Abstract
Yersinia enterocolitica is able to efficiently invade Peyer's patches with the aid of invasin, an outer member protein involved in the attachment and invasion of M cells. Invasin is encoded by inv, which is positively regulated by RovA in both Y. enterocolitica and Yersinia pseudotuberculosis while negatively regulated by YmoA in Y. enterocolitica and H-NS in Y. pseudotuberculosis. In this study we present data indicating H-NS and RovA bind directly and specifically to the inv promoter of Y. enterocolitica. We also show that RovA and H-NS from Y. enterocolitica bind to a similar region of the inv promoter and suggest they compete for binding sites. This is similar to recently published data from Y. pseudotuberculosis, revealing a potentially conserved mechanism of inv regulation between Y. enterocolitica and Y. pseudotuberculosis. Furthermore, we present data suggesting H-NS and YmoA form a repression complex on the inv promoter, with H-NS providing the binding specificity and YmoA interacting with H-NS to form a repression complex. We also demonstrate that deletion of the predicted H-NS binding region relieves the requirement for RovA-dependent transcription of the inv promoter, consistent with RovA acting as a derepressor of H-NS-mediated repression. Levels of H-NS and YmoA are similar between 26°C and 37°C, suggesting that the H-NS/YmoA repression complex is present at both temperatures, while the levels of rovA transcript are low at 37°C and high at 26°C, leading to expression of inv at 26°C. Expression of RovA at 37°C results in transcription of inv and production of invasin. Data presented here support a model of inv regulation where the level of RovA within the cell governs inv expression. As RovA levels increase, RovA can successfully compete for binding to the inv promoter with the H-NS/YmoA complex, resulting in derepression of inv transcription.
Yersinia enterocolitica is a gram-negative human pathogen capable of colonizing the gastrointestinal tract. Y. enterocolitica is normally acquired through ingestion of contaminated food or water, with swine serving as a major reservoir for human pathogenic strains (4). Invasin is an outer membrane protein found on the surface of Y. enterocolitica and Yersinia pseudotuberculosis (23, 24, 38, 47) that is responsible for binding to β1-integrins (24) on the apical surface of M cells and initiating uptake of the organism (7, 48). Migration through these cells leads to the accumulation of bacteria in the underlying lymphoid tissue (Peyer's patches) and spread to the mesenteric lymph nodes (6, 18, 20, 48). Once Y. enterocolitica establishes an infection, it is able to cause a variety of syndromes, including enterocolitis, mesenteric lymphadenitis, and terminal ileitis (4).
While a considerable amount of work has demonstrated a role for invasin during Yersinia infection, less is understood about the mechanisms controlling the expression of the gene encoding invasin (inv). Expression of inv in Y. enterocolitica responds to both temperature and pH (46). inv expression is higher at 26°C than 37°C during in vitro growth, with maximal expression occurring during late logarithmic to early stationary phase; however, expression of inv at 37°C can be restored to levels comparable to 26°C by adjusting the pH of the medium to 5.5 (46). Presence of invasin has also been shown in the Peyer's patches of infected animals during infection (46, 50). Two regulators have been identified through genetic screens that affect the expression of inv: RovA (41, 50), which is required for inv expression, and YmoA (12), which is involved in the negative regulation of inv.
RovA is a member of the MarR/SlyA family of regulators and is present in all three pathogenic species of Yersinia. Members of this family are widely distributed in nature, with at least 336 putative members in 45 species of bacteria and 13 species of archaea present in the Clusters of Orthologous Groups database (http://www.ncbi.nlm.nih.gov/COG/). Members of this family regulate a wide variety of biological processes, including antibiotic resistance (MarR in Escherichia coli) (15), antimicrobial agents (Rap in Serratia marcescens), and virulence (SlyA in Salmonella enterica serovar Typhimurium) (5, 10, 26, 29). In many cases the mechanism by which these proteins repress or activate gene expression is unknown. However, SlyA appears to activate hlyE (also known as clyA or sheA) transcription in E. coli by competing for binding to the hlyE promoter with the negative regulator H-NS (60). Specifically, SlyA binds to two sites within the hlyE promoter that overlap with H-NS binding site I, suggesting that binding of SlyA may block H-NS binding and relieve H-NS-mediated repression (60). Recently, a similar mechanism was proposed for the regulation of inv in Y. enterocolitica (11, 12) and further explored in Y. pseudotuberculosis (21). In Y. pseudotuberculosis, RovA and H-NS bind to multiple overlapping sites within the inv promoter, indicating that like SlyA, RovA interactions with the inv promoter may inhibit H-NS binding and quell H-NS-mediated repression (21).
YmoA is a member of a growing family of proteins that have homology to the N-terminal dimerization domain of H-NS (30). Studies with the YmoA homolog Hha in E. coli revealed that Hha interacts with H-NS to negatively regulate the expression of the hly operon of plasmid pHly152 (31, 43). The mechanism of repression is not completely understood, but it is believed that H-NS provides DNA binding specificity and Hha interacts with H-NS to form a higher-order complex to regulate hly expression (31, 43). YmoA has been shown to interact with H-NS (42), leading us to hypothesize that YmoA may act in a similar fashion to repress inv expression indirectly by forming a complex with H-NS on the inv promoter.
While RovA is required for expression of inv in both Y. enterocolitica and Y. pseudotuberculosis (41, 50), it is unclear if it acts on the Y. enterocolitica inv promoter in the same manner as reported for Y. pseudotuberculosis; two lines of evidence indicate that the regulatory mechanism of inv may differ between the two species. First, restoration of inv expression at 37°C by changes in medium pH has not been observed in Y. pseudotuberculosis (41). Secondly, comparison of the two promoters reveals significant sequence differences between Y. enterocolitica and Y. pseudotuberculosis. Based on the differences in regulation and sequence divergence in the promoter regions of the two species, our goals in this work were as follows: (i) to determine if H-NS was involved in the negative regulation of inv in Y. enterocolitica; (ii) to further investigate the mechanism by which RovA activates inv transcription in Y. enterocolitica; and (iii) to better define the mechanism of YmoA-mediated repression of the inv promoter in Y. enterocolitica.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The “v” designation refers to strains harboring the virulence plasmid; “c” refers to strains that have been cured of the virulence plasmid. Bacterial cultures were grown in Luria-Bertani (LB) broth at 26°C or 37°C as indicated. When appropriate, antibiotics were used at the following concentrations: chloramphenicol, 25 μg ml−1 (for growth of E. coli) and 12.5 μg ml−1 (for growth of Y. enterocolitica); ampicillin, 100 μg ml−1; kanamycin, 50 μg ml−1; nalidixic acid, 20 μg ml−1; spectinomycin, 50 μg ml−1; streptomycin, 50 μg ml−1; tetracycline, 15 μg ml−1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Source |
|---|---|---|
| Y. enterocolitica strainsa | ||
| JB580c | Serogroup O:8 Nalr ΔyenR(R− M+) | 27 |
| JB41v | inv′-′phoA inv+ Nalr Cmr | 2 |
| MBL014 | inv′-′lacZ inv+ Nalr Cmr | This work |
| MBL015 | ΔrovA inv′-′lacZ inv+ Nalr Cmr | This work |
| YVM567c | inv′-′phoA inv+ymoB::mTn5Kn2 Nalr Cmr | This work |
| YVM927c | ΔrovA Cmr | This work |
| E. coli strains | ||
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3), a λ prophage carrying the T7 RNA polymerase gene | 55 |
| MC4100 | F−araD139 Δ(lacIOPZYA)U169 rpsL thiA | 14 |
| RO949 | MC1000 hns205::Tn10 Tetr | R. Osuna |
| VM1303 | MC4100 hns205::Tn10 Tetr | This work |
| Plasmids | ||
| pBAD33 | Expression vector containing PBAD promoter, Cmr | 19 |
| pBADrovA:knr | pBAD33 containing Knr cassette in the SphI site and rovA, Cmr | This work |
| pWKS30:StrSpec | pWKS30 with a Str/Spec cassette in the HindIII site of the polylinker, Ampr Strr Specr | Lab collection |
| pELL21 | pWKS30::StrSpec carrying ΔymoBymoA and upstream promoter region | 12 |
| pET-24+ | Overexpression vector with T7 promoter and C-terminal His tag | Novagen |
| pETrovA-his | RBS plus rovA open reading frame cloned into pET | This work |
| pEThns-his | RBS plus hns open reading frame cloned into pET | This work |
| PGEX-6P-1 | Overexpression vector with tac promoter and cleavable N-terminal GST tag | Pharmacia |
| pGST-ymoA | ymoA open reading frame cloned into pGex | This work |
| phns | pWKS30::StrSpec carrying hns and upstream promoter region | This work |
| pROBE-gfp[LVA] | gfp with LVA tag to decrease its half-life, Knr | 39 |
| pinv16 | pROBE-gfp[LVA], containing inv promoter fragment −305 to +146 | This work |
| pinv29 | pROBE-gfp[LVA], containing inv promoter fragment −276 to +146 | This work |
| pinv30 | pROBE-gfp[LVA], containing inv promoter fragment −239 to +146 | This work |
| pinv31 | pROBE-gfp[LVA], containing inv promoter fragment −190 to +146 | This work |
| pinv17 | pROBE-gfp[LVA], containing inv promoter fragment −158 to +146 | This work |
| pinv12 | pROBE-gfp[LVA], containing inv promoter fragment −69 to +146 | This work |
All Y. enterocolitica strains listed are derivatives of JB580v.
