Abstract
Bacterial populations produce dormant persister cells that are resistant to killing by all antibiotics currently in use, a phenomenon known as multidrug tolerance (MDT). Persisters are phenotypic variants of the wild type and are largely responsible for MDT of biofilms and stationary populations. We recently showed that a hipBA toxin/antitoxin locus is part of the MDT mechanism in Escherichia coli. In an effort to find additional MDT genes, an E. coli expression library was selected for increased survival to ampicillin. A clone with increased persister production was isolated and was found to overexpress the gene for the conserved aerobic sn-glycerol-3-phosphate dehydrogenase GlpD. The GlpD overexpression strain showed increased tolerance to ampicillin and ofloxacin, while a strain with glpD deleted had a decreased level of persisters in the stationary state. This suggests that GlpD is a component of the MDT mechanism. Further genetic studies of mutants affected in pathways involved in sn-glycerol-3-phosphate metabolism have led to the identification of two additional multidrug tolerance loci, glpABC, the anaerobic sn-glycerol-3-phosphate dehydrogenase, and plsB, an sn-glycerol-3-phosphate acyltransferase.
Persisters are specialized cells that exhibit multidrug tolerance (MDT), an ability to survive killing by antibiotics acting against unrelated targets (8, 20, 29, 38, 44, 47, 48). Tolerance is mechanistically distinct from resistance, which acts to prevent the antibiotic from inhibiting the target. We have suggested that tolerance results from antibiotic targets being blocked in a benign way by MDT proteins (29). Bactericidal antibiotics kill not by merely inhibiting a function but also by corrupting the target. For example, β-lactam antibiotics inhibit cross-linking of peptidoglycan, causing autolysis, aminoglycosides create toxic misfolded peptides by interrupting translation, and fluoroquinolones inhibit the ligase function of topoisomerase, resulting in DNA strand breaks (39, 41, 46).
In previous work, we isolated persisters from a population of Escherichia coli lysed with ampicillin and obtained an expression profile of these cells (29). The profile showed downregulation of proteins involved in energy production and nonessential functions such as flagellar synthesis, suggesting that persisters are dormant cells. The expression profile was also consistent with the finding that persisters formed by a hipA7 mutant mutant strain (an allele conferring high persistence) of E. coli are nongrowing (or slow-growing) cells (4). The profile pointed to proteins that may be responsible for dormancy: RMF, a stationary-state inhibitor of translation, SulA, an inhibitor of septation, and toxin-antitoxin (TA) module elements RelBE, DinJ, and MazEF (7, 12, 51). We have shown that expression of RelE, a “toxin” that causes reversible stasis by inhibiting translation, strongly increases tolerance to antibiotics (29). As with RelE, expression of the putative HipA toxin increased tolerance as well, but unlike RelE, deletion of the hipBA locus caused a sharp, 10- to 100-fold decrease in persister production in stationary-state and biofilm cultures (29). This suggested that hipA is the first bona fide persister/MDT gene. Deletion of other potential candidates, such as rmf, relE, or mazF, had no discernible phenotype in persister production, possibly due to the high degree of redundancy of these elements. In E. coli there are at least 10 TA modules, and there are >60 in Mycobacterium tuberculosis (23). We have suggested that the term “toxin” is a misnomer and that the function of chromosomal TA modules like hipBA is to protect the population from lethal factors by creating a subpopulation of multidrug-tolerant, dormant persister cells.
Persisters are not created in early exponential phase but rise to ∼1% in a stationary population (28, 44). This suggests that the major role of persisters is to protect the population from extinction when it is in a nongrowing, surviving mode. The presence of persisters, especially in biofilms protected from the immune system, presents a serious human health problem by creating multidrug-tolerant infections (16, 32). In Western countries, biofilms are responsible for 65% of all human infections (33). Finding the mechanism of tolerance will provide an understanding of an important microbial survival strategy and may lead to the development of much-needed antipersister therapies.
In this study, we report the identification of three additional genes involved in MDT: plsB, glpD, and glpABC. PlsB, the glycerol-3-phosphate acetyltransferase, is an essential and conserved persister protein that is a potential target for antipersister therapy.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids are listed in Table 1. Wild-type E. coli K-12 EMG2 (3, 14) was obtained from the E. coli Genetic Stock Center (EGSC 4401) and used as the parental strain. Cells were cultured in Luria-Bertani (LB; 10 g tryptone, 5 g yeast extract, 10 g sodium chloride; Fisher Scientific) medium with aeration at 37°C unless otherwise indicated. When required, the medium was supplemented with antibiotics at the following final concentrations: ampicillin (Sigma), 100 μg/ml; chloramphenicol (Sigma), 30 to 50 μg/ml; kanamycin (Fisher), 50 μg/ml; ofloxacin (Sigma), 5 μg/ml; and ciprofloxacin (Sigma), 5 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Parent strain or plasmid | Reference or construction details |
|---|---|---|---|
| Strains | |||
| EMG2 | (Wild type) K-12 F+ λ+ | 1 | |
| KLE416 | ΔglpD::FRT | EMG2 | This study |
