Abstract
Pantoea stewartii subsp. stewartii is a bacterial pathogen of corn. Its pathogenicity depends on the translocation of effector proteins into host cells by the Hrp type III secretion system. We previously showed by genetic analysis that the HrpX sensor kinase and the HrpY response regulator are at the head of a complex cascade of regulators controlling hrp/hrc secretion and wts effector genes. This cascade also includes the HrpS response regulator and the HrpL alternative sigma factor. These regulators are shared among many important plant pathogens in the genera Pantoea, Erwinia, and Pseudomonas. In this study, we dissect the regulatory elements in the hrpS promoter region, using genetic and biochemical approaches, and show how it integrates various environmental signals, only some of which are dependent on phosphorylation of HrpY. Primer extension located the transcriptional start site of hrpS at a σ70 promoter 601 bp upstream of the open reading frame. Electrophoretic mobility shift assays and DNase I footprinting analysis demonstrated that HrpY binds to conserved regulatory elements immediately adjacent to this promoter, and its binding affinity was increased by phosphorylation at D57. A consensus sequence for the two direct repeats bound by HrpY is proposed. Deletion analysis of the promoter region revealed that both the HrpY binding site and additional sequences farther upstream, including a putative integration host factor binding site, are required for hrpS expression. This finding suggests that other unknown regulatory proteins may act cooperatively with HrpY.
The hrp/hrc genes of gram-negative, phytopathogenic bacteria encode type III secretion systems that deliver effector proteins into host cells. These proteins may elicit cell death or suppress host defenses. They are required for infection and colonization of plants by all biotrophic pathogens of the genera Pseudomonas, Ralstonia, Xanthomonas, Erwinia, and Pantoea (3). They also play a role in rapid establishment of necrotrophic, soft-rot bacteria in the genus Erwinia, where enzymatic tissue disintegration is delayed by hrp mutations (33). Pantoea stewartii subsp. stewartii (synonym, Erwinia stewartii) causes vascular wilting and leaf blight of sweet corn and maize. This pathogen carries a group I hrp gene cluster (1, 7, 8, 11), similar to those in Pseudomonas, Erwinia, and other Pantoea species. The group I hrp gene clusters are regulated by the HrpS enhancer-binding protein and the HrpL extracytoplasmic function alternative sigma factor (12, 25, 36-39). P. stewartii hrp genes are also controlled by the HrpX/HrpY two-component regulatory system, which is also present in Pantoea agglomerans pv. gypsophilae, Erwinia amylovora, Erwinia carotovora subsp. carotovora, and Erwinia chrysanthemi (19, 25, 27, 35, 39). In erwinias and pantoeas, the hrpL-hrpXY-hrpS genes have the same map order, and the corresponding proteins share greater than 80% similarity at the amino acid level (25). However, in P. stewartii, a large remnant of an insertion sequence (IS) element is present between hrpS and its promoter. In all cases where genetic analysis has been performed, the HrpY response regulator appears to act upstream of HrpL in the regulatory cascade and is absolutely required for pathogenicity (25, 27, 35). In Pantoea spp. (25, 27) and E. chrysanthemi (39), HrpY has been shown by genetic analysis to regulate hrpS, but its direct target gene(s) in E. amylovora is not yet clear.
HrpY has sequence similarity to the FixJ class of response regulators (25), which includes Escherichia coli NarL and UhpA and Bacillus subtilis DegU. Members of this class share a typical modular organization (31), with a conserved N-terminal receiver domain, a flexible linker, and a C-terminal domain that contains a LuxR/GerR superfamily DNA-binding motif. The N-terminal receiver domain of P. stewartii HrpY contains three conserved aspartyl residues (D11, D12, and D57). By homology to other response regulators and protein modeling (22), D57 is expected to be the phosphorylation site. In agreement with this prediction, conservative and structurally neutral amino acid substitutions at this position, such as D57N and D57A, abolish virulence when the hrpY allele is expressed in single copy from the chromosome (25). Interestingly, the same mutant proteins are functional when expressed from a low-copy-number plasmid. Using a genetic approach, we previously showed that P. stewartii HrpY activates expression of hrpS and autoregulates the hrpXY operon (25). Moreover, the hrpS promoter (PhrpS) region appears to be a key control point for the integration of various signaling pathways that modulate spatiotemporal hrp gene expression in response to metabolic status, pH, and osmotic pressure (22). In addition, HrpS initiates a novel autoregulatory loop caused by readthrough transcription from hrpL into hrpXY (24). In contrast, the hrpXY operon is constitutively expressed at a moderate level and appears to be involved in transducing only a few of these signals (23). Although HrpX is required for full hrp gene expression in inducing medium, hrpX mutants are still quite virulent on corn, in contrast to hrpY mutants that are totally nonpathogenic. This suggests that cross talk among related sensor kinases or acetyl phosphate occurs in planta. Together, these findings open the possibility that other regulatory factors also modulate hrpS transcription, either by direct regulation of PhrpS or via phosphorylation of HrpY. It is interesting that each phytopathogenic bacterium appears to respond somewhat differently to external signals and growth conditions (19, 25, 27, 35, 36, 39), so that regulation of hrpS by specific and global regulators may be fine-tuned for each pathogen's niche. For this reason, it is important to understand how various regulatory molecules, especially HrpY, interact with PhrpS. At this point, a direct, physical interaction between HrpY and hrpS regulatory sequences has not been demonstrated in any species.
In this study, we show that only some of the multiple signals controlling the hrpS promoter are mediated by HrpY. We characterized the hrpS promoter by locating its transcription start site, identifying the region required for HrpY-dependent regulation, and determining the sequences to which HrpY binds. We also showed that HrpY has a single phosphorylation site and that phosphorylation increases its binding affinity to PhrpS. Our results further suggest that other unidentified regulatory factors may act in concert with HrpY to control hrpS expression.
MATERIALS AND METHODS
Strains, growth conditions, and pathogenicity tests.
E. coli and P. stewartii strains and plasmids used in this study are listed in Table 1. Luria-Bertani (LB) broth and agar were routinely used for growing bacteria. To induce hrp genes in P. stewartii strains, a minimal inducing medium at pH 5.5 (IM5.5) (25) with or without the addition of casamino acids was used. For experiments testing various hrp-repressing conditions, IM5.5 was modified in the following ways: alkalinized with HEPES to pH 8.0, amended with 250 mM NaCl, amended with 2 mM nicotinic acid, used with 55 mM citrate as the sole carbon source, or used with 1% tryptone as the sole carbon and nitrogen sources. Liquid cultures were grown in flasks or tubes with shaking at 200 rpm at 37°C for E. coli or 28°C for P. stewartii. When appropriate, antibiotics were supplied at the following concentrations: ampicillin, 200 μg ml−1; kanamycin, 50 μg ml−1; tetracycline, 20 μg ml−1; and chloramphenicol, 34 μg ml−1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant phenotype and genotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| BL21(DE3)/pLysS | F−ompT hsdSB (r−Bm−B) gal dcm (DE3) carrying pLysS | Novagen |
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 endA1 recA1 deoR Δ(ara,leu)7697 araD139 galU galK nupG rpsL λ− | Invitrogen |
| HB101 | F−thi-1 hsd20 (r−Bm−B) sup E44 recA13 ara-14 leuB6 proA2 lacY1 rpsL20 (Smr) xyl-5 mtl-1 | 5 |
| Sφ200Rif | metB strA purB Δ(agg-uidA-man) Rifr | 35 |
| P. stewarti subsp. stewartiii strains | ||
| DC283 | SS104 wild-type; Wts+, HR+, Nalr | 7 |
| DM064 | DC283 hrpY1296::Tn5 (Kanr) | 11 |
| DM733 | DC283 hrpY64::Tn5 hrpS1::Tn3HoHoI (Kanr Ampr) | This study |
| MEX1 | DC283 hrpS1::Tn3HoHo (Ampr) | 11 |
| MM254 | DC283 hrpY[D57N] | 25 |
| Plasmids | ||
| pBluescript KS and SK(+) | ColE1 αlacZ (Apr) | Stratagene |
| pET15b | ColE1; T7lac; poly-His tag expression vector (Apr) | Novagen |
| pPL6GUSC | pLAFR6 derivative carrying a promoterless uidA gene (Tcr) | 17 |
| pRK415 | IncP αlacZ (Tcr) | 16 |
| pDM2912 | pPL6GUSC with a +629 to −231 hrpSΔ(−111 to −64) PCR fragment fused to uidA | This study |
| pMM46 | pBluescript KS(+) with a 0.7-kb HindIII hrpY PCR fragment, transcribed from Plac | 25 |
| pMM50 | pPL6GUSC with a +994 to −317 hrpS PCR fragment fused to uidA | 25 |
| pMM52 | pRK415 with the 0.7-kb BamHI-EcoRI hrpY+ fragment from pMM46, transcribed from Plac | 25 |
| pMM57 | pBluescript KS(+) with a 0.7-kb HindIII hrpY[D57A] spliced overlap extension-PCR fragment | 25 |
| pMM58 | pBluescript SK(+) with a 5.3-kb insert containing hrpL+, hrpXY+, and hrpS+ | 25 |
| pMM74 | pRK415 with the 0.7-kb HindIII hrpY[D57A] fragment from pMM57; transcribed from Plac | This study |
| pMM92 | pBluescript KS(+) with a 0.7-kb HindIII hrpY[D57N] SOE-PCR fragment | 25 |
| pMM118 | pRK415 with a 0.7-kb HindIII hrpY[D57N] fragment from pMM92 | This study |
| pMM221 | pET15b with a 0.7-kb NdeI hrpY PCR fragment | This study |
| pMM222 | pET15b with a 0.7-kb NdeI hrpY[D57N] PCR fragment | This study |
| pMM396 | pPL6GUSC with a +629 to −165 hrpS PCR fragment fused to uidA | This study |
| pMM393 | pPL6GUSC with a +629 to −90 hrpS PCR fragment fused to uidA | This study |
| pMM400 | pPL6GUSC with a +629 to −231 hrpS PCR fragment fused to uidA | This study |
| pMM403 | pPL6GUSC with a +629 to −51 hrpS PCR fragment fused to uidA | This study |
| pMM1001 | pDM2945 with DR1 site from −99 to −90 deleted | This study |
| pMM1002 | pDM2945 with DR2 site from −78 to −69 deleted | This study |
| pMM1100 | pDM2945 with IHF site from −191 to −179 deleted | This study |
| pMM1101 | pDM2945 with a 5-bp deletion from −144 to −140 | This study |
| pMM1102 | pDM2945 with a 5-bp deletion from −139 to −135 | This study |
| pMM1103 | pDM2945 with a 10-bp deletion from −144 to −135 | This study |
| pDM2912 | pPL6GUS with pMM400 BamHI insert carrying a DR1-DR2 deletion from −121 to −70 | This study |
| pDM2932 | pPL6GUS with pMM400 BamHI insert carrying a DR1-DR2 deletion from −99 to −69 | This study |
| pDM2945 | pUC19 with the pMM400 BamHI insert carrying the +629 to −231 hrpS region | This study |
| pDM2949 | pPL6GUS with pMM1001 BamHI insert | This study |
| pDM2953 | pPL6GUS with pMM1002 BamHI insert | This study |
| pDM2958 | pPL6GUS with pMM1100 BamHI insert | This study |
| pDM2960 | pPL6GUS with pMM1101 BamHI insert | This study |
| pDM2963 | pPL6GUS with pMM1102 BamHI insert | This study |
| pDM2965 | pPL6GUS with pMM1103 BamHI insert | This study |
Construction of plasmid-borne PhrpS gene fusions.