Strain and plasmid construction.
Primers used in this study are listed in Table 2. To construct an hns mutant in E. coli strain MC4100, the mutant allele hns205::Tn10 was introduced by P1 transduction from strain RO949.
TABLE 2.
Primers
| Primer | Sequence (5′-3′)a |
|---|---|
| RovAFRBSR1 | CGGAATTCGGTAGTTATGCTAGCACGC |
| RovAxhoR3His | GGTCTCGAGCTTACTTTGTAGTTGAATAATGTTTCTCTCAAGC |
| H-NSFnotHis | ATAAGAATGCGGCCGCCAGCAGGAAATCATCCAG |
| H-NSRR1His | CGGAATTCCACTCTATTATTATCCAGAC |
| YmoAgstFR1 | GGAATTCATGACAAAAACTGACTACCT |
| YmoAgstRxho | CCGCTCGAGCTATTTCACATGTTGCCATA |
| InvgfpH1F | CGGATCCGATAAGTGAAGGAGCAAC |
| InvgfpR1R | GGAATTCCTAATTATTTTAGTCACAGTTAGC |
| H-NsccxbaF | TGCTCTAGATGATATGATCAAGGCTCGCC |
| H-NsccnotR | ATAAGAATGCGGCCGCGAACATTAACCAGGTTAGAGCG |
| Inv500F | GATAAGTGAAGGAGCAAC |
| Inv300F | AGTCTATTTTATGAGACGAATG |
| Inv150F | GTAAGTTCTGCTATTCTATTG |
| Inv300R | CATTCGTCTCATAAAATAGACT |
| Inv200R | GCAATGACAAATGAAATGC |
| Inv150R | CAATAGAATAGCAGAACTTAC |
| Inv3R | TATTAGCCTGCTAATTATTTTAGTCACAGTTAGCG |
| Inv194F | GCAGAGAGTCTATTTTATGAGACGAATGTAAAC |
| Inv34R | AAAATTAAAAATAACAGCAATGACAAATGAAATGC |
Restriction sites are indicated by underlining.
To generate C-terminal histidine (His)-tagged recombinant RovA and H-NS, the genes were ligated into pET-24(+) (Novagen). The genes were amplified using primers RovAFRBSR1 and RovAXhoR3His for rovA and H-NSFnotHis and H-NSRR1His for hns. Both products contained the native ribosome binding site (RBS) and were digested with the appropriate restriction enzymes and ligated into pET-24(+). To generate N-terminal glutathione S-transferase (GST)-tagged YmoA, ymoA was amplified by PCR with primers ymoAgstFR1 and ymoAgstRXho and cloned into pGEX-6P-1 (Pharmacia). Plasmids provA-his, phns-his, and pGST-ymoA were checked by DNA sequencing to ensure no errors had been introduced by PCR. Plasmid pinv-gfp was constructed by amplifying the inv promoter using primers invgfpH1F and invgfpR1R, generating a ∼500-bp fragment that was digested and ligated into pROBE-gfp(LVA) (39). inv promoter truncations were subcloned from previously constructed plasmids pAJD-16, pAJD-29, pAJD-30, pAJD-31, pAJD17, and pAJD-12 by digesting with XbaI/NruI. The digested fragments were then ligated into the XbaI/SmaI sites in pROBE-gfp(LVA) (39), generating plasmids pinv-gfp-16, pinv-gfp-29, pinv-gfp-30, pinv-gfp-31, pinv-gfp-17, and pinv-gfp-12, respectively. To construct an H-NS-complementing plasmid, hns was amplified using primers H-NSccXbaF and H-NSccNotR. The resulting fragment was digested and ligated into pWKS30:StrSpec, generating phns.
Purification of RovA-His, H-NS-His, and YmoA.
Overnight cultures of BL21(DE3) containing either provA-his, phns-his, or pGST-ymoA were diluted 1/200 and allowed to grow at 37°C until an optical density at 600 nm (OD600) of 0.5 was reached. To induce protein expression, isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 1 mM and growth was continued for 5 h. Cells were harvested by centrifugation at 7,000 rpm for 30 min; cell pellets were resuspended in 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole pH 8.0 for His tag purification or 10 mM Na2HPO4, 1.8 mM KH2PO4, 140 mM NaCl, 2.7 mM KCl pH 7.3 for purification of GST-YmoA and frozen at −80°C. The cell suspension was removed from −80°C and allowed to thaw at room temperature. The samples were sonicated five times for 30 s with a 1-min incubation on ice between sonication steps. To isolate the soluble fraction, the samples were centrifuged at 10,000 × g for 30 min and transferred to a clean 50-ml conical tube. Five ml of cleared lysate was added to 0.5 ml of presettled Ni-nitrilotriacetic acid agarose (QIAGEN) or glutathione-Sepharose 4B (Amersham Biosciences) in 1-ml polypropylene columns (QIAGEN) and allowed to flow over the column at room temperature. For purification of His-tagged proteins, the column was then washed with 5 ml of 50 mM NaH2PO4, 300 mM NaCl, 5% glycerol, 1% Triton X-100, 20 mM imidazole pH 8.0, followed by 5 ml of 50 mM NaH2PO4, 1 M NaCl, 20 mM imidazole pH 8.0. Excess salt was removed by washing the column with 5 ml of 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole pH 8.0. The wash procedure was the same for GST-YmoA purification except that the base buffer was 10 mM Na2HPO4, 1.8 mM KH2PO4, 140 mM NaCl, 2.7 mM KCl pH 7.3. The His-tagged proteins were eluted from the column in 2 ml of 50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole pH 8.0 or 50 mM Tris-HCl, 10 mM reduced glutathione pH 8.0 for GST-YmoA purification. To remove excess imidazole, protein samples were run over PD-10 desalting columns (Amersham) and exchanged into buffer containing 50 mM NaH2PO4, 300 mM NaCl, 15% sucrose pH 8.0; 50-μl aliquots were stored at −80°C. To remove the GST tag from YmoA, purified protein was added to PD-10 desalting columns (Amersham) and exchanged into 10 mM Na2HPO4, 1.8 mM KH2PO4, 140 mM NaCl, 2.7 mM KCl, pH 7.3. This was applied to a new 1-ml glutathione-Sepharose 4B column and allowed to bind at 4°C. The column was equilibrated with 10 ml of 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, pH 7.0. Eighty units of PreScission protease (Amersham) was diluted with 1 ml of 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, pH 7.0, and mixed with glutathione-Sepharose 4B containing the bound GST-YmoA. This mixture was incubated at 4°C for 4 h, and then YmoA was collected from the column by washing with 1 ml of 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, pH 7.0. Purified YmoA was concentrated and exchanged into buffer containing 20 mM Tris-HCl, 5% glycerol, 20 mM NaCl, 5 mM MgCl2. For all proteins, the concentration was determined by using the Bradford protein assay (Bio-Rad).
EMSA.
Primers used to generate inv promoter fragments used in electrophoretic mobility gel shift assay (EMSA) experiments are listed in Table 2. An internal 500-bp fragment of the lacZ gene was used as a nonspecific control. EMSA experiments were performed at room temperature in binding buffer containing 20 mM Tris-HCl, 15% glycerol, 50 mM NaCl, 5 mM MgCl2, and 1 μg of salmon sperm DNA. Random 32P-labeled DNA was generated by PCR with [α-32P]dATP and added to protein to a final activity of 3,000 cpm in 10-μl reaction mixtures. The reaction mixtures were incubated at room temperature for 15 min and then run on a 3.5%, 5%, or 8% polyacrylamide gel, depending on the size of the fragment. For the competition experiments, RovA was added and allowed to bind for 5 min before the addition of H-NS. The reaction mixtures were then incubated for 10 min before loading onto a 5% polyacrylamide gel. Competition experiments were preformed in binding buffer containing 20 mM Tris-HCl, 15% glycerol, 80 mM NaCl, 5 mM MgCl2, 100 ng of salmon sperm DNA, and 1 μg of bovine serum albumin.
