Abstract
Natural genetic transformation in Streptococcus pneumoniae entails transcriptional activation of at least two sets of genes. One set of genes, activated by the competence-specific response regulator ComE, is involved in initiating competence, whereas a second set is activated by the competence-specific alternative sigma factor ComX and functions in DNA uptake and recombination. Here we report an initial characterization of CoiA, a ComX-dependent gene product that is induced during competence and is required for transformation. CoiA is widely conserved among gram-positive bacteria, and in streptococci, the entire coiA locus composed of four genes is conserved. By use of immunoblot assay, we show that, similar to its message, CoiA protein is transient, appearing at 10 min and largely disappearing by 30 min post-competence induction. Using complementation analysis, we establish that coiA is the only gene of this induced locus needed for transformability. We find no indication of CoiA having a role in regulating competence. Finally, using 32P- and 3H-labeled donor DNA, we demonstrate that a coiA mutant can internalize normal amounts of donor DNA compared to the wild-type strain but is unable to process it into viable transformants, suggesting a role for CoiA after DNA uptake, either in DNA processing or recombination.
Natural genetic transformation was first observed in Streptococcus pneumoniae by Griffith (13). Since then, similar abilities to incorporate exogenous fragments of donor DNA into the bacterial genome have been reported in more than 40 species (28). The state in which naturally transformable species take up external DNA and alter their genetic makeup is termed competence. In S. pneumoniae, competence for genetic transformation is a transient, cell density-dependent state which is activated through quorum sensing (55) mediated by a peptide pheromone, CSP (competence-stimulating peptide) (14). A 42-amino-acid prepeptide encoded by comC (43) is exported outside the cell through a dedicated transporter ComAB (17, 18). During export, it is matured into the 17-amino-acid active peptide CSP-1 that is responsible for activating the histidine kinase ComD, which upon phosphorylation is thought to activate its cognate response regulator ComE (9, 43). Activated ComE recognizes a direct repeat sequence in the promoter region of several genes (59) and activates the first wave of gene expression (early genes) in response to CSP (1, 15, 41, 45, 46, 48). These early genes include the comAB and comCDE loci and duplicate genes encoding a competence-specific alternative sigma factor, ComX (27, 30). ComX associates with the core RNA polymerase for recognition of a noncanonical promoter, the combox, and activation of a second wave of gene expression (late genes), which produces proteins required for DNA uptake and recombination processes (3, 7, 27, 41, 44).
Several late genes have been assigned roles in DNA binding, uptake, or recombination. Based on mutant phenotypes and on homologies to Bacillus subtilis counterparts, protein products of the cfl, cel, ccl, and cgl loci (comF, comE, comC, and comG in B. subtilis, respectively), for example, have been proposed to form the membrane pore for the entry of donor DNA (5; reviewed in reference 20). Although it is known that donor DNA is cut endonucleolytically upon binding to competent cells and is reduced to a single strand during transport into the cell (19, 20, 21, 22, 23, 25, 34, 37, 47), less is understood about its fate once it is inside the cell in a single-stranded state. Two cytoplasmic products of late genes, an Ssb and RecA, have been implicated in binding the single-stranded DNA and in promoting its search for a homologous partner (4, 35, 39, 41). Recently, a third late competence gene product, DprA, has been proposed to play a role in DNA protection, in parallel to RecA. In dprA or recA mutants, donor DNA is taken up but is degraded, completely failing to recombine (4, 5).
While the transcription of many competence-inducible genes is strongly regulated, the expression pattern of the corresponding protein products during competence development is known only for ComX (30) and ComE (59), both products of early genes. The rapid shutoff of competence has been proposed to be due to the degradation of ComX (11, 46), and late gene mRNA loss does indeed precede the decay of competence; however, a corresponding disappearance or degradation of late competence proteins has not been investigated.
coiA (competence inducible A) was identified as a competence-specific gene in a screen of a lacZ transcriptional fusion library (43), which was created using the insertion vector pEVP3 (10). Disruption of the coiA locus reduces transformability (43). Recently, DNA microarrays revealed directly that coiA mRNA was indeed competence specific, that its expression depended on the competence-specific alternative sigma factor ComX, and that it has a late gene expression profile (46). Its promoter region contains the combox sequence, TACGAATA, typical of late genes (7, 43). Together, these features establish coiA as a late competence gene, whose transcription increases at competence about 16- to 60-fold.
Although coiA was among the first genes to be identified as expressed specifically at competence and is required for efficient transformation, nothing is yet known about its role in transformation. Here we report that CoiA is a transient competence-associated protein, that the temporal expression patterns of early and late genes are unaffected by coiA mutation, and that coiA mutants are defective in DNA processing at a stage after DNA uptake.
MATERIALS AND METHODS
Strains, media, and transformation.
S. pneumoniae strains and primers used in this study are listed in Tables 1 and 2, respectively. DNA from strain CP1500 served as the donor for transformation assays, DNA from CPM6 (27) was the template for PCRs, the JANUS cassette (53) was the source for the kanamycin resistance marker, and pR412 was the source for the spectinomycin resistance gene, aad9 (31). Oligonucleotides listed in Table 2 were obtained from QIAGEN, Inc. (Valencia, CA).
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype and description | Source or referencea |
|---|---|---|
| CP1016 | nov-1 str-1 ery-2 vlt | 40 |
| CP1250 | hex malM511 str-1 bgl-1, Rx derivative, low β-galactosidase background | 43 |
| CPM3 | CP1250, comX1′ΩpEVP3 ′comX1+ | 27 |
| CPM7 | CP1250, ssb2′ΩpEVP3 ′ssb2+ | 27 |
| CP1335 | CP1250, ΔSP0979-81::Spcr | This work |
| CP1389 | CP1250, ΔdprA::Kanr | This work |
| CP1500 | hex nov-r1 bry-r str-1 ery-r1 ery-r2 Novr Smr Emr Brr | 8 |
| CP1548 | CP1250, cglA′ΩpEVP3 ′cglA | 27 |
| CP1649 | CP1250, comA′ΩpEVP3 ′comA | 27 |
| CP1721 | CP1250, comWΩpEVP3 comW+ | 54 |
| CP1793 | CP1250, ΔcoiA::Kanr | This work |
| CP1795 | CP1250, ssb2′ΩpEVP3 ′ssb2+ ΔcoiA::Kanr | CP1793 × CPM7 |
| CP1796 | CP1250, comX1′ΩpEVP3 ′comX1+ ΔcoiA::Kanr | CP1793 × CPM3 |
| CP1800 | CP1250, cglA′ΩpEVP3 ′cglA ΔcoiA::Kanr | CP1793 × CP1548 |
| CP1806 | CP1250, comCΩpEVP3 comC+ | This work |
| CP1807 | CP1250, comCΩpEVP3 comC+ ΔcoiA::Kanr | CP1793 × CP1806 |
| CP1808 | CP1250, comCΩpEVP3 comC+ ΔdprA::Kanr | CP1389 × CP1806 |
| CP1824 | CP1250, coiA′ΩpEVP3-coiA-C-His6 tag | This work |
| CP1847 | CP1250, comA′ΩpEVP3 ′comA ΔcoiA::Kanr | CP1793 × CP1649 |
| CP1848 | CP1250, comWΩpEVP3 comW+ ΔcoiA::Kanr | CP1793 × CP1721 |
| CP1870 | CP1250, Prafaga::coiA::Spc::rafE | This work |
| CP1871 | CP1250, Prafaga::coiA-C term-His6::Spc::rafE | This work |
| CP1873 | CP1250, Prafaga::coiA::Spc::rafE ΔcoiA::Kanr | CP1793 × CP1870 |
| CP1874 | CP1250, Prafaga::coiA-C term-His6::Spc::rafE ΔcoiA::Kanr | CP1793 × CP1871 |
Construction of a strain by transformation between two different mutants is shown as DNA × recipient.
