules, often carboxylate-containing intermediates in primary metabolism. They consist of a conserved N-terminal helix-turn-helix DNA-binding domain and a variable C-terminal-effector-binding/oligomerization domain (25, 44). Previous screens for developmental mutants of streptomycetes have led to the identification of three other genes encoding members of the GntR family: whiH (46); dasR (48), and SCO7168 (49).
FIG. 2.
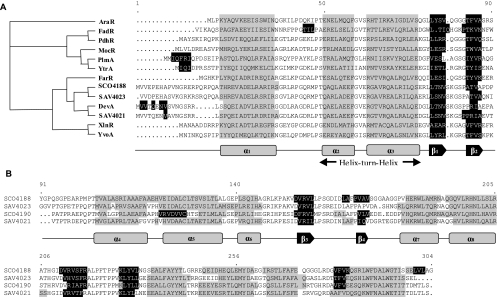
In silico analysis of the DevA/GntR subfamily. Transcription factor abbreviations are as indicated in Table 2. A. Cladogram built from multiple alignment of the helix-turn-helix motifs (alpha-helices two and three) and structure-based alignment of the DNA-binding domain of the selected GntR members. DevA members appear in a common clade with HutC/GntR members, suggesting, for both subfamilies, a common ancestor for their DNA-binding domain. B. Structure-based alignment of the effector-binding domain of DevA subfamily members, highlighting a novel type of secondary-structure topology for the GntR superfamily.
DevA defines a new subfamily of the GntR family.
The GntR family contains four major subfamilies (FadR, HutC, MocR, and YtrA) and two minor subfamilies (AraR and PlmA), recognized by secondary-structure prediction and phylogenetic analysis (34, 44). BLAST results indicated that high levels of similarity to the GntR family are limited to the N-terminal region of DevA, coinciding with the DNA-binding domain (Fig. 2A). The GntR-like proteins that are most similar to DevA in the N-terminal domain belong to the HutC subfamily, but the C-terminal domain, usually associated with effector binding and oligomerization, is not similar to those of other members of the GntR family, and secondary-structure predictions imply a new topology fused to the GntR-type DNA-binding domain (Fig. 2B). Additional phylogenetic analysis, secondary-structure predictions (Fig. 2), and BLAST results confirmed that DevA and its relatives (SCO4188, and their orthologues in Streptomyces avermitilis, SAV4021, and SAV4023) form a new GntR subfamily.
Immediately upstream of devA is a gene encoding a small (62-amino-acid) hypothetical protein (SCO4189) with no known homologues. The next two genes upstream of devA, SCO4187 and SCO4188, appear to represent a duplication event; SCO4187 is similar to SCO4189 (51.1% identity), and SCO4188 is similar to DevA (57.6% identity) and is also a member of the new DevA subfamily of GntR regulators (see above). Downstream from and overlapping devA is a gene encoding a 237-amino-acid putative hydrolase (SCO4191; devB) (Fig. 1), with a Pfam match to a poorly defined hydrolase family (PF00702) which includes proteins with wide-ranging functions, including phosphatases and haloacid-dehalogenases. BLAST and PSI-BLAST searching revealed resemblances to many proposed phosphatases, with the closest match being a 235-amino-acid putative phosphatase from Neisseria meningitidis (EMBL accession no. AL162756; 29.5% identity).
This cluster of genes and their organization are conserved in Streptomyces avermitilis and Streptomyces scabies (except for the SCO4187 homologue), but the cluster could not be found in other sequenced actinomycete genomes (Mycobacterium tuberculosis, Mycobacterium leprae, Corynebacterium glutamicum, and Nocardia farcinica) by BLAST searching. The absence of these genes in the unicellular and hyphal but nonsporulating actinomycetes is consistent with the role of this gene in aerial hyphal formation and sporulation in streptomycetes.
Disruption of devA causes the formation of rare, aberrant aerial hyphae.