Northern blot assays.
To examine the levels of hns mRNA in JB41c and YVM567c, the strains were grown overnight at 37°C, subcultured to an OD600 of 0.2, and allowed to grow at 37°C to an OD600 of 2.0; RNA was harvested using the RiboPure-Bacteria extraction kit (Ambion). To determine the levels of inv, rovA, hns, and ymoA mRNA, overnight cultures were grown at either 26°C or 37°C and subcultured to an OD600 of 0.2, and samples were harvested at early log (0.5 to 1.0 OD600), middle log (1.1 to 2.5 OD600), and late log (2.6 to 3.5 OD600); RNA was harvested using the RiboPure-Bacteria extraction kit (Ambion). Equal amounts of purified RNA were run on a 1% agarose gel containing 1× morpholineethanesulfonic acid and 6.7% formaldehyde and transferred overnight to a nitrocellulose membrane. The membranes were hybridized overnight at 42°C with either 32P-labeled inv, rovA, hns, or ymoA probes consisting of the genes' corresponding open reading frames and exposed to an imaging screen. The image was scanned on an FLA-5000 phosphorimager and analyzed using Image Gauge (version 4.2; Fujifilm).
Green fluorescent protein assays.
Strains were grown in triplicate overnight in 2 ml of LB at 26°C with aeration, and the levels of fluorescence intensity were determined as previously described (12).
AP assays.
Strains were grown in triplicate overnight in 2 ml of LB at 37°C with aeration. Alkaline phosphatase (AP) activity was measured according to previously described methods (32).
β-Galactosidase assays.
Cultures were grown in triplicate overnight in 2 ml of LB broth at 37°C or 26°C containing 0.002, 0.0002, 0.00002, 0.000002, or 0% arabinose. β-Galactosidase activity was measured according to previously described methods (37).
Western blot assays.
One OD equivalent of cells was harvested and pelleted for 3 min at 13,000 rpm; the resulting pellets were resuspended in 50 μl Laemmli buffer containing 10% β-mercaptoethanol. A 0.1 OD equivalent was loading onto a 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and run at 200 V for 1 h. Proteins were transferred to a nitrocellulose membrane, and RovA was detected using a polyclonal anti-RovA antibody (Covance) at a 1:1,000 dilution. H-NS was detected using a 1:2,000 dilution of anti-H-NS antibody kindly supplied by Yeong-Jae Seok, Seoul National University (53). YmoA was detected using a 1:1,000 dilution of anti-YmoA antibody kindly supplied by Gregory V. Plano (25).
RESULTS
hns silences expression of the inv promoter.
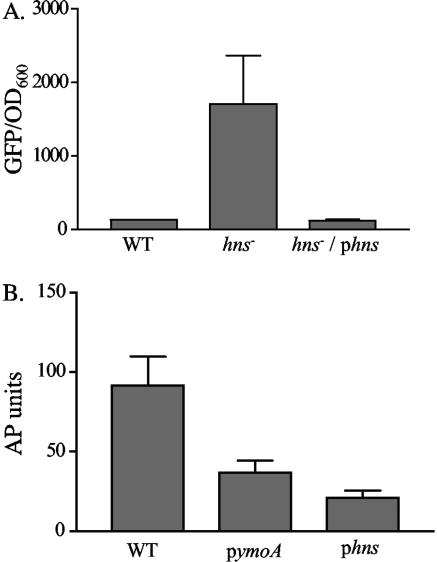
To determine if hns was involved in the negative regulation of inv, we initially attempted to inactivate hns in Y. enterocolitica. Several attempts to generate an hns mutant were unsuccessful, suggesting that hns is essential for viability of Y. enterocolitica strain JB580. Similar results have been reported for Y. pseudotuberculosis (21) and were also found for ymoA, a negative regulator of inv, in Y. enterocolitica strain JB580 (12). However, hns has been successfully inactivated in E. coli, and we tested the effect of hns on inv expression in this surrogate background. Plasmid pinv-gfp, containing an inv-gfp transcriptional fusion, was transformed into wild-type (WT) E. coli MC4100 and an isogenic hns mutant of MC4100 (VM1303). These strains were grown overnight at 26°C and assayed for expression of inv-gfp. MC4100 and MC4100/pinv-gfp both demonstrated similar levels of fluorescence, indicating that inv-gfp is expressed at low levels in WT E. coli, while a mutation of hns resulted in a ∼7-fold increase in inv-gfp expression (Fig. 1A). Complementation of the hns mutant with hns from Y. enterocolitica restored the level of inv-gfp expression to that of MC4100 harboring pinv-gfp. These data suggest that Y. enterocolitica H-NS is able to repress transcription of inv.
FIG. 1.
Effect of hns on the expression of inv. (A) Cultures of MC4100 containing pinv-gfp and VM1303 (an hns mutant) containing pinv-gfp or pinv-gfp and phns were grown overnight in triplicate at 26°C. Fluorescence was calculated by dividing average fluorescence by the OD600. WT refers to MC4100 containing pinv-gfp, hns− refers to VM1303 containing pinv-gfp, and hns−/phns refers to VM1303 containing pinv-gfp and phns. (B) Cultures of JB41v containing the plasmids phns, pymoA (pEll21), or pWKS30strspec were grown overnight in triplicate at 26°C and assayed for expression of inv-phoA. WT refers to JB41v containing pWKS30strspec as a vector control.
To determine if H-NS is able to repress inv expression in Y. enterocolitica, the inv-phoA reporter stain (JB41c) was transformed with either phns or the vector control (pWKS30:Strspec) and assayed for expression of inv-phoA (Fig. 1B). The strain containing pWKS30:StrSpec exhibited high levels of AP activity as expected when cultured at 26°C. When phns was present, a decrease in AP activity was observed. Supplying excess amounts of a second negative regulator of inv, YmoA, in trans also resulted in a reduction of inv expression in Y. enterocolitica (Fig. 1B). These data suggest that H-NS negatively regulates expression of Y. enterocolitica inv and strengthen previous data showing that YmoA negatively regulates the expression of inv.
RovA and H-NS interact specifically with the inv promoter.
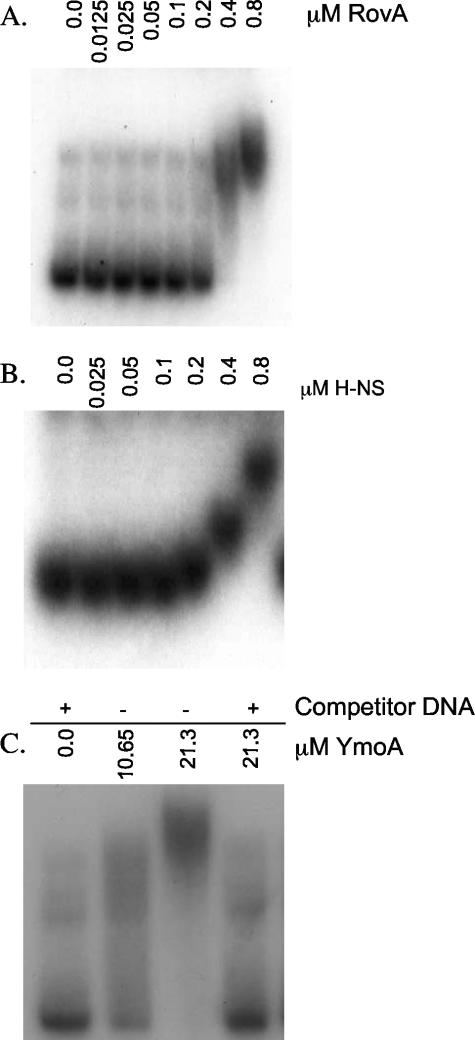
To gain insight into the interaction of RovA, H-NS, and YmoA with the inv promoter, EMSA experiments were performed. Recombinant C-terminal His-tagged RovA and H-NS were generated and purified as described in Materials and Methods. Purified YmoA was obtained by purifying an N-terminal GST fusion protein that was later cleaved to yield a protein with a six-amino-acid N-terminal extension. We chose to use an N-terminal cleavable GST tag because previous work done with E. coli Hha showed that a C-terminal His tag rendered the protein unable to interact with H-NS (42). Purified proteins were incubated with a random 32P-labeled inv fragment (−344 to +146) and 1 μg of salmon sperm DNA as nonspecific competitor. Both RovA and H-NS bound the inv promoter in the presence of excess competitor DNA (Fig. 2A and B). Binding specificity of RovA was demonstrated by an inability of the recombinant protein to bind the ysaE promoter from Y. enterocolitica under the same conditions tested for inv (data not shown). H-NS binding was unaffected by excess nonspecific competitor DNA, but H-NS did not bind 32P-labeled inv probe in the presence of excess unlabeled inv promoter, indicating specific binding to the inv promoter (data not shown). Unlike RovA and H-NS, YmoA was unable to bind the inv promoter in the presence of nonspecific competitor DNA, even at elevated concentrations of purified protein (Fig. 2C). YmoA was able to bind DNA in the absence of nonspecific competitor DNA (Fig. 2C), consistent with previous reports for Hha, which revealed no binding specificity for Hha to the hly operon of plasmid pHly152 (31, 43). Hha and YmoA both have been shown to interact with H-NS, raising the possibility that our YmoA preparations are contaminated with H-NS (42). However, our data show that there is little contaminating H-NS protein in our YmoA preparations, because there is no binding to the inv promoter in the presence of nonspecific competitor DNA. These data indicate that RovA and H-NS bind specifically to the inv promoter while suggesting that YmoA does not.