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence | Location or use |
|---|---|---|
| BVD26 | 5′-AAACGGGAGTCTATCAAACGTCGTGAGCAA | Upstream of coiA |
| BVD27 | 5′-ATGGATCCTGAATTCCCTCCTTTTCTATATCAT | In coiA |
| BVD28 | 5′-ATGGGCCCGAATAGAAAGGATGGAGGAATCTAA | In coiA |
| BVD29 | 5′-GTAGACATCGTACATCTTGAGATCTGAAAT | Downstream of coiA |
| BVD52 | 5′-ATGGGCCCCTAGTGGATCCCCCGTTTGATTTT | In spc cassette |
| BVD53 | 5′-ATATGCATTGCAGATTTTACATGATCCCCCATGTTG | In spc cassette |
| BVD83 | 5′-ATAGATCTCCAGCTTGTGGAGGCCAGCTCCATTTG | In coiA |
| BVD84 | 5′-AATATGCATTAGTGATGGTGATGGTGATGGTATTCTACCATATTTTTCAAGAAATATTGCTGATAAAAG | In coiA/C-term His6 extension |
| BVD106 | 5′-ATGGGCCCGATTCCTCCATCCTTTCTATTCTACC | In SP0979 |
| BVD107 | 5′-ATATGCATTCAAACGAATAGTCCAAATCAATGAG | In SP0981 |
| BVD108 | 5′-AGTCCTGCTCTGGTGGTGGTGGACGAAATG | In aga gene |
| BVD109 | 5′-ATGGTACCATATCATAGTTTTCTAAAAATATACTGTCTACTC | In aga gene |
| BVD110 | 5′-ATGGTACCTAGAAAAGGAGGGAATTCATGTTTG | In coiA |
| BVD111 | 5′-ATGGGCCCTTTCTATTCTACCATATTTTTC | In coiA |
| BVD112 | 5′-ATGGGCCCTCAATGGTGATGGTGATGGTGTTCTACCATATTTTTCAAGAAATATTG | In coiA/C-term His6 extension |
| BVD113 | 5′-ATATGCATCTGACCCCAAAAGTTAGATTTATTC | In rafE |
| BVD114 | 5′-CGGTAAAGCCAATCGCATTC | In rafE |
| BVD121 | 5′-GTAAATCAGGTATTTGAGTCTTAAAACTTGTTTTC | Sequencing of junction fragments |
| BVD122 | 5′-AAGGAGCGTTTGACTCGTCTACAGCAAGG | Sequencing of junction fragments |
| DAM301 | 5′-CGCGCAAGCTGGGGATCCG | In kan cassette |
| DAM302 | 5′-ACGTGGGCCCTAGGTACTAAAACAATTCATCCAGTAA | In kan cassette |
| DAM563 | 5′-GATAGAGGCGATAAGCATGGCACATAGTAA | Upstream of dprA |
| DAM564 | 5′-ATGGGCCCTGCCATCATTTGATTCAAGAAG | In dprA |
| DAM565 | 5′-GGATCCATAACGGCTGGATTACGGCAACCT | In dprA |
| DAM566 | 5′-GATTGGGAACTCGCTTGCGTCCTATGACTGA | Downstream of dprA |
Pneumococcal strains were routinely cultured in 18-mm tubes in casein hydrolysate yeast extract medium (CAT) (38), supplemented with 10 mM HCl to prevent endogenous competence induction. Cells were grown to an optical density at 550 nm (OD550) of 0.08 to 0.1 (Spectronic 20D+) at 37°C and then held on ice until needed. For competence induction, the culture was further supplemented with bovine serum albumin (0.2%), CaCl2 (0.005%), and CSP-1 (250 ng ml−1) obtained from Chiron Mimitopes (Raleigh, NC) (14) and incubated at 37°C for an additional 10 min. Competent cultures were exposed to the donor DNA at the indicated concentrations at 30°C for 1 h for DNA uptake and recombination. Serial dilutions of the transformed cultures were plated in 60-mm-diameter petri dishes with a 4-layer (3-ml each) technique for phenotypic development (38). Final concentrations of antibiotics for the selection of recombinant strains were as follows: chloramphenicol, 2.5 μg ml−1; kanamycin, 200 μg ml−1; spectinomycin, 100 μg ml−1.
Construction of insertion-deletion mutants.
Construction of new mutations used the strategy described by Lau et al. (26). To construct a coiA deletion mutant (CP1793), PCR was employed to amplify two DNA segments (approximately 1,000 bp) flanking the coiA gene using primer pairs BVD26-27 and BVD28-29. The resulting DNA segments, coiA up and coiA dn had terminal BamHI site and ApaI sites, respectively. The kanamycin cassette was amplified with corresponding BamHI and ApaI sites using primers DAM301 and DAM302. These fragments were digested with the respective restriction enzymes, purified using the QIAGEN PCR purification kit, ligated, and transformed directly into the wild-type strain, CP1250. Transformants were selected with kanamycin and checked for the correct structure by amplifying both new junction fragments. One coiA mutation was further purified by backcross into the wild-type strain CP1250 and isolation of a single clone, named CP1793. The dprA and SP0979-81 (Table 1) mutations were constructed similarly, and their structures were confirmed by demonstrating the presence of new junction fragments.
Construction of C-terminal CoiA-His-tagged strain.
coiA-His was amplified using HiFi Platinum Taq polymerase (Invitrogen) with primers BVD83 and BVD84 (Table 2). The PCR product had a 5′ BglII site, six new His codons, and a 3′ NsiI site. The amplified product was digested, purified, and inserted into the integrative vector, pEVP3, between the BglII and NsiI sites. The resulting plasmid, pBD04 in Escherichia coli host strain DH5α, was extracted, purified, and transformed into CP1250 to create strain CP1824 (Pcin::coiA-C-term His6). The CoiA-His insert in pBD04 was sequenced to confirm the absence of any point mutations.
Construction of CoiA overexpression strains.
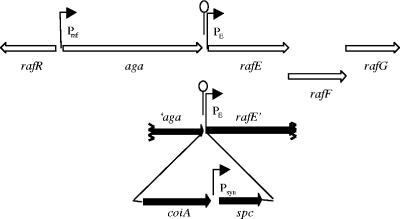
CP1870 (PcincoiA+, Praf::coiA) was constructed using the PCR amplification-ligation strategy (see Fig. 4). Briefly, four DNA fragments (′aga, coiA, spc, and rafE′) were amplified using the primers described in Table 2, with mutually compatible restriction enzyme sites. Digestion, purification, and ligation of the four DNA fragments assembled a four-piece cassette ′aga::coiA::spc::rafE′ which was transformed into CP1250. Several independent Spcr colonies were analyzed for all three new junctions using PCR. From 1 of 3 clones with the correct structure, the mutation was back-crossed into strain CP1250 to purify it further. Several resulting single colony clones were sequenced to confirm the absence of any point mutations in the coiA gene. One strain with the desired coiA sequence was named CP1870. CP1873 (PcincoiA null, Praf::coiA) was constructed by transforming the coiA mutation (from CP1793) into strain CP1870 and selecting a single transformant for further work.
