Abstract
The cytoplasmic membrane protein DsbD keeps the periplasmic disulfide isomerase DsbC reduced, using the cytoplasmic reducing power of thioredoxin. DsbD contains three domains, each containing two reactive cysteines. One membrane-embedded domain, DsbDβ, transfers electrons from thioredoxin to the carboxy-terminal thioredoxin-like periplasmic domain DsbDγ. To evaluate the role of conserved amino acid residues in DsbDβ in the electron transfer process, we substituted alanines for each of 19 conserved amino acid residues and assessed the in vivo redox states of DsbC and DsbD. The mutant DsbDs of 11 mutants which caused defects in DsbC reduction showed relatively oxidized redox states. To analyze the redox state of each DsbD domain, we constructed a thrombin-cleavable DsbD (DsbDTH) from which we could generate all three domains as separate polypeptide chains by thrombin treatment in vitro. We divided the mutants with strong defects into two classes. The first mutant class consists of mutant DsbDβ proteins that cannot receive electrons from cytoplasmic thioredoxin, resulting in a DsbD that has all six of its cysteines disulfide bonded. The second mutant class represents proteins in which the transfer of electrons from DsbDβ to DsbDγ appears to be blocked. This class includes the mutant with the most clear-cut defect, P284A. We relate the properties of the mutants to the positions of the amino acids in the structure of DsbD and discuss mechanisms that would interfere with the electron transfer process.
Extracytoplasmic compartments, including the external milieu, are oxidative environments in both eukaryotic and prokaryotic cells. Many proteins in these environments contain disulfide bonds as stable parts of their structures. The formation of structural disulfide bonds depends on enzymes present in certain of the extracytoplasmic compartments. In the periplasm of Escherichia coli, the enzyme DsbA acts as an oxidant to promote disulfide bond formation in secreted or membrane proteins via a thiol-disulfide exchange reaction (1). However, the formation of disulfide bonds in protein substrates by DsbA is not perfect, and occasionally nonnative pairs of disulfide bonds are introduced into target proteins. This “misoxidation” occurs most commonly in those proteins which in their native states contain disulfide bonds between cysteines that follow nonsequentially in the amino acid sequence (3). Thus, bacteria (and eukaryotes as well) require a second enzyme, a periplasmic protein disulfide isomerase, which can correct nonnative disulfide bonds in target proteins. In E. coli, this enzyme is DsbC (15, 22, 30).
DsbC, which also exhibits chaperone activity (4), is thought to recognize proteins that contain nonnative disulfide bonds because they are misfolded. These bonds are attacked by one of the active-site cysteines of DsbC, presumably leading to an intermediate, mixed disulfide between DsbC and the substrate. The rearrangement of disulfide bonds in the substrate can occur either by resolution of the mixed disulfide, involving attack of another cysteine located in the substrate, or by reduction of the incorrect disulfide bond and further interaction with DsbA (29). The latter process would result in the oxidation of DsbC. The reduced state of DsbC is necessary for its activity. Its cysteines are maintained in the reduced state by the membrane protein DsbD, which transfers electrons across the cytoplasmic membrane from cytoplasmic thioredoxin-1 (Trx1) (5, 25). Both DsbA and DsbC, along with one of the periplasmic domains of DsbD, are members of the thioredoxin family of proteins.
DsbD is composed of three domains, each containing two reactive cysteines. The α domain has an immunoglobulin-like fold; the β domain is composed of eight transmembrane (TM) segments; and the γ domain exhibits a thioredoxin-like structure. The electron transfer pathway from Trx1 to DsbC through DsbD (β→γ→α domain) was determined by detecting the mixed disulfide complexes, Trx1-DsbDβ and DsbDα-DsbC, and by assessing the oxidation state of each domain when one of the other domains was missing (11). Our studies showed that in the absence of electron transfer from thioredoxin the DsbDβ domain is found with its two cysteines joined in a disulfide bond (11, 12). Thus, DsbD presents the first case of a protein that uses redox-active cysteines to transfer electrons across a membrane and that (at one stage of its activity) contains a disulfide bond in a membrane-embedded domain.
DsbD also contributes indirectly to the reduction of apocytochrome c, which is necessary for its incorporation of a heme. The direct substrate of DsbD in the cytochrome c assembly pathway is CcmG, which is a periplasmic protein containing a thioredoxin fold tethered to the cytoplasmic membrane (2). Bioinformatic analysis has, so far, revealed three classes of DsbD-like proteins in nature: the E. coli type, COG4233-containing DsbD, and CcdA. The first two classes of proteins have two periplasmic domains in addition to the TM domain and differ mainly in the nature of their amino-terminal periplasmic domain. CcdA is a homologue of DsbDβ that in most instances has six TM segments rather than the eight TM segments found in the other two classes (13, 17).
A central question in the study of DsbD function is how its central membrane-embedded domain (DsbDβ) transfers electrons via its two cysteines across the cytoplasmic membrane and how these cysteines are able to interact with thioredoxin-like molecules on both sides of the membrane. We have shown that the two cysteines of DsbDβ can form a disulfide bond which is accessible for reduction by thioredoxin in the cytoplasm (11). When the disulfide bond is reduced, at least one of the cysteines is still accessible to oxidized thioredoxin and both are accessible to a high-molecular-weight (5-kDa) alkylating agent from the cytoplasmic side, not the periplasmic side (12). Interestingly, the two reactive cysteines are accessible from the periplasmic side to a low-molecular-weight (0.5-kDa) membrane-impermeable alkylating agent (S.-H. Cho, A. Porat, and J. Beckwith, unpublished results). The remaining and important questions to be resolved are the following. Which amino acid residues in addition to the two cysteines are important for the functioning of DsbDβ? How do the mutations affect the functionality? How are the electrons transferred to the β domain from thioredoxin and then from the β domain to the periplasmic domain? Does DsbD undergo significant conformational changes that allow the two cysteines to interact alternately with cytoplasmic and periplasmic proteins? Is there a channel in the membrane that allows these interactions?
In this paper, we begin to address the first two questions by changing each of 19 conserved amino acids of DsbD to alanine and observing the in vivo redox states of DsbC and DsbD. Some of the mutations result in defects in DsbC reduction, showing the protein in relatively oxidized redox states compared to that of the wild type. Strikingly, one of the mutations, P284A, alters DsbD such that, while the β domain remains largely in the reduced state, the α and γ domains are oxidized, in contrast to their states in a wild-type strain. We show that P284A DsbDβ still interacts with cytoplasmic thioredoxin. Our results suggest that this proline is important for the formation of the disulfide bond in the β domain or for the β and γ domains to interact efficiently. For three other mutants (P162A, P289A, and G316A), it appears that either a change of redox potential or reduced affinity of the β domain for the γ domain is caused by these mutations.
MATERIALS AND METHODS
Strains and media.