FIG. 2.
Ability of RovA, H-NS, and YmoA to interact with inv. Randomly 32P-labeled inv (−344 to +146) was incubated with purified RovA, H-NS, or YmoA at room temperature for 15 min and run on a 3.5% or 5% nondenaturing polyacrylamide gel. (A) RovA; (B) H-NS. In panels A and B, 1 μg of salmon sperm was included in all of the reaction mixtures. (C) YmoA. The + indicates the presence of 1 μg of salmon sperm; − indicates the absence of salmon sperm. The concentration of protein is shown across the top of the gel (in μM).
RovA and H-NS bind similar sites on the inv promoter.
RovA and H-NS bind to overlapping binding sites within the inv promoter of Y. pseudotuberculosis (21). These binding regions display considerable sequence differences in Y. enterocolitica, and we were unsure whether these differences affected RovA or H-NS binding in Y. enterocolitica (Fig. 3). To identify regions bound by RovA and H-NS, a set of fragments (Table 3) representing the inv promoter were generated by PCR and incubated with purified protein. EMSA experiments were performed as described above, and both RovA and H-NS demonstrated preferential binding to the same inv promoter fragments (Table 3). The fragments bound indicated that the binding sites are located between bp −194 and −10 relative to the +1 transcriptional start site. To confirm that RovA and H-NS bound to this region, a PCR product encompassing bp −194 to −34 was generated and the ability of the two proteins to bind this product was determined (Table 3). EMSA experiments revealed that RovA and H-NS independently bind this region of the inv promoter, raising the possibility that RovA and H-NS bind overlapping sites within the inv promoter of Y. enterocolitica.
FIG. 3.
Alignment of inv promoters. Shown is an alignment of the RovA and H-NS binding region of inv from Y. enterocolitica and Y. pseudotuberculosis. Conserved bases are shaded in black, RovA binding sites are underlined in black, and H-NS bindings sites are indicated by a gray underline. The RovA and H-NS binding sites are based on DNase I protection data from Y. pseudotuberculosis (21). The start of transcription, marked +1, was previously determined (41, 46).
TABLE 3.
Binding to inv fragmentsa
| Fragment | 5′ primer | 3′ primer | Region | RovA | H-NS |
|---|---|---|---|---|---|
| A | Inv500F | Inv3R | −343 to +146 | + | + |
| B | Inv300F | Inv3R | −193 to +146 | + | + |
| C | Inv150F | Inv3R | −19 to +146 | − | − |
| D | Inv500F | Inv150R | −343 to +2 | + | + |
| E | Inv500F | Inv200R | −343 to −49 | + | + |
| F | Inv500F | Inv300R | −343 to −171 | − | − |
| G | Inv194F | Inv34R | −194 to −34 | + | + |
+ indicates binding; − indicates no binding.
H-NS and RovA compete for binding sites on the inv promoter.
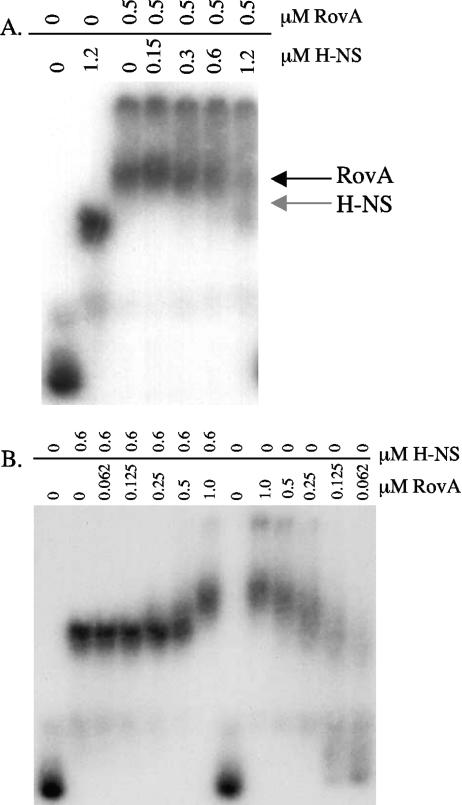
As described previously and recently shown by DNase I footprinting in Y. pseudotuberculosis (21), RovA and H-NS bind to overlapping sites within the inv promoter. These data suggest that RovA and H-NS compete for binding to the inv promoter. To test this directly, EMSA experiments were used to determine if RovA and H-NS were able to replace one another once bound to inv (−194 to −34). RovA (0.5 μM) and H-NS (0.6 μM) were added in separate reaction mixtures at a concentration required to completely shift inv. To replace the prebound protein, increasing amounts of H-NS or RovA were added to the reaction mixtures (Fig. 4A and B). The data indicate that H-NS was able to displace RovA once bound to the inv promoter, as increasing concentrations of H-NS produced a mobility shift identical to the mobility shifts seen with H-NS incubated alone with inv (Fig. 4A). Similarly, increasing concentrations of RovA were able to displace prebound H-NS from the inv promoter, producing a shift identical to the mobility shifts seen with RovA incubated alone with inv (Fig. 4B). In both experiments, increasing amounts of RovA or H-NS were required to displace the prebound protein, suggesting that RovA and H-NS compete for binding sites on the inv promoter. We do not believe that the two proteins are able to bind simultaneously, as no cooperative binding (i.e., an additional shifted species) was observed at lower concentrations of protein known to shift the inv promoter (Fig. 4A and B). These data indicate that RovA and H-NS compete for binding sites within the inv promoter.
FIG. 4.
Competition between RovA and H-NS for the inv promoter. (A) Randomly 32P-labeled inv (−194 to −34) was incubated for 5 min with purified RovA (0.5 μM), shown across the top of the gel. To these reaction mixtures increasing concentrations of H-NS were added, as shown across the top of the gel (in μM), and incubated for 10 min. Samples were run on a 5% nondenaturing polyacrylamide gel. (B) Randomly 32P-labeled inv (−194 to −34) was incubated for 5 min with purified H-NS (0.6 μM), shown across the top of the gel. To these reaction mixtures increasing concentrations of RovA were added, as shown across the top of the gel (in μM), and incubated for 10 min. Samples were run on a 5% nondenaturing polyacrylamide gel.
hns expression is unaffected in the ymoB::Tn10 mutant.