| E4152 | EcoK r− m− McrBC− Mrr− Dam+ Dcm+ carrying pACYC184 | Er2420 | NEB |
| JM109 | e14− (McrA−) recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 Δ(lac-proAB) (F′ traD36 proAB lacIqZΔM15) | Stratagene | |
| HM22 | zde-264::Tn10 dapA6 hipA7 | HM21 | 39 |
| KL312 | ΔhipBA::FRT | EMG2 | 31 |
| KLE401 | JM109 carrying pACYC184 | JM109 | This study |
| KLE414 | EMG2 carrying pBADglpD | EMG2 | This study |
| KLE413 | ΔhipBA::FRT ΔglpD::kan | KL312 | This study |
| KLE418 | ΔglpABC::kan | EMG2 | This study |
| KLE421 | EMG2 carrying pBADcontrol | EMG2 | This study |
| KLE443 | EMG2 carrying pBAD3-2 | EMG2 | This study |
| KLE4481 | ΔmgsA::FRT | EMG2 | This study |
| KLE449 | ΔgloA::kan | EMG2 | This study |
| MV1703 | ΔglpR::FRT | EMG2 | This study |
| MV1723 | ΔglpR::FRT ΔglpD::kan | MV1703 | P1 from KOJW3389 |
| CAG12164 | malF3089::Tn10 | C. Gross | C. Gross |
| KLE456 | ΔdgkA::kan malF3089::Tn10 | KOJW4002 | P1 from CAG1216 |
| BB26-36 | plsB26 plsX50 glpD3 glpR2 phoA8 StrsrelA1 spoT1 tonA22 T2R pit-10 HfrC | Strain 8 | 22 |
| KLE459 | BB26-36 malF3089::Tn10 | BB26-36 | P1 from KLE456 |
| KLE456 | malF3089::Tn10 | EMG2 | P1 from KLE459 |
| KLE457 | malF3089::Tn10 plsB26 | EMG2 | P1 from KLE459 |
| TJS2 | malT::Tn10 glpR12 | T. J. Larson | |
| MV1714 | malT::Tn10 | EMG2 | P1 from TJS2 |
| MV1715 | malT::Tn10 glpR12 | EMG2 | P1 from TJS2 |
| MV1718 | malT::Tn10 ΔglpD::kan | MV1714 | P1 from KLE416 |
| MV1719 | malT::Tn10 glpR12 ΔglpD::kan | MV1715 | P1 from KLE416 |
| KLE458 | ΔglpQ::kan | EMG2 | P1 from KOJW2233 |
| KOJW3414 | ΔugpQ::kan | BW25113 | 4 |
| KLE459 | ΔugpQ::kan | EMG2 | P1 from KOJW3414 |
| KOJW4323 | ΔmdoB::kan | BW25113 | 4 |
| KLE460 | ΔmdoB::kan | EMG2 | P1 from KOJW4323 |
| KOJW1037 | ΔmdoH::kan | BW25113 | 4 |
| KLE461 | ΔmdoH::kan | EMG2 | P1 from KOJW1037 |
| KOJW3897 | ΔglpK::kan | BW25113 | 4 |
| KLE462 | ΔglpK::kan | EMG2 | This study |
| KLE463 | ΔglpD::FRT ΔglpK::kan | KLE416 | P1 from KOJW3897 |
| Plasmids | |||
| pKD13 | Template plasmid aphA | 19 | |
| pKD46 | λ Red recombinase expression plasmid bla | 19 | |
| pACYC184 | p15A cat tet | NEB | |
| pBAD33 | P15 cat ara pBAD | 14 | |
| pBAD3035 | P15 cat ara pBAD::relE | pBAD33 | 13 |
| pBADglpD | relE removed from MCS with SphI and XbaI and replaced with glpD in those same sites | pBAD3035 | This study |
| pBAD3-2 | Site-directed mutant, Gly15Arg | pBADglpD | This study |
Recombinant DNA procedures.
PCR amplification was performed using Taq DNA polymerase (New England BioLabs [NEB]) except when PCR products were used for cloning. In the latter case, Pfu DNA polymerase (Stratagene) was used. Sequencing was performed by the Forsyth sequencing facility, Boston, MA, or Seqwright, Houston, TX.
Expression library construction.
Genomic DNA from EMG2 was purified using Promega's Wizard genomic DNA purification kit and partially digested with Sau3aI. The digest was then separated on an agarose gel, and the region corresponding to 3- to 10-kb fragments was excised. Fragments were ligated into the BamHI (NEB) site of pACYC184 (NEB E4152S). Chemically competent JM109 cells (Stratagene) were transformed with the genomic library, plated on LB agar with 50 μg/ml chloramphenicol, and cultured overnight at 37°C. Approximately 4,500 of the resulting colonies were lifted into 4 ml LB broth with 8% dimethyl sulfoxide and frozen at −80°C in 50-μl aliquots.
Selection procedure and clone validation.
The genomic library was cultured in LB medium to exponential phase and selected for survival upon exposure to ampicillin (100 μg/ml). Survival was compared to the negative control containing an empty vector in strain KLE401. A KLE401 culture spiked with a high-persistence hipA7 mutant strain, HM22, at a 1:10,000 ratio served as a positive control for the selection procedure. Cultures of HM22, KLE401, or the genomic library were first grown for 18 h to stationary phase. For the first round of selection, the stationary-phase negative control culture was diluted 1:100 into LB medium and the genomic library was diluted 1:100 in 100 ml LB with 75 μg/ml diaminopimelic acid (no chloramphenicol added) in a 1-liter flask and grown for 3 h. The positive control contained KLE401 diluted 1:100 and HM22 diluted 1:10,000 and was grown under identical conditions. After 3 hours the cultures were sampled and initial cell density was determined by diluting and plating. Ampicillin was then added to a final concentration of 100 μg/ml. After 3 h of exposure to ampicillin, the cells were centrifuged at 10,000 × g for 10 min at 10°C. The pellet was resuspended in 12.5 ml LB, sampled for surviving cells, and grown overnight in a 125-ml flask. The sample of surviving cells was plated on LB, and the positive control was plated on LB with chloramphenicol. One-milliliter samples of each culture were frozen in 20% glycerol after each round of selection. Each subsequent round of selection was initiated by diluting the previously selected overnight culture 1:100 in 100 ml LB followed by 3 h of growth, killing by ampicillin, and recovery procedures. After the fourth sequential selection, individual clones from the library were picked and grown for plasmid isolation. The plasmids were transformed into a clean JM109 background and tested for high-persistence phenotype under conditions corresponding to a single selection round. Unique high-persistence clones were identified by restriction digestion using SspI (NEB). Unique clones were sequenced using primers to plasmid pACYC184: LpACYC184 (5′-GTCACTATGGCGTGCTGCTA-3′) and RpACYC184 (5′-GGTGATGTCGGCGATATAGG-3′).