FIG. 4.
Construction of a raffinose-regulated ectopic copy of coiA. Open arrows, five central genes of the raffinose utilization locus (50). The four-part ligation product (′aga, coiA, spc, and rafE′) was transformed into CP1250 to construct CP1870 (PcincoiA+ and Praf::coiA+). Gray bars, regions targeting homologous recombination; black arrows, coiA and the Spcr cassette inserted in the raffinose locus; Praf, promoter controlling aga gene; PE, promoter controlling rafE operon; Psyn, promoter driving expression of the spc cassette; stem-loop, transcriptional terminator.
A similar strategy was employed for constructing strains CP1871 and CP1874 (Table 1) using primers described in Table 2.
Assay of the transformation phenotype.
To determine the kinetics of competence induction for mutants and wild-type strains, parallel cultures were grown in CAT as described above at 37°C. At an OD550 of 0.08 to 0.1, cultures were chilled on ice and diluted 10-fold in 10 ml cold CAT supplemented with bovine serum albumin, CaCl2, and HCl at the mentioned concentrations. Diluted cultures (1.5 ml) were transferred to sterile microcentrifuge tubes in a 37°C block. After 10 min at 37°C, all of the cultures were induced to competence in parallel by adding CSP-1 (250 ng ml−1). At 5-min intervals, 0.15 ml of cells was mixed with 10 ng of 5MC donor DNA and incubated at 30°C for 5 min, and DNA uptake was terminated by transferring 0.015 ml into 1.5 ml of CAT containing DNase I (40 μg ml−1) and MgCl2 (10 mM). After 60 min at 30°C, the entire sample was plated in CAT agar to determine Novr transformants.
Beta-galactosidase activity assay.
S. pneumoniae strains carrying the lacZ reporter gene were grown and induced to competence as described above. Samples (0.5 ml) were harvested before CSP treatment (“0 min” time point) and at the indicated time intervals after the addition of CSP. Samples were collected on ice, mixed with a 1/3 volume of 2% Triton X-100 (final, 0.5%), allowed to lyse at 37°C for 10 min, and chilled until assay. The lysates were mixed with 1.5 ml of ONPG reaction mix (1.6 mg o-nitrophenyl-β-d-galactopyranoside [ONPG], 20 mM sodium phosphate [16.4 mM Na2HPO4 and 3.6 mM NaH2PO4, pH 7.5], 20 mM NaOH, 2 mM MgCl2 and 90 mM β-mercaptoethanol), and incubated at 37°C until yellow color developed. The reaction was stopped by adding 1 ml of 1 M Na2CO3. The absorbance at 420 nm was determined, and β-galactosidase activity was calculated using the following relation: Miller units (MU) = (1,000 × OD420)/(incubation time in min × culture sample volume in ml × culture OD550 at addition of CSP). The “0-min” time sample was used as the blank.
Immunoblot analysis.
S. pneumoniae strains CP1824 and CP1874 were grown in 400 ml of CAT to an OD550 of 0.08 to 0.1. A 25-ml “0-min” sample was harvested just before CSP addition, after which successive 25-ml samples were harvested every 2.5 min. Samples were cooled rapidly in a 1-liter prechilled stainless steel beaker and stored in 30-ml corex glass tubes on ice. Cells were collected by centrifugation at 8,000 × g for 15 min at 4°C, resuspended in 0.2 ml of 0.1% sodium dodecyl sulfate, and lysed for 5 min at 95°C. A sample of the lysate (10 μl) was used to determine the total protein concentration using Bio-Rad's Bradford protein assay reagent. Two sodium dodecyl sulfate-polyacrylamide gel electrophoresis minigels (10% Tris-HCl [Bio-Rad]) were developed in parallel in Tris-glycine buffer at 85 V until the dye reached the bottom of the gels. After the gels were washed twice in water, one gel was stained with Bio-Rad's BioSafe Coomassie. The other gel, a polyvinylidene difluoride membrane (Hybond-P; Amersham, Piscataway, NJ), fiber supports, and blotting papers were equilibrated in the transfer buffer (48 mM Tris, 39 mM glycine, pH 9.2) for at least 10 min before being assembled together in an electrotransfer unit (Bio-Rad). After transfer at 40 V for 90 min at 4°C, the membrane was washed twice for 15 min with 25 ml TBST (20 mM Tris-HCl, pH 7.6, 140 mM NaCl, and 0.1% Tween 20) buffer before the membrane was blocked overnight at 4°C with 30 ml TBST containing 5% nonfat dry milk (Bio-Rad, Hercules, CA). The polyvinylidene difluoride membrane was then rinsed twice with 25 ml TBST and probed for 2 h with anti-His6 primary antibody (RDI, Inc.) (1:4,000 in 20 ml TBST containing 1% nonfat dry milk) at room temperature. The membrane was washed thoroughly with TBST for a total time of 1 h with 6 25-ml buffer changes at room temperature. The membrane was then probed with horseradish peroxidase (HRP)-conjugated secondary antibody (Amersham Biosciences, Piscataway, NJ) (1:20,000 in 20 ml TBST containing 1% nonfat dry milk) for 2 h at room temperature. After secondary probing, the membrane was again washed thoroughly in TBST to remove all unbound antibody, and bands were visualized using Amersham's ECL plus immunoblotting detection system and X-OMAT imaging films (Kodak, Rochester, NY).
Preparation of 3H-labeled DNA.
CP1016 was grown in 10 ml CAT containing 0.5 mCi of [3H]thymidine (specific activity, 20 to 40 Ci mmol−1; MP Biomedicals) for 6.5 generations to an OD550 of 0.2. The culture was chilled, harvested, and then processed for DNA extraction as recommended by the QIAGEN Genomic DNA handbook (QIAGEN, Inc., Valencia, CA). Yield of the DNA from such preparations was approximately 12 to 15 μg, with a specific activity of 1.0 to 1.5 × 106 cpm μg−1.
DNA uptake assay.
CP1250, CP1793, and CP1389 were grown in 10 ml CAT at 37°C to an OD550 of 0.08 to 0.1. Cells were chilled if required and returned to 37°C before CSP treatment. After competence induction, 1-ml cultures were exposed to 40 ng of CP1016 chromosomal DNA (approximately 0.6 × 105 cpm). For determining transformation, after 5 min of DNA uptake, 0.15 ml of cells was mixed with 1.5 ml of CAT containing DNase I as described above, incubated at 30°C for 60 min, and then plated in soft agar plates (measure of transformation). For measuring the DNA uptake stages, the remaining culture was incubated at 30°C for 30 min and centrifuged at 10,000 rpm at 4°C for 2 min. Supernatant recovered from the spin was mixed with a 1/4 volume of 50% trichloroacetic acid (TCA), incubated on ice for at least 10 min, and centrifuged at 10,000 rpm for 10 min. A portion of the resulting TCA-soluble supernatant (0.1 ml) was mixed with 0.4 ml of water and 10 ml of EcoLite scintillation fluid for measuring radioactivity (measure of degradation). The pellet obtained from the first spin was washed once without resuspension (21), resuspended in 0.1 ml of CAT containing 40 μg ml−1 DNase I, and incubated for 10 min at 30°C. Cells were centrifuged, and the supernatant containing the DNase I accessible product was mixed with 10 ml EcoLite scintillation fluid (MP Biomedicals) (measure of DNA binding). The cell pellet from the last spin was washed once with 1 ml of cold CAT, resuspended in 0.1 ml SEDS buffer (52), lysed at 37°C for 10 min, and mixed with 0.4 ml water and 10 ml EcoLite scintillation fluid for determining DNA uptake.