Isolation and manipulation of recombinant DNA molecules used standard procedures (4) or the various product manufacturers' instructions. Plasmids were introduced into P. stewartii by conjugation or transformation as previously described (7, 25). Fragments containing deletions of the 5′ regulatory region of hrpS were produced by PCR using the primers given in Table S1 in the supplemental material. Oligonucleotide primers were synthesized at Integrated DNA Technologies. All PCR products were generated with 15 to 20 cycles of amplification using Pfu DNA polymerase (Clontech) and 500 ng of plasmid pMM58 as a template. Gel-purified products were digested with BamHI and ligated into pPL6GUSC to create transcriptional fusions to a uidA (β-glucuronidase [GUS]) reporter gene. In particular, the 5′ hrpS regulatory fragments from −241 to +618, −175 to +618, −100 to +618, and −61 to +628 were amplified with the primer pairs SF3509B/SR4368B, SF3575B/SR4368B, SF3650B/SR4368B, and SF3689B/SR4368B to produce plasmids pMM400, pMM396, pMM393, and pMM403, respectively. (Unless otherwise noted, coordinates here and elsewhere are relative to the transcriptional start of hrpS.) PhrpS gene fusions with deletions between −70 and −121 and between −99 and −69 were constructed by spliced overlap extension-PCR (2) using primer pairs SF3509B/SRS1 plus SR4368B/SFS2 and SF3509B/SR2 plus SR4368B/SF3 to form plasmids pDM2912 and pDM2932, respectively. Small deletions in the PhrpS region were introduced by oligonucleotide site-directed mutagenesis as follows. About 100 ng of plasmid pDM2945, a pUC19 derivative carrying the pMM400 insert, was methylated in vitro with SssI methylase by following the manufacturer's instructions (New England Biolabs). The partially overlapping primer pairs M1/MR1 (deleting nucleotides [nt] −191 to −179), M2/MR2 (deleting nt −144 to −140), M3/MR3 (deleting nt −139 to −135), M4/MR4 (deleting nt −144 to −135), M5/MR5 (deleting nt −99 to −90), and M6/MR6 (deleting nt −78 to −69) were annealed to pDM2945 DNA, and PCRs were performed using Herculase DNA polymerase (Stratagene) according to the manufacturer's instructions. Unmethylated linear PCR products were transformed into McrBC-proficient E. coli hosts such as DH5α. The recombinant plasmids were screened by DNA sequencing to identify deletion mutants. The resulting plasmids pMM1100, pMM1101, pMM1102, pMM1103, pMM1001, and pMM1002 were digested with BamHI, and the inserts were subcloned into pPL6GUS to produce plasmids pDM2958, pDM2960, pDM2963, pDM2965, pDM2949, and pDM2953, respectively. Plasmids pMM74 and pMM118, carrying the hrpY D57A and D57N alleles, were generated by subcloning hrpY from plasmids pMM57 and pMM92 into pRK415 for expression from Plac.
Construction of hrpY plasmids for protein expression and purification.
hrpY was amplified by PCR using Pfu DNA polymerase (Clontech), primers ET-YF-NdeI and ET-YR-NdeI (see Table S1 in the supplemental material) and plasmids pMM46 hrpY+ or pMM92 hrpY(D57N) as DNA templates (25). Primer ET-YF-NdeI introduced an NdeI site at the start codon of hrpY and maintained the reading frame with the vector's N-terminal His6 tag; primer ET-YR-NdeI introduced three stop codons downstream of the hrpY open reading frame (ORF). The 670-bp PCR fragments, spanning either hrpY+ or hrpY(D57N), were gel purified, digested with NdeI, and ligated into an NdeI-linearized pET-15b vector (Novagen). The resulting plasmids, pMM221 and pMM222, carried hrpY and hrpY(D57N), respectively, fused to an N-terminal His6 tag and expressed from a T7lac IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter. The inserts in plasmids pMM221 and pMM222 were sequenced to confirm that they did not contain PCR errors.
Purification of His6-HrpY proteins.
Plasmids pMM221 and pMM222 were transformed by electroporation into E. coli BL21(DE3) carrying pLysS. For protein expression, bacteria were grown in 1 liter of LB broth until the culture reached an A600 of 0.8. IPTG was added to a final concentration of 1 mM, and the culture was grown at 15°C for 18 h until an A600 reached 1.4. Cells were harvested by centrifugation and resuspended in 35 ml of ice-cold lysis buffer (10 mM imidazole, 500 mM NaCl, 1 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride [PMSF], 20 mM Tris-HCl, pH 7.9 at 4°C), containing 50 μl of protease inhibitor cocktail (Sigma). The cell suspension was incubated in 0.1 mg ml−1 lysozyme for 20 min on ice and then passed two times through a 40-K French pressure cell (Thermo-IEC) at 10,000 lb/in2. The lysate was centrifuged twice at 20,000 × g for 30 min at 4°C. Column chromatography was performed at 4°C. The supernatant was then applied to a 1.5-ml Ni2+-iminodiacetic acid column packed using His-Bind resin (Novagen), prepared according to the manufacturer's instructions. The column was washed with 10 column volumes of lysis buffer and 10 column volumes of washing buffer (40 mM imidazole, 500 mM NaCl, 1 mM MgCl2, 0.5 mM PMSF, 20 mM Tris-HCl, pH 7.9 at 4°C). The target His6-HrpY proteins were eluted with a step gradient of 100 mM and 250 mM imidazole buffers (100 mM or 250 mM imidazole, 500 mM NaCl, 20 mM Tris-HCl, pH 7.9 at 4°C, 1 mM MgCl2, 0.5 mM PMSF). Fractions containing higher concentrations of the 26.1-kDa recombinant protein, as assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), were pooled and dialyzed overnight against storage buffer (67 mM potassium glutamate, 125 mM HEPES, pH 7.9 at 4°C, 0.5 mM dithiothreitol [DTT], 0.5 mM PMSF, 20% glycerol). The protein was further purified by anion exchange chromatography in a Q-Sepharose (Amersham-Pharmacia) column after dialysis against buffer A (20 mM Tris-HCl, pH 8.0 at 4°C, 25 mM NaCl, 0.25 mM EDTA, 0.2 mM PMSF) prepared according to the manufacturer's instructions. Proteins were eluted in a linear buffer gradient prepared by mixing buffer A with buffer B (20 mM Tris-HCl, pH 8.0 at 4°C, 500 mM NaCl, 0.25 mM EDTA, 0.2 mM PMSF). Fractions containing highly purified His6-HrpY were pooled, and the buffer was exchanged for 1× storage buffer (10 mM Tris-HCl, pH 7.9, 125 mM NaCl, 1 mM DTT, 3 mM MgCl2, 38% glycerol) by repeated centrifugation, according to the manufacturer's instructions, in a Vivaspin 15R ultrafiltration tube (Viva Science) with a molecular mass cutoff of 10 kDa or by Sephadex G25-HiTrap Desalt chromatography (Amersham). Protein concentration was determined by the dye-binding method of Bradford (6). The degree of purification was estimated by Coomassie brilliant blue staining after SDS-PAGE (18).