Construction of a GlpD overexpression vector and glpD mutagenesis.
glpD was cloned into the multicloning site of pBAD33 (13) to create pBADglpD. The gene was amplified from K-12 genomic DNA using primers xbaIglpD (5′-GCTCTAGAGGAGTGAAACGATGGAAACCAAAGATCTGATTGT-3′ with XbaI site [italics], Shine-Dalgarno [underlined], and start codon [bold]) and sphIglpD (5′-AAAAGCATGCAATTTACGACGCCAGCGATA-3′). This vector was transformed into EMG2 to create strain KLE414. A glpD point mutant was created by mutating glycine 15 to an arginine in the flavin adenine dinucleotide (FAD) binding site. The codon was mutated from GGT to AGG using primers g43a/t45gglpD (5′-GGGCGGCATCAATAGGGCTGGTATCGCGGC-3′) and ASg43a/t45glpD (5′-GCCGCGATACCAGCCCTATTGATGCCGCCC-3′). The mutagenesis was done using Stratagene's QuikChange site-directed mutagenesis kit, and the resulting plasmid, pBAD3-2, was transformed into EMG2, creating strain KLE443. The resulting mutant's sequence was confirmed.
l-α-Glycerophosphate dehydrogenase assay.
l-α-Glycerophosphate dehydrogenase activity was measured in crude cell extracts using a procedure adapted from Lin et al. (34). Stationary cultures were diluted 1:1,000 in 20 ml LB containing 50 μg/ml chloramphenicol and 0.2% arabinose and grown until the cell density reached ∼2 × 107 CFU/ml. The cells were pelleted at 10,000 × g for 10 min at 4°C, washed in 25 ml deionized H2O, and resuspended in 0.5 ml 0.1 M Tris-HCl, pH 7.4. The cells were disrupted using an ultrasonic homogenizer (model 150V/T; Biologics, Inc.) with the power set at 40 and pulsed at 30% for 4 min in an ice-H2O bath. Glycerophosphate dehydrogenase activity was measured by following reduction of 1-(4,5-dimethylthiazol-2-yl)-3,5-diphenylformazan (MTT) into formazan absorbing at 550 nm. Five μl of freshly prepared crude cell extract was added to 3 ml of the reaction buffer containing 0.01 M KCN, 0.33 mM phenazine methosulfate in phosphate-buffered saline (PBS), pH 7.5, 89 μM MTT in 1× PBS, pH 7.5, and 1× PBS, pH 7.5, at 25°C. The reaction was started by adding 0.1 ml of 1 M dl-α-glycerophosphate. The total protein concentration of the samples was determined using the Pierce Biotechnology Micro BCA protein assay reagent kit.
G3P determination.
Glycerol-3-phosphate (G3P) was determined by a procedure adapted from references 25 and 42. One milliliter of stationary-phase cells (or 10 ml of a 1:10 dilution of stationary-phase cultures in fresh LB medium) was washed with the same volume of room temperature PBS, pH 7, and resuspended in 60 μl ice-cold water. Ten microliters of this suspension was mixed with 40 μl water and heated for 15 min at 95°C to quantify the protein concentration using the Bradford reagent (Sigma) following the manufacturer's instructions. Four microliters of aldehyde-free formic acid (Fisher) was added to the remainder of the cell suspension (final concentration, 1.4 M) and then incubated on ice for 1 h. The suspension was centrifuged, and the supernatant was moved to a clean tube and neutralized with 30 μl of 5 M K2CO3 (final concentration, 1.8 M). The enzymatic assay for detecting glycerol-3-phosphate was based on the conversion of NAD to NADH by glycerol-3-phosphate dehydrogenase (Fisher) by measuring NADH fluorescence at 445 nm (excitation, 340 nm) with a Molecular Dynamics fluorescent plate reader. Five microliters of cell lysate was added to 143.5 μl of a glycine-hydrazine buffer (1 M glycine, 0.4 M hydrazine sulfate, 1 M NaOH, 5 mM disodium-EDTA) containing also 2 mg/ml β-NAD and was blanked after a 10-min incubation at 37°C. Then, 1.5 μl glycerol-3-phosphate dehydrogenase (0.1 mg/ml final concentration; Fisher) was added and the measurement was taken at 445 nm (excitation, 340 nm) after a 15-min incubation at 37°C. A standard curve was prepared using glycerol-3-phosphate (Fisher). The concentration of glycerol-3-phosphate was expressed per total protein concentration of each sample.
MIC assay.
The MIC of each antibiotic was determined by the standard NCCLS broth microdilution method (2).
Multidrug tolerance assay.
Persistence was measured by determining survival upon exposure upon exposure to 100 μg/ml ampicillin, 5 μg/ml ofloxacin, or 5 μg/ml ciprofloxacin in exponential- and stationary-phase cultures.