Incorporation of [32P]dATP in a large 11-kb donor DNA amplicon.
An 11-kb 32P-labeled PCR product was prepared using HiFidelity Taq polymerase (Invitrogen), primer pairs BVD81 and BVD82, and 5MC as template DNA. The PCR (50 μl) was carried out in the presence of 125 μCi of [32P]dATP (Amersham BioSciences) according to the manufacturer's recommendations.
Fate of labeled DNA.
CP1806, CP1807, and CP1808 were cultured in 10 ml CAT to an OD550 of 0.08 at 37°C and chilled until required. After the cultures were rewarmed to 30°C and induced to competence at 30°C for 10 min, 4 ml of cells was exposed to 150 ng of a [32P]dATP-labeled 11-kb PCR product for 20 min at 30°C. Cells were incubated further with DNase I (40 μg ml−1) at 30°C for 60 min, chilled for 5 min, diluted fivefold with cold CAT containing 5 mM EDTA, and centrifuged at 10,000 rpm for 10 min at 4°C. The resulting cell pellet was washed once with 1 ml of cold CAT containing EDTA and lysed at 37°C for 5 min in 0.4 ml SEDS containing 70 μg ml−1 RNase A. Chromosomal DNA from the lysed cells was extracted using the phenol-chloroform method (51), precipitated with ethanol, and dissolved in 30 μl of warm 10 mM Tris-Cl buffer, pH 8.5. The extracted DNA was digested with EcoRI and BstBI according to the manufacturer's recommendations and fractionated at 65 V in a 1% agarose gel (24 by 20 cm) for 20 h at room temperature. The gel was stained in ethidium bromide to confirm equal loadings, dried in vacuo at 80°C for 4 h, and exposed to a phosphorimager screen for about 36 h. The radiolabeled pattern was visualized using the PhosphorImager SI (Molecular Dynamics, Sunnyvale, CA).
RESULTS
coiA is a ComX-dependent late gene with transient protein expression.
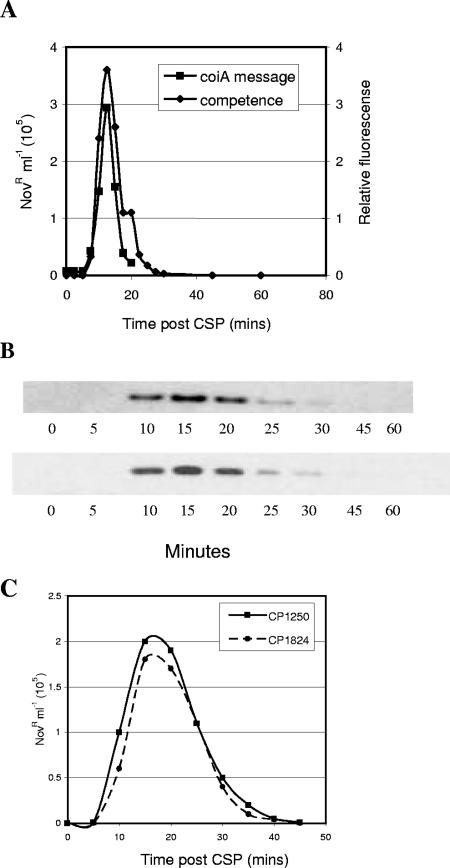
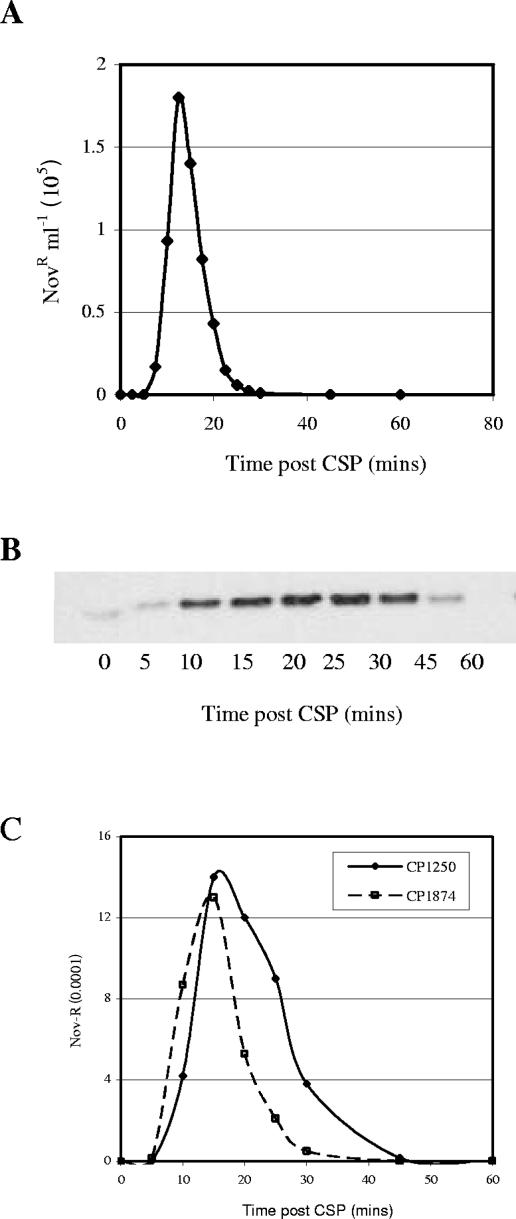
To ask whether the competence-specific transcription of coiA is reflected by changes in the level of protein, we created a coiA gene encoding an epitope-tagged protein (CoiA-His6) and showed that transformability of the recombinant strain was not affected by this alteration in CoiA structure (Fig. 1). Immunoblotting using a His6-specific antiserum (anti-His6) revealed that the amount of CoiA was low in noncompetent cells. The protein was first detectable 10 min after CSP treatment, was maximal at 15 min, and disappeared soon after 20 min (Fig. 1). This is the first time that the temporal protein expression profile for a late competence gene product has been determined during competence. The only other competence protein expression data available are for early competence gene products. ComE is stable (59), but ComX is unstable, with a temporal pattern similar to that of CoiA (30).
FIG. 1.
Competence-dependent expression of CoiA. (A) Kinetics of competence development of CoiA-His6 mutant in CP1824 (⧫) and transcriptional profile of coiA in CP1250 (▪). (B) CoiA-His6 protein levels were determined in strain CP1824 in two replicate experiments. (C) Comparison of the transformation phenotype between CP1824 and CP1250. Details for the transformation assay and immunoblot analysis can be found in the Materials and Methods. Details for the coiA mRNA can be found in the report from Peterson et al. (46).