The strains used in this study are listed in Table 1. E. coli cells were grown in NZ medium (18) at 37°C. When needed, 200 μg/ml of ampicillin and 100 μg/ml of spectinomycin were added. Strains were grown overnight, diluted to a ratio of 1:100 in 5 ml NZ medium, and grown to an optical density at 600 nm of 0.1. Expression of the genes was induced for 1 h by adding appropriate concentrations of isopropyl-β-d-thiogalactopyranoside (IPTG) when using pAM238 or by adding 0.2% l-arabinose when using pBAD18.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or feature | Source or reference |
|---|---|---|
| Strain | ||
| MC1000 | araD139(araABC-leu)7679 galU galK Δ(lac)X74 rpsL thi | Laboratory collection |
| FED126 | MC1000 ΔdsbD | 25 |
| FED513 | FED126 trxB::Kan | 11 |
| FED163 | FED126 ΔtrxA trxC::Tn10 | F. Katzen, unpublished data |
| FED184 | FED163 trxB::Kan | F. Katzen, unpublished |
| SEN44 | MC1000 F′ lacIqZ+Y+ | This study |
| SEN45 | FED126 F′ lacIqZ+Y+ | This study |
| Plasmid | ||
| pAM238 | lac promoter, pSC101-based, spectinomycin | 6 |
| pAD18 | Arabinose regulator, pBR-based, ampicillin | 8 |
| pSC01 | pAM238 with DsbD | This study |
| pSC01-1 | pSC01G155A | This study |
| pSC01-2 | pSC01P162A | This study |
| pSC01-3 | pSC01P166A | This study |
| pSC01-4 | pSC01P169A | This study |
| pSC01-5 | pSC01Y194A | This study |
| pSC01-6 | pSC01M198A | This study |
| pSC01-7 | pSC01Y202A | This study |
| pSC01-8 | pSC01G206A | This study |
| pSC01-9 | pSC01G275A | This study |
| pSC01-10 | pSC01P284A | This study |
| pSC01-11 | pSC01P289A | This study |
| pSC01-12 | pSC01L290A | This study |
| pSC01-13 | pSC01L294A | This study |
| pSC01-14 | pSC01G316A | This study |
| pSC01-15 | pSC01P320A | This study |
| pSC01-16 | pSC01G336A | This study |
| pSC01-17 | pSC01W338A | This study |
| pSC01-18 | pSC01M339A | This study |
| pSC01-19 | pSC01G347A | This study |
| pSC49 | pAM238 with DsbD derivative containing thrombin cleavage site and c-Myc epitope (DsbDTH) | This study |
| pSC49-10 | pSC49P284A | This study |
| pSC51 | pBAD18 with DsbDTH | This study |
| pSC51-2 | pSC51P162A | This study |
| pSC51-10 | pSC51P284A | This study |
| pSC51-11 | pSC51P289A | This study |
| pSC51-14 | pSC51G316A | This study |
| pAP06 | pBAD18 with DsbDss-3HA-DsbDβ-c-Myc | A. Porat, unpublished data |
DNA manipulation.
Standard techniques were used for cloning, analysis of DNA, and PCR (20). All the dsbD sequences in the plasmids in this paper were verified by DNA sequencing by the Micro Core facility of the Department of Microbiology and Molecular Genetics, Harvard Medical School. pFK093 containing intact dsbD was digested with KpnI and PstI to obtain the DNA to be inserted (11). To construct pSC01, this DNA was ligated with the vector pAM238, previously digested with the same enzymes. The primers for site-directed mutagenesis for the alanine substitution were generated using a standard method, and the template was the pSC01 plasmid. Site-directed mutagenesis was performed using Pfu Turbo (Stratagene) according to the manufacturer's protocol. For insertion of the thrombin cleavage sequence and c-Myc epitope, mutagenesis was performed in the same way. The DsbD sequences of 128-Ala-Pro-Gln-Pro-Val-Ser-133 and 444-Leu-Val-Glu-Ala-447 were replaced by the sequence Leu-Val-Pro-Arg-Gly-Ser, with the thrombin cleavage sequences inserted between the domains (α to β and β to γ). These changes were accomplished by mutagenesis, using pSC01 as a template and the primers DsbD-th-8 (5′-GAAGTGGTCGCCAACAACGCACTGGTTCCGCGTGGATCCGTTCCGCAGCAAGAGCAGCCC-3′, a forward primer) and DsbD-th-6 (5′-GTTAAATCAGGCGCTCGTTCCGCGTGGATCCAAAGGCAAACCGGTGATG-3′, a forward primer), respectively. The Lys-Leu-Ile-Ser-Glu-Glu-Asp-Leu sequence was inserted after the Gln in the DsbD sequence of 138-Glu-Gln-Pro-140, which resulted in expression of the c-Myc epitope after the thrombin cleavage sequence between the α and β domains. This change was accomplished by mutagenesis, using the primer DsbD-139-Myc (5′- GTTCCGCAGCAAGAGCAGAAACTGATTTCTGAAGAGGATCTGCCCACCGCGCAATTGCCC-3′, a forward primer) after constructing pSC01, which contains the nucleotide sequence that encodes the thrombin cleavage sites. This construct was designated as pSC49. pSC49-10 was constructed by site-directed mutagenesis, using pSC49 as a template. To move these constructs into pBAD18, pSC49 and pSC49-10 were digested with PstI and KpnI to obtain the DNAs to be inserted. These DNAs were ligated with the vector pBAD18, previously digested with the same enzymes. Those constructs were designated as pSC51 and pSC51-10, respectively. From pSC51 as a template, pSC51-2, pSC51-11, and pSC51-14 were constructed by site-directed mutagenesis. The pAP06 plasmid encodes the DsbDβ domain, which has a signal sequence of DsbD and the three-hemagglutinin (HA) epitope in its N terminus and the c-Myc epitope in its C terminus. It was kindly donated by Amir Porat.
The above insertion sites for the thrombin cleavage sequence and the c-Myc epitope were selected from among several sites that we tested. According to the analysis of sequence alignment of E. coli type DsbD, the linking region between the α and β domains is not conserved and is relatively long to modify and accommodate some sequences, but the linking region between the β and γ domains is shorter than that between the α and β domains. Each site we inserted between the α and β domains was very efficiently cleaved by thrombin, whereas the efficiency of cleavage between the β and γ domains was variable no matter where the sequence was inserted. However, the site described above that is located in the loop region between helix α1 and beta sheet β2 in the N-terminal portion of the γ domain was efficiently cleaved (14; data not shown).
Sequence analyses.
The E. coli type DsbD sequences, which were retrieved from the sequences used in our previous paper (13), were used to select the conserved amino acid residues. The selection was done using Clustal X to perform sequence alignment (26), using the default parameters. To compare the sequences with those of CcdA and DsbD containing COG4233 (17), the sequence alignment was performed in the same way, using Clustal X. Four strains in each group were chosen randomly from among those that had evolved quite distantly (13). Membrane topology was predicted using HMMTOP (http://www.enzim.hu/hmmtop/), a membrane protein topology prediction program (28). The predicted topology was visualized using TMRPres2D (http://bioinformatics.biol.uoa.gr/TMRPres2D/), which automates the creation of graphical images and/or models of TM proteins (23).
Antibodies.
Anti-c-Myc (A-14) rabbit polyclonal antiserum was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.). The anti-DsbDγ, anti-DsbC, and anti-DsbDα antibodies were previously described (10, 11, 25).
Thiol-redox state analyses.
To determine the in vivo redox states of the proteins, free thiols were acid trapped by trichloroacetic acid (TCA) and alkylated with the high-molecular-mass reagent 4-acetamido-4′maleimidylstilbene-2,2′-disulfonic acid (AMS) (Molecular Probes) as previously described (25). When indicated, dithiothreitol (DTT) was added to the cells at a final concentration of 50 mM before AMS alkylation.
Redox analysis of each domain in DsbDTH.
To digest the proteins at the thrombin cleavage sites, 1 ml of both nonreduced and reduced AMS-alkylated cell extracts were prepared in the same way as described in the preceding paragraph. TCA was added to the samples again to a final concentration of 10%, and the mixture was washed twice with acetone. These second extracts were solubilized in 100 μl of thrombin cleavage buffer containing 50 mM Tris-HCl (pH 8.4), 150 mM NaCl, 2.5 mM CaCl2, 10% glycerol, and 0.1% n-dodecyl-β-d-maltoside (Calbiochem) and were subjected to sonication. Bovine thrombin (10 U; Amersham) was added, and the samples were incubated overnight at room temperature. After centrifugation, sodium dodecyl sulfate (SDS) was added to the supernatants at a final concentration of 2%, and the supernatants were subjected to Western blotting, using the appropriate antibodies. For comparison, 10 mM DTT was added in the same samples before loading onto the gel.