Stationary-phase cultures were produced by inoculating 3 ml LB with a 1-μl aliquot of frozen stock into 17- by 100-mm polypropylene tubes and growth for at least 16 h. The initial cell concentration was determined by sampling 10 μl and serially diluting and plating on LB agar. Cell survival was tested by diluting the culture 1:10 into fresh medium containing antibiotics and sampling after 6 h of shaking at 37°C.
Late-exponential-phase cultures were produced by first diluting a stationary-phase culture 1:1,000 in LB medium (12.5 μl in 125 ml). These cells were then shaken with aeration at 37°C for 5 h. The initial cell concentration was determined by sampling 10 μl and serially diluting and plating on LB agar. One-milliliter aliquots of the late-exponential-phase cells were exposed to antibiotics for 3 h. After 3 h, samples were removed, serially diluted, and plated on LB agar.
Strain construction.
The strains were constructed as previously described by homologous recombination using a PCR product that deleted the entire gene and stably replaced it with a selectable marker (18). Primers were designed with 50 bp upstream and downstream of the gene plus the complementary sequence for amplification of the kanamycin resistance cassette of pKD13. Primers used for creating and verifying knockouts will be made available on request. Some deletions were transduced from other backgrounds into EMG2 using P1 (Table 1).
RESULTS
Selecting a genomic library for persister genes.
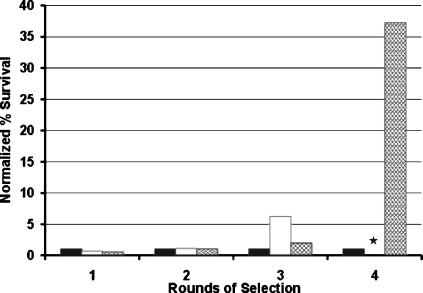
A genomic library was prepared by cloning partially digested DNA from strain EMG2 into pACYC184 and transformation into E. coli JM109. For selection, cells were cultured in LB medium to exponential phase and ampicillin was added at 100 μg/ml. At this concentration, ampicillin effectively lyses regular cells and only persisters survive (6, 29). E. coli does not form mutants resistant to such a high dose of ampicillin, ensuring that any surviving cells are persisters. An HM22 hipA7 strain that produces 1,000-fold-more persisters (1%) in exponential phase than the wild type was used as a control to probe the selection procedure. hipA7 cells were mixed with the wild type at a 1:10,000 ratio and subjected to repeated rounds of selection by exposing cells to ampicillin and then culturing surviving cells. Plating on LB agar and LB agar with tetracycline allowed for separately counting colonies of the KLE401 and hipA7 cells, correspondingly. After three cycles, the surviving cells were HM22 (no chloramphenicol-resistant KLE401 cells were detected), showing that this method can effectively select for cells with an increased probability of persister formation (Fig. 1). Cells from the recombinant library were subjected to a similar selection, and after four rounds of enrichment, 4.0 × 106 CFU/ml out of the 2.4 × 107 CFU/ml of logarithmically growing culture of library clones survived, while only 5 × 105 CFU/ml of 3.2 × 107 CFU/ml of the negative control survived. After normalization to the initial cell concentration of the negative control, the survival of the library was 37 times longer than the control with an empty vector (Fig. 1).
FIG. 1.
Selection of an E. coli overexpression clone library for increased persister production. Ampicillin (100 μg/ml) was added to exponential-phase cells in a growth medium, and after 3 h of incubation surviving cells were collected, washed, and regrown for a subsequent round of selection. The percent survival is normalized to the amount of killing of the negative control on each day. Black bars, negative control (KLE401); white bars, positive control (KLE401) and high-persistence mutant HM22 added at 1:10,000 before round 1; hatched bars, expression library. The level of persisters for the positive control was not determined on day 4; this is indicated by a star.
Plasmids prepared from six of the clones present after the fourth selection were retransformed into wild-type cells, and exponential-phase cells were tested for tolerance to ampicillin and ofloxacin in a dose-response experiment (data not shown). Four of the six clones survived better than the wild-type control. Clone 4-1 showed the strongest increase in tolerance and was examined further.
Analysis of GlpD involvement in persister formation.
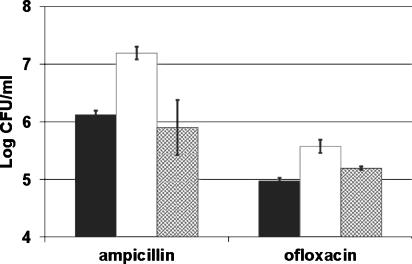
The recombinant plasmid of clone 4-1 had a 6.3-kb insert in the pACYC184 vector. Sequencing of the insert showed that it contained two genes, glpD and yzgL. GlpD is an inner membrane-associated sn-glycerol-3-phosphate dehydrogenase, and yzgL is a conserved hypothetical protein (45). The glpD gene was PCR amplified, cloned into the pBAD33 vector under the arabinose promoter, and transformed into wild-type KLE401 cells. Cells carrying the pBADglpD plasmid expressing glpD showed increased tolerance to ampicillin and ciprofloxacin in late exponential phase (Fig. 2).
FIG. 2.
Levels of persistence to ampicillin or ofloxacin in late exponential phase. Cells were diluted 1:1,000 and induced with 0.2% arabinose at time zero. After 5 h; samples were removed and tested for their multidrug tolerance to 100 μg/ml ampicillin or 5 μg/ml ofloxacin. The starting cell number for each was 1 × 109 CFU/ml. Black bars, wild-type control (KLE421); white bars, overexpression of GlpD (KLE414); hatched bars, overexpression of inactive GlpD (KLE443). These data are the averages of three independent experiments, and the error bars correspond to the standard deviations of the means.