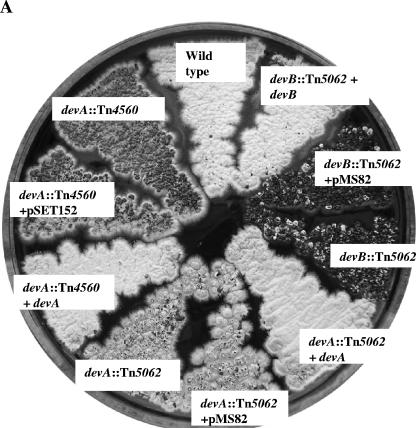
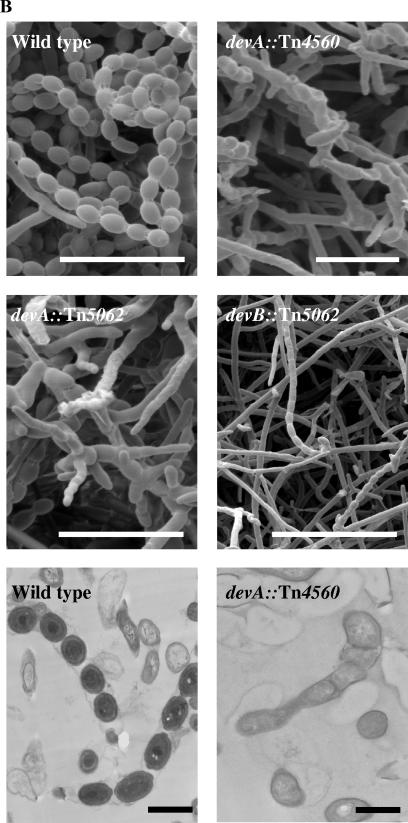
On plates of R5, colonies of J3101 (devA::Tn4560) produced only rare aerial hyphae, mainly confined to the colony edges, which, even on prolonged incubation, remained white (Fig. 3A). SEM of J3101 revealed few aerial hyphae, with spore chains shorter than wild type (<10 spores per chain). Septum formation was irregular, resulting in aberrantly shaped spores (Fig. 3B). TEM confirmed the SEM observations and revealed that in addition to misplaced septa, the compartment walls lacked the thickening observed in wild-type spores (Fig. 3B), and the normal condensation of the chromosome was not apparent, although DAPI (4′,6′-diamidino-2-phenylindole) staining confirmed the presence of DNA in each spore compartment (data not shown).
FIG.3.
A. Effect of devA and devB disruptions on colony appearance and complementation of mutants. Strains were grown on R5 medium. B. Electron microscopy images of devA and devB mutants. Colonies were grown on R5 medium, except for J3104 (devB::Tn5062), which was grown on minimal medium with mannitol. SEM bar = 10 μm; TEM bar = 1 μm.
To confirm that the phenotype of J3101 was caused by the insertion of Tn4560 into devA, a second devA mutant was constructed in the M600 background by using a derivative of cosmid D66 carrying Tn5062 (an apramycin-resistant derivative of Tn5) inserted into devA (at nucleotide 4598766). This cosmid derivative (a kind gift of Paul Herron and Paul Dyson), generated using the in vitro transposition method of Bishop et al. (6), was introduced into S. coelicolor by conjugation from E. coli. Mutants exhibiting the double-crossover phenotype (apramycin resistant, kanamycin sensitive) were confirmed by Southern hybridization (not shown), and one was designated J3102. On R5 medium, colonies of J3102 (devA::Tn5062) (Fig. 3A) exhibited an appearance similar to that of J3101 (devA::Tn4560). SEM analysis revealed short, sparse aerial hyphae, with the characteristic aberrant septation associated with J3101 (Fig. 3B). TEM examination of J3102 also revealed short spore chains, lacking the cell wall thickening associated with wild-type spores and showing the aberrant septa formation and apparent lack of chromosome condensation observed in J3101 (data not shown).