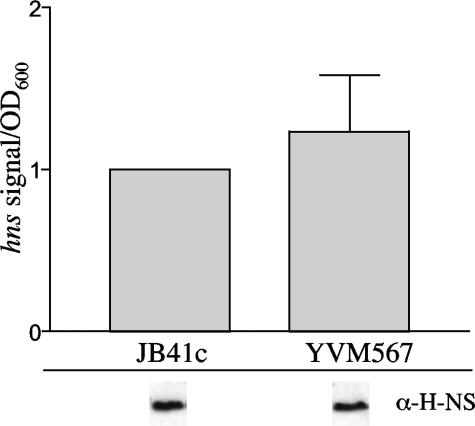
Due to the essential nature of YmoA and H-NS for growth of Y. enterocolitica, we have been unable to successfully inactivate either gene. This has made it difficult to address the individual contribution of YmoA and H-NS in the regulation of inv. However, a previously characterized transposon insertion mutation of ymoB (YVM567c), the gene upstream of ymoA, exhibits a strong polar effect on ymoA expression (12). Strain YVM567c demonstrates a drastically reduced level of ymoA transcription (12), which leads to an increase in the expression of inv at 37°C. This increase in inv expression is not due to elevated levels of rovA transcription (12) or increased levels of RovA protein, as equivalent amounts of RovA are present in the parental strain (JB41c) and YVM567c (data not shown). Another possibility for the increase in inv expression at 37°C in YVM567c is a decrease in the levels of hns transcript, leading to a reduction in H-NS bound to the inv promoter and higher levels of inv expression at 37°C. This would correlate with recently published data suggesting that Hha positively regulates hns expression in E. coli (45). To determine if YmoA affects hns expression, RNA was harvested from JB41c and YVM567c cultures grown with aeration at 37°C to an OD600 of 2.0, and levels of hns transcript were compared (Fig. 5). Northern blot analysis demonstrated no detectable difference in the expression of hns in the ymoA mutant. To determine if the expression levels of hns correlated with levels of H-NS protein, Western blot assays were performed. Similar levels of H-NS protein were detected in the two strains, suggesting that the levels of H-NS protein were unaffected in strain YVM567c (Fig. 5). Our result is different from the published work on the previously mentioned hha mutation in E. coli that showed a reduction in hns levels (45). This could be due to the fact that YVM567c still expresses low levels of ymoA, which could be enough to sustain wild-type expression of hns, could reflect differences between YmoA and Hha, or could reflect differences between E. coli and Y. enterocolitica. These data suggest that the increased expression of inv at 37°C in the ymoA mutant is not due to a reduction in both H-NS and YmoA but that the reduction in ymoA expression alone is responsible for the elevated levels of inv expression.
FIG. 5.
Levels of hns in YVM567c. JB41c (WT) and YVM567c (ymoB::Tn10) were grown overnight at 37°C, and cultures were then subcultured to an OD600 of 0.2 and grown to an OD600 of 2.0. Transcriptional levels of hns were determined by Northern blotting. Levels of H-NS in the two strains are shown below the graph from samples grown in the same manner as for the Northern blot assay. Equal OD equivalents were loaded for each strain for the Western analysis, and data shown are from the same gel.
Interaction of YmoA and H-NS leads to the formation of a repression complex.
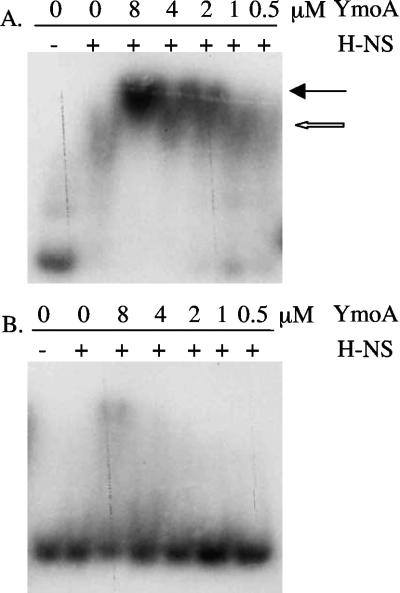
While it has been demonstrated that YmoA influences the expression of inv, it appears that this is not by binding specifically to the inv promoter. It has been shown that the YmoA homolog Hha regulates the hly operon of plasmid pHly152 through specific interactions with H-NS (43). In pull-down experiments, YmoA has also previously been shown to interact with H-NS (42). We therefore hypothesized that YmoA may regulate inv expression through a similar mechanism by interacting with H-NS at the inv promoter. To determine if YmoA and H-NS form a complex in the presence of inv, EMSA experiments were used to determine if migration of the H-NS-inv complex was altered by addition of YmoA to the reaction. Purified YmoA and H-NS were incubated with 32P-labeled inv promoter (−344 to +146), and mobility of the labeled DNA was determined on a nondenaturing polyacrylamide gel (Fig. 6A). We observed that addition of YmoA to the reaction resulted in slightly slower migration of the H-NS-inv complex, suggesting an interaction of YmoA with H-NS. To show that the H-NS/YmoA complex was specific for the inv promoter, the same assay was performed using an internal ∼500-bp fragment of lacZ (Fig. 6B). The data show that H-NS was unable to bind the lacZ fragment under the conditions tested and that very little H-NS/YmoA complex formed with the lacZ fragment under these conditions, indicating that the H-NS/YmoA complex is specific for inv. The ability to shift lacZ was only seen at the highest concentrations of YmoA and most likely represents nonspecific binding by YmoA to lacZ, as YmoA slightly shifts inv and lacZ when incubated alone at the highest concentration tested in this experiment (data not shown). Nevertheless, H-NS and YmoA still formed a complex with the inv promoter at lower concentrations of YmoA, at which there is an absence of nonspecific shifting. The H-NS/YmoA complex, like the H-NS/Hha complex, is also stable in the presence of excess competitor DNA (data not shown). These data indicate that YmoA and H-NS are able to form a complex in the presence of the inv promoter.
FIG. 6.
Ability of H-NS and YmoA to form a repression complex on the inv promoter. (A) Randomly 32P-labeled inv (−344 to +146) was incubated with purified H-NS and YmoA at room temperature for 15 min and run on a 5% nondenaturing polyacrylamide gel. (B) Randomly 32P-labeled lacZ was incubated with purified H-NS and YmoA at room temperature for 15 min and run on a 5% nondenaturing polyacrylamide gel. The concentration of YmoA is shown across the top of the gel (in μM), and H-NS was added at a constant concentration of 0.1 μM. The open arrow indicates an H-NS-inv complex, with the closed arrow indicating an H-NS/YmoA-inv complex.
RovA acts as a derepressor of H-NS/YmoA-mediated repression of the inv promoter.
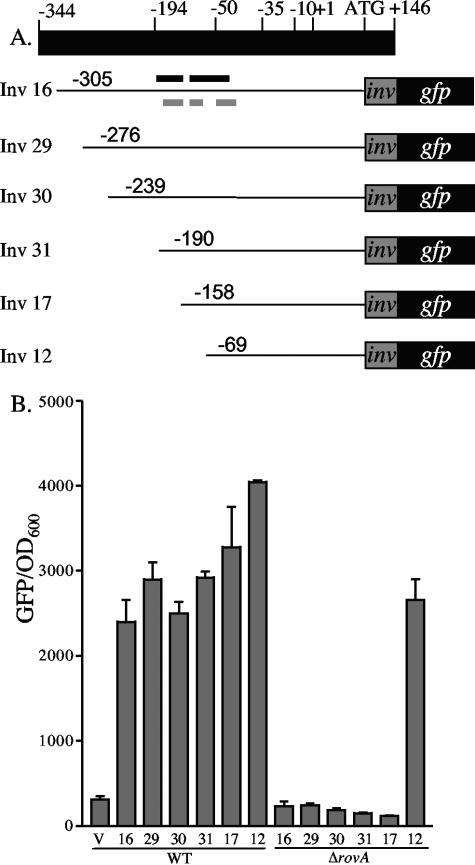
While RovA is required for the expression of inv, it is unclear if regulation is mediated by RovA activating inv expression or through a mechanism of derepression by limiting the availability of H-NS/YmoA binding sites in the inv promoter. We hypothesized that if RovA is not required for activation then loss of H-NS/YmoA binding to the inv promoter would not alter expression of inv in the absence of RovA. To test this hypothesis, a series of inv promoter truncations fused to gfp were generated (Fig. 7A) and transformed into WT Y. enterocolitica (JB580c) and ΔrovA (YVM927c). Cultures were inoculated and incubated with aeration overnight at 26°C and assayed for inv-gfp expression (Fig. 7B). All promoter truncations showed high levels of inv-gfp expression in WT Y. enterocolitica. In the ΔrovA mutant, five of the six truncations demonstrated no expression of inv-gfp, but the shortest truncation (pinv12), which lacks two of the three predicted H-NS binding sites (21), showed robust expression, indicating RovA is not required to activate inv expression. EMSA experiments revealed that H-NS was still able to bind this fragment; however, we hypothesize that without the other H-NS binding sites upstream, efficient repression does not occur (data not shown).
FIG. 7.