MS.
Protein molecular mass measurements were performed at the Ohio State University Campus Chemical Instrument Center-Mass Spectrometry Facility by electrospray ionization (ESI) mass spectrometry (MS) using a Micromass Q-TOF II (quadrupole-time of flight) mass spectrometer equipped with an orthogonal nanospray source (Z-spray) operated in positive ion mode. Tandem MS of [Glu]-fibrinopeptide B was used for mass calibration in a calibration range of m/z 100 to 2,000. Salt buffers from the protein samples were cleaned using manual syringe protein traps from Michrom BioResources. Desalted protein samples were prepared in a solution containing 50% acetonitrile-0.1% formic acid and infused into the nanospray source at a rate of 0.5 to 1.0 ml min−1. Optimal conditions were a capillary voltage of 3,000 V, a source temperature of 110°C, and a cone voltage of 60 V. Quadrupole 1 was set to optimally pass ions from m/z 100 to 2,000, and all ions transmitted into the pusher region of the TOF analyzer were scanned over the m/z range with a 1-s integration time. Data were acquired in the continuum mode until acceptable averaged data were obtained (10 to 15 min). ESI data were deconvoluted using MaxEnt I (Micromass).
In vitro phosphorylation of HrpY.
For enzymatic phosphorylation, 15 μl of a mixture containing 25 μM His6-HrpY, 75 μM Salmonella enterica BarA198 (obtained from M. Teplitski) (32), 40 μM [γ32-P]ATP (10 Ci/mmol; NEN) and 0.1 mM cold ATP in phosphorylation buffer was incubated at 25°C for 90 min. Reactions were terminated with 5 μl of 4× stop buffer (125 mM Tris-HCl, pH 6.8, 8 mM EDTA, 4% SDS, 8% β-mercaptoethanol, 20% glycerol, 0.02% bromophenol blue), separated by SDS-PAGE (12% monomer gel), and visualized by Coomassie brilliant blue staining (18). Gels were dried under vacuum at 60°C for 30 min. The radiolabeled proteins were visualized with a storage phosphor screen, analyzed on a Molecular Dynamics Storm-840 PhosphorImager, and quantified with ImageQuant.
EMSA.
DNA fragments used as probes for electrophoretic mobility shift assays (EMSA) were made by PCR using plasmid pMM58, which carries the hrpL, hrpXY, and hrpS genes (25), as a template. Six probes (A, B, C, C1, C2, and C3) were designed to amplify various fragments of the hrpS regulatory region located upstream of the IS-like element. Primers SF3509 and SR3845 were used for amplification of fragment A (327 bp), SF3509 and SR3575 for fragment B (76 bp), SF3575 and SR3645 for fragment C (261 bp), SF3575 and SR3650 for fragment C1 (76 bp), SF3650 and SR3689 for fragment C2 (86 bp), and SF3689 and SR3845 for fragment C3 (101 bp). PCR products were purified from agarose or acrylamide gels and quantified by the ethidium bromide staining spot method (4). DNA fragments were labeled at the 5′ ends by incubation with T4 polynucleotide kinase (USB) and [γ-32P]ATP (3,000 Ci/mmol; NEN). Unincorporated nucleotides were removed by Sephadex G-25 spin chromatography (4), and labeled DNA was diluted to 10,000 cpm μl−1 (Cerenkov counts). DNA binding reactions (20 μl) contained 15 to 50 fmol of 32P-labeled DNA probe, various amounts of His6-HrpY, 200 ng of acetylated bovine serum albumin (NEB), and 1 μg of poly(dI-dC) (Roche) in binding buffer (125 mM potassium glutamate, 125 mM HEPES, pH 7.9, 75 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA, 0.2 mM DTT). Unlabeled specific competitor DNA was added before the addition of His6-HrpY. The reactions were incubated at 25°C for 30 to 40 min. Reaction mixtures were loaded, without loading buffer, onto 6% acrylamide nondenaturing gels in a vertical gel apparatus (model V16; Waterman), run at 200 V for 2.0 to 2.5 h. Gel and running buffers were 1× TBE (97 mM Tris-HCl, 90 mM borate, 3 mM EDTA, pH 8.0). The gels were dried under vacuum at 80°C, and the radioactive fragments were visualized either by autoradiography at −80°C with Kodak MS films or by using a storage phosphorimager for quantification.
DNase I footprinting.
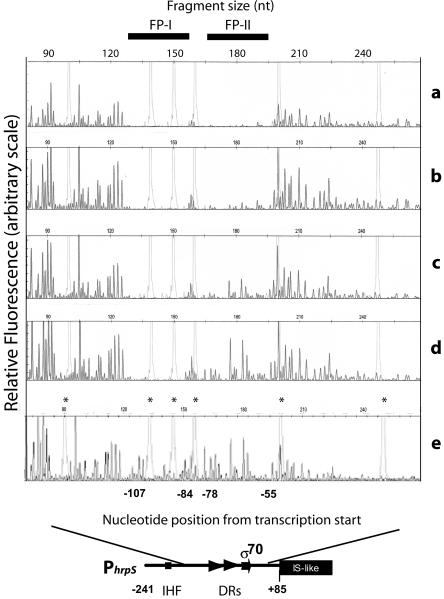
DNase I footprinting to map the physical contacts between HrpY and the hrpS promoter region was performed using fluorescently labeled DNA and an automated sequencer to resolve the digestion products, as described by Zianni et al. (40). Primers SF3509 and SR3845, which delimit hrpS fragment C, were synthesized as 6-carboxyfluorescein (6-FAM) 5′-labeled oligonucleotides by Integrated DNA Technologies. Fragment C was amplified as a singly end-labeled PCR product using plasmid pMM58 as a template. PCR products were purified by gel electrophoresis and quantified by UV spectrophotometry. The labeled probes (45 ng) were incubated with amounts of His6-HrpY protein ranging from 0 to 40 μM in binding buffer (150 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 8% glycerol in 10 mM Tris-HCl, pH 8.0). Nuclease digestion was performed with 0.0025 Kunitz units of DNase I (Worthington Biochemicals) per 20-μl reaction mixture for 5 min. The reaction was stopped by the addition of 0.25 M EDTA and extracted with phenol-chloroform-isoamyl alcohol (25:24:1). The DNA fragments were purified in a QIAGEN spin column. Sequencing reactions were performed using a Thermo Sequenase Dye Primer Manual Cycle sequencing kit from USB according to the manufacturer's instructions. Reactions contained 200 ng of pMM58 plasmid DNA template and 20 pmol of 6-FAM-labeled SF3509 primer. Cycling conditions consisted of a 30-s denaturation at 95°C, 30-s annealing step at 50°C, and 60-s extension at 72°C, for a total of 60 cycles. A 0.1-μl aliquot of Genescan-500 LIZ size standard (Applied Biosystems) was combined with either 1 μl of sequencing product or 5 μl of DNase I digestion products in a 10-μl final volume. Samples were loaded onto a 3730 DNA analyzer (Applied Biosystems) for electrophoresis and detection. Electropherograms were aligned using GeneMapper 3.5 (Applied Biosystems).
GUS enzyme assays.
GUS activity was assayed fluorometrically using 4-methyl-umbelliferyl-β-d-glucuronide as described by Jefferson (15), but the assays were scaled down to fit microtiter plates and analyzed using a Victor 1420-2 multilabel reader (PE Applied Biosystems). Net GUS activity of each strain was corrected for the basal fluorescence of P. stewartii DC283 carrying pPL6GUSC without an insert. Specific activity was expressed in GUS units (1 unit is defined as 1 pmol of 4-methyl-umbelliferone min−1 per unit of optical density at 600 nm ml−1 of culture at 37°C).
Primer extension, DNA sequencing, and bioinformatics.
DNA-free total RNA was isolated from 25 ml of overnight cultures of bacteria grown in IM5.5 liquid medium using an RNA Wizard prep kit (Promega) as per the manufacturer's instructions. RNA was precipitated by adding two volumes of isopropanol, and the precipitate was washed in 70% ethanol. The RNA pellet was resuspended in 50 μl of diethypyrocarbonate-treated water and treated with DNase I (Invitrogen) according to the manufacturer's instructions. Analysis of the 5′ ends of the hrpS mRNA transcripts was performed by primer extension using 6-FAM-labeled primers. A total of 100 pmol of primer 6-FAM-SR3735 was annealed to 50 μg of RNA purified by using a Wizard Total RNA kit (Promega). Synthesis of cDNA was performed using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. DNA sequencing was performed with the same primers used for cDNA synthesis by following the procedure used for DNA footprinting. Denatured single-stranded DNAs were analyzed in an ABI 3770 capillary electrophoresis sequencer. DNA bending ability was predicted using Bend-It (26). Promoter and DNA binding site predictions were determined with BPROM (http://www.softberry.com/), PromScan (http://molbiol-tools.ca/mtoolwww-cgi/promscan.cgi), and SEQSCAN (http://www.bmb.psu.edu/seqscan/).
Nucleotide sequence accession number.
The updated sequence of the P. stewartii hrp cluster has been deposited in the GenBank database under accession number AF282857.