RESULTS
In vivo redox states of DsbC and DsbD in the strains expressing 19 mutant DsbDs.
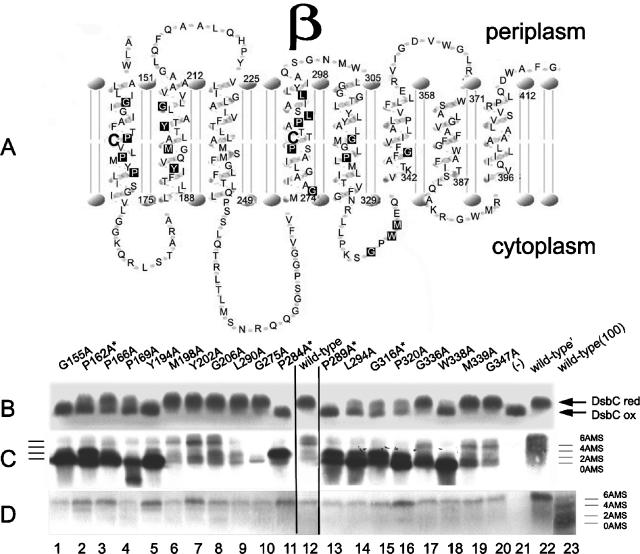
To select the conserved amino acids in DsbDβ to be changed, we compared the sequence from E. coli with those from other organisms containing E. coli type DsbD, using the alignment program Clustal X. Comparison of the sequences led to the selection of 19 amino acid residues to be changed. We performed the analyses again, using all three classes of DsbD-like proteins, including COG4233-containing DsbD and CcdA proteins (Fig. 1). Gly155, Pro162, Pro166, Gly275, Pro284, Leu290, Leu294, Gly316, Pro320, and Gly347 are conserved among most of the three types. Tyr194, Met198, Tyr202, Gly206, and Pro289 are conserved only among the E. coli types. In the case of Pro289, the non-E. coli DsbD types contain a proline that is shifted by one amino acid residue toward the N terminus. Gly336, Trp338, and Met339 are not conserved among CcdA homologues. Pro169 is not conserved among the COG4233 proteins.
FIG. 1.
Sequence alignments of membrane-embedded domains of CcdA and DsbD. The strains having CcdA, DsbD containing COG4233, or DsbD are denoted with a superscript a, b, or c, respectively. The residue number of E. coli DsbD was calculated from the mature DsbD. Rho_cap represents Rhodobacter capsulatus; the accession number of CcdA is AAF26218, available from the ERGO-Light Database of Integrated Genomics (http://www.ergo-light.com/). Str_coe represents Streptomyces coelicolor A3(2); the DDBJ/EMBL/GenBank accession number is NP_628639. Oce_ihe represents Oceanobacillus iheyensis HTE831, accession number NP_692598; Met_maz represents Methanosarcina mazei Goe1, accession number NP_634265; Glo_vio represents Gloeobacter violaceus, accession number NP_926909; Cau_cre represents Caulobacter crescentus CB15, accession number NP_419036; Chl_pne represents Chlamydophila pneumoniae TW-183, accession number NP_877086; Sal_ent represents Salmonella enterica subsp. enterica serovar Typhimurium, accession number NP_455609; Esc_col represents Escherichia coli K12, accession number NP_418559; Nit_eur represents Nitrosomonas europaea, accession numbers ATCC 19718 and NP_842384; Pse_aer represents Pseudomonas aeruginosa PAO1, accession number AAG05866; and Cam_jej represents Campylobacter jejuni subsp. jejuni, accession numbers NCTC 11168 and NP_281786.
The positions of these amino acid residues in TM segments were predicted using the HMMTOP (http://www.enzim.hu/hmmtop/) membrane protein topology prediction program, as shown in Fig. 2A. The topology is roughly similar to that proposed previously based on alkaline phosphatase fusions (25). The first group of mutations described above is located mainly in TM segments 1, 4, and 5, the second group mainly in TM segment 2, and the last group in the loop region between TM segments 5 and 6. We replaced each of these residues with alanine, using site-directed mutagenesis. Alanine-scanning mutagenesis is used because alanine generally causes little structural change, due to its small size and because it has no charge.
FIG. 2.
Membrane topology of the membrane-embedded domain (β) of DsbD and in vivo mutational effects. (A) The topology of DsbDβ was predicted using HMMTOP. The two reactive cysteines are represented by the large boldface letters, and the conserved amino acid residues, which are substituted with alanines, are represented as letters inside black rectangles. The residue numbers represent the start or end of the TM helixes. (B and C) The in vivo redox states of DsbC (B) and DsbD (C) in the strains expressing 19 mutant DsbDs. Expression of the genes was induced by 20 μM IPTG, harvested at mid-log phase, precipitated with TCA, and subjected to AMS alkylation. Proteins were separated by SDS-PAGE and visualized by Western blotting, using antibodies against DsbC (B) and DsbDα (C). DsbC and DsbD that are reduced become alkylated and display a higher molecular weight than the oxidized forms. (D) Reduced redox states of DsbD in the strains expressing 19 mutant DsbDs. Expression of the genes was induced by 20 μM IPTG, harvested at mid-log phase, reduced by 50 mM DTT, precipitated with TCA, and subjected to AMS alkylation. Proteins were separated by SDS-PAGE and visualized by Western blotting, using an antibody against DsbDα. For lane 23 [wild-type (100)], expression of the genes was induced with 100 μM IPTG and processed in the same way except that there was no reduction with DTT. Wild-type′ is the strain which retains the chromosomal dsbD gene. The strain background used was SEN45 (the dsbD mutant) in all lanes except lane 22 (SEN44). The following plasmids were used: pSC01-1 (G155A, lane 1), pSC01-2 (P162A, lane 2), pSC01-3 (P166A, lane 3), pSC01-4 (P169A, lane 4), pSC01-5 (Y194A, lane 5), pSC01-6 (M198A, lane 6), pSC01-7 (Y202A, lane 7), pSC01-8 (G206A, lane 8), pSC01-12 (L290A, lane 9), pSC01-9 (G275A, lane 10), pSC01-10 (P284A, lane 11), pSC01-11 (P289A, lane 13), pSC01-13 (L294A, lane 14), pSC01-14 (G316A, lane 15), pSC01-15 (P320A, lane 16), pSC01-16 (G336A, lane 17), pSC01-17 (W338A, lane 18), pSC01-18 (M339A, lane 19), pSC01-19 (G347A, lane 20), pAM238 [(−), lane 21, and wild-type′, lane 22], and pSC01 (wild-type, lane 12, and wild-type (100), lane 23). P162A, P284A, P289A, and G316A are marked by superscript stars because they were analyzed further for Fig. 5.
To assess the effects of each mutation on the ability of DsbD to act as a reductant of DsbC, we first determined the in vivo redox states of the active-site cysteines of DsbC. These cysteines are in the reduced state in the strains expressing DsbD and in the oxidized state in the absence of DsbD (19). The oxidation of DsbC is believed to be due to its activity as a reductase of substrates misoxidized by DsbA (29). We were concerned that high expression levels of the mutant DsbD proteins might obscure the effects of the mutations and allow reduction of DsbC by the newly synthesized reduced DsbD. Accordingly, we used a low-copy plasmid expressing DsbD from the lac promoter and induced expression to levels comparable to that for wild-type chromosomally located DsbD, using appropriate concentrations of IPTG.