We have previously shown that persisters are not formed in the early exponential phase of E. coli growth but instead increase in mid- to late-exponential-phase growing cells (28). In agreement with this finding, a time course experiment showed the persister level in GlpD-expressing cells increased faster than the wild type carrying the control vector (data not shown), and in late exponential phase (5 h of growth), this strain formed 10 times more persisters (1.6 × 107 CFU) than the wild type carrying the control vector (1.3 × 106 CFU) (Fig. 2). The persister level of the GlpD-producing strain was four times higher (an average of 3.7 × 105 CFU) when tested with ofloxacin, an inhibitor of DNA gyrase/topoisomerase that is capable of killing nongrowing cells, compared to the wild type carrying the control vector (an average of 9.3 × 104 CFU) (Fig. 2). The survival rate observed in the experiment is different from that in the original selection. We believe this is a result of different culture conditions as well as differences in time of exposure to the antibiotics. Variations in persister levels have also been observed by others studying the persistence phenomenon (30, 48).
Cells overexpressing glpD had a similar growth rate compared to wild type, and their MICs to ampicillin and ofloxacin were unchanged (6.25 μg/ml and 0.1 μg/ml, respectively). This confirms that while expression of GlpD increases the frequency of persisters, it does not affect resistance.
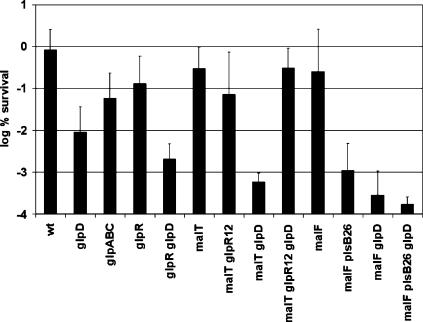
To test persister formation in cells lacking GlpD, a precise glpD deletion was created by allelic replacement, producing strain KLE416. This strain was unable to grow on a medium containing glycerol as a sole carbon source and grew on glucose minimal medium, confirming the ΔglpD phenotype (34). Exponential-phase cells of KLE416 did not show an apparent change in the level of persisters. However, the persister level in KLE416 decreased by 20-fold in undiluted (data not shown) and by 55-fold in diluted stationary state compared to the wild type (Fig. 3). This is similar to our previous observations with ΔhipA cells, which have a decreased level of persisters but only in stationary state (29). Note that the level of persisters at stationary state is ∼1% for wild-type cells (compared to 10−6 to 10−4 in early exponential phase), indicating that they have a significantly more important role in a nongrowing population. Knocking out glpD eliminates the bulk of stationary-state persisters, suggesting that glpD is essential in MDT cells.
FIG. 3.
Survival of mutants affected in putative persister genes. Cells were grown to stationary phase and diluted 1:10 in fresh medium (final concentration, ∼3 × 108 CFU/ml) containing 5 μg/ml ciprofloxacin. After 6 h of exposure, cells were washed, diluted, and spot plated. The wild type is EMG2, and all mutations were made in this background. These data are the averages of the log percent survival of at least three independent experiments, and the error bars correspond to the standard deviations of the means.
It was important to learn whether the effect of GlpD on persister formation was due to its catalytic activity or, rather, to the physical presence of this protein. Site-directed mutagenesis was used to replace a conserved arginine in the glycine-rich FAD-binding region with glycine. The complete loss of glycerol-3-phosphate dehydrogenase activity in the mutant was confirmed using a colorimetric assay for G3P (data not shown). The strain carrying plasmid pBAD3-2 with the mutant glpD gene had the same level of persisters as the wild type carrying the control vector in exponential phase (data not shown) and in late exponential phase (an average of 1.3 × 105 CFU versus 0.8 × 105 CFU) (Fig. 2). This result links persistence to the G3P dehydrogenase activity of GlpD.
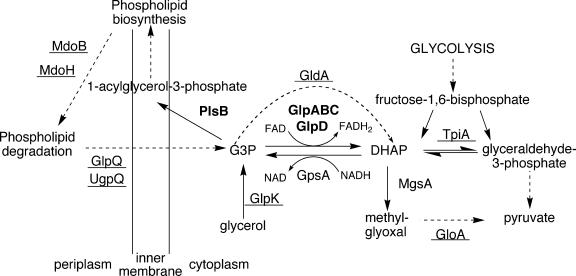
To further analyze the role of GlpD in persister formation, we considered the role of GlpD in E. coli metabolism (Fig. 4). GlpD and its paralog, GlpA, are involved in the enzymatic conversion of dihydroxyacetone phosphate (DHAP) into G3P (15, 26). It seemed logical to test both for their roles in persister formation. Under aerobic conditions, ΔglpABC (KLE418) had a lower persister level than the wild type but it was still higher than ΔglpD (KLE416) (Fig. 3). The effect of the loss of glpABC on persister formation was not large, and we hesitate to draw far-reaching conclusions from this. However, the behavior of ΔglpD and ΔglpABC, coupled with the absence of a phenotype in a strain overexpressing a nonfunctional GlpD, implicates dehydrogenation of G3P in persister formation.
FIG. 4.
Role of glycerol-3-phosphate in E. coli metabolism. Enzyme names in bold indicate involvement in persistence. Underlined enzyme names indicate there was no detectable change in persistence in a deletion mutant.