RESULTS
HrpY-dependent and HrpY-independent signals control hrpS expression.
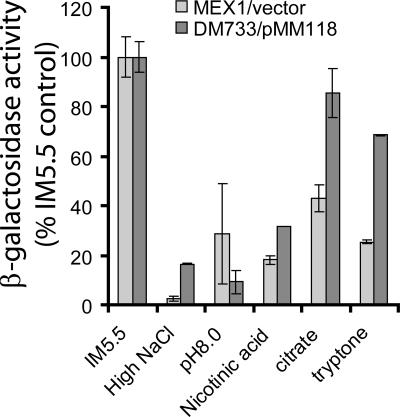
In a previous study (25), we demonstrated that hrpXY expression was constitutive and that hrpS was the first gene in the Hrp regulatory cascade to be repressed by high osmolarity, high pH, and the presence of nicotinic acid, since these signals could all be overridden by ectopic expression of hrpS. In this study, we used a similar approach to determine which of these environmental signals might be mediated via the phosphorylation of HrpY. We tested the ability of ectopic overexpression of hrpY(D57N) to override the repression of a hrpS::lacZ fusion by each signal (Fig. 1). The D57N allele of HrpY was used to bypass the effects of phosphorylation of HrpY by any particular sensor kinase and/or acetyl phosphate. We therefore compared the effect of each signal on the hrpS::lacZ fusions in strains MEX1 hrpY+ and DM733(pMM118). The latter strain was derived from MEX1 and contains the same hrpS::lacZ fusion along with an hrpY::Tn5 mutation. By introducing pMM118 into DM733, we simply replaced the chromosomal hrpY+ gene of MEX1 with a plasmid-borne hrpY(D57N) allele, which was previously shown to activate PhrpS when overexpressed (25). As shown in Fig. 1, the repression by alkaline pH (pH 8) was not overridden by pMM118, and the repression by high osmolarity (250 mM NaCl) or nicotinic acid was only slightly reduced, whereas the repression by citrate or tryptone was significantly alleviated (Fig. 1). This suggests that HrpY is phosphorylated primarily in response to metabolic changes during growth on Krebs cycle intermediates and/or complex nitrogen sources. These results indicate that both HrpY-dependent and HrpY-independent mechanisms regulate PhrpS and further emphasize the pivotal role of this region in integrating responses to a range of environmental conditions.
FIG. 1.
The effect of environmental stimuli on hrpS-lacZ expression and suppression of these effects by ectopic overexpression of hrpY(D57N). The P. stewartii strain MEX1 hrpS-lacZ and its derivative DM733 hrpY::Tn5 hrpS-lacZ carrying pMM118 Plac-hrpY(D57N) were grown to an A600 nm of 0.6 to 0.8. Expression of hrpS-lacZ was measured using a fluorometric β-galactosidase assay, and the data are shown as a percentage of the respective vector control strain grown in IM5.5. Media are as follows: IM5.5, standard IM lacking casamino acids with 10 mM sucrose as the C-source; High NaCl, IM5.5 with 250 mM NaCl; pH 8.0, IM buffered with HEPES to pH 8.0; nicotinic acid, IM5.5 with 2 mM nicotinic acid; citrate, IM5.5 with 10 mM trisodium citrate as a sole C source; tryptone, IM5.5 with 0.125% tryptone as the sole source of C and N. Error bars indicate standard deviations. Data are from a representative experiment with three replicates.
Organization of the hrpS promoter region.
In order to determine the elements required for HrpY-dependent regulation of hrpS, we carried out a more complete analysis of the hrpS 5′ untranslated region (UTR). In our previous study of this region (25), the smallest fragment with HrpY-dependent promoter activity extended 928 bp upstream from the ORF. This fragment included a 483-bp remnant of an IS element that is located just 7 bp upstream of the hrpS ribosomal binding site, which in turn is only a few bases from the start codon of hrpS. The remnant IS element does not appear to encode any proteins, but computer analysis (not shown) suggests that it could form a strong hairpin structure (ΔG° of −149.2 kcal/mol). At present, the regulatory significance of this feature is unknown, and it will not be addressed in this study.
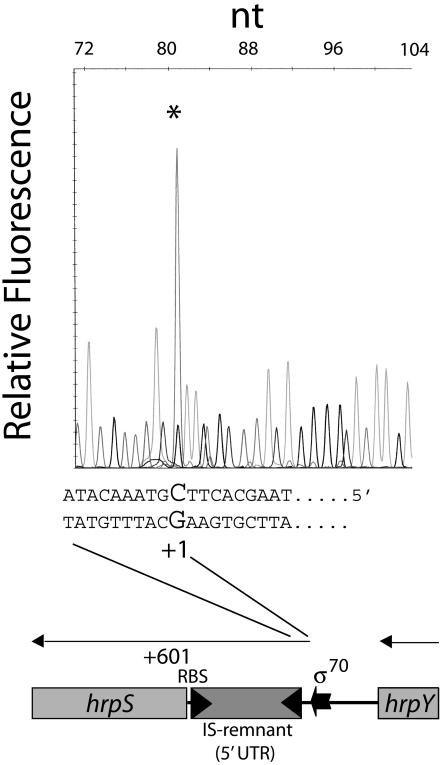
Primer extension was used to determine the 5′ end of the hrpS mRNA transcript. Given the large size of the 5′ hrpS region, we used five primers spanning the entire hrpS 5′ region (see Table S1 in the supplemental material). Only one of the primers resulted in a clear extension product. Using the 6-FAM-labeled SR3835 primer under Hrp-inducing growth conditions, the hrpS mRNA starting site was located at the cytosine base 602 bp upstream of the hrpS start codon (Fig. 2). A putative σ70 promoter (TTATCT-N17-TCTTAT) centered at −29 bp from the transcription start (see Fig. S1 in the supplemental material) was identified using PromScan (score, 70). The DNA sequences of the hrpS 5′ regions from E. amylovora, P. stewartii, P. agglomerans pv. gypsophilae, E. carotovora subsp. carotovora, E. carotovora subsp. atroseptica, and E. chrysanthemi were aligned by ClustalW (not shown) in order to identify common motifs. Given that these species exhibit similar HrpY-dependent regulation of the hrpS gene and that the 5′ regions are very similar, we predicted that common regulatory elements might be present. The regions chosen for analysis included nucleotides +1 to −241 from the transcriptional start in P. stewartii and the corresponding sequences in the hrpY-hrpS intergenic regions of various erwinia group species starting from the 3′ UTR of hrpY. (P. stewartii is the only species that has an IS-like element upstream of hrpS). The P. stewartii hrpS 5′ region appears to be most closely related to that of P. agglomerans pv. gypsophilae, with E. amylovora being the second most similar species. The pectolytic Erwinia spp. grouped separately at higher genetic distances upon phylogenetic analysis (not shown). The P. stewartii and P. agglomerans pv. gypsophilae 5′ hrpS gene sequences exhibited 69.8% identity to each other over the entire 232 bp, whereas P. stewartii and E. carotovora subsp. atroseptica had only 39.6% identity. Two short direct repeats (DRs) were centered at −74 and −95 (AAATCCTTAC-N11-AATTCCTTAC) (Fig. S1). These were present in all the erwinias, but were most conserved in the pantoeas. A potential integration host factor (IHF) binding site at −191 to −179 (TTTCAACAGGTTA; consensus, WATCAA-N4-TTR; score, 74) was identified using PromScan with the E. coli K12 consensus matrix (Fig. S1). IHF belongs to a class of DNA bending proteins that are able to impose strong curvatures in A+T-rich DNA (30). IHF binding sites are associated with many different kinds of promoters and are usually located between 60 and 150 bp upstream, which is fairly close to the location of the site found by PromScan. The curvature propensity and bending ability of the P. stewartii hrpS promoter region was therefore analyzed using the Bend-It algorithm. A high propensity for bending (>6° per helical turn) was predicted between −170 and −70 bp, with two peaks of intrinsic curvature located around positions −139 and −157 bp.
FIG. 2.
Primer extension analysis of the hrpS promoter. Total RNA from DC283 cells grown in IM5.5 was annealed with 6-FAM-labeled primer SR3745 complementary to the 5′ UTR of hrpS and extended as described in Material and Methods. Dideoxy sequencing ladders were generated for the hrpS 5′ UTR with the same primer and pMM56 DNA as a template. Reactions were appropriately diluted and run in a capillary electrophoresis sequencer to correctly estimate the size and position of the 6-FAM-labeled primer extension product (asterisk). The signal from each chromatographic peak is reported as relative fluorescence units. The bases corresponding to the transcription start on the coding strand are indicated, with the transcription start (+1) shown in a larger font. Map coordinates are relative to the transcription start site.