J3101 (devA::Tn4560) was fully complemented by pIJ6970 (Fig. 3A), a pSET152 derivative carrying a 1.5-kb fragment including devA and its promoter region (Fig. 1). Introduction of pSET152 alone, which integrates site specifically into the S. coelicolor chromosome at the phage φC31 attB site, had no effect on the morphological phenotype of J3101 (Fig. 3A). The devA::Tn5062 mutant (J3102) was also complemented by the same fragment, introduced on the integrating vector pMS82 (data not shown).
Disruption of devB causes two alternative conditional developmental phenotypes.
In the same way, a disruption mutant of devB (SCO4191; putative phosphatase) was constructed using a derivative of cosmid D66 carrying Tn5062 inserted into devB (at nucleotide 4599827) (a kind gift of Paul Herron and Paul Dyson). The resulting strain was confirmed by Southern hybridization and designated J3104. On R5 medium, J3104 colonies were bald (Fig. 3A). However, production of nonsporulating aerial hyphae could be restored by growth on minimal medium with mannitol as a carbon source; SEM examination showed that although aerial hyphae were present at approximately wild-type levels, sporulation septation was absent (Fig. 3B). Thus, the devB mutant has two distinct developmental phenotypes, depending on medium conditions.
The devB mutant (J3104) was not complemented by pIJ6972, a pMS82 derivative containing a 1.3-kb fragment carrying devB and the 3′ end of devA (Table 1; Fig. 1). However, it was complemented by pIJ6976, a pMS82 derivative containing a 2-kb fragment carrying the devA promoter region, devA, and devB (Table 1). Colonies of the resulting strain appeared wild type on all media and could be seen by phase-contrast microscopy to sporulate abundantly. The devB mutant was also complemented by a pMS82 derivative (pIJ6979) containing a fragment carrying an in-frame devA deletion, such that the devA promoter reads directly into devB (Fig. 1 and 3A). These data suggest cotranscription of devA and devB from the same promoter.
DevA negatively regulates its own transcription.
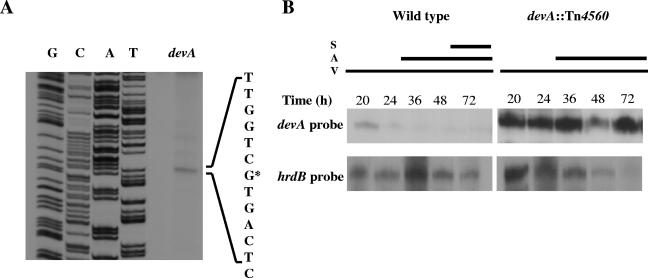
S1 nuclease protection analysis identified a single devA promoter. High-resolution mapping (Fig. 4A) localized the transcription start point to the first “G” of the most likely translational start codon (GTG at nucleotide 4598634), suggesting that the devA mRNA is leaderless, a frequent feature of Streptomyces transcripts (28). Indeed, whiH, encoding another GntR-type transcriptional repressor involved in S. coelicolor development, also has a leaderless transcript (46). RNA was isolated from surface-grown cultures of S. coelicolor M600 and J3101 (devA) throughout the growth cycle, providing samples when vegetative mycelium, aerial mycelium, and spores were present in the cultures or at equivalent time points. The devA transcript was present in M600 at the earliest time point tested (20 h) but was barely detectable at subsequent time points during differentiation (Fig. 4B). In contrast, transcription was markedly upregulated in the 20-h time point in J3101 (devA), and the devAB transcript was equally abundant at all subsequent time points (Fig. 4B). These observations are consistent with a role for DevA as a negative transcriptional autoregulator, similar to that of other GntR family transcriptional regulators (17, 18, 36, 42, 43, 46).
FIG. 4.