DsbC is a homodimeric protein in which each monomer contains four cysteines, two of them its redox-active cysteines and the other two forming a structural disulfide bond. Only the disulfide bond formed by the reactive cysteines can be reduced by DsbD (30). We assessed the redox states of DsbC, using the alkylating agent AMS. AMS causes a 0.5-kDa molecular mass shift of the protein bands during SDS-polyacrylamide gel electrophoresis (PAGE) when it alkylates a cysteine in a protein. Figure 2B shows the in vivo redox states of DsbC determined by AMS alkylation for 19 mutants. The absence (Fig. 2B, lane 21) or presence (Fig. 2B, lanes 12 and 22) of wild-type DsbD expressed from a plasmid or chromosome resulted in accumulation of oxidized or reduced DsbC, respectively. Expression of the G155A, Y194A, P284A, P289A, and W338A DsbDs caused a strong defect in DsbC reduction (Fig. 2B, lanes 1, 5, 11, 13, and 18). We observed a partial defect in DsbC reduction in strains expressing P162A, P166A, P169A, L294A, G316A, and P320A DsbDs (Fig. 2B, lanes 2 to 4 and 14 to 16). The other mutant DsbDs behaved nearly like wild-type DsbD, showing DsbC mostly in the reduced state (Fig. 2B, lanes 6 to 10, 17, 19, and 20).
We also used AMS alkylation to determine the redox state of the mutant DsbDs. While such analysis can indicate the overall oxidation state of DsbD, it does not directly provide information on the redox state of each of the three domains of the protein. Since all three domains have two cysteines, we cannot distinguish which of the domains are oxidized or reduced, except in the case of a fully oxidized or fully reduced protein. Nevertheless, we can determine how many cysteines of the protein can be alkylated by AMS. This analysis could have been complicated by the fact that the wild-type DsbD expressed from the chromosome has eight cysteines, one in the signal sequence that does not appear in the mature protein and one, not essential for DsbD activity, at amino acid residue 282 in the mature sequence (25). We replaced these cysteines with alanines in the plasmid-expressed wild-type and all mutant DsbDs used in the following experiments. If DsbD is expressed from the chromosome, it will always be so indicated in the following section. Since the chromosomal copy of the gene in one of our control strains still contains Cys282, it shows an apparently lower mobility of the ladder of bands in lane 22 (chromosomal DsbD) compared with those expressed from the plasmids in lane 12 of Fig. 2C.
We determined the in vivo redox states of DsbD from the 19 mutants, using AMS alkylation (Fig. 2C). Western blotting was performed, using an antibody against the DsbDα domain. We note here and discuss below the peculiar behavior of this antibody, which appears to react much more effectively with oxidized DsbDα than with its reduced form (11). This property of the antibody results in much fainter bands of reduced than of oxidized DsbD on Western blots. Thus, in considering the data presented in Fig. 2C, it should be noted that any mutant accumulating significant amounts of DsbD with its α domain oxidized will show more-intense bands on Western blots.
Expression of wild-type DsbD from either a plasmid or the chromosome revealed a ladder of several bands (Fig. 2C, lanes 12 and 22) which were not present in cells lacking DsbD (Fig. 2C, lane 21). The M198A, Y202A, G206A, L290A, G275A, G336A, M339A, and G347A DsbDs (Fig. 2C, lanes 6 to 10, 17, 19, and 20) showed ladders of bands comparable in intensity to that of the wild-type DsbD. Thus, these mutants show no defects in electron transfer through the DsbD-DsbC pathway. In contrast, the G155A, P166A, P169A, Y194A, and W338A DsbDs (Fig. 2C, lanes 1, 3 to 5, and 18) showed much more intense bands than that of the wild type, in each case corresponding to the lowest band (0 AMS). This band corresponds to fully oxidized DsbD (see below). With P169A DsbD, there was a significant amount of a band that may be a degradation product. The remainder of the mutant DsbDs (Fig. 2C, lanes 2, 11, and 13 to 16) also showed much more intense bands than that of the wild-type, corresponding either to the second-lowest band (2 AMS) or to a mix of the lowest and second-lowest bands (0 and 2 AMS).
The mutant DsbDs showing a different pattern from wild-type DsbD must be present in a fully or partially oxidized state, since they appear to express species that have not been alkylated at all or alkylated in only one domain. The properties of these mutant DsbDs are consistent with the finding that these same mutants are the ones that cause strong or partial defects in DsbC reduction (Fig. 2B and C, lanes 1 to 5, 11, 13 to 16, and 18). The five fully oxidized mutant DsbDs appear to have a defect in receiving electrons from thioredoxin, as this is the first step in which electrons are transferred to DsbD (Fig. 2C, lanes 1, 3 to 5, and 18). The remainder appear to have a defect in transferring electrons between the DsbDβ and DsbDγ domains. Perhaps the most interesting of these mutants, because it has the strongest effects, is P284A, as this mutant DsbD protein is trapped in a band (2 AMS) corresponding to a protein with only two of its six cysteines reduced (Fig. 2C, lane 11).
As noted above, we observed a significantly higher band intensity for the mutant DsbDs accumulating in the most-oxidized states (Fig. 2C, lanes 1 to 5, 11, and 13 to 18) compared to those accumulating the more-reduced forms of the proteins (Fig. 2C, lanes 6 to 10, 19, and 20). This difference could be due to expression levels or to decreased binding affinity of the anti-DsbDα antibody for the reduced α domain, at least in its alkylated form. If the latter were the case, reduction of the more-oxidized proteins should show an apparent reduction in amounts of the protein on Western blots, as assessed with anti-DsbDα antibody. To estimate the expression level of each mutant protein as well as to confirm that the most-shifted bands during SDS-PAGE were in the most-reduced state, we reduced extracts with DTT and then treated them with AMS (Fig. 2D). The overall band intensity was substantially lowered by DTT in all of the samples showing relatively oxidized states of DsbD (Fig. 2D, lanes 1 to 5, 11, and 13 to 18), such that there now appeared to be no difference in the “expression” levels of the mutant DsbDs. We conclude that the main reason for the difference in band intensity between the mutant DsbDs in Fig. 2C is the decreased affinity of the anti-DsbDα antibody for the reduced α domain compared to that for the oxidized form of this domain. Although P162A, P166A, Y194A, Y202A, P284A, and P320A DsbDs (Fig. 2D, lanes 2, 3, 5, 7, 11, and 16) showed somewhat higher expression levels compared with the wild-type DsbD and other mutant DsbDs, only the Y202A mutant protein is able to reduce DsbC to a level comparable to that of the wild-type DsbD. The uppermost band (6 AMS) from the nonreduced wild-type sample (Fig. 2D, lane 23) coincided with the reduced and alkylated bands from the wild-type and mutant DsbDs.
A useful version of DsbD containing two thrombin cleavage sites and a c-Myc epitope sequence is functional.