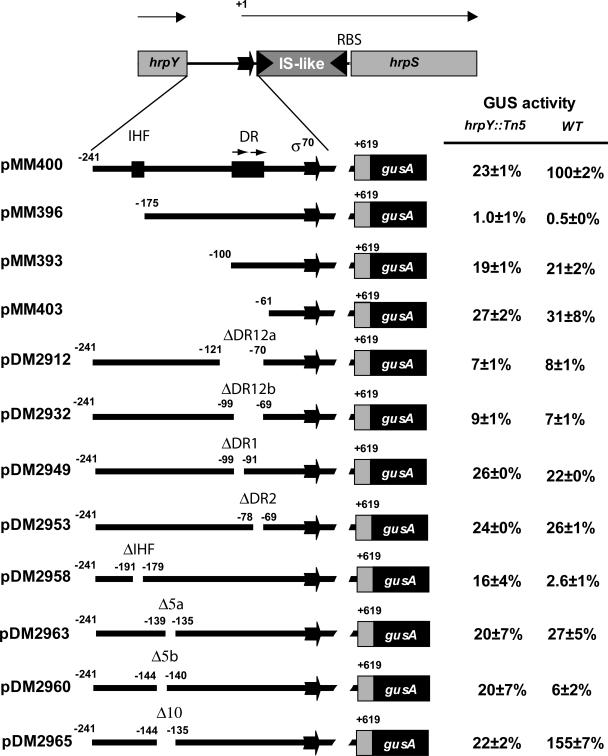
Regions of PhrpS required for activation by HrpY.
The importance of various sequences in the hrpS promoter region was genetically tested by deletion analysis using plasmid-borne transcriptional fusions (Fig. 3). These plasmids were mobilized into P. stewartii wild-type and hrpY::Tn5 strains and assayed for expression of PhrpS-uidA after growth in IM5.5 Hrp-inducing medium. The reporter fusion in plasmid pMM400, containing the entire hrpS 5′ regulatory region, required HrpY for full activation (Fig. 3), consistent with our previous findings for a similar construct. This fusion was used for comparison with all the other derivatives. Removal of the region from −241 to −175 in pMM396 totally abolished PhrpS-uidA expression. This deletion removed the putative IHF site. When just the IHF element was ablated in pMM2958, the requirement for this site was confirmed, but in this case the residual activity was somewhat higher in the hrpY background. Further deletion of the 5′ region up to, but not including, the two DRs (−241 to −100 in pMM393) (Fig. 3) restored low basal activity (ca. 20% wild-type levels), but the expression was HrpY independent, suggesting that elements upstream of the DRs are required for HrpY-dependent regulation of hrpS. A similar result was observed for pMM403, which extended the deletion to −61 but still contained the putative σ70 promoter (Fig. 3).
FIG. 3.
Deletion analysis of the hrpS regulatory region. The various hrpS promoter fragments carrying deletions from the 5′ end or internal deletions were cloned into plasmid pPL6GUSC to create PhrpS-uidA (gusA) transcriptional gene fusions. All fusions included the 493-bp IS element remnant in the 5′ UTR and a portion of the hrpS ORF. GUS activity was determined for each plasmid in wild-type strain DC283 (296 ± 5 GUS units), the hrpY::Tn5 mutant DM064, and the hrpY(D57N) mutant MM254 after growth in IM5.5 for 16 h. Data are expressed as percent activity compared to the wild-type control fusion in DC283. Data are from a representative experiment with four independent replicates per strain. GUS activities are reported as pmol of 4-methyl-umbelliferone min−1 per unit of optical density at 600 nm ± standard deviation. The coordinates shown above the genetic map are relative to the hrpS transcriptional start site. The σ70 promoter is indicated. RBS indicates the ribosomal binding site; DR12 is the region spanned by the two DRs described in the legend of Fig. 2; and Δ5a, Δ5b, and Δ10 are deletions of 5 bp, 5 bp, and 10 bp, respectively.
To test the importance of the DRs, just this region was deleted (ΔDR12, −121 to −70 in pDM2912). This resulted in a strong reduction in PhrpS-uidA expression in both genetic backgrounds to levels about 8% that of the pMM400 control fusion (Fig. 3). We then made a smaller deletion (ΔDR12, −99 to −69 in pDM2932) that included only the two DR sites and the 11 bp separating them (approximately one DNA helix turn), and this construct exhibited the same very low level of expression. To estimate the relative contribution of individual repeats, we then individually deleted each DR. The resulting fusions (ΔDR1, −99 to −90 in pDM2949, and ΔDR2, −78 to −69 in pDM2958) showed twice as much activity as the ΔDR12 fusions, but their expression was still only 22 to 26% of the pMM400 control (Fig. 3). This result suggests that these two repeats contribute in a cooperative way to the expression of hrpS.
The above findings are consistent with a model whereby both sequences next to the hrpS promoter and other elements much farther upstream are required for full activation of PhrpS. The strong reduction in expression observed upon deletion of the putative IHF site also suggests the involvement of DNA bending, which might bring an unknown coactivator, binding near the IHF site, into contact with HrpY. This model predicts that small progressive deletions, changing the phase of the DNA helix, will lead to incorrect topology of the binding elements for regulatory factors. To test this hypothesis, we separately created two different 5-bp deletions in the region halfway between the IHF element and the DR region (−144 to −140 in pDM2960 and −139 to −135 in pDM2963). Each deletion was expected to shift the phase of the helix by half a turn. Both mutations had a large negative effect on PhrpS expression (Fig. 3). In contrast, an in-phase 10-bp deletion spanning both sites (−144 to −135 in pDM2965) did not decrease PhrpS expression but, instead, seemed to stimulate it above wild-type levels (Fig. 3). These findings are consistent with the existence of factors that require a correct DNA topology to bind to upstream elements and activate the hrpS promoter.
Purification of recombinant HrpY.
To begin addressing questions concerning the biochemistry of the interaction of HrpY with its target promoter, we purified the wild-type HrpY protein and a nonphosphorylatable D57N mutant version of it. His6-HrpY and His6-HrpY(D57N) were overexpressed using the pET-15b vector. To show that the His6 tag did not affect biological activity, the his6-hrpY gene was subcloned into pRK415. Although this recombinant plasmid had a mild adverse effect on the growth of P. stewartii transconjugants, it complemented a hrpY null mutant for virulence (data not shown). We therefore decided to use the His6-tagged form of the protein for further studies due to the convenience of purification. Induction of the hrpY and hrpY(D57N) genes in plasmids pMM221 and pMM222 in E. coli BL21(DE3/pLysS) resulted in the production of 26-kDa polypeptides, which were not present in uninduced cells. (For His6-HrpY see Fig. S2, lane 2 versus lane 1, in the supplemental material; His6-HrpY[D57N] is not shown). Fig. S2 in the supplemental material shows recovery of His6-HrpY after various steps in its purification by immobilized metal affinity chromatography using step-elution gradients. Highly purified His6-HrpY was recovered from the 136 to 250 mM NaCl fractions after anion exchange chromatography. In all cases, purity was >95%, and background kinase activity was not visible by autoradiography of protein samples incubated with [γ-32P]ATP. The molecular weight of the polypeptides was confirmed by Q-TOF ESI MS. For the His6-HrpY sample, the major protein species had a molecular mass of 26,051 ± 58 Da, whereas His6-HrpY(D57N) had a mass of 26,122 ± 76 Da, both in close agreement with the expected mass of 26.1 kDa. Finally, Western blot analysis using anti-His6 monoclonal antibody confirmed the identity of the recombinant polypeptides (not shown).
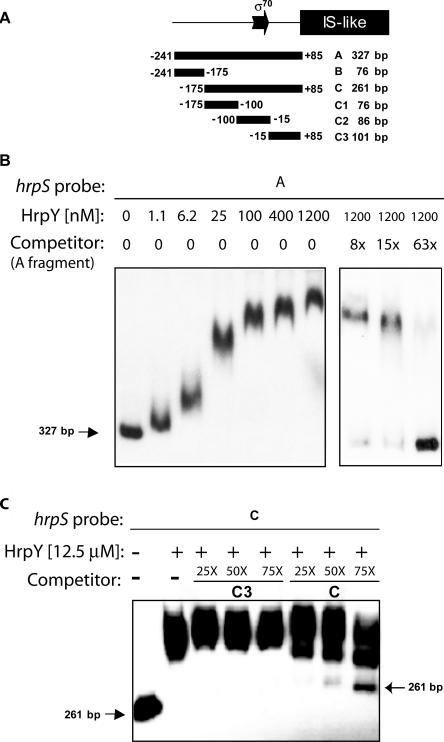
In vitro binding of HrpY to the hrpS regulatory region.
The binding of HrpY to various DNA fragments spanning the hrpS promoter and regulatory region located between the IS-like element and hrpY was studied by EMSAs. These fragments were synthesized by PCR and end-labeled with 32P. Their map positions are shown in Fig. 4A. In an initial experiment using the 327-bp fragment A (+85 to −241) as a probe, increasing concentrations of unphosphorylated His6-HrpY (≤1.2 μM) were incubated with 20 fmol of 32P-labeled fragment A DNA for 40 min, and then the protein-DNA complexes were separated from the free probe by electrophoresis in native 6% acrylamide gels (Fig. 4B). All reactions were performed in a large molar excess of the nonspecific competitor poly(dI-dC). At 1.2 μM, HrpY maximally retarded the mobility of fragment A. When lower concentrations of HrpY were used, complexes of intermediate mobility were observed migrating between the free probe and the retarded probe. The intermediate complexes formed wavy bands and may have been produced upon dissociation of the binding complex at lower HrpY concentrations. This has been reported in other systems (9). Similar results were obtained with the His6-HrpY(D57N) variant (data not shown). In a specificity control, the binding of HrpY to fragment A was dramatically reduced by competition with a 63-fold molar excess of cold fragment A (Fig. 4B). These results demonstrate specific binding of His6-HrpY to the full-length hrpS promoter region.