Effect of promoter truncations on the expression of inv-gfp in WT and ΔrovA strains. (A) Schematic of the inv promoter with the predicted −35, −10, and +1 sites, along with the start codon (+107) and fusion junction (+170). The predicted RovA binding sites (black lines) from Y. pseudotuberculosis are shown above inv16, and the predicted H-NS binding sites (gray lines) are shown below (21). (B) JB580 (WT) and YVM927 (ΔrovA) strains were transformed with the inv-gfp promoter truncations and grown overnight at 26°C. Fluorescence was calculated by dividing the average fluorescence by OD600. V indicates the pROBE-gfp[LVA], containing no promoter, and the numbers below the bars represent the different inv-gfp truncations shown in panel A. Shown below the numbers are the strains in which the truncations were tested.
Expression levels of H-NS and YmoA are similar at 26°C and 37°C.
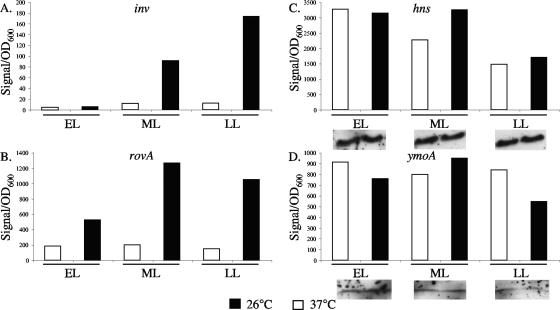
inv expression has been shown to increase when cultures are grown at 26°C and repressed when grown at 37°C (41, 46); similar results have also been shown for the expression of rovA (41). However, the expression levels of hns and ymoA have yet to be investigated at 26°C and 37°C in Y. enterocolitica. To test if changes in hns and ymoA expression were involved in the regulation of inv between 26°C and 37°C, levels of hns and ymoA were determined using Northern blotting. Samples from early, middle, and late log phases of growth at 26°C and 37°C were harvested and assayed for inv, rovA, ymoA, and hns expression. As expected, expression of inv at 37°C was low and increased from early to late log phases of growth at 26°C (Fig. 8A). A similar pattern of expression was observed for rovA, with low expression at 37°C and increasing expression as growth progressed at 26°C (Fig. 8B). The expression levels of hns and ymoA were similar between 26°C and 37°C at early, middle, and late log phases of growth (Fig. 8C and D). To determine if the expression data correlated with levels of protein, Western blot assays were done on H-NS and YmoA (Fig. 8C and D). These data suggest that the expression and the levels of H-NS and YmoA remain relatively constant between 26°C and 37°C and the increased expression of inv at 26°C is not associated with a decrease in H-NS or YmoA relative to 37°C.
FIG. 8.
Expression levels of inv, rovA, ymoA, and hns at early, middle, and late log phases of growth. (A) inv; (B) rovA; (C) hns; (D) ymoA. Cultures of WT bacteria were grown overnight and subcultured to an OD600 of 0.2. Approximately 4 OD600 of cells were taken at early log (0.5 to 1.0 OD600), middle log (1.1 to 2.5), and late log (2.6 to 3.5) phases of growth, and the RNA was extracted. Three biological replicates were performed, but because of high signal variability between replicates, representative graphs are shown for inv, rovA, ymoA, and hns. The trends shown here were the same for all of the replicates, and the expression levels of hns and ymoA between 26°C and 37°C never differed by more than twofold. Corresponding levels of H-NS and YmoA, determined by Western analysis as described in Materials and Methods, are indicated below their graphs. For both H-NS and YmoA, the inserts were taken from the same gel and the same exposure time.
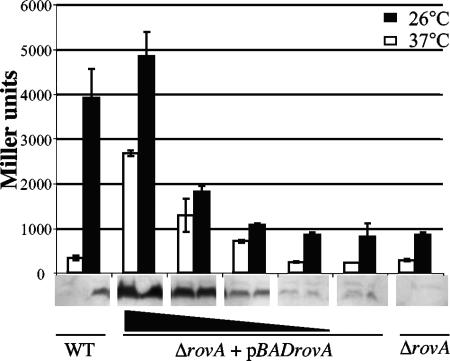
Increasing the levels of RovA at 37°C leads to expression of inv.
Based on our previous observations, we reasoned that the levels of RovA are a critical component for controlling the expression of inv, and increasing the expression of rovA under conditions where rovA is normally repressed (37°C at neutral pH) would activate inv transcription. To test this hypothesis, strain MBL015, a rovA mutant containing an inv-lacZ chromosomal fusion and an arabinose-inducible rovA construct (pBADrovA), was grown overnight at 26°C and 37°C in the presence of increasing concentrations of arabinose and assayed for inv-lacZ expression and protein levels of RovA (Fig. 9). The expression of inv-lacZ showed a dose-dependent increase in inv expression as the levels of RovA increased at both 26°C and 37°C (Fig. 9). The presence of invasin was also detected by Western blotting of cells grown at 37°C and 26°C and showed similar results (data not shown). To control for effects of arabinose on the expression of inv, strain MBL014 (containing an inv-lacZ chromosomal fusion) was grown in the presence of the same concentrations of arabinose used in the previous experiments, and no effect on the expression of inv at 26°C or 37°C was observed (data not shown). The expression of inv was also unaffected in strain MBL015 harboring the vector control (data not shown). These data indicate that increasing the levels of RovA under conditions of inv repression activate expression of inv, supporting a model where the levels of RovA in the cell are a key determinant of expression levels of inv.
FIG. 9.
Expression of inv at 37°C by induction of rovA. Cultures were grown overnight at 26°C or 37°C in triplicate and assayed for inv-lacZ expression and relative levels of RovA, which are shown below the graph. WT and ΔrovA strains are shown on the ends of the graph, and the ΔrovA+pBADrovA mutant is shown in the middle; increasing concentrations of arabinose (0.000002, 0.00002, 0.0002, and 0.002%) are indicated by the black triangle.
DISCUSSION
Invasin is expressed by both Y. pseudotuberculosis and Y. enterocolitica and is an important virulence factor involved in attachment and invasion of M cells (7, 18, 20, 23, 33, 38). Considerable work has been done to understand interactions between invasin and host ligands (7, 24) and the role of invasin in invasion of the host (6, 48); however, the mechanism of inv regulation has only recently begun to emerge.
The regulators RovA and H-NS previously have been reported to bind to the inv promoter in Y. pseudotuberculosis (21, 41), and here we demonstrate that these two regulators bind to a similar region within the inv promoter of Y. enterocolitica, suggesting a conserved mechanism of inv regulation between the two species. DNase I protection assays in Y. pseudotuberculosis revealed that the binding sites for RovA and H-NS overlap (21). We also showed that RovA and H-NS are unable to cooperatively bind to the inv promoter and that they compete with each other for binding to inv. Like H-NS, Y. pseudotuberculosis RovA also binds AT-rich regions of DNA (21), and we suspect that this will hold true in Y. enterocolitica, as the promoter fragments bound by RovA and H-NS also contain AT-rich segments. However, H-NS is believed to recognize DNA conformation (9, 61) rather than a consensus sequence. Recently, we identified additional genes regulated by RovA in Y. enterocolitica and Yersinia pestis and are currently analyzing the promoters to determine if a consensus sequence may be present that is not obvious from examination of the inv promoters alone (J. Cathelyn, D. W. Ellison, and V. L. Miller, unpublished data).
While RovA is required for expression from the native inv promoter, we demonstrated that truncation of the promoter and loss of putative H-NS binding sites results in inv expression in the absence of RovA. Our current interpretation of these results is that RovA mediates inv expression by competing for binding to the promoter with H-NS and counteracting H-NS-mediated repression. This is similar to the mechanism proposed for derepression of the hlyE promoter by SlyA (60) and is supported by data from Heroven et al. that show inv expression increases in an hns mutant E. coli but expression is not enhanced by addition of RovA to the mutant (21). While RovA is not required for the expression of inv, recent data from in vitro transcription assays indicate that RovA is able to enhance transcription of inv by interaction with RNA polymerase (57). This is similar to the regulation of tcp in Vibrio cholerae, where ToxT counteracts H-NS repression and enhances transcription through stimulation of RNA polymerase (44, 62). The identification of other genes regulated by RovA poses the question of whether or not RovA regulates expression of these targets in the same fashion as it does with inv.