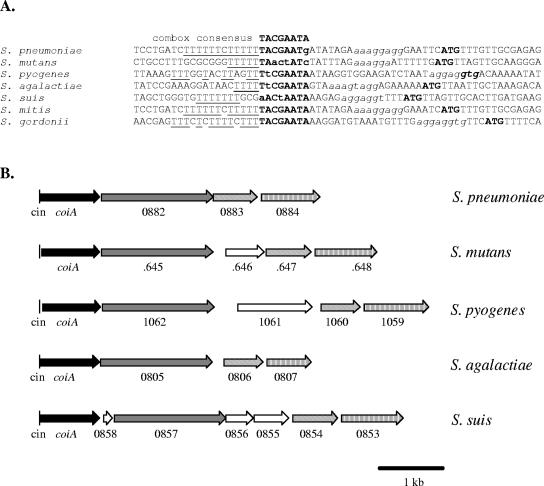
CoiA is a widely conserved protein of unknown function.
CoiA is widely conserved in gram-positive bacteria (Table 3), and the entire coiA locus (see below) is preserved within the streptococci (Fig. 2B). Interestingly, the streptococcal coiA homologs are also associated with a combox promoter sequence (Fig. 2), suggesting a common functional role for these proteins, perhaps in DNA transformation. Curiously, CoiA is also invariably associated with a common downstream neighbor annotated as an oligopeptidase (data not shown). The B. subtilis homolog of CoiA, YjbF, has also been reported to have a promoter sequence specifically regulated by the Bacillus competence regulator ComK (6). Although CoiA is so widely conserved, nothing is known about the function of any of its homologs.
TABLE 3.
Homologs of CoiA
| Organisma | % Identityb | % Similarity | Functional annotationc |
|---|---|---|---|
| Streptococcus pneumoniae | 100 | 100 | Competence protein CoiA |
| Streptococcus mitis | 89 | 92 | Competence protein CoiA |
| Streptococcus gordonii | 54 | 72 | Competence protein |
| Streptococcus suis | 50 | 68 | Competence protein |
| Streptococcus agalactiae | 49 | 66 | Similar to hypothetical transcription factor |
| Streptococcus mutans | 46 | 66 | Putative competence protein/transcription factor |
| Lactococcus lactis | 44 | 64 | Competence protein CoiA |
| Lactococcus lactis | 43 | 62 | Transcription factor |
| Lactococcus lactis | 42 | 62 | Oligopeptidase PepF2 |
| Streptococcus pyogenes | 41 | 61 | Putative transcription factor |
| Staphylococcus aureus | 40 | 59 | Hypothetical protein |
| Bacillus halodurans | 37 | 56 | Unknown conserved protein |
| Bacillus anthracis | 34 | 57 | Hypothetical protein |
| Bacillus thuringiensis | 34 | 57 | Possible competence protein |
| Enterococcus faecalis | 34 | 57 | Competence protein, putative |
| Bacillus cereus | 33 | 54 | Putative competence protein/transcription factor |
| Bacillus subtilis | 33 | 49 | Hypothetical protein |
| Lactobacillus plantarum | 33 | 46 | Competence protein |
| Bacillus licheniformis | 31 | 49 | Conserved protein |
| Pediococcus pentosaceus | 30 | 50 | Competence protein |
| Lactobacillus acidophilus | 30 | 45 | Competence protein |
| Listeria monocytogenes | 29 | 50 | Putative competence protein |
| Staphylococcus haemolyticus | 29 | 45 | Hypothetical protein |
| Listeria innocua | 28 | 49 | Putative competence protein |
| Lactobacillus johnsonii | 27 | 48 | Hypothetical protein |
| Staphylococcus saprophyticus | 26 | 46 | Putative competence protein |
| Enterococcus faecium | 26 | 45 | Competence CoiA-like |
| Lactobacillus gasseri | 25 | 49 | Competence protein |
Species in the NCBI database with homology to CoiA.
Proportion of identical residues in full-length alignment with S. pneumoniae CoiA.
Functional annotations are as described in the NCBI database.
FIG. 2.
Sequence conservation at coiA locus in streptococci. (A) Alignment of combox sequence in the promoter of coiA homologs of several streptococci. A consensus combox sequence is displayed at the top; putative combox sequences are highlighted in boldface type, with nonconserved nucleotides in lowercase type; adjacent thymine residues (7, 44) are underlined; pneumococcal ribosome binding sites, aaaggaggtg, are in lowercase italic type (2, 25); and predicted translation start sites (ATG/GTG) are shown in boldface type. (B) Comparison of coiA loci among five streptococci. Similar ORF patterns represent functional similarity. For S. pneumoniae, the three coiA downstream genes annotated by Hoskins et al. (16) are group B oligopeptidase (spr0882), probable methyltransferase (spr0883), and peptidylprolyl isomerase (spr0884), respectively. cin, combox promoter; open arrows, ORFs not displaying functional similarity.
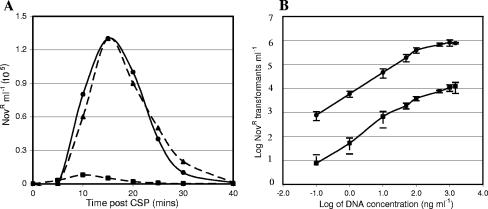
coiA deficiency reduces transformability.
To characterize the role of CoiA in transformation, a deletion mutant (CP1793) was created by replacing the coiA gene with a kanamycin resistance marker (Kanr) using the strategy described in Materials and Methods. The transformation efficiency of CP1793 was severely reduced compared to its wild-type parent, CP1250 (Fig. 3A). To determine whether this defect was dependent on donor DNA concentration, the assay was repeated over a wide range of donor DNA concentrations. As shown in Fig. 3B, the low transformability of the coiA mutant was consistent over a wide range of DNA concentrations and is consistent with the phenotype observed earlier for several coiA insertion-duplication mutants (43).
FIG. 3.
Reduced transformability of a coiA mutant. (A) Competence of CP1250 (•; wild type), CP1793 (▪; coiA null), and CP1335 (▴; SP0979-0981−) determined at 5-min intervals. (B) Comparison of transformation of CP1793 (▪; coiA null), and CP1250 (•; wild type) after incubation in the presence of different amounts of donor DNA for 30 min at 37°C. Error bars indicate standard errors of results from three independent experiments.
Loss of CoiA is responsible for the observed transformation defect.
coiA is the first gene of a 4-open reading frame (ORF) cluster of CSP-inducible late genes, spr0881 to spr0884 (Fig. 2B) (46). Expression of its upstream neighbor gene, spr0880, is invariant in response to CSP, and a terminator was identified between spr0884 and spr0885 using the GeSTer algorithm (46, 57). This arrangement suggests that a single CSP-induced mRNA may encompass coiA and 3 adjacent genes. The observed transformation defect in CP1793 might in principle be due to either the loss of CoiA or a polar effect of the mutation on the expression of downstream neighbors. We took two approaches to distinguish these possibilities. First, the three downstream genes were deleted by being replaced with a spectinomycin (Spcr) cassette while leaving coiA intact. As the resulting mutant, CP1335, was fully transformable, the phenotype of CP1793 could not be replicated by disruption of these linked genes (Fig. 3A).