Transcriptional analysis of devA. A. High-resolution S1 nuclease mapping of the devA transcription start point. Lanes G, C, A, and T represent a dideoxy sequencing ladder generated using the same oligonucleotide used to generate the S1 nuclease mapping probe. The most likely transcription start point is indicated by the asterisk. B. S1 nuclease protection analysis of devA transcription during development of S. coelicolor M600 and J3101 (devA::Tn4560) on R5 medium. The time points at which mycelium was harvested for RNA isolation and the presence of vegetative mycelium (V), aerial mycelium (A), and spores (S) , as judged by microscopic examination, are shown.
devA is transcribed in vegetative hyphae.
The devA mutant phenotype and the temporal association of devA transcription with vegetative growth in the wild type prompted examination of the spatial localization of devA transcription in differentiating colonies. The promoter region of devA was cloned into pIJ8660 (50), creating a transcriptional fusion to an enhanced version of the green fluorescent protein (EGFP) gene. The resulting plasmid, pIJ6974, was introduced into wild-type S. coelicolor (M600) and J3101 (devA). In M600, fluorescence was barely detectable at 20 h, when only vegetative hyphae were present (Fig. 5, left panel). At later time points expression was undetectable by confocal microscopy (Fig. 5, middle panel). In contrast, in the devA null mutant (J3101) the EGFP gene was abundantly expressed in vegetative, aerial hyphae and spores (Fig. 5, right panel). These data showed that devA was transcribed in substrate hyphae in the wild type and were again consistent with negative regulation of the devA promoter by DevA itself.
FIG. 5.
Localization of devA activity by using the reporter EGFP gene. Confocal microscope images of wild-type (M600) and J3101 (devA::Tn4560) carrying pIJ6974 (devAp-egfp) are shown. Left panel, M600 containing devA::egfp promoter fusion at 20 h of growth; middle panel, wild type (M600) containing devA::egfp promoter fusion at 36 h of growth; right panel, devA (J3101) containing devA::egfp promoter fusion at 36 h of growth.
DevA binds the devA promoter region in vitro.
Since the transcriptional data suggested that devA was autoregulatory, it seemed likely that there was a DevA-binding site in the devA promoter, an arrangement demonstrated for the GntR-like regulators DasR (45), FarR (42), and AraR (36). To address this possibility, we investigated the ability of DevA to bind a 444-bp fragment encompassing the SCO4189-devA intergenic region, using a gel mobility shift assay. A crude cell extract of E. coli expressing His6-DevA retarded the devA promoter to a defined position (Fig. 6, lane A), whereas an equivalent extract of E. coli carrying the parent vector (pET15b) did not (data not shown). Furthermore, purified His6-DevA retarded the labeled fragment to the same position. A DNA fragment internal to the devA coding sequence, added as a negative control to each assay, was not bound by His6-DevA (Fig. 6).
DISCUSSION
DevA defines a new subfamily of GntR regulators.
The amino acid sequence of the helix-turn-helix motif places DevA within the large HTH GntR superfamily, which has been subdivided on the basis of heterogeneity observed in the C-terminal-effector-binding/oligomerization domain (44). DevA defines the seventh GntR subfamily to be identified, following the four major subfamilies FadR, HutC, MocR, and YtrA and the two minor subfamilies AraR and PlmA. There are currently six members of the DevA subfamily, i.e., DevA and SCO4188 in S. coelicolor and their orthologues in S. avermitilis and S. scabies. BLAST and PSI-BLAST searching of fragments of the C termini of all DevA members revealed no conserved domains/folds or local gene context that would give insights into the type of effector molecule modulating the DNA-binding affinity of DevA, as was possible for GntR of Bacillus subtilis (17) and FadR of E. coli (12). Currently there are 1,300 known members of the family (2; www.sanger.ac.uk/Software/Pfam/). The S. coelicolor genome contains 57 GntR-like regulators (2, 3), with representatives of all the subfamilies identified by Rigali et al. (45) except the PlmA and AraR subfamilies, which are confined to cyanobacteria and firmicutes, respectively (34, 36, 44).
Does DevA respond to the metabolic state of the hyphae?