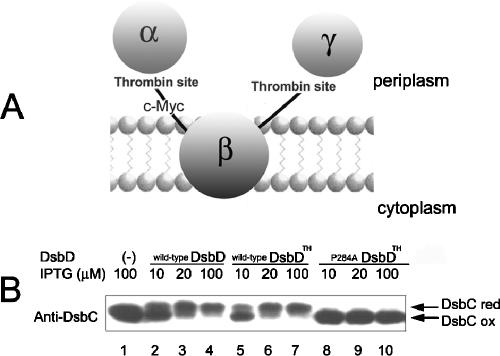
We have presumed that the fastest-moving bands (0 AMS) from the mutant DsbDs in Fig. 2C represent DsbDs in which all three domains are oxidized and can therefore not be alkylated. In addition, according to the above interpretation, we suspected that the second-lowest band (2 AMS) from the mutant DsbDs showing relatively oxidized states might contain the α domain as one of its reduced domains (Fig. 2C, lanes 2, 11, and 13 to 16). We wished to confirm these deductions and, in general, determine the redox state of each domain of the mutant DsbDs. To do this, we constructed a derivative of the dsbD gene that allowed us to separate the three domains. This derivative contains two thrombin cleavage sites, one between the α and β domains and the other between the β and γ domains. In addition, the protein contains a c-Myc epitope sequence after the thrombin cleavage site between the α and β domains to allow detection of the β domain (Fig. 3A). This construct allows us to release all three domains of DsbD by thrombin cleavage and to detect the DsbDβ domain with an antibody against c-Myc. (We have antibodies against the DsbDα and γ domains.) We have given this useful construct of DsbD the name DsbDTH (TH = thrombin).
FIG. 3.
Verification of the function of DsbDTH. (A) Scheme showing DsbDTH, DsbD containing the thrombin cleavage site between the domains, and the c-Myc epitope after the thrombin cleavage site between the α and β domains. (B) The in vivo redox states of DsbC were assessed in the strains expressing wild-type DsbD, wild-type DsbDTH, and P284A DsbDTH using AMS alkylation. In lane 1, SEN45 (the dsbD mutant) containing only the vector (pAM238) was used. In the other lanes, the cells express wild-type DsbD (pSC01, lanes 2 to 4), wild-type DsbDTH (pSC49, lanes 5 to 7), and P284A DsbDTH (pSC49-10, lanes 8 to 10).
To verify the functionality of DsbDTH, we assessed the in vivo redox states of DsbC, using wild-type DsbD, wild-type DsbDTH, and P284A DsbDTH. Expression of wild-type DsbD and wild-type DsbDTH with increasing concentrations of IPTG resulted in increasing reductions of DsbC (Fig. 3B, lanes 2 to 7). In contrast, the expression of P284A DsbDTH in the absence of wild-type DsbD caused a significant defect in DsbC reduction (Fig. 3B, lanes 1 and 8 to 10), thus behaving like the original P284A DsbD. These results indicate that DsbDTH is functional and behaves like normal DsbD.
Thioredoxins are the main and direct oxidants of DsbDβ in a thioredoxin reductase-deficient strain.
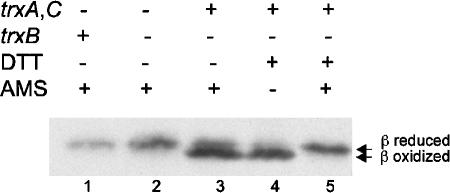
In subsequent sections, we used an approach that allowed us to assess the ability of thioredoxin and the DsbDβ domain to interact. This was done by studying the properties of DsbDβ in a strain missing thioredoxin reductase (trxB). We have shown previously that cytoplasmic thioredoxins can act as oxidants of alkaline phosphatase expressed in the cytoplasm of a trxB mutant (24). We also showed that DsbDβ could be oxidized in a trxB strain and, further, that a mixed disulfide complex between C285A DsbDβ and Trx1 was detected in this background (11). We have considered three explanations for the oxidation of DsbDβ in a trxB strain. First, the absence of reducing potential in the thioredoxin pathway results in failure to reduce DsbDβ that is spontaneously oxidized. Second, oxidized thioredoxins can act positively to promote disulfide bond formation in DsbDβ. Third, other unidentified substrates that can be reduced by thioredoxin reductase could exist and act as the main oxidants in a trxB strain. To distinguish among these possibilities, we expressed DsbDβ in mutants missing Trx1 and Trx2 (trxA and trxC) with or without expression of thioredoxin reductase and assessed the redox states of DsbDβ. DsbDβ was not oxidized in this background regardless of thioredoxin reductase expression (Fig. 4). From these and previous data, we can exclude the first and last possibilities and conclude that the presence of oxidized thioredoxins is required for the oxidation of DsbDβ, a reversal of the normal reaction.
FIG. 4.
In vivo redox state of DsbDβ in thioredoxins or thioredoxin reductase-deficient strain. The samples in lanes 1 to 3 were precipitated with TCA and subjected to AMS alkylation. The samples in lane 4 and 5 were harvested without TCA precipitation, reduced by 50 mM DTT, and precipitated with TCA, followed by AMS alkylation for the sample in lane 5. Western blotting was performed, using an antibody against c-Myc. The strain backgrounds used were FED163 (the trxA, trxC, and dsbD mutant, lane 1), FED184 (the trxA, trxB, trxC, and dsbD mutant, lane 2), and FED513 (the trxB and dsbD mutant, lanes 3 to 5).
In vivo redox state of each domain in wild-type, P284A, P162A, P289A, and G316A DsbDTH.
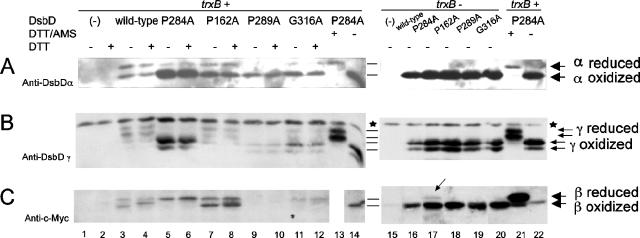
We used the DsbDTH construct described above to further define the defects in electron transfer of the mutants in conserved amino acids of DsbD. We chose those mutants that exhibited the clearest defects, including P284A (the strongest), P162A, P289A, and G316A. For these experiments, we switched to a higher-copy plasmid (pBAD18) for expressing the constructs, since the low expression levels of the plasmid used in the earlier experiments, in addition to the decreased yield after solubilization of the denatured protein in thrombin-cleavage buffer, made it difficult to detect all of the domains after thrombin treatment of extracts. When mutant DsbDTHs are expressed from pBAD18 and the extracts are prepared and treated with thrombin, we can readily detect the three DsbD domains on Western blots. Unexpectedly, both the oxidized and reduced states of the thrombin-released γ domains appeared as two bands each (Fig. 5B). The double band may be due to an additional cleavage near the introduced thrombin site. Nevertheless, the redox states of the γ domains of the wild-type and all the tested mutant DsbDs could clearly be determined (Fig. 5B). This additional cleavage, which might have been thought to result in two molecular-weight versions of the released β domains, did not do so; interpretation of the redox states of the β domains was also straightforward (Fig. 5C).
FIG. 5.
In vivo redox state of each domain in wild-type, P284A, P162A, P289A, and G316A DsbD. Proteins were detected by Western blotting, using antibodies against DsbDα (A), DsbDγ (B), and c-Myc (C). The samples in lanes 1 to 12 and 15 to 20 were precipitated with TCA and subjected to AMS alkylation. The samples in lanes 13, 14, 21, and 22 were harvested without TCA precipitation, subjected to reduction by 50 mM DTT, and precipitated with TCA. The samples in lanes 13 and 21 were subjected to AMS alkylation. The extracts were subjected to thrombin cleavage. The samples in the even-numbered lanes from 1 to 12 were subjected to reduction by 10 mM DTT before being loaded onto the gel. The strain backgrounds used were FED126 (the dsbD mutant, lanes 1 to 14, 21, 22) and FED513 (the dsbD and trxB mutant, lanes 15 to 20). The following plasmids were used: pBAD18 (−, lanes 1, 2, and 15), pSC51 (wild-type, lanes 3, 4, and 16), pSC51-10 (P284A, lanes 5, 6, 13, 14, 17, 21, and 22), pSC51-2 (P162A, lanes 7, 8, and 18), pSC51-11 (P289A, lanes 9, 10, and 19), and pSC51-14 (G316A, lanes 11, 12, and 20). Stars indicate nonspecific bands detected by anti-DsbDγ antibody, and the arrow in lane 17 indicates a small amount of the reduced β domain. The experiments were performed three times, and the results were reproducible.