Some strains tested for their survival were created using cotransduction with a selectable marker. malT::Tn10 and malF::Tn10 were both used in the experiments detailed in Fig. 3. Note that these mutations alone had no effect on the level of persistence when compared to the wild type. However, both malT::Tn10 and malF::Tn10 mutations somewhat exaggerated the effect of a glpD mutation in a wild-type background. This result points to the possible involvement of other metabolites, like maltose, in the persistence phenomenon.
Another potential player in persister formation is GpsA, an enzyme that preferentially catalyzes the reaction from DHAP to G3P (Fig. 4) (17, 26). However, our inability to construct a strain with gpsA deleted suggests it is an essential protein.
G3P has a prominent position in E. coli metabolism and is linked to several pathways that could potentially affect persister formation. We were particularly interested in the methylglyoxal (MG) pathway, which produces MG, an intermediate that causes cell stasis (1). MG synthesis is a glycolytic bypass that leads to pyruvate via DHAP and MG (Fig. 4) (1, 11, 22, 36). DHAP is the product of GlpD-catalyzed dehydrogenation of G3P, and one would expect that an increase in DHAP would lead to an increase in MG, which could in turn produce dormancy. However, persister levels were not affected in strains with a deletion of mgsA (KLE4481), the enzyme converting DHAP into MG (data not shown), nor were they affected in a strain with a deletion of gloA (KLE449), an enzyme that catalyzes the detoxification of MG to pyruvate (36) (data not shown). These results indicate that MG is not involved in persister formation.
Deletion of glpD leads to at least two important changes: it abolishes the conversion of G3P to DHAP under aerobic conditions, resulting in an increase in the intracellular concentration of G3P, and this in turn leads to derepression of the glp regulon through the binding of G3P to the GlpR repressor (52). We sought to assess these two effects (high G3P concentration and induction of glp regulon) of the glpD mutation separately. We reasoned that we could eliminate the derepression of the glp regulon by taking advantage of an allele of the glpR repressor gene, glpR12, that codes for a repressor that is insensitive to G3P concentration (52). In this mutant strain, the glp regulon cannot be derepressed. We compared persister levels in control strains and in a ΔglpD glpR12 double mutant (MV1715 in the wild-type background, MV1718 in the KLE416 background), as well as a ΔglpDΔglpR double mutant (MV1703 in the wild-type background, MV1723 in the KLE416 background). We observed that each glpR mutation alone did not affect multidrug tolerance compared to the isogenic wild type (Fig. 3). As expected, a double deletion of glpD and glpR (where high G3P cannot further derepress the constitutive expression of the regulon) had tolerance levels similar to the ΔglpD background. On the other hand, when the ΔglpD mutant was combined with a glpR12 noninducible allele, it lost its phenotype and had wild-type levels of persistence (Fig. 3). These results show that both the induction of the glp regulon and high levels of G3P are needed for the observed effect on persistence.
G3P is the precursor of the backbone of phospholipid biosynthesis (Fig. 4) (26). It seemed possible that depletion of G3P would cause an inhibition of phospholipid biosynthesis and result in a larger subpopulation of multidrug-tolerant, dormant cells. The PlsB enzyme that uses G3P to produce 1-acyl-G3P is essential, and we could not analyze the behavior of a null mutant. However, a strain with a mutant PlsB enzyme with a higher Km has been described (plsB26), and we used it to examine persister formation (24). The plsB26 strain grew normally and showed the same MICs to ampicillin and ofloxacin as an isogenic wild-type control (data not shown). Interestingly, the plsB26 strain produced considerably fewer persisters than either the wild type or the ΔglpD strain (Fig. 3). This result was unexpected, as a decrease in PlsB function would diminish the rate of phospholipid synthesis, which could result in dormancy and increase the level of persisters.
Testing of additional members of the glp regulon and components of metabolism connected to G3P showed no effect on persister formation (data not shown). Thus, strains with deletion of tpiA (40), mdoB (27), mdoH (27), glpQ (31), ugpQ (31), gldA (5, 50), glpK (35), glpF (35), glpX (19), and glpT (35) had no phenotype when their survival in the presence of ofloxacin was tested in the stationary state. This mutational analysis essentially completes the known pathways linked to GlpD and G3P.
Intracellular G3P concentration.
It seemed interesting to learn whether the intracellular level of G3P correlated with persister production. G3P concentration was therefore measured in a variety of strains described in the previous sections.
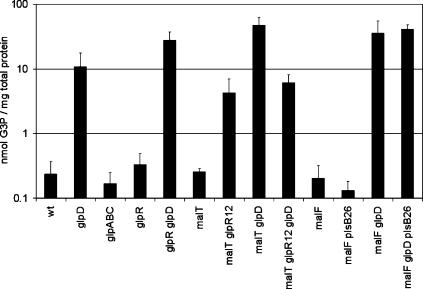
G3P concentration in stationary phase was low in the wild type and, as expected, much higher (50-fold) in the glpD strain (Fig. 5). A mutation in glpD increased the G3P level in all backgrounds except when combined with glpR12, the noninducible GlpR repressor (Fig. 5). In a glpR12 background, glpD itself cannot be induced above its basal level and therefore does not dehydrogenate a significant amount of G3P. Hence, the G3P level is higher in a glpR12 background than in the wild type, but not as high as in ΔglpD. G3P is a substrate of PlsB, but the decrease in PlsB activity (plsB26 allele) did not lead to an increase in G3P concentration (Fig. 5). This result shows that the partially functional plsB26 allele was not affecting persistence by elevating G3P.
FIG. 5.
Intracellular G3P concentration in stationary phase in strains involved in G3P metabolism. These data are the averages of at least four independent experiments, and the error bars correspond to the standard deviations of the means.