In a complementary approach, we asked if the transformation defect of the ΔcoiA::Kan mutant could be complemented by providing an ectopic copy of the coiA gene alone. The aga promoter Praf (PA in reference 50), regulated by the products of the rafRS genes, is responsive to raffinose (50). coiA was amplified using PCR, linked to a spectinomycin (Spcr) marker, and inserted in the raffinose utilization locus, downstream of the aga gene (Fig. 4). The resulting strain, CP1870, had two copies of coiA, the native copy retained under the control of its combox promoter (PcincoiA), and the ectopic copy retained under the control of the raffinose promoter (Praf::coiA). To study the complementation of the ΔcoiA mutation, the native copy of the gene was replaced with a Kanr marker by crossing CP1793 with CP1870. The resulting strain, CP1873 (Praf::coiA, ΔPcincoiA), was treated with synthetic CSP-1, and the ectopic copy of coiA was induced by adding raffinose at a level known to be sufficient to induce maximum levels of aga expression (29). The presence of 100 μM raffinose did not have an adverse effect on competence development in the wild-type control strain CP1250; however, as shown in Table 4, induction of the Praf copy of coiA in CP1873 rescued the transformation defect caused by deletion of CoiA (compare CP1793). From the consistent results of the two approaches, we conclude that the transformation defect observed in strain CP1793 is due to the loss of the coiA gene alone and not to polar effects of the coiA mutation.
TABLE 4.
Complementation of coiA deletion by raffinose-controlled coiA
| Strain | Genotype | No. of Novr transformants (105 cells ml−1) witha:
|
|
|---|---|---|---|
| No raffinose | 100 μM raffinoseb | ||
| CP1250 | PcincoiA+ Prafaga | 1.64 | 1.63 |
| CP1793 | PcincoiA null Prafaga | 0.01 | 0.01 |
| CP1870 | PcincoiA+ Praf::coiA+ | 1.23 | 0.87 |
| CP1873 | PcincoiA null Praf::coiA+ | 0.12 | 0.99 |
Transformation was assayed by exposing 1 ml of CSP-treated cells to 5MC donor DNA.
When used, 100 μM raffinose was supplied at the same time as CSP.
coiA deletion or overexpression does not alter competence regulation in S. pneumoniae.
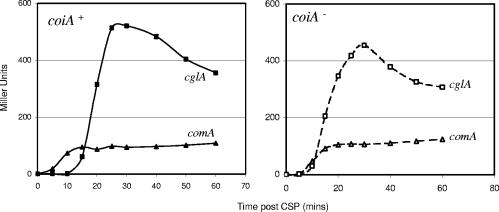
Most CoiA homologs in the NCBI database are annotated as proteins with unknown function or as having a function in competence (based on the function previously assigned to CoiA) (43). The only other function assigned to a CoiA homolog is that of transcription factor, made in the case of a gene in Lactococcus lactis, and based on a 15% identity and 39% similarity to the tomato transcription factor Vsf-1 (42, 49, 56). Although this similarity is weak, we asked if mutation of coiA in pneumococcus caused a general disruption in the CSP response of early or late competence genes by making use of the integrative reporter vector pEVP3 to analyze the expression of four early (comA, comC, comX, and comW) and three late (cglA, celB, and ssbB) competence genes. Upon CSP induction, the expression of these representative genes was unaltered in the coiA background compared to the corresponding wild-type strains (Fig. 5; data shown only for comA and cglA).
FIG. 5.
Lack of effect of ΔcoiA mutation on early and late competence gene expression. LacZ activity was determined at indicated times after competence induction in wild-type (left) and coiA-deficient (right) backgrounds. ▴, comA; ▪, cglA.
Transcriptional regulators are sometimes found to alter expression of target genes when overexpressed. To explore this possibility for CoiA, the recombinant S. pneumoniae strain CP1874 (PcincoiA null and PrafcoiA-His6) was induced to competence, while CoiA-His6 was overexpressed by induction using 100 μM raffinose. A 16-fold overexpression of CoiA (data not shown) did not affect transformation levels in the recombinant strain, although we did observe a small shift in the timing of competence (Fig. 6). Thus, it seems unlikely that CoiA may act as a general transcriptional regulator of competence genes.
FIG. 6.
Competence development in the presence of overexpressed CoiA. (A) Transformation of CP1874 was determined at 2.5-min intervals after adding CSP-1 to a culture grown for 1 generation in 100 μM raffinose. (B) CoiA-His6 protein levels determined by immunoblot analysis in extracts prepared at 5-min intervals. (C) Transformation phenotypes of CP1874 and CP1250, respectively.
coiA-deficient mutants take up normal amounts of donor DNA.
As a coiA mutant exhibited a severe defect in transformability but did not appear to have a regulatory defect, we sought to determine whether it was affected in DNA uptake steps of transformation. The coiA mutant was compared with the wild-type and with a dprA mutant strain (CP1389) constructed by deletion replacement. Consistent with the previous report (4, 5), this dprA mutant had a severe transformation defect (0.0001% of wild-type; data not shown) but took up comparable amounts of DNA and produced amounts of TCA-soluble extracellular products similar to the wild type. The coiA mutant also exhibited DNA uptake properties similar to the dprA mutant. Although there was a >95% defect in transformability in a coiA mutant, it took up DNA and produced TCA-soluble products in amounts similar to the wild-type and dprA mutant strains (Table 5). Thus, the coiA mutant, CP1793, was similar to the dprA mutant in two ways: it was efficient in taking up 3H-labeled donor DNA and it produced few viable transformants.
TABLE 5.
DNA uptake and processing in coiA mutant
| Strain | DNA (cpm ml−1) at processing stagea:
|
Transformation (Novr cells ml−1) | ||
|---|---|---|---|---|
| Degradationb | Bindingc | Uptaked | ||
| CP1250 (wild type) | 1,060 ± 37 | 1,140 ± 30 | 600 ± 27 | 1,900 ± 450 |
| CP1793 (coiA null) | 880 ± 88 | 900 ± 61 | 440 ± 4 | 53 ± 30 |
| CP1389 (dprA null) | 990 ± 38 | 890 ± 99 | 600 ± 30 | 0.0001 |
Cells were exposed to CSP and 105 cpm DNA for 30 min at 30°C. Data are means ± standard errors of results from three independent experiments.
Acid-soluble extracellular products.
DNase-susceptible bound DNA.
DNase-resistant bound DNA.
In an independent approach to trace the fate of donor DNA in the coiA mutant, a strategy described previously (4, 33) was used with some modifications. An 11-kb PCR product including the gyrB point mutation nov-1, conferring Novr, was amplified in the presence of [32P]dATP for use as a donor. The 32P label was used to distinguish nucleotides incorporated by recombination specifically at the gyrB locus from material distributed more broadly into the chromosome via degradation and DNA synthesis. Both coiA and dprA mutants were able to take up the donor DNA, as seen from the uniform presence of label in many restriction fragments (data not shown). While in the wild-type strain, CP1250, intensely labeled bands at specific positions reflected homologous recombination, such bands were absent in the coiA and dprA mutants, demonstrating that both mutants failed to integrate donor genes into the resident genome. The nonspecific bands observed on the gel were the result of degradation of the labeled DNA and incorporation of the label during DNA synthesis (33, 58). This interpretation was strengthened by repeating the experiment in the presence of an inhibitor of DNA synthesis, HpUra (33, 58). For the wild-type, use of HpUra limited the incorporation of label to the bands corresponding to homologous recombination, while for the coiA and dprA mutants, no labeled bands were detected (data not shown). Together, these results suggest that, as in dprA mutants, donor DNA is taken up in a coiA mutant, but subsequent processing is blocked at some step leading to recombination.