GntR and many of its relatives, including WhiH, PdhR, AraR, and FarR, are autoregulatory, and some bind operator sequences comprising inverted repeats in their own promoters (17, 18, 36, 42, 43, 46). The spatial and temporal deregulation of devA transcription seen in a devA mutant suggested a direct negative autoregulatory mechanism, a possibility confirmed by the ability of purified DevA to bind and retard its own promoter region in vitro.
Our data raise the possibility that DevA is responsive to the physiological state of the hyphae, repressing transcription in the absence of a specific effector metabolite. Further, because devA and devB are cotranscribed, DevA, through autoregulation, also controls expression of the putative phosphatase/hydrolase DevB, suggesting that a phosphorylated metabolic intermediate or signaling molecule might be processed by DevB upon derepression of the devAB operon. In E. coli, in response to long-chain fatty acids, FadR brings about global changes in gene expression via the transcriptional repressor IclR (11). By analogy, perhaps DevA could act as a repressor of another repressor involved in the development of aerial hyphae, giving DevA an overall positive role in development. However, at least one member of the GntR family (FadR) can also act as a transcriptional activator in the absence of its effector molecule (11-13, 26, 27). DevA is the fourth GntR-like transcription factor shown to be involved in sporulation in streptomycetes. WhiH and SCO7168 belong to the FadR subfamily, and DasR belongs to the HutC subfamily. The involvement of members of three separate subfamilies of GntR-like regulators, each likely to respond to a different effector molecule, reinforces the possible influence of primary metabolism on Streptomyces development and provides a potential focus for future studies.
Acknowledgments
We thank Grant Calder for assistance with the confocal microscope, Paul Herron and Paul Dyson for the gift of in vitro-mutagenized cosmids, and David Hopwood, Keith Chater, Matt Hutchings, Bertolt Gust, and Marie Elliot for helpful discussion and comments on the manuscript.
This work was funded by BBSRC grant 208/EGH16080 and by a grant-in-aid to the John Innes Centre from the BBSRC.
REFERENCES
- 1.Aínsa, J. A., H. D. Parry, and K. F. Chater. 1999. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 34:607-619. [DOI] [PubMed] [Google Scholar]
- 2.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley, S. D., K. F. Chater, A-M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M.-H. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 4.Bibb, M. J., V. Molle, and M. J. Buttner. 2000. σBldN, an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2). J. Bacteriol. 182:4606-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 6.Bishop, A., S. Fielding, P. J. Dyson, and P. R. Herron. 2003. Concerted mutagenesis of a streptomycete genome: a link between osmoadaptation, development and antibiotic production. Genome Res. 14:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 8.Chater, K. F., and S. Horinouchi. 2003. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48:9-15. [DOI] [PubMed] [Google Scholar]
- 9.Chung, S.-T. 1987. Tn4556, a 6.8-kilobase-pair transposable element of Streptomyces fradiae. J. Bacteriol. 169:4436-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiRusso, C. C., and T. Nyström. 1998. The fats of Escherichia coli during infancy and old age: regulation by global regulators, alarmones and lipid intermediates. Mol. Microbiol. 27:1-8. [DOI] [PubMed] [Google Scholar]
- 12.DiRusso, C. C., A. K. Metzger, and T. L. Heimert. 1993. Regulation of transcription of genes required for fatty acid transport and unsaturated fatty acid biosynthesis in Escherichia coli by FadR. Mol. Microbiol. 7:311-322. [DOI] [PubMed] [Google Scholar]
- 13.DiRusso, C. C., T. L. Heimert, and A. K. Metzger. 1992. Characterization of FadR, a global transcriptional regulator of fatty acid metabolism in Escherichia coli. Interaction with the fadB promoter is prevented by long chain fatty acyl coenzyme A. J. Biol. Chem. 267:8685-8691. [PubMed] [Google Scholar]
- 14.Elliot, M. A., and N. J. Talbot. 2004. Building filaments in the air: aerial morphogenesis in bacteria and fungi. Curr. Opin. Microbiol. 7:594-601. [DOI] [PubMed] [Google Scholar]
- 15.Elliot, M. A., M. J. Bibb, M. J. Buttner, and B. K. Leskiw. 2001. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol. Microbiol. 40:257-269. [DOI] [PubMed] [Google Scholar]
- 16.Fowler, K. 2002. Transposon mutagenesis of Streptomyces coelicolor A3(2). Ph.D thesis. University of East Anglia, Norwich, United Kingdom.