In the AMS-trapping experiments with full-length DsbD described earlier, the P284A mutant exhibited a band only in the second-lowest position (2 AMS) from the bottom (Fig. 2C, lane 11). After the P284A version of DsbDTH was cleaved with thrombin, the Western blots show that the β domain remained reduced while the α and γ domains remained largely oxidized (Fig. 5, lanes 5 and 6). At the same time, the α and γ domains of wild-type DsbDTH were largely in the reduced state while about half of the β domain was in the oxidized state (Fig. 5, lanes 3 and 4). This conclusion takes into account the higher affinity of anti-DsbDα antibody for the oxidized α domain, as mentioned above. These results show that the “2 AMS” band was the result of alkylation of a reduced β domain of a DsbD that was oxidized in its other two domains.
Thus, P284A exhibits a strong defect in electron transfer from the β domain to the γ domain. We can conceive of three explanations for the defect in this mutant. First, the alteration may result in a conformational change in DsbDβ such that the orientation of or distance between the thiols of the two cysteines disfavors the formation of the disulfide bond in DsbDβ. Then the transfer of electrons from a reduced β domain to DsbDγ would be less likely to occur. Second, the reduced state of the cysteines in the β domain of P284A may be stabilized compared to that of the wild type, reflecting an increase in the redox potential of the β domain, again making the reduction of DsbDγ by DsbDβ less favored. Third, a conformational alteration of the β domain may have reduced its affinity for the γ domain so that it does not interact effectively with the γ domain and thus cannot transfer electrons to it.
We were able to test the first explanation for the defects of the P284A mutation: inability to form a disulfide bond in the mutant β domain. In a trxB strain, wild-type DsbDβ is oxidized by oxidized thioredoxins (Fig. 4). Therefore, we assessed the redox state of the β domain of the P284A DsbD in a trxB strain. We found that the β domain of P284A DsbD accumulates largely in the oxidized form, with a disulfide bond between its two redox-active cysteines (Fig. 5C, lane 17), like the β domain of the wild-type DsbD (Fig. 5C, lane 16). This result indicates that P284A DsbD is still capable of forming a disulfide bond in its β domain, making unlikely the explanation for its properties that invokes a conformational change interfering with the formation of a disulfide bond. Rather, the defect is more likely due either to a higher redox potential of the mutant β domain than that of the wild type or to the lack of interaction of it with the γ domain. We did observe that the P284A DsbDβ domain in the trxB strain has a slight resistance to oxidation compared to the wild-type and other mutant DsbDs (Fig. 5C, lane 17). This incomplete oxidation could be due to lowered affinity of the β domain for thioredoxins or to a higher redox potential of the β domain.
The remaining three mutants, P162A, P289A, and G316A, exhibited two bands in the first- and second-lowest positions (0 and 2 AMS) (Fig. 2C, lanes 2, 13, and 15), indicating an earlier defect in the electron transfer pathway. With P162A DsbDTH, there was an increase in the amounts of oxidized forms of the periplasmic domains compared to those for the wild-type, but the β domain showed redox states similar to those of the wild-type (Fig. 5, lanes 7 and 8). In a trxB strain, where oxidized thioredoxins accumulate, all the domains were in the oxidized states, indicating that the interaction of the β domain with thioredoxins was not impaired (Fig. 5, lane 18). We can conceive of two explanations for the defect in this mutant. First, although the β domains remain in redox states similar to that of the wild type and the impairment of the electron transfer to the periplasmic domains was smaller than that of P284A (Fig. 5A and B, compare lanes 5 and 6 with lanes 7 and 8), a small change of redox potential—either higher or lower—could decrease the efficiency of electron transfer from the β domain to the γ domain. Second, a conformational alteration of the β domain may partially reduce its affinity for the γ domain.
With P289A DsbDTH, there may be degradation of the mutant protein; we could not detect the β domain but did detect small amounts of periplasmic domains which were largely in the oxidized state (Fig. 5, lanes 9 and 10). In a trxB strain, all of the domains exhibited amounts comparable to those of wild-type and other mutant DsbDs and were in the oxidized state. Thus, the interaction of the β domain with thioredoxins was not impaired, and the oxidized form appeared more stable than the reduced one (Fig. 5, lane 19). We suggest similar explanations for a redox potential change or reduced affinity of the β domain for the γ domain.
While the β domain of G316A DsbDTH is largely in the reduced state, it shows defects in its electron transfer to the γ domain (not as significant as the defect of P284A DsbDβ) (Fig. 5, lanes 11 and 12). In a trxB strain, all domains were in the oxidized states (Fig. 5, lane 20). The mutation appears to cause a similar defect to that of P284A. The same explanations we have suggested for P284A could apply to G316A.
The efficiency in electron transfer from the β domains of the wild-type and the mutant DsbDs to the periplasmic domains exhibited the following order: wild-type > P162A > G316A > P289A > P284A (Fig. 5A and B, lanes 3 to 12). This order of efficiency of electron transfer among the wild-type and mutant DsbDs correlated with the ability to reduce DsbC that was determined in conditions of low expression (Fig. 2B, lanes 2, 11, 12, 13, and 15) (wild-type > G316A ≥ P162A > P289A ≥ P284A). The consistency of the results in the two analyses suggests that the results obtained in the analysis of the cleaved domain are representative of the actual redox states of each domain.
The even-numbered lanes between lanes 1 and 12 in Fig. 5 were included in the hopes of observing mixed disulfide complexes between the domains of the wild-type and mutant DsbDs. Any such complexes should disappear upon reduction by DTT and release the component domains to add to the bands representing the free domain. We could detect unidentified DTT-sensitive bands (data not shown). If mixed disulfide complexes existed, their amounts would not be high because the amounts of the domains from the odd- and even-numbered lanes showed almost no differences. In lane 4 and lane 8 in Fig. 5C, a small band shift and broadening of the oxidized β domain, respectively, were observed by reduction during SDS-PAGE, which might be due to a conformational change of the β domain upon reduction during SDS-PAGE.
DISCUSSION
In this paper, we present a mutational analysis of DsbDβ, a membrane-embedded domain of DsbD that promotes transfer of electrons from the cytoplasm to the periplasm via two redox-active cysteines. We have converted each of the 19 most-conserved amino acid residues of this domain into alanines and assessed their effects on the redox state of DsbC, a substrate that is reduced by DsbD. Eleven of these mutations cause a decrease in the reduction of DsbC. All 19 mutations were further studied to determine their effects on the redox state of DsbD itself. We used AMS alkylation and Western blotting to permit distinction between oxidized and reduced forms of DsbD. The 11 mutants showed a decrease in reduction of DsbD, accumulating forms of the protein with varying degrees of oxidation. For the purposes of this discussion, we have separated the mutants into two classes. We have characterized several of these mutants in more detail, including P284A, the mutant from one of these classes with the strongest effects.