DISCUSSION
As coiA is part of a four-gene competence-inducible operon, the transformation defect reported by Pestova and Morrison (44) for coiA insertion-duplication mutants cannot be directly associated with the disruption of coiA. Indeed, Lacks (24) proposed that the products of all four genes act in cell wall remodeling during development of competence. To identify the specific genes responsible for the defect in transformation, insertion-deletion mutations in the coiA locus were constructed. These mutations in combination with complementation analysis established coiA as the only gene of this locus required for transformation. A similar organization of competence-inducible operons has also been described before (46): Peterson et al. reported that for seven competence-inducible operons, only one of the more proximal genes was needed for transformation, while the downstream genes were dispensable for the process.
Genetic transformation in S. pneumoniae involves induction and shutoff of competence. It was previously observed that induction of this transient process was accompanied by a shift in the expression of many cellular proteins (35). Recent genomic surveys demonstrated a coherent activation of approximately 124 genes during competence induction (12, 46). In contrast, competence shutoff has only been associated with the disappearance of the messages of these induced genes. It has been proposed that one or more gene products under the transcriptional regulation of ComX may be responsible for competence shutoff (27, 46, 54). However, it is not known what change(s) in protein level or activity cause the rapid decay in the ability of competent cells to take up or process DNA. Since the accumulation of CoiA protein was transient and followed kinetics parallel to that of competence, its loss may contribute to the loss of competence. Protein expression profiles are known for only two other competence genes, both in the early gene category: ComE is stable throughout competence (59) and ComX is unstable (30). If other late gene products also follow expression patterns similar to CoiA and ComX, disappearance of these competence-specific proteins could explain the shutoff of competence.
The rapid disappearance of ComX has been attributed to its degradation by the protease ClpEP (54). Additionally, the above study suggested that this degradation can be prevented by an antiprotease activity of ComW. As the level of CoiA achieved using transcription controlled by the Praf promoter was strongly elevated in competent cells (Fig. 6), it is possible that CoiA is regulated similarly to ComX by the activities of ClpEP and/or ComW and that the stability of CoiA may depend on the presence of ComW or on other posttranscriptional regulation.
A homolog of CoiA is annotated in the NCBI database as a transcriptional regulator, based on a weak similarity (15% identity and 33% similarity) of this CoiA homolog in Lactococcus lactis to the transcriptional factor Vsf-1 (42, 56). In addition, our unpublished results (B. Desai and D. Morrison) demonstrate the presence of a C2H2 type of zinc (Zn) finger motif in the N terminus of CoiA. These types of Zn finger motifs are extremely rare in prokaryotes but are abundant in eukaryotes and are involved in DNA recognition. Since CoiA possesses this Zn finger motif and as one of the CoiA homologs was possibly related to a transcriptional regulator, we sought tp determine whether CoiA has a role in regulating competence. Deletion of coiA did not block the expression of any of four early or three late competence genes. In addition, approximately 16-fold overexpression of CoiA did not have any affect on the level of competence. Thus, there is no direct indication of any role for CoiA as a regulator of competence.
As CoiA did not have a major role in regulating competence, we sought to determine whether CoiA had a more direct role in DNA processing. Among 81 late competence-induced proteins, only 15 are known to be required for transformation. Most of these 15 proteins are membrane proteins involved in DNA binding or degradation (5, 20). Little is known about the proteins interacting with donor DNA after internalization. Two ComX-regulated proteins, DprA and RecA, are essential for transformation but not for DNA uptake (5). In both mutants, donor DNA is immediately degraded after entry into the cell, implicating them in DNA protection (4). Another competence-induced protein is implicated in forming a protective complex with a single-stranded donor (35, 36, 39). Finally, a noninduced protein, MmsA (RecG), is known to be important for DNA processing (32). As coiA mutants exhibited a severe defect in transformability but could internalize normal amounts of donor DNA, CoiA also appears to act in a post-DNA uptake step. Once internalized, DNA is known to undergo several stages of processing: protection, eclipse complex formation, and heteroduplex formation/recombination. It will be of interest to determine how far along this pathway single-stranded donor DNA progresses in a coiA mutant.
Acknowledgments
This work was supported in part by the U.S. National Science Foundation (MCB-0110311).
We are grateful to Jean-Pierre Claverys for providing plasmids pR410 and pR412. We thank Haiying Li for assistance in constructing the raffinose inducible systems and Sharmistha Dev for assistance in transformation assays.
REFERENCES
- 1.Alloing, G., B. Martin, C. Granadel, and J. P. Claverys. 1998. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29:75-83. [DOI] [PubMed] [Google Scholar]
- 2.Bacot, C. M., and R. H. Reeves. 1991. Novel tRNA gene organization in the 16S-23S intergenic spacer of the Streptococcus pneumoniae rRNA gene cluster. J. Bacteriol. 173:4234-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartilson, M., A. Marra, J. Christine, J. S. Asundi, W. P. Schneider, and A. E. Hromockyj. 2001. Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence stimulatory peptide. Mol. Microbiol. 39:126-135. [DOI] [PubMed] [Google Scholar]
- 4.Bergè, M., I. Mortier-Barrière, B. Martin, and J. P. Claverys. 2003. Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol. Microbiol. 50:527-536. [DOI] [PubMed] [Google Scholar]
- 5.Bergè, M., M. Moscoso, M. Prudhomme, B. Martin, and J. P. Claverys. 2002. Uptake of transforming DNA in gram-positive bacteria: a view from Streptococcus pneumoniae. Mol. Microbiol. 45:411-421. [DOI] [PubMed] [Google Scholar]
- 6.Berka, R. M., J. Hahn, M. Albano, I. Draskovic, M. Persuh, X. Cui, A. Sloma, W. Widner, and D. Dubnau. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, E. A., S. Y. Choi, and H. R. Masure. 1998. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol. Microbiol. 27:929-939. [DOI] [PubMed] [Google Scholar]
- 8.Cato, A., Jr., and W. R. Guild. 1968. Transformation and DNA size. I. Activity of fragments of defined size and a fit to a random double cross-over model. J. Mol. Biol. 37:157-178. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, Q., E. A. Campbell, A. M. Naughton, S. Johnson, and H. R. Masure. 1997. The com locus controls genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 23:683-692. [DOI] [PubMed] [Google Scholar]
- 10.Claverys, J. P., A. Dintilhac, E. V. Pestova, B. Martin, and D. A. Morrison. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123-128. [DOI] [PubMed] [Google Scholar]
- 11.Claverys, J. P., and L. S. Håvarstein. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7:d1798-d1814. [DOI] [PubMed] [Google Scholar]
- 12.Dagkessamanskaia, A., M. Moscoso, V. Henard, S. Guiral, K. Overweg, M. Reuter, B. Martin, J. Wells, and J. P. Claverys. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51:1071-1086. [DOI] [PubMed] [Google Scholar]
- 13.Griffith, F. 1928. The significance of pneumococcal types. J. Hyg. 27:113-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Håvarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Håvarstein, L. S., and D. A. Morrison. 1999. Quorum sensing and peptide pheromones in streptococcal competence for genetic transformation, p. 9-46. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 16.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hui, F. M., and D. A. Morrison. 1991. Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J. Bacteriol. 173:372-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hui, F. M., L. Zhou, and D. A. Morrison. 1995. Competence for genetic transformation in Streptococcus pneumoniae: organization of a regulatory locus with homology to two lactococcin A secretion genes. Gene 153:25-31. [DOI] [PubMed] [Google Scholar]
- 19.Lacks, S. 1979. Uptake of circular deoxyribonucleic acid and mechanism of deoxyribonucleic acid transport in genetic transformation of Streptococcus pneumoniae. J. Bacteriol. 138:404-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lacks, S. 2004. Transformation, p. 89-115. In E. I. Tuomanen, T. J. Mitchell, D. A. Morrison, and B. G. Spratt (ed.), The pneumococcus. ASM press, Washington, D.C.