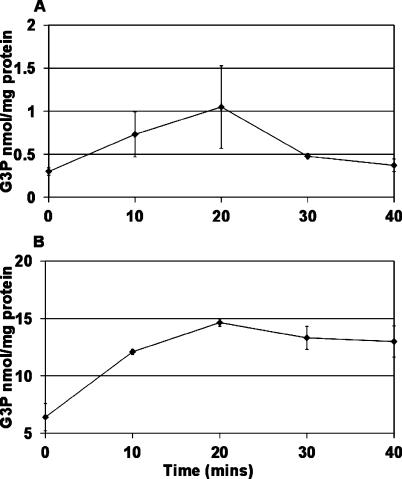
Persisters resuscitate from their dormant state when a stationary culture is inoculated into fresh medium. We therefore wanted to know how G3P levels change under these conditions. Upon inoculation of stationary-phase cells into fresh LB medium, the G3P level of both wild-type and ΔglpD strains increased significantly (Fig. 6). In the wild type and in the ΔglpD strain, the G3P concentration peaked at 20 min after the reinoculation. This transient increase was dramatic in the wild type, where it dropped to the basal level by 40 min. The decrease in ΔglpD cells was much slower. This result is consistent with high G3P levels correlating with the low persister frequency typically observed in exponential phase and in the ΔglpD mutant.
FIG. 6.
Changes in G3P following dilution of cells in fresh medium. (A) G3P concentration in the wild type; (B) G3P concentration in the glpD deletion strain, KLE416. These data are the averages of three independent experiments, and the error bars correspond to the standard deviations of the means.
DISCUSSION
Approaches to the study of persisters.
Persisters have a clear and striking phenotype, surviving treatment with high doses of bactericidal antibiotics. This should have made it easy to identify persister genes by straightforward genetic approaches. Unexpectedly, finding persister genes proved to be a formidable challenge. Our previous research, building on the work of others (9, 10, 19, 21, 30, 37, 38, 43, 49), led to the identification of the first persister gene, HipA, a component of the hipBA toxin/antitoxin module (29). We found that a strain with hipBA deleted had a considerably lower level of persisters, but only at stationary state. Previous studies that examined the role of this locus in persister formation reported no phenotype of the deletion, but those experiments were restricted to exponential-phase cells (9, 37). Homologs of hipA are present in a number of gram-negative bacteria, but its conservation is poor. At the same time, all species, including gram-positive bacteria, form persisters irrespective of whether or not they carry hipBA (29, 32). For example, Pseudomonas aeruginosa, the model organism for biofilm studies, forms highly tolerant persisters (44) but does not have a hipBA locus. These observations suggested that there are different ways by which persisters may be formed, and additional genes responsible for persister formation are yet to be discovered. A gene profile of isolated persisters indicated overexpression of several toxin/antitoxin modules (MazEF, RelBE, and DinJ/YafQ), suggesting a general theme in the mechanism of persister formation (29). In principle, overexpression of any “toxin” that can reversibly inhibit a target function (such as MazF or RelE, which block translation by cleaving mRNAs) can send the cell into a dormant, multidrug-tolerant state. Expression of either HipA (21, 29) or RelE (29) indeed increases the level of surviving persisters, but only a hipA deletion produced a marked decrease in the persister level (29). Persisters are relatively abundant in stationary state, making up 1% of cells, and deletion of hipA decreases them by 10- to 100-fold. Genes responsible for the remaining persisters are yet to be discovered.
One of the standard approaches to finding a function is by screening transposon insertion libraries, which does not work well for functions encoded by redundant genes. Indeed, our attempts to find persister genes by this method have not been successful. An alternative method is to identify genes by screening/selecting a library cloned into an expression vector for gain of function. In this case, even a weak contributor to a multigene function can be identified when overexpressed. However, this approach is problematic for persisters as well, since overproduction of many (especially membrane) proteins leads to misfolded toxic products that can stop cell growth and will create an artifact emulating a dormant state. In order to make this approach work, we decided to use mild overexpression by cloning a library into a low-copy vector, using native promoters for expression, and introducing a growth step between rounds of selection by a bactericidal antibiotic. In this manner, any cells that grew considerably slower than the wild type would be selected against. This procedure enriched the population in cells with elevated persister production. A clone with consistent persister overproduction carried the glpD gene and was examined in detail in this study.
G3P and persister formation.
Cells overexpressing GlpD, the sn-glycerol-3-phosphate dehydrogenase (35), had an elevated level of persisters tolerant to both ampicillin and ofloxacin. GlpD and GlpA are sn-glycerol-3-phosphate dehydrogenases which are primarily involved in utilization of glycerol as a carbon source. Glycerol is passively transported into the cell by GlpF and phosphorylated by GlpK, generating sn-glycerol-3-phosphate. GlpD and GlpA move G3P into the central metabolism by converting it into the glycolysis metabolite DHAP, using FAD as a cofactor. DHAP can then be isomerized by TpiA to glyceraldehyde-3-phosphate. GlpD is active under aerobic conditions, while GlpA requires GlpB and GlpC to complete the same reaction under anaerobic conditions. DHAP is converted to G3P by GpsA, known as the biosynthetic glycerol-3-phosphate dehydrogenase. In the absence of glycerol in the medium, this enzyme is essential for production of G3P, the precursor to phospholipid biosynthesis. GpsA uses NADH as a cofactor and is not structurally related to GlpD or GlpA. G3P is also generated from the breakdown of phospholipids by the periplasmic enzymes MdoB and MdoH. This G3P is then transported back into the cell by GlpQ, UgpQ, or GlpT.