FIG. 4.
EMSA of the HrpY-PhrpS interaction. (A) Map of the probes used. The hrpS promoter region was amplified by PCR from plasmid pMM58 to produce fragments A, B, C, C1, C2, and C3. Coordinates are relative to the transcription start site, and sizes are reported in base pairs. The fragments shown were used in electrophoretic mobility shift experiments as either radiolabeled probes or cold specific competitors. (B) Binding of HrpY to fragment A and specific competition titration. Increasing concentrations of HrpY (0 to 1.2 μM range) were incubated with 25 fmol of 32P-labeled fragment A. The molar excesses of the unlabeled specific competitor are indicated above the lanes (0×, 8×, 15×, and 63×). The size of the free probe in base pairs is shown. (C) Competition analysis of the binding of HrpY to hrpS fragment C. HrpY was incubated with 32P-labeled DNA fragment C with and without competing unlabeled DNA fragments at various molar excesses (fragment C was used as the unlabeled specific competitor while fragment C3, which does not bind HrpY, was the nonspecific competitor). Unbound probe C in lane 8 does not match the size of unbound probe C in lane 1 due to gel migration distortion.
To further locate the HrpY-binding sites within fragment A, probes carrying smaller portions of this region were synthesized by PCR and end-labeled with 32P (Fig. 4A). Binding assays were performed with probe B (−175 to −241) containing sequences upstream of the region with high bending ability, probe C1 (−100 to −175) carrying the IHF binding site, probe C2 (−15 to −100) containing the two DRs and the σ70 promoter, probe C3 (+85 to −15) containing the 5′ UTR upstream of the IS-like element, and probe C (+85 to −175) spanning C1, C2, and C3. In each assay, 10 to 15 fmol of probe was incubated with 4.7 μM or 12.5 μM HrpY with or without cold specific competitor DNA. HrpY formed high-affinity complexes with fragment C, similar to what was observed for probe A (Fig. 4C). Fragments B, C1, and C3 were not bound by HrpY, but weak binding was observed to fragment C2 (data not shown). Subsequent footprinting experiments, described below, revealed that HrpY protected a region that was mostly contained in fragment C2 but extended 8 bp into fragment C1. To further demonstrate the specificity of the binding of HrpY to fragment C, excess unlabeled fragment C3 was incubated with HrpY and 32P-labeled probe C (Fig. 4C). Up to 75-fold molar excess of unlabeled fragment C3, acting here as an nonspecific competitor, did not block the binding of HrpY to probe C, whereas binding was increasingly reduced in the control assay using cold fragment C (Fig. 4C).
DNase I footprinting of the hrpS regulatory region.
The precise HrpY-binding sequences were identified by testing the ability of HrpY to protect against DNase I cleavage of the hrpS promoter. A fragment with the −175 to + 85 region of PhrpS (Fig. 4A, fragment C), which was fluorescently 5′ labeled with 6-FAM, was incubated with increasing amounts of His6-HrpY and then treated with DNase I. The digestion patterns were analyzed by capillary electrophoresis using an automated sequencer (Fig. 5). Two regions between −107 and −84 (FP-I) and between −78 and −55 (FP-II) were protected, which was indicated by disappearing nucleotide peaks as the HrpY concentration increased. The bases in FP-I were protected at a lower concentration of protein than FP-II, implying higher binding specificity for that region. These footprinting results further demonstrate that the binding of HrpY to PhrpS described in the gel shift experiments is specific to a precise region of the promoter localized just upstream of the −35 to −10 promoter box. DNase I footprinting did not reveal any binding elsewhere in the PhrpS region (Fig. 5 and data not shown). The two footprinted regions span the direct repeats described above, which are conserved to various degrees in Erwinia spp., and correspond to the region shown by deletion analysis to be essential for HrpY-dependent regulation of hrpS (Fig. 3).
FIG. 5.
DNase I footprinting of the HrpY binding sites. DNase I digestion reactions were prepared and analyzed by capillary electrophoresis in an ABI 3770 sequencer as described in Materials and Methods. The fluorescence intensity of the 6-FAM-labeled fragments is shown on the y axis of each electropherogram, fragment sizes are given along the top in nucleotides as determined by comparison to the internal molecular weight standards (light gray peaks marked by an asterisk in the bottom electropherogram), and the coordinates of the peaks (bases) relative to the transcription start site are given along the bottom. Solid bars at the top (labeled as FP-I and FP-II) indicate PhrpS regions protected by His6-HrpY. Four electropherograms from reactions with decreasing amounts of His6-HrpY are shown: 80 μM (a) 8 μM (b), 2 μM (c), and 0.5 μM (d) HrpY. The bovine serum albumin-only control is also shown (e).
In vitro phosphorylation of HrpY and the role of D57.
Response regulators are usually phosphorylated at a single aspartyl residue in the receiver domain, although secondary phosphorylation sites are sometimes present (10). The tertiary structure of HrpY was modeled using a homology approach based on the experimental crystallographic data from NarL and Spo0F (not shown). Our model predicted that three conserved aspartyl residues (D11, D12, and D57) at the top of a β-strand form the catalytic triad that is conserved in response regulator receivers. As shown for other response regulators (31), D57 is the likely phosphorylation site for HrpY, given its exposed position at the top of a β-strand. Substitutions at this site usually affect function but not structure.
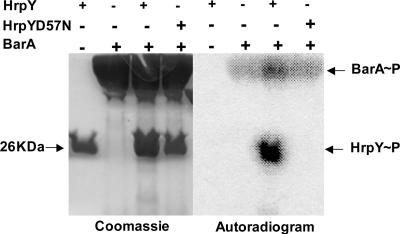
To directly test whether or not D57 is the sole phosphorylation site in HrpY, we developed an in vitro system to phosphorylate HrpY. The putative cognate sensor kinase for HrpY is probably HrpX, because it is encoded by the same operon and nonpolar, null mutations in hrpX decrease both hrp gene expression in IM5.5 medium and virulence (25). However, our attempts to purify either full-length or truncated active HrpX for in vitro transphosphorylation assays repeatedly failed (data not shown). Alternatively, we used recombinant S. enterica BarA198 sensor kinase (a truncated version of BarA engineered with an N-terminal his6 tag) for our experiments. In S. enterica, BarA transphosphorylates the SirA response regulator, which belongs to the same class of regulators (FixJ) as HrpY. Figure 6 (lanes 1 and 5) shows that purified HrpY alone did not catalyze an autophosphorylation reaction in the presence of [γ-32P]ATP. On the other hand, BarA198 autophosphorylated in presence of [γ-32P]ATP (lanes 2 and 6), and BarA198∼P efficiently transferred 32Pi to HrpY (lanes 3 and 7). The HrpY/BarA198 complex appeared more active than BarA198 alone in catalyzing the autophosphorylation reaction, at least under the conditions used here. Finally, the HrpY(D57N) mutant protein was not phosphorylated by BarA198 and [γ-32P]ATP (lanes 4 and 8), demonstrating that D57 is probably the only phosphorylation site in HrpY efficiently recognized by this kinase. Furthermore, this is consistent with our previous finding that D57A and D57N mutant alleles abolish hrp gene expression and virulence when present in single copy (25).
FIG. 6.
In vitro transphosphorylation of HrpY. Reaction mixtures contained [γ-32P]ATP, S. enterica BarA198 sensor kinase, and either HrpY or HrpY(D57N). Reactions were separated by SDS-PAGE and visualized by Coomassie brilliant blue staining (left) or PhosphorImager scanning (right). Negative controls consisted of HrpY and HrpY(D57N) incubated in the presence of [γ-32P]ATP without BarA.
Effect of phosphorylation on DNA binding affinity.
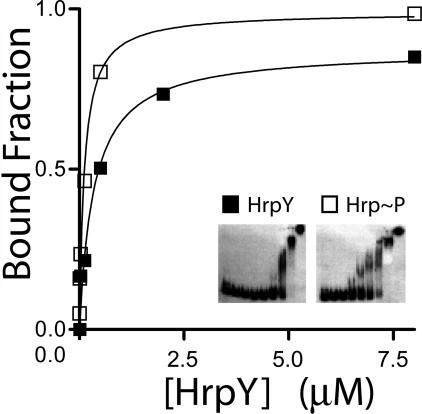
The effect of phosphorylation on the affinity of HrpY for its target promoter was analyzed by gel shift assays. Fragment C (Fig. 4) was used as a probe for EMSAs. His6-HrpY was phosphorylated either by BarA198 or by 50 mM carbamoyl phosphate, which is a small phosphodonor molecule. Increasing concentrations of phosphorylated or unphosphorylated HrpY (from 5 nM to 80 μM) were incubated with a constant amount of labeled fragment C at nanomolar concentrations. Protein-DNA complexes were separated on a 6% acrylamide gel, and the bound fraction was measured by quantifying the amount of free probe with a phosphorimager scanner and comparing it to the no-protein controls (Fig. 7). The difference was used to calculate the bound fraction. Data were plotted against protein concentrations, the binding isotherm was fitted to a hyperbolic binding model (Fig. 7), and the KD (dissociation constant) was calculated. When HrpY was phosphorylated by BarA198, the KD for binding to probe C was decreased ca. eightfold (from 8 μM to 1.2 μM), which is similar to values obtained for other response regulators of this class. Similar changes in KD values were obtained using carbamoyl phosphate, implying that small phosphodonors can also activate HrpY (data not shown). The observed KD showed variability between batches of purified protein or between stored stocks, perhaps due to inactivation, but the effect of phosphorylation was consistent. These results indicate that in vitro phosphorylation of HrpY can increase its affinity for target sites in the hrpS promoter region.