The first class includes six mutants: the P284A mutant (2AMS), which has the most clear-cut effects, and the P162A, P289A, L294A, G316A, and P320A mutants, which show a mixture of two forms of DsbD (0 and 2 AMS). Proline 284 is a well-conserved amino acid residue among the homologues of DsbD (Fig. 1) and is located just before the more-carboxy terminus of the two cysteines in the β domain, Cys285. We determined which domains of the P284A DsbD were altered in their redox state, using a specially constructed derivative of DsbD (DsbDTH) which permits us to separate the three domains. We showed that the P284A strain, in contrast to the wild-type strain, maintains its DsbDβ domain mainly in the reduced state and accumulates the oxidized forms of DsbDα and γ. We ruled out the possibility that the mutant DsbDβ domain is so structurally altered that its cysteines cannot form a disulfide bond; that bond does appear in a genetic background lacking thioredoxin reductase. In this background, oxidized thioredoxins accumulate and carry out the reverse of the normal reaction between it and DsbD, promoting disulfide bond formation in the reduced DsbDβ. This finding also shows that the mutant DsbD is still capable of interacting effectively with thioredoxins. Altogether, these results indicate that P284A DsbDβ transfers its electrons to DsbDγ very inefficiently. This defect is consistent with the presumed role of the redox-active Cys285 adjacent to Pro284. Of the two cysteines in DsbDβ, Cys163 has been shown to interact directly with Trx1, forming a mixed disulfide complex. Thus, Cys285, which is adjacent to Pro284, is thought to interact with DsbDγ.
Either the P284A β domain may be so structurally altered that it cannot interact effectively with the γ domain or the redox potential of its cysteines may be increased to the point that they cannot act efficiently as a reductant of γ. Significant changes in the redox potential of thioredoxin-like proteins with single amino acid substitutions have been observed previously. For example, the redox potential of thioredoxin-fold proteins is altered when the middle dipeptide of their Cys-Xaa-Xaa-Cys motif is changed (7, 16). In the case of DsbA, the wild-type (Cys-Pro-His-Cys) equilibrium constant, Kox, measured with glutathione, is 0.1 mM, while one mutant (Cys-Pro-Pro-Cys) has a Kox of 200 mM, close to that of E. coli Trx1 (Kox, about 2 M) (7).
The P162A, P289A, L294A, G316A, and P320A mutants show a mixture of two forms of DsbD, the fully oxidized band (0 AMS) and the band with four of its six cysteines oxidized (2 AMS). The latter band found in these mutants must represent a species in which one of the three domains of DsbD is in the reduced state (Fig. 2C, lanes 2 and 13 to 16). We exclude the possibility that much of the α domain is in the reduced state because significant amounts of it would be detected, due to the sensitivity of the anti-DsbDα antibody. Using the DsbDTH construct, we analyzed the redox state of each domain for three of these DsbD mutants, P162A, P289A, and G316A. This was done to determine the nature of the 2 AMS bands and to see whether it was possible to detect mixed disulfide complexes between the domains of DsbD. We could obtain no evidence that these amino acid changes resulted in the accumulation of such complexes. P162A DsbD maintained its β domain in the same redox state as that of the wild type but accumulated a small amount of the oxidized forms of DsbDα and γ. The periplasmic domains of P289A DsbD were largely in the oxidized states, although some portion of the reduced protein might be degraded as a result of instability. G316A DsbD showed characteristics similar to those of P284A DsbD, but its β domain could transfer electrons to the periplasmic domains better than that of P284A. It is possible that a small fraction of its β domain becomes oxidized after electron transfer, although we could not observe this, due to the faint bands on the Western blot. Considering the experiment using DsbDTH, the bands (2 AMS) from these mutant DsbDs (Fig. 2C, lanes 2, 13, and 15) might contain a significant portion of the reduced β domain.
The interactions between the β domains and thioredoxin in these three mutant DsbDs and thioredoxins were not impaired, but their β domains have defects in electron transfer to their γ domains. The amino acid changes in the mutants L294A and P320A are closest to Cys285, at least in the primary sequence, while P162A is next to Cys163, which ultimately is disulfide bonded to Cys285. The simplest explanation for the properties of these mutants is that they, like P284A, are defective in the reduction of DsbDγ by DsbDβ, probably due to their reduced affinity for DsbDγ or to a changed redox potential. Although Pro162 is located just before Cys163, it seems important for the interaction of the β domain with DsbDγ rather than thioredoxins. This property may be related to its location closer to the periplasmic side of the membrane than Cys163 (Fig. 2A). Further interpretation of these results will require a deeper understanding of the relative kinetics of each of the electron transfer steps in this pathway.
The mutant proteins in the second class appear to affect the interaction of DsbDβ with thioredoxin. This class, which includes G155A, P166A, P169A, Y194A, and W338A, showed mainly a form of DsbD that was the fastest-moving band on gels. This band almost certainly represents the most-oxidized redox state, with all of its six cysteines disulfide bonded (0 AMS) (Fig. 2C, lanes 1, 3 to 5, and 18). This conclusion is based on our analysis of the redox states of the separated domains of the P284A DsbD, which confirmed our conclusion that the band we labeled 2AMS has one domain (the β domain) in the reduced state and, therefore, only two cysteines that can be alkylated by AMS (Fig. 2C, lane 11). Establishing the redox state of this band, which was the next-to-fastest-moving DsbD band, allows us to be more certain of the redox state (number of reduced cysteines) of the mutant DsbDs in this second class. As with P284A, there are different explanations for the properties of these mutants. They may have altered the redox potential of the β domain so that it is now more stable in the oxidized state and cannot readily receive electrons from thioredoxins. Alternatively, the affinity of the β domain for thioredoxins may be decreased by these amino acid substitutions so that the β domain is not effectively reduced by thioredoxins. Of these mutants, G155A and Y194A show very strong defects in DsbC reduction, indicating that they manifest very strong defects in one of the above steps. The others may retain a small amount of reductive activity, allowing some DsbC reduction. The finding of several mutants that show DsbD overwhelmingly in the oxidized state but still have substantial amounts of reduced DsbC suggests that DsbD may be in substantial excess in relation to the need for reduction of DsbC.
According to the topological analysis of DsbD, most of the amino acid changes in the mutant DsbDs in the second class are located in the cytoplasm (Trp338) or near the redox-active Cys163. Cys163 interacts directly with Trx1, forming a mixed disulfide complex (11). Given the properties of these mutants and their locations in the protein, we suggest that the defects they exhibit are defects in the interaction of Cys163 with thioredoxin.
A number of the conserved residues in DsbD and its homologues are prolines. Proline residues in the middle of a TM helix can distort α-helix geometry, due to the loss of a backbone hydrogen bond, and can introduce flexibility in the helix that may result in substantial kinks (21). These proline-induced hinges may have a direct role in gating in some membrane proteins (9, 27). We have proposed that DsbD may operate by a gated-channel mechanism in transferring electrons across the cytoplasmic membrane (17). The many conserved prolines in DsbD may have an important function in gating, explaining the significant effects of the several proline mutants.
The newly generated construct of DsbD containing two thrombin cleavage sites and a c-Myc epitope (DsbDTH) behaves functionally like the wild-type DsbD in vivo and has provided an improved means of carrying out redox analysis on DsbD and its mutant derivatives. An advantage of using this construct over the previously described system in which the three domains of DsbD were expressed as separate constructs is that the latter approach did not result in fully reconstituted activity in vivo. This new cleavable construct may allow detection of in vivo mixed disulfide complexes between the domains of DsbD that we failed to detect previously using the three-piece system (11). It may be possible to detect a mixed disulfide α to γ complex or β to γ complex produced in vivo by performing thrombin cleavage of the domains, followed by reduction of disulfide bonding and identification of complex components using two-dimensional SDS-PAGE. Introduction of mutations of a redox-active cysteine into these domains may cause an increase in the yield of the mixed disulfide complexes, as has been seen previously (11), and allow determination of which cysteines are involved in these linkages.