The absolute numbers of cells that survive antibiotic treatment may change from day to day but, as has been shown by others, the relative numbers tend to stay the same (30, 48). This variability may reflect the stochastic nature of persister formation. An upregulation of glpD has also been seen in a transcriptional profile of isolated persister cells (D. Shah and K. Lewis, BMC Microbiol., in press). This change in survival was not related to growth rate, and an unchanged MIC indicated that the increased survival is not a result of increased resistance (the ability to grow in the presence of elevated amounts of antibiotic) to ampicillin or ofloxacin. The wild-type growth rate of the overexpressing strain indicates the lack of apparent pleiotropic effects, such as a deficiency in phospholipid synthesis, or the creation of a metabolic futile cycle.
Deletion of glpD did not affect tolerance in exponential phase but eliminated the majority of persisters in stationary state. This is similar to the behavior of ΔhipA cells, which only show a decrease in persisters at stationary state (29). In E. coli, persisters are not created at early exponential phase, then they sharply increase as the density of the culture rises and reach 1% at stationary state. It seems logical that persisters become more prominent in a nongrowing stationary population, where survival (as opposed to growth) is the defining feature. The prominent effect of GlpD on persister formation at stationary state suggests that glpD is an important persister gene.
Deletion of glpABC, the operon encoding a second G3P dehydrogenase and accessory proteins that function primarily under anaerobic conditions, also lowered the persister fraction, but less so than ΔglpD alone. This underscores the difficulties in identifying constituents of a redundant function.
GlpD could be affecting either persister formation or resuscitation. The persistence levels of the ΔglpD deletion strain were lower than the wild type under all conditions tested. However, the difference was consistently more pronounced in diluted stationary-state cells (55-fold) than in undiluted stationary-state cells (25-fold). This increased sensitivity was paralleled by an increase in G3P concentration when stationary-state cells were diluted into fresh medium. An increase in G3P may facilitate resuscitation of persisters from a dormant state. When resuscitated, a cell would become as sensitive as the bulk of the population. This possibility is supported by our observation of wild-type cells undergoing a spike in G3P concentration when diluted into fresh medium (Fig. 6).
glpD is part of the glp regulon, consisting of five operons, all controlled by the GlpR repressor. Deletion of glpD leads to an increase in G3P, the inducer of the regulon, which in turn derepresses all glp operons through G3P binding to GlpR. In order to separate these two effects of glpD mutation (G3P increase and glp regulon derepression), we constructed strains with G3P-insensitive GlpR (the glp regulon cannot be induced) and with glpR deleted (constitutive expression of the glp regulon). While inactivation of glpD reduced persistence by the same amount in wild-type and ΔglpR backgrounds, it had no effect when combined with a noninducible glpR allele. This suggests that derepression of the glp regulon is essential for the observed effect of the ΔglpD mutation. However, constitutive expression of the glp regulon alone is not sufficient because the deletion of glpR does not lead to the same drop in persistence as does deletion of glpD (Fig. 3). The effect of glpD therefore is the combination of both high intracellular G3P levels and the derepression of GlpR-regulated genes.
We considered additional components linked to G3P or the Glp regulon for a possible role in persister formation. Deletions of tpiA (40), mdoB (27), mdoH (27), glpQ (31), ugpQ (31), gldA (5, 50), glpK (35), glpF (35), glpX (19), and glpT (35) had no effect on persister formation (Fig. 4), with one exception—a mutant attenuated in an essential plsB gene had a considerably decreased level of persister formation. The plsB mutant had a low level of G3P, similar to the wild type. We suggest, therefore, that the PlsB effect on persistence is downstream of G3P.
In conclusion, we identified three possible components affecting persister formation: G3P level, the glp regulon, and PlsB. The exact relationship between these components is currently under investigation.
REFERENCES
- 1.Ackerman, R. S., N. R. Cozzarelli, and W. Epstein. 1974. Accumulation of toxic concentrations of methylglyoxal by wild-type Escherichia coli K-12. J. Bacteriol. 119:357-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, J. M. 2001. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48(Suppl. 1):5-16. [DOI] [PubMed] [Google Scholar]
- 3.Bachmann, B. J. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36:525-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. [DOI] [PubMed] [Google Scholar]
- 5.Bell, R. M., and J. E. Cronan. 1975. Mutants of Escherichia coli defective in membrane phospholipid synthesis, phenotypic suppression of sn-glycerol-3-phosphate acyltransferase Km mutants by loss of feedback inhibition of biosynthetic sn-glycerol-3-phosphate dehydrogenase. J. Biol. Chem. 250:7153-7158. [PubMed] [Google Scholar]
- 6.Bergstrom, S., and S. Normark. 1979. Beta-lactam resistance in clinical isolates of Escherichia coli caused by elevated production of the ampC-mediated chromosomal beta-lactamase. Antimicrob. Agents Chemother. 16:427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi, E., and J. Lutkenhaus. 1993. Cell-division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 175:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bigger, J. W. 1944. Treatment of staphylococcal infections with penicillin. Lancet ii:497-500. [Google Scholar]
- 9.Black, D. S., B. Irwin, and H. S. Moyed. 1994. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 176:4081-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black, D. S., A. J. Kelly, M. J. Mardis, and H. S. Moyed. 1991. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 173:5732-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booth, I. R., G. P. Ferguson, S. Miller, C. Li, B. Gunasekera, and S. Kinghorn. 2003. Bacterial production of methylglyoxal: a survival strategy or death by misadventure? Biochem. Soc. Trans. 31:1406-1408. [DOI] [PubMed] [