- 17.Fujita, Y., and T. Fujita. 1987. The gluconate operon gnt of Bacillus subtilis encodes its own transcriptional negative regulator. Proc. Natl. Acad. Sci. USA 84:4524-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita, Y., and Y. Miwa. 1989. Identification of an operator sequence for the Bacillus subtilis gnt operon. J. Biol. Chem. 264:4201-4206. [PubMed] [Google Scholar]
- 19.Gehring, A. M., J. R. Nodwell, S. M. Beverley, and R. Losick. 2000. Genomewide insertional mutagenesis in Streptomyces coelicolor reveals additional genes involved in morphological differentiation. Proc. Natl. Acad. Sci. USA 97:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gehring, A. M., N. J. Yoo, and R. Losick. 2001. An RNA polymerase sigma factor that blocks morphological differentiation by Streptomyces coelicolor A3(2). J. Bacteriol. 183:5991-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gehring, A. M., S. T. Wang, D. B. Kearns, N. Y. Storer, and R. Losick. 2004. Novel genes that influence development in Streptomyces coelicolor. J. Bacteriol. 186:3570-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregory, M. A., R. Till, and M. C. M. Smith. 2003. Integration site for Streptomyces phage ϕBT1 and development of site-specific integrating vectors. J. Bacteriol. 185:5320-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. Gene replacement by PCR targeting in Streptomyces and its use to identify a protein domain involved in the biosynthesis of the sesquiterpene odour geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gust, B., G. Chandra, D. Jakimowicz, Y. Tian, C. J. Bruton, and K. F. Chater. 2004. λ Red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54:107-128. [DOI] [PubMed] [Google Scholar]
- 25.Haydon, D. J., and J. R. Guest. 1991. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 63:291-295. [DOI] [PubMed] [Google Scholar]
- 26.Henry, M. F., and J. E. Cronan. 1991. Escherichia coli transcription factor that both activates fatty acid synthesis and represses fatty acid degradation. J. Mol. Biol. 222:843-849. [DOI] [PubMed] [Google Scholar]
- 27.Henry, M. F., and J. E. Cronan. 1992. A new mechanism of transcriptional regulation: release of an activator triggered by small molecule binding. Cell 70:671-679. [DOI] [PubMed] [Google Scholar]
- 28.Janssen, G. R. 1993. Eubacterial, archaebacterial and Eukaryotic genes that encode leaderless mRNA, p. 59-67. In R. H. Baltz, G. D. Hegeman, and P. L. Skatrud (ed.), Industrial microorganisms: basic and applied molecular genetics. American Society for Microbiology, Washington, D.C.
- 29.Karandikar, A., G. P. Sharples, and G. Hobbs. 1997. Differentiation of Streptomyces coelicolor A3(2) under nitrate-limited conditions. Microbiology 143:3581-3590. [DOI] [PubMed] [Google Scholar]
- 30.Kelemen, G. H., and M. J. Buttner. 1998. Initiation of aerial mycelium formation in Streptomyces. Curr. Opin. Microbiol. 1:656-662. [DOI] [PubMed] [Google Scholar]
- 31.Kelemen, G. H., G. L. Brown, J. Kormanec, L. Potûcková, K. F. Chater, and M. J. Buttner. 1996. The positions of the sigma-factor genes, whiG and sigF, in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2). Mol. Microbiol. 21:593-603. [DOI] [PubMed] [Google Scholar]
- 32.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces Genetics. John Innes Foundation, Norwich, United Kingdom.