FIG. 7.
Effect of phosphorylation on the in vitro binding affinity of HrpY to PhrpS DNA. The binding isotherm plot for HrpY is shown. Increasing concentrations of HrpY or HrpY∼P (from 1 nM to 80 μM), prepared upon incubation with a 2:1 molar ratio of kinase:response regulator for 30 min, were mixed with an average of 40 fmol of 32P-labeled probe DNA at room temperature. Unphosphorylated HrpY samples contained the sensor kinase, but ATP was omitted from the reaction. The positions of the free probes and complexes and their intensities were determined by exposing the dried gel to a phosphor storage screen (inset). The signal was quantified with ImageQuant software. The bound fraction was plotted against protein concentration and fitted to a hyperbolic model using GraphPad Prism.
DISCUSSION
We previously reported that P. stewartii hrpS was controlled by the HrpX/HrpY two-component regulatory system and that the hrpS promoter responded to multiple environmental signals (25). Although hrpY was required for hrpS expression, mutations in hrpX had only a minor effect on virulence, even though they decreased PhrpS expression sixfold in IM5.5. In this study, we showed that PhrpS is activated by direct interaction with HrpY at specific regions containing 10-bp DRs and that it is also repressed by HrpY-independent mechanisms of gene regulation. Alkaline pH and high osmolarity, two signals that repress hrp gene expression, occur in planta during the late stages of pathogenesis, when the host tissues have been extensively damaged. These conditions may serve to turn off the type III secretion system when it is no longer needed. The fact that HrpY overexpression cannot bypass these repressing signals indicates that they act independently of HrpY and that their perception does not involve HrpX or other mechanisms of phosphorylating HrpY (e.g., alternate kinases or small phosphodonors). To make sure of this we employed a mutant HrpY protein that was confirmed in vitro to be nonphosphorylatable. In contrast, overexpression of HrpY was able to override the repression caused by organic acids and complex carbon and nitrogen sources, such as citrate and tryptone. Although we do not know what other mechanisms might phosphorylate HrpY or the nature of the ligands sensed by HrpX, it is noteworthy that HrpX is cytoplasmic and contains two tandem PAS domains (25). Such domains are commonly involved in monitoring redox potential, oxygen, small ligands, and the energy level inside cells, so it is possible that the HrpX/HrpY system monitors changes in redox potential during growth on organic acids and complex carbon or nitrogen sources.
An initial objective in our characterization of the hrpS promoter region was to locate the DNA sequences needed for activation by HrpY. Using deletion analysis, we demonstrated that at least two sites in the long 5′ hrpS regulatory region are simultaneously required for HrpY-dependent activation of PhrpS. The first of these sites is just a few bases upstream of a σ70 promoter box, which is over 600 bp from the ORF. HrpY binds to this region in vitro with two footprints of 24 and 16 bp, which span the two 10-bp DRs (Fig. 6). The second site (contained within fragment B) (Fig. 4) is located farther upstream (−175 to −241) and does not bind HrpY (Fig. 4), even when it is phosphorylated (data not shown). Deletion of this second region rendered the hrpS-uidA fusion in pMM396 totally inactive in both wild-type and hrpY backgrounds (Fig. 3). This deletion included a likely IHF binding site at −191 to −179, and precise deletion of this element led to a dramatic decrease in hrpS expression in both genetic backgrounds, which was comparable to that of pMM396 (Fig. 3). To genetically test the notion that upstream binding of IHF and/or other factors may be important for DNA looping and downstream HrpY-dependent expression of hrpS, we altered the DNA helix phase by introducing 5-bp or 10-bp deletions (corresponding to one half or one full helical turn) into a region midway between the IHF consensus element and the HrpY binding site. Consistent with a requirement for DNA looping, both 5-bp deletions reduced hrpS expression to levels comparable to those of hrpY null mutants, whereas the 10-bp deletion spanning the same sites actually increased hrpS expression. At this point, we have no biochemical proof that any factor, other than HrpY, directly binds the hrpS promoter region, but our preliminary finding that crude extracts from hrpY null mutants still retard fragment A in gel shift experiments (M. Merighi and D. Coplin, unpublished data) is consistent with this hypothesis.
As mentioned above, the region proximal to the hrpS promoter, which was extensively characterized by deletion analysis, EMSA, and DNase I footprinting, includes two 10-bp DRs (AAATCCTTAC-N11-AATTCCTTAC; consensus, AAWCCTTAC). These are conserved in P. agglomerans pv. gypsophilae, E. amylovora, E. carotovora subsp. atroseptica, and E. chrysanthemi (data not shown). Similar A+T-rich binding sites have been found for other FixJ class regulators, e.g., RAAAYY for E. coli UhpA (9) and TACYNMT for E. coli NarL (34). Complex binding patterns at multiple sites have likewise been observed for NarL and UhpA and also for E. coli OmpR, a “winged helix” class regulator. NarL dimers (21) bind, in a cooperative and hierarchical manner, to four heptamers located in front of the fndG operon and to eight heptamers in front of the narG operon, (20). UhpA binds as a dimer to inverted repeats in the uhpT promoter, which were mapped by iron chelated hydroxy-radical footprinting (28). On the other hand, asymmetrical OmpR dimers bind, in a cooperative fashion, to multiple 10-bp sites arranged in tandem (13). At this point, we do not know whether HrpY binds as a dimer or as an oligomeric complex. The relatively large footprinted region that we found suggests that the latter may occur. Interestingly, the distal footprint (FP-I) seems to show an apparently higher affinity for HrpY than the proximal footprint (FP-II), so that activating site occupancy may differ at various levels of HrpY phosphorylation in vivo. A simplified working model for hrpS regulation therefore involves HrpY binding at the proximal site, possibly in two different steps, and then acting cooperatively with an unidentified coactivator(s) that binds at the distal site (−175 to −241; fragment B). The DNA-bending introduced by IHF would then facilitate the interaction of this coactivator with HrpY. This would explain why loss of the IHF site in pDM2958 did not have as dramatic effect on the basal level of PhrpS expression in the hrpY background (24% to 19%) as did deletion of fragment B in pMM396 (24% to 0.7%). The restoration of the basal level by further deletion of the region from −175 to −100 in pMM393 could mean that a corepressor could bind between HrpY and the putative coactivator.
Another objective of this study was to explore the role of phosphorylation in modulating the activity of HrpY as a DNA-binding protein. Computer modeling predicted that D57 is the most likely phosphorylation site in HrpY (22). The function of this site was initially tested using hrpY alleles with conservative or neutral substitutions at residue D57. We observed the ability of hrpY(D57A) or hrpY(D57N) to complement hrpY null mutants for virulence when ectopically expressed from plasmids (25). On the other hand, substitutions at D57 abolished virulence when expressed in single copy from the chromosome. Such copy number effects are not uncommon for response regulators of this class (9). One frequent explanation is that phosphorylation enhances binding to the target promoters, but, at high concentrations, the unphosphorylated response regulator may still bind to and form active complexes on the target DNA. Another common hypothesis is that increased expression of the response regulator may lead to cross talk with other sensor kinases. The first explanation seems more likely for HrpY. Indeed, expression of an hrpS::lacZ fusion was only partially stimulated by the hrpY(D57N) and hrpY(D57A) alleles when they were ectopically expressed (data not shown). In protein phosphorylation experiments using the heterologous mutant kinase BarA198 (Fig. 7), the HrpY(D57N) protein was not phosphorylated, which provided further evidence that D57 is the sole phosphorylation site. Finally, our in vitro EMSA experiments showed that phosphorylation increased the affinity of HrpY for the hrpS promoter by severalfold. The reduction in hrpS activation by the hrpY(D57N) and hrpY(D57A) alleles, therefore, could simply be due to their decreased ability to bind at the hrpS promoter to the same extent as HrpY∼P. Since we found that the proximal binding site, FP-II, had a lower affinity for HrpY, we speculate that, at normal concentrations of HrpY, it may only be occupied when HrpY is completely phosphorylated and that binding to it may be important for full contact and activation of the RNA polymerase holoenzyme at the weak σ70 promoter.
Although HrpY needs to be phosphorylated in order to activate PhrpS at normal physiological levels, the HrpX sensor kinase does not appear to be absolutely required for hrp gene expression (25); so we proposed above that an alternate sensor kinase or small phosphodonors may phosphorylate HrpY during infection. HrpX was purified as a recombinant protein, but it did not show kinase activity under the conditions tested (22). Usually, there is a certain degree of specificity between response regulators and their cognate kinases in order to achieve proper regulation (14), but cross talk is a known phenomenon. It is therefore interesting that we were able to use a truncated, mutant form of the S. enterica BarA sensor kinase for this purpose, since this enzyme is reportedly quite specific for its cognate regulator (29). BarA/SirA orthologs, such as GacA/GacS, are found in many γ-proteobacteria, including the genera Pseudomonas, Pantoea, and Erwinia. Moreover, these regulators have important roles in regulating the virulence of salmonellae, erwinias, and pseudomonads. A recent BLAST search of the P. stewartii draft genome sequence (http://www.hgsc.bcm.tmc.edu/projects/microbial/Pstewartii/) confirmed the presence of a barA/gacS ortholog. Consequently, we are currently investigating the possibility that this ortholog may function as an alternate kinase in place of HrpX under certain environmental conditions.