We have also shown for the first time in Y. enterocolitica that an H-NS/YmoA complex is involved in the negative regulation of gene expression. Although YmoA has the ability to bind to the inv promoter, this interaction does not appear to be specific, as addition of nonspecific competitor DNA prevents binding. Our data suggest that YmoA interacts with H-NS at the inv promoter to form a negative regulatory complex. However, YmoA may still interact with DNA to stabilize the repression complex or provide further specificity in promoter binding within the H-NS/YmoA complex. Furthermore, interactions between YmoA homologs and H-NS may be a conserved regulatory mechanism, as a similar regulatory scheme has been proposed in E. coli for hly regulation by Hha (43). In the future it will be interesting to determine if the H-NS/YmoA complex represses other genes regulated by RovA in a similar manner. The regulation of rovA in Y. pseudotuberculosis shows similar regulation to inv. It is positively autoregulated by RovA and negatively regulated by H-NS through competition for binding sites within the rovA promoter (21, 41). These data provide evidence that RovA and H-NS may regulate multiple genes in a similar manner.
The regulation of hns and ymoA in response to temperature had not previously been investigated in Y. enterocolitica. Previously, YmoA was shown to be present at both 26°C and 37°C in Y. pestis. However, YmoA was shown to be degraded more rapidly at 37°C than at 26°C in Y. pestis (25). Our data indicate little difference in the steady-state expression or protein levels of H-NS and YmoA at 26°C versus 37°C in Y. enterocolitica. This led us to hypothesize that the repression complex forms at both 26°C and 37°C and expression of RovA at 26°C would relieve repression of inv, presumably by competing for binding with H-NS. Therefore, the activation of inv expression is largely dependent on the concentration of RovA in the cell and its ability to compete for binding with the H-NS/YmoA complex. To test this hypothesis, we expressed RovA under conditions where inv is normally repressed (37°C) and we were able to activate the expression of inv, lending support for our model. These results are similar to data for VirB from Shigella flexneri that show that under H-NS-repressed conditions increasing the levels of VirB leads to activation of transcription of VirB-dependent promoters (3). While the concentration of RovA in the cell has been shown to be important for inv regulation, the factors that control the RovA concentration within the cell remain an open question in Y. enterocolitica.
Nucleoid proteins have been shown to repress transcription of genes through several different mechanisms (for reviews, see references 1 and 35). Here we have demonstrated that H-NS preferentially binds to a region of the inv promoter just upstream of the −35 to −10 region, similar to results observed in Y. pseudotuberculosis using DNase I footprinting (21). This led us to propose three possible models for H-NS-mediated repression of inv: (i) the upstream region of inv could provide a high-affinity H-NS binding site, allowing nucleation of H-NS and recruitment of YmoA to form a higher-order repression complex that blocks RNA polymerase from binding to the promoter (51). (ii) Formation of an H-NS/YmoA repression complex upstream of the −35 sequence could cause silencing of the promoter by locally constraining negative supercoils (16, 17, 40, 58). (iii) The H-NS/YmoA complex could trap RNA polymerase in an open complex that is incapable of proceeding into the elongation phase (52, 53). While none of these can be ruled out, the repression of inv could involve more than one of the mentioned mechanisms.
Temperature has been shown to be involved in the regulation of many virulence genes (for reviews, see references 22, 28, 34, and 36). H-NS is involved in silencing a number of virulence genes at ambient temperature, and this is released when organisms are shifted to 37°C (49, 56, 59). This is thought to be due to structural changes in the DNA that reduce the ability of H-NS to repress transcription (13). Thermoregulation of inv is unique in that inv is highly expressed at 26°C and repressed by an H-NS/YmoA complex at 37°C, whereas other YmoA-regulated genes are expressed primarily at 37°C. It is interesting that YmoA is involved in the repression of genes at both 26°C and 37°C in Y. enterocolitica (8, 12). The ability of H-NS to form tetramers is important for its function, and recent research has shown that as the growth temperature decreases so does the ability of H-NS to form tetramers (54). The reduced ability of H-NS to form tetramers at lower temperature could have an impact on the regulation of inv. However, further investigation is needed to determine what role a reduction in H-NS tetramer formation may have on inv transcription. Temperature has also been shown to affect the structure of DNA. Changes in DNA bending and/or supercoiling could have an effect on the regulation of inv, as these structural changes can affect binding by regulatory proteins such as H-NS. It will be interesting in the future to investigate the influence that temperature may have on the structure of the inv promoter and how these changes affect inv regulation.
This work has demonstrated that the regulation of inv in Y. enterocolitica and Y. pseudotuberculosis appears to be mechanistically conserved, with RovA acting primarily as a depressor of inv by competing with H-NS for binding sites within the inv promoter. We have also furthered the understanding of inv regulation by showing that inv is repressed by an H-NS/YmoA protein complex. We have determined that the concentration of RovA within the cell is critical for the activation of inv and presented a model where the H-NS/YmoA complex is able to repress the transcription of inv under conditions of low RovA concentration, but under conditions of increased RovA concentration RovA is able to successfully compete with the H-NS/YmoA complex and activate transcription of inv. While the levels of RovA within the cell are clearly important for activation of inv, there are other factors that may play a significant role in the regulation of inv. These include changes in DNA structure, changes in the oligomeric state of H-NS, and interaction with RNA polymerase and other proteins that have yet to be identified. Understanding the interactions between H-NS, YmoA, and RovA may provide insight into the global regulation of other potential virulence factors.
Acknowledgments
This study was supported by National Institutes of Health grant AI52167 awarded to V.L.M.
We thank Robert Osuna for the gift of strain RO949 and Mollie Winfield and Eduardo Groisman for supplying P1 phage and strain MC4100. The H-NS antibody was generously supplied by Yeong-Jae Seok, and the YmoA antibody was kindly supplied by Gregory V. Plano. We also thank Matthew Lawrenz and Joseph Vogel for critical review of the manuscript and Peter Chivers for helpful discussion and suggestions.
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Badger, J. L., and V. L. Miller. 1998. Expression of invasin and motility are coordinately regulated in Yersinia enterocolitica. J. Bacteriol. 180:793-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beloin, C., and C. J. Dorman. 2003. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 47:825-838. [DOI] [PubMed] [Google Scholar]
- 4.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchmeier, N., S. Bossie, C. Y. Chen, F. C. Fang, D. G. Guiney, and S. J. Libby. 1997. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect. Immun. 65:3725-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter, P. B. 1975. Pathogenicity of Yersinia enterocolitica for mice. Infect. Immun. 11:164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, M. A., B. H. Hirst, and M. A. Jepson. 1998. M-cell surface beta1 integrin expression and invasin-mediated targeting of Yersinia pseudotuberculosis to mouse Peyer's patch M cells. Infect. Immun. 66:1237-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelis, G. R., C. Sluiters, I. Delor, D. Geib, K. Kaniga, C. Lambert de Rouvroit, M. P. Sory, J. C. Vanooteghem, and T. Michiels. 1991. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 5:1023-1034. [DOI] [PubMed] [Google Scholar]
- 9.Dame, R. T., C. Wyman, and N. Goosen. 2001. Structural basis for preferential binding of H-NS to curved DNA. Biochimie 83:231-234. [DOI] [PubMed] [Google Scholar]
- 10.Daniels, J. J., I. B. Autenrieth, A. Ludwig, and W. Goebel. 1996. The gene slyA of Salmonella typhimurium is required for destruction of M cells and intracellular survival but not for invasion or colonization of the murine small intestine. Infect. Immun. 64:5075-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellison, D. W., M. B. Lawrenz, and V. L. Miller. 2004. Invasin and beyond: regulation of Yersinia virulence by RovA. Trends Microbiol. 12:296-300. [DOI] [PubMed] [Google Scholar]