- 33.Kodani, S., M. E. Hudson, M. C. Durrant, M. J. Buttner, J. R. Nodwell, and J. M. Willey. 2004. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 101:11448-11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, M. H., M. Scherer, S. Rigali, and J. W. Golden. 2003. PlmA, a new member of the GntR family, has plasmid maintenance functions in Anabaena sp. strain PCC 7120. J. Bacteriol. 185:4315-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molle, V., W. J. Palframan, K. C. Findlay, and M. J. Buttner. 2000. WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 182:1286-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mota, L. J., P. Tavares, and I. Sá-Nogueira. 1999. Mode of action of AraR, the key regulator of l-arabinose metabolism in Bacillus subtilis. Mol. Microbiol. 33:476-489. [DOI] [PubMed] [Google Scholar]
- 37.Nodwell, J. R., K. McGovern, and R. Losick. 1996. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol. 22:881-893. [DOI] [PubMed] [Google Scholar]
- 38.Nodwell, J. R., M. Yang, D. Kuo, and R. Losick. 1999. Extracellular complementation and the identification of additional genes involved in aerial mycelium formation in Streptomyces coelicolor. Genetics 151:569-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Connor, T. J., P. Kanellis, and J. R. Nodwell. 2002. The ramC gene is required for morphogenesis in Streptomyces coelicolor and expressed in a cell type-specific manner under the direct control of RamR. Mol. Microbiol. 45:45-57. [DOI] [PubMed] [Google Scholar]
- 40.Paget, M. S. B., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor, σE, is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pope, M. K., B. D. Green, and J. Westpheling. 1996. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell-cell signalling. Mol. Microbiol. 19:747-756. [DOI] [PubMed] [Google Scholar]
- 42.Quail, M. A., C. E. Dempsey, and J. R. Guest. 1994. Identification of a fatty acyl responsive regulator (FarR) in Escherichia coli. FEBS Lett. 356:183-187. [DOI] [PubMed] [Google Scholar]
- 43.Quail, M. A., D. J. Haydon, and J. R. Guest. 1994. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol. Microbiol. 12:95-104. [DOI] [PubMed] [Google Scholar]
- 44.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507-12515. [DOI] [PubMed] [Google Scholar]
- 45.Rigali, S., M. Schlicht, P. A. Hoskisson, H. Northaft, M. Merzbacher, B. Joris, and F. Titgemeyer. 2004. Extending the classification of bacterial transcription factors beyond the helix-turn-helix motif as an alternative approach to discover new cis/trans relationships. Nucleic Acids Res. 32:3418-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryding, N. J., G. H. Kelemen, C. A. Whatling, K. Flärdh, M. J. Buttner, and K. F. Chater. 1998. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 29:343-357. [DOI] [PubMed] [Google Scholar]
- 47.Ryding, N. J., M. J. Bibb, V. Molle, K. C. Findlay, K. F. Chater, and M. J. Buttner. 1999. New sporulation loci in Streptomyces coelicolor A3(2). J. Bacteriol. 181:5419-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seo, J. W., Y. Ohnishi, A. Hirata, and S. Horinouchi. 2002. ATP-binding cassette transport system involved in regulation of morphological differentiation in response to glucose in Streptomyces griseus. J. Bacteriol. 184:91-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sprusansky, O., L. Zhou, S. Jordan, J. White, and J. Westpheling. 2003. Identification of three new genes involved in morphogenesis and antibiotic production in Streptomyces coelicolor. J. Bacteriol. 185:6147-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun, J., G. H. Kelemen, J. M. Fernandez-Abalos, and M. J. Bibb. 1999. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145:2221-2227. [DOI] [PubMed] [Google Scholar]
- 51.Willey, J., J. Schwedock, and R. Losick. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 7:895-903. [DOI] [PubMed] [Google Scholar]