- 21.Lacks, S., B. Greenberg, and M. Neuberger. 1974. Role of a deoxyribonuclease in the genetic transformation of Diplococcus pneumoniae. Proc. Natl. Acad. Sci. USA 71:2305-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacks, S., B. Greenberg, and M. Neuberger. 1975. Identification of a deoxyribonuclease implicated in genetic transformation of Diplococcus pneumoniae. J. Bacteriol. 123:222-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacks, S., and B. Greenberg. 1976. Single-strand breakage on binding of DNA to cells in the genetic transformation of Diplococcus pneumoniae. J. Mol. Biol. 101:255-275. [DOI] [PubMed] [Google Scholar]
- 24.Lacks, S. A. 1999. DNA uptake by transformable bacteria, p. 136-168. In J. K. Broome-Smith, S. Baumberg, C. J. Stirling, and F. B. Ward (ed.), Transport of molecules across microbial membranes. Cambridge University Press, Cambridge, United Kingdom.
- 25.Lacks, S. A., S. Ayalew, A. G. de la Campa, and B. Greenberg. 2000. Regulation of competence for genetic transformation in Streptococcus pneumoniae: expression of dpnA, a late competence gene encoding a DNA methyltransferase of the DpnII restriction system. Mol. Microbiol. 35:1089-1098. [DOI] [PubMed] [Google Scholar]
- 26.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 27.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo, P., H. Li, and D. A. Morrison. 2003. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol. Microbiol. 50:623-633. [DOI] [PubMed] [Google Scholar]
- 30.Luo, P., and D. A. Morrison. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185:349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, B., M. Prudhomme, G. Alloing, C. Granadel, and J. P. Claverys. 2000. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol. Microbiol. 38:867-878. [DOI] [PubMed] [Google Scholar]
- 32.Martin, B., G. J. Sharples, O. Humbert, R. G. Lloyd, and J. P. Claverys. 1996. The mmsA locus of Streptococcus pneumoniae encodes a RecG-like protein involved in DNA repair and in three-strand recombination. Mol. Microbiol. 19:1035-1045. [DOI] [PubMed] [Google Scholar]
- 33.Méjean, V., and J. P. Claverys. 1984. Use of a cloned DNA fragment to analyze the fate of donor DNA in transformation of Streptococcus pneumoniae. J. Bacteriol. 158:1175-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Méjean, V., and J. P. Claverys. 1988. Polarity of DNA entry in transformation of Streptococcus pneumoniae. Mol. Gen. Genet. 213:444-448. [DOI] [PubMed] [Google Scholar]
- 35.Morrison, D. A., and M. F. Baker. 1979. Competence for genetic transformation in pneumococcus depends on synthesis of a small set of proteins. Nature 282:215-217. [DOI] [PubMed] [Google Scholar]
- 36.Morrison, D. A., M. Baker, and B. Mannarelli. 1979. A protein component of the pneumococcal eclipse complex, p. 43-52. In S. W. Glover and L. O. Butler (ed.), Transformation 1978. Cotswold Press Ltd., Oxford, United Kingdom.
- 37.Morrison, D. A., and W. R. Guild. 1973. Breakage prior to entry of donor DNA in Pneumococcus transformation. Biochim. Biophys. Acta 299:545-556. [DOI] [PubMed] [Google Scholar]
- 38.Morrison, D. A., S. A. Lacks, W. R. Guild, and J. M. Hageman. 1983. Isolation and characterization of three new classes of transformation-deficient mutants of Streptococcus pneumoniae that are defective in DNA transport and genetic recombination. J. Bacteriol. 156:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrison, D. A., and B. Mannarelli. 1979. Transformation in pneumococcus: nuclease resistance of deoxyribonucleic acid in the eclipse complex. J. Bacteriol. 140:655-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morrison, D. A., M. C. Trombe, M. K. Hayden, G. A. Waszak, and J. D. Chen. 1984. Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAMβ1. J. Bacteriol. 159:870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mortier-Barrière, I., A. de Saizieu, J. P. Claverys, and B. Martin. 1998. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol. Microbiol. 27:159-170. [DOI] [PubMed] [Google Scholar]
- 42.Nardi, M., P. Renault, and V. Monnet. 1997. Duplication of the pepF gene and shuffling of DNA fragments on the lactose plasmid of Lactococcus lactis. J. Bacteriol. 179:4164-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pestova, E. V., L. S. Håvarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 44.Pestova, E. V., and D. A. Morrison. 1998. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J. Bacteriol. 180:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peterson, S., R. T. Cline, H. Tettelin, V. Sharov, and D. A. Morrison. 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol. 182:6192-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051-1070. [DOI] [PubMed] [Google Scholar]
- 47.Puyet, A., B. Greenberg, and S. A. Lacks. 1990. Genetic and structural characterization of endA. A membrane-bound nuclease required for transformation of Streptococcus pneumoniae. J. Mol. Biol. 213:727-738. [DOI] [PubMed] [Google Scholar]
- 48.Rimini, R., B. Jansson, G. Feger, T. C. Roberts, M. de Francesco, A. Gozzi, F. Faggioni, E. Domenici, D. M. Wallace, N. Frandsen, and A. Polissi. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 36:1279-1292. [DOI] [PubMed] [Google Scholar]
- 49.Ringli, C., and B. Keller. 1998. Specific interaction of the tomato bZIP transcription factor VSF-1 with a non-palindromic DNA sequence that controls vascular gene expression. Plant Mol. Biol. 37:977-988. [DOI] [PubMed] [Google Scholar]
- 50.Rosenow, C., M. Maniar, and J. Trias. 1999. Regulation of the alpha-galactosidase activity in Streptococcus pneumoniae: characterization of the raffinose utilization system. Genome Res. 9:1189-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Shoemaker, N. B., and W. R. Guild. 1972. Kinetics of integration of transforming DNA in pneumococcus. Proc. Natl. Acad. Sci. USA 69:3331-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sung, C. K., H. Li, J. P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sung, C. K., and D. A. Morrison. 2005. Two distinct functions of ComW in stabilization and activation of the alternative sigma factor ComX in Streptococcus pneumoniae. J. Bacteriol. 187:3052-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomasz, A., and R. D. Hotchkiss. 1964. Regulation of the transformability of pneumococcal cultures by macromolecular cell products. Proc. Natl. Acad. Sci. USA 51:480-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Torres-Schumann, S., C. Ringli, D. Heierli, N. Amrhein, and B. Keller. 1996. In vitro binding of the tomato bZIP transcriptional activator VSF-1 to a regulatory element that controls xylem-specific gene expression. Plant J. 9:283-296. [DOI] [PubMed] [Google Scholar]
- 57.Unniraman, S., R. Prakash, and V. Nagaraja. 2002. Conserved economics of transcription termination in eubacteria. Nucleic Acids Res. 30:675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vijayakumar, M. N., and D. A. Morrison. 1983. Fate of DNA in eclipse complex during genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 156:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ween, O., P. Gaustad, and L. S. Håvarstein. 1999. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33:817-827. [DOI] [PubMed] [Google Scholar]