Future studies will include additional determination of the redox state of each domain of the mutants with the strongest effects, measurement of the redox potentials of the purified β domains, and determination of the Km values of interactions between mutant β domains and either γ domains or thioredoxins. Suppressor analysis of the P284A mutation may reveal mutations affecting affinity properties or redox potential, depending on the nature of the original defect. Ultimately, interpretation of many of the findings will depend on a combination of the mutational/biochemical analysis with knowledge of the three-dimensional structure of DsbD and changes in that structure that take place during the electron transfer process.
We note that several of the remainder of the mutants, which had no measurable effect on DsbD activity or DsbC reduction, lie in the second TM of DsbD and that their conservation does not extend beyond the group that just includes homologues with structures similar to E. coli DsbD. The CcdA homologues and the COG4233 family of homologues do not contain these conserved residues. These differences may reflect different mechanisms of transfer to substrates, such as the homologues of CcmG.
Acknowledgments
We gratefully acknowledge Federico Katzen and Amir Porat for providing many strains and plasmids. We also thank members of the Beckwith laboratory, particularly Hiroshi Kadokura, Markus Eser, and Mehmet Berkman, for helpful discussions. J.B. is an American Cancer Society Research Professor.
This study was supported by NIH grant GM55090 to J.B.
REFERENCES
- 1.Bardwell, J. C. A., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581-589. [DOI] [PubMed] [Google Scholar]
- 2.Beckett, C. S., J. A. Loughman, K. A. Karberg, G. M. Donato, W. E. Goldman, and R. G. Kranz. 2000. Four genes are required for the system II cytochrome c biogenesis pathway in Bordetella pertussis, a unique bacterial model. Mol. Microbiol. 38:465-481. [DOI] [PubMed] [Google Scholar]
- 3.Berkmen, M., D. Boyd, and J. Beckwith. 2005. The nonconsecutive disulfide bond of Escherichia coli phytase (AppA) renders it dependent on the protein-disulfide isomerase, DsbC. J. Biol. Chem. 280:11387-11394. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J., J. L. Song, S. Zhang, Y. Wang, D. F. Cui, and C. C. Wang. 1999. Chaperone activity of DsbC. J. Biol. Chem. 274:19601-19605. [DOI] [PubMed] [Google Scholar]
- 5.Chung, J., T. Chen, and D. Missiakas. 2000. Transfer of electrons across the cytoplasmic membrane by DsbD, a membrane protein involved in thiol-disulphide exchange and protein folding in the bacterial periplasm. Mol. Microbiol. 35:1099-1109. [DOI] [PubMed] [Google Scholar]
- 6.Gil, D., and J. P. Bouche. 1991. ColE1-type vectors with fully repressible replication. Gene 105:17-22. [DOI] [PubMed] [Google Scholar]
- 7.Grauschopf, U., J. R. Winther, P. Korber, T. Zander, P. Dallinger, and J. C. A. Bardwell. 1995. Why is DsbA such an oxidizing disulfide catalyst? Cell 83:947-955. [DOI] [PubMed] [Google Scholar]
- 8.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin, T., L. Peng, T. Mirshahi, T. Rohacs, K. W. Chan, R. Sanchez, and D. E. Logothetis. 2002. The βγ subunits of G proteins gate a K+ channel by pivoted bending of a transmembrane segment. Mol. Cell 10:469-481. [DOI] [PubMed] [Google Scholar]
- 10.Joly, J. C., and J. R. Swartz. 1997. In vitro and in vivo redox states of the Escherichia coli periplasmic oxidoreductases DsbA and DsbC. Biochemistry 36:10067-10072. [DOI] [PubMed] [Google Scholar]
- 11.Katzen, F., and J. Beckwith. 2000. Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell 103:769-779. [DOI] [PubMed] [Google Scholar]
- 12.Katzen, F., and J. Beckwith. 2003. Role and location of the unusual redox-active cysteines in the hydrophobic domain of the transmembrane electron transporter DsbD. Proc. Natl. Acad. Sci. USA 100:10471-10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katzen, F., M. Deshmukh, F. Daldal, and J. Beckwith. 2002. Evolutionary domain fusion expanded the substrate specificity of the transmembrane electron transporter DsbD. EMBO J. 21:3960-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, J. H., S. J. Kim, D. G. Jeong, J. H. Son, and S. E. Ryu. 2003. Crystal structure of DsbDγ reveals the mechanism of redox potential shift and substrate specificity. FEBS Lett. 543:164-169. [DOI] [PubMed] [Google Scholar]
- 15.Missiakas, D., C. Georgopoulos, and S. Raina. 1994. The Escherichia coli dsbC (xprA) gene encodes a periplasmic protein involved in disulfide bond formation. EMBO J. 13:2013-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mossner, E., M. Huber-Wunderlich, A. Rietsch, J. Beckwith, R. Glockshuber, and F. Aslund. 1999. Importance of redox potential for the in vivo function of the cytoplasmic disulfide reductant thioredoxin from Escherichia coli. J. Biol. Chem. 274:25254-25259. [DOI] [PubMed] [Google Scholar]
- 17.Porat, A., S.-H. Cho, and J. Beckwith. 2004. The unusual transmembrane electron transporter DsbD and its homologues: a bacterial family of disulfide reductases. Res. Microbiol. 155:617-622. [DOI] [PubMed] [Google Scholar]
- 18.Rietsch, A., D. Belin, N. Martin, and J. Beckwith. 1996. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:13048-13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rietsch, A., P. Bessette, G. Georgiou, and J. Beckwith. 1997. Reduction of the periplasmic disulfide isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J. Bacteriol. 179:6602-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Senes, A., D. E. Engel, and W. F. DeGrado. 2004. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 14:465-479. [DOI] [PubMed] [Google Scholar]
- 22.Shevchik, V. E., G. Condemine, and J. Robert-Baudouy. 1994. Characterization of DsbC, a periplasmic protein of Erwinia chrysanthemi and Escherichia coli with disulfide isomerase activity. EMBO J. 13:2007-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spyropoulos, I. C., T. D. Liakopoulos, P. G. Bagos, and S. J. Hamodrakas. 2004. TMRPres2D: high quality visual representation of transmembrane protein models. Bioinformatics 20:3258-3260. [DOI] [PubMed] [Google Scholar]
- 24.Stewart, E. J., F. Aslund, and J. Beckwith. 1998. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. EMBO J. 17:5543-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart, E. J., F. Katzen, and J. Beckwith. 1999. Six conserved cysteines of the membrane protein DsbD are required for the transfer of electrons from the cytoplasm to the periplasm of Escherichia coli. EMBO J. 18:5963-5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tieleman, D. P., I. H. Shrivastava, M. R. Ulmschneider, and M. S. P. Sansom. 2001. Proline-induced hinges in transmembrane helices: possible roles in ion channel gating. Proteins 44:63-72. [DOI] [PubMed] [Google Scholar]
- 28.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 29.Walker, K. W., and H. F. Gilbert. 1999. Scanning and escape during protein-disulfide isomerase-assisted protein folding. J. Biol. Chem. 272:8845-8848. [DOI] [PubMed] [Google Scholar]
- 30.Zapun, A., D. Missiakas, S. Raina, and T. E. Creighton. 1995. Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry 34:5075-5089. [DOI] [PubMed] [Google Scholar]