Supplementary Material
Acknowledgments
This work was supported by the U.S. Department of Agriculture, Cooperative State Research, Education and Extension Service, under agreement number 2002-35319-11562. Additional salaries and research support were provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University. M. M. was supported by a Presidential Fellowship from The Ohio State University.
Purified S. enterica BarA198 protein was a gift from Max Teplitski and Brian Ahmer.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahmad, M., D. R. Majerczak, S. Pike, M. E. Hoyos, A. Novacky, and D. L. Coplin. 2001. Biological activity of harpin produced by Pantoea stewartii subsp. stewartii. Mol. Plant-Microbe Interact. 14:1223-1234. [DOI] [PubMed] [Google Scholar]
- 2.Aiyar, A., and J. Leis. 1993. Modification of the megaprimer method of PCR mutagenesis: improved amplification of the final product. BioTechniques 14:366-369. [PubMed] [Google Scholar]
- 3.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology, vol. 1. John Wiley & Sons, New York, N.Y.
- 5.Boyer, H. W., and Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Coplin, D. L., R. D. Frederick, D. R. Majerczak, and E. S. Haas. 1986. Molecular cloning of virulence genes from Erwinia stewartii. J. Bacteriol. 168:619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coplin, D. L., R. D. Frederick, D. R. Majerczak, and L. D. Tuttle. 1992. Characterization of a gene cluster that specifies pathogenicity in Erwinia stewartii. Mol. Plant-Microbe Interact. 5:81-88. [Google Scholar]
- 9.Dahl, J. L., B.-Y. Wei, and R. J. Kadner. 1997. Protein phosphorylation affects binding of the Escherichia coli transcription activator UhpA to the uhpT promoter. J. Biol. Chem. 272:1910-1919. [DOI] [PubMed] [Google Scholar]
- 10.Delgado, J., S. Forst, S. Harlocker, and M. Inouye. 1993. Identification of a phosphorylation site and functional analysis of conserved aspartic acid residues of OmpR, a transcriptional activator for ompF and ompC in Escherichia coli. Mol. Microbiol. 10:1037-1047. [DOI] [PubMed] [Google Scholar]
- 11.Frederick, R. D., M. Ahmad, D. R. Majerczak, A. S. Arroyo-Rodriguez, S. Manulis, and D. L. Coplin. 2001. Genetic organization of the Pantoea stewartii subsp. stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Mol. Plant-Microbe. Interact. 14:1213-1222. [DOI] [PubMed] [Google Scholar]
- 12.Frederick, R. D., D. R. Majerczak, and D. L. Coplin. 1993. Erwinia stewartii WtsA, a positive regulator of pathogenicity gene expression, is similar to Pseudomonas syringae pv. phaseolicola HrpS. Mol. Microbiol. 9:477-485. [DOI] [PubMed] [Google Scholar]
- 13.Harrison-McMonagle, P., N. Denissova, E. Martinez-Hackert, R. H. Ebright, and A. M. Stock. 1999. Orientation of OmpR monomers within an OmpR:DNA complex determined by DNA affinity cleaving. J. Mol. Biol. 285:555-566. [DOI] [PubMed] [Google Scholar]
- 14.Hoch, J. A., and K. I. Varughese. 2001. Keeping signals straight in phosphorelay signal transduction. J. Bacteriol. 183:4941-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jefferson, R. A. 1987. Assaying chimeric genes in plants: the GUS gene fusion system. Plant. Mol. Biol. Rep. 5:387-405. [Google Scholar]
- 16.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 17.Knoop, V., B. Staskawicz, and U. Bonas. 1991. Expression of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria is not under the control of hrp genes and is independent of plant factors. J. Bacteriol. 173:7142-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lehtimaki, S., A. Rantakari, J. Routtu, A. Tuikkala, J. Li, O. Virtaharju, E. T. Palva, M. Romantschuk, and H. T. Saarilahti. 2003. Characterization of the hrp pathogenicity cluster of Erwinia carotovora subsp. carotovora: high basal level expression in a mutant is associated with reduced virulence. Mol. Genet. Genomics 270:263-272. [DOI] [PubMed] [Google Scholar]
- 20.Li, J., S. Kustu, and V. Stewart. 1994. In vitro interaction of nitrate-responsive regulatory protein NarL with DNA target sequences in the fdnG, narG, narK and frdA operon control regions of Escherichia coli K-12. J. Mol. Biol. 241:150-165. [DOI] [PubMed] [Google Scholar]
- 21.Maris, A. E., M. R. Sawaya, M. Kaczor-Grzeskowiak, M. R. Jarvis, S. M. Bearson, M. L. Kopka, I. Schroder, R. P. Gunsalus, and R. E. Dickerson. 2002. Dimerization allows DNA target site recognition by the NarL response regulator. Nat. Struct. Biol. 9:771-778. [DOI] [PubMed] [Google Scholar]
- 22.Merighi, M. 2003. Ph.D. dissertation. Ohio State University, Columbus, Ohio.
- 23.Merighi, M., D. R. Majerczak, and D. L. Coplin. 2001. The hrp genes of Pantoea stewartii are regulated by a complex system that senses environmental signals. p. 201-204 In S. H. De Boer (ed.), Plant pathogenic bacteria. Proceedings of the 10th International Conference on Plant Pathogenic Bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 24.Merighi, M., D. R. Majerczak, and D. L. Coplin. 2005. A novel transcriptional autoregulatory loop enhances expression of the Pantoea stewartii subsp. stewartii Hrp type III secretion system. FEMS Microbiol. Lett. 243:479-487. [DOI] [PubMed] [Google Scholar]
- 25.Merighi, M., D. R. Majerczak, E. H. Stover, and D. L. Coplin. 2003. The HrpX/HrpY two-component system activates hrpS expression, the first step in the regulatory cascade controlling the Hrp regulon in Pantoea stewartii subsp. stewartii. Mol. Plant-Microbe. Interact. 16:238-248. [DOI] [PubMed] [Google Scholar]
- 26.Munteanu, M. G., K. Vlahovicek, S. Parthasaraty, I. Simon, and S. Pongor. 1998. Rod models of DNA: sequence-dependent anisotropic elastic modeling of local bending phenomena. Trends Biochem. Sci. 23:341-346. [DOI] [PubMed] [Google Scholar]
- 27.Nizan-Koren, R., S. Manulis, H. Mor, N. M. Iraki, and I. Barash. 2003. The regulatory cascade that activates the Hrp regulon in Erwinia herbicola pv. gypsophilae. Mol. Plant-Microbe. Interact. 16:249-260. [DOI] [PubMed] [Google Scholar]
- 28.Olekhnovich, I. N., and R. J. Kadner. 2002. Mutational scanning and affinity cleavage analysis of UhpA-binding sites in the Escherichia coli uhpT promoter. J. Bacteriol. 184:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pernestig, A. K., O. Melefors, and D. Georgellis. 2001. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem. 276:225-231. [DOI] [PubMed] [Google Scholar]
- 30.Stenzel, T. T., P. Patel, and D. Bastia. 1987. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell 49:709-717. [DOI] [PubMed] [Google Scholar]
- 31.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 32.Teplitski, M., R. I. Goodier, and B. M. Ahmer. 2003. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J. Bacteriol. 185:7257-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toth, I. A., and R. J. Birch. 2005. Rotting softly and stealthily. Curr. Opin. Plant Biol. 8:424-429. [DOI] [PubMed] [Google Scholar]
- 34.Tyson, K. L., J. A. Cole, and S. J. Busby. 1994. Nitrite and nitrate regulation at the promoters of two Escherichia coli operons encoding nitrite reductase: identification of common target heptamers for both NarP- and NarL-dependent regulation. Mol. Microbiol. 13:1045-1055. [DOI] [PubMed] [Google Scholar]
- 35.Wei, Z. M., J. F. Kim, and S. V. Beer. 2000. Regulation of hrp genes and type III protein secretion in Erwinia amylovora by HrpX/HrpY, a novel two-component system, and HrpS. Mol. Plant-Microbe. Interact. 13:1251-1262. [DOI] [PubMed] [Google Scholar]
- 36.Xiao, Y., S. Heu, J. Yi, Y. Lu, and S. W. Hutcheson. 1994. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J. Bacteriol. 176:1025-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao, Y., and S. W. Hutcheson. 1994. A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J. Bacteriol. 176:3089-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao, Y., Y. Lu, S. Heu, and S. W. Hutcheson. 1992. Organization and environmental regulation of the Pseudomonas syringae pv. syringae 61 hrp cluster. J. Bacteriol. 174:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yap, M. N., C. H. Yang, J. D. Barak, C. E. Jahn, and A. O. Charkowski. 2005. The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J. Bacteriol. 187:639-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zianni, M., K. Tessanne, M. Merighi, R. Laguna, and F. R. Tabita. 2006. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 17:103-113. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.