- 12.Ellison, D. W., B. Young, K. Nelson, and V. L. Miller. 2003. YmoA negatively regulates expression of invasin from Yersinia enterocolitica. J. Bacteriol. 185:7153-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falconi, M., B. Colonna, G. Prosseda, G. Micheli, and C. O. Gualerzi. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 17:7033-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming, T. P., M. S. Nahlik, and M. A. McIntosh. 1983. Regulation of enterobactin iron transport in Escherichia coli: characterization of ent::Mu Δ(Apr lac) operon fusions. J. Bacteriol. 156:1171-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.George, A. M., and S. B. Levy. 1983. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J. Bacteriol. 155:541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goransson, M., B. Sonden, P. Nilsson, B. Dagberg, K. Forsman, K. Emanuelsson, and B. E. Uhlin. 1990. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature 344:682-685. [DOI] [PubMed] [Google Scholar]
- 17.Gowrishankar, J., and D. Manna. 1996. How is osmotic regulation of transcription of the Escherichia coli proU operon achieved? A review and a model. Genetica 97:363-378. [DOI] [PubMed] [Google Scholar]
- 18.Grutzkau, A., C. Hanski, H. Hahn, and E. O. Riecken. 1990. Involvement of M cells in the bacterial invasion of Peyer's patches: a common mechanism shared by Yersinia enterocolitica and other enteroinvasive bacteria. Gut 31:1011-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanski, C., U. Kutschka, H. P. Schmoranzer, M. Naumann, A. Stallmach, H. Hahn, H. Menge, and E. O. Riecken. 1989. Immunohistochemical and electron microscopic study of interaction of Yersinia enterocolitica serotype O8 with intestinal mucosa during experimental enteritis. Infect. Immun. 57:673-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heroven, A. K., G. Nagel, H. J. Tran, S. Parr, and P. Dersch. 2004. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol. Microbiol. 53:871-888. [DOI] [PubMed] [Google Scholar]
- 22.Hurme, R., and M. Rhen. 1998. Temperature sensing in bacterial gene regulation—what it all boils down to. Mol. Microbiol. 30:1-6. [DOI] [PubMed] [Google Scholar]
- 23.Isberg, R. R., and S. Falkow. 1985. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature 317:262-264. [DOI] [PubMed] [Google Scholar]
- 24.Isberg, R. R., and J. M. Leong. 1990. Multiple beta 1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell 60:861-871. [DOI] [PubMed] [Google Scholar]
- 25.Jackson, M. W., E. Silva-Herzog, and G. V. Plano. 2004. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol. Microbiol. 54:1364-1378. [DOI] [PubMed] [Google Scholar]
- 26.Kaneko, A., M. Mita, K. Sekiya, H. Matsui, K. Kawahara, and H. Danbara. 2002. Association of a regulatory gene, slyA with a mouse virulence of Salmonella serovar Choleraesuis. Microbiol. Immunol. 46:109-113. [DOI] [PubMed] [Google Scholar]
- 27.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R-M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 28.Konkel, M. E., and K. Tilly. 2000. Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2:157-166. [DOI] [PubMed] [Google Scholar]
- 29.Libby, S. J., W. Goebel, A. Ludwig, N. Buchmeier, F. Bowe, F. C. Fang, D. G. Guiney, J. G. Songer, and F. Heffron. 1994. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc. Natl. Acad. Sci. USA 91:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madrid, C., J. M. Nieto, and A. Juarez. 2002. Role of the Hha/YmoA family of proteins in the thermoregulation of the expression of virulence factors. Int. J. Med. Microbiol. 291:425-432. [DOI] [PubMed] [Google Scholar]
- 31.Madrid, C., J. M. Nieto, S. Paytubi, M. Falconi, C. O. Gualerzi, and A. Juarez. 2002. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J. Bacteriol. 184:5058-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marra, A., and R. R. Isberg. 1996. Analysis of the role of invasin during Yersinia pseudotuberculosis infection of mice. Ann. N. Y. Acad. Sci. 797:290-292. [DOI] [PubMed] [Google Scholar]
- 34.Maurelli, A. T. 1989. Temperature regulation of virulence genes in pathogenic bacteria: a general strategy for human pathogens? Microb. Pathog. 7:1-10. [DOI] [PubMed] [Google Scholar]
- 35.McLeod, S. M., and R. C. Johnson. 2001. Control of transcription by nucleoid proteins. Curr. Opin. Microbiol. 4:152-159. [DOI] [PubMed] [Google Scholar]
- 36.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Miller, V. L., and S. Falkow. 1988. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun. 56:1242-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, W. G., J. H. Leveau, and S. E. Lindow. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant-Microbe Interact. 13:1243-1250. [DOI] [PubMed] [Google Scholar]
- 40.Mukerji, M., and S. Mahadevan. 1997. Characterization of the negative elements involved in silencing the bgl operon of Escherichia coli: possible roles for DNA gyrase, H-NS, and CRP-cAMP in regulation. Mol. Microbiol. 24:617-627. [DOI] [PubMed] [Google Scholar]
- 41.Nagel, G., A. Lahrz, and P. Dersch. 2001. Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol. Microbiol. 41:1249-1269. [DOI] [PubMed] [Google Scholar]
- 42.Nieto, J. M., C. Madrid, E. Miquelay, J. L. Parra, S. Rodriguez, and A. Juarez. 2002. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J. Bacteriol. 184:629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nieto, J. M., C. Madrid, A. Prenafeta, E. Miquelay, C. Balsalobre, M. Carrascal, and A. Juarez. 2000. Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS. Mol. Gen. Genet. 263:349-358. [DOI] [PubMed] [Google Scholar]
- 44.Nye, M. B., J. D. Pfau, K. Skorupski, and R. K. Taylor. 2000. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J. Bacteriol. 182:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paytubi, S., C. Madrid, N. Forns, J. M. Nieto, C. Balsalobre, B. E. Uhlin, and A. Juarez. 2004. YdgT, the Hha paralogue in Escherichia coli, forms heteromeric complexes with H-NS and StpA. Mol. Microbiol. 54:251-263. [DOI] [PubMed] [Google Scholar]
- 46.Pepe, J. C., J. L. Badger, and V. L. Miller. 1994. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol. Microbiol. 11:123-135. [DOI] [PubMed] [Google Scholar]
- 47.Pepe, J. C., and V. L. Miller. 1990. The Yersinia enterocolitica inv gene product is an outer membrane protein that shares epitopes with Yersinia pseudotuberculosis invasin. J. Bacteriol. 172:3780-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pepe, J. C., and V. L. Miller. 1993. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc. Natl. Acad. Sci. USA 90:6473-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prosseda, G., P. A. Fradiani, M. Di Lorenzo, M. Falconi, G. Micheli, M. Casalino, M. Nicoletti, and B. Colonna. 1998. A role for H-NS in the regulation of the virF gene of Shigella and enteroinvasive Escherichia coli. Res. Microbiol. 149:15-25. [DOI] [PubMed] [Google Scholar]
- 50.Revell, P. A., and V. L. Miller. 2000. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol. Microbiol. 35:677-685. [DOI] [PubMed] [Google Scholar]
- 51.Rimsky, S., F. Zuber, M. Buckle, and H. Buc. 2001. A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 42:1311-1323. [DOI] [PubMed] [Google Scholar]
- 52.Schroder, O., and R. Wagner. 2000. The bacterial DNA-binding protein H-NS represses ribosomal RNA transcription by trapping RNA polymerase in the initiation complex. J. Mol. Biol. 298:737-748. [DOI] [PubMed] [Google Scholar]
- 53.Shin, M., M. Song, J. H. Rhee, Y. Hong, Y. J. Kim, Y. J. Seok, K. S. Ha, S. H. Jung, and H. E. Choy. 2005. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Eσ70 as a cofactor for looping. Genes Dev. 19:2388-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stella, S., M. Falconi, M. Lammi, C. O. Gualerzi, and C. L. Pon. 2006. Environmental control of the in vivo oligomerization of nucleoid protein H-NS. J Mol. Biol. 355:169-174. [DOI] [PubMed] [Google Scholar]
- 55.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 56.Trachman, J. D., and M. Yasmin. 2004. Thermo-osmoregulation of heat-labile enterotoxin expression by Escherichia coli. Curr. Microbiol. 49:353-360. [DOI] [PubMed] [Google Scholar]
- 57.Tran, H. J., A. K. Heroven, L. Winkler, T. Spreter, B. Beatrix, and P. Dersch. 2005. Analysis of RovA, a transcriptional regulator of Yersinia pseudotuberculosis virulence that acts through antirepression and direct transcriptional activation. J. Biol. Chem. 51:42423-42432. [DOI] [PubMed] [Google Scholar]
- 58.Tupper, A. E., T. A. Owen-Hughes, D. W. Ussery, D. S. Santos, D. J. Ferguson, J. M. Sidebotham, J. C. Hinton, and C. F. Higgins. 1994. The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J. 13:258-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Umanski, T., I. Rosenshine, and D. Friedberg. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148:2735-2744. [DOI] [PubMed] [Google Scholar]
- 60.Wyborn, N. R., M. R. Stapleton, V. A. Norte, R. E. Roberts, J. Grafton, and J. Green. 2004. Regulation of Escherichia coli hemolysin E expression by H-NS and Salmonella SlyA. J. Bacteriol. 186:1620-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamada, H., S. Muramatsu, and T. Mizuno. 1990. An Escherichia coli protein that preferentially binds to sharply curved DNA. J. Biochem. (Tokyo) 108:420-425. [DOI] [PubMed] [Google Scholar]
- 62.Yu, R. R., and V. J. DiRita. 2002. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol. Microbiol. 43:119-134. [DOI] [PubMed] [Google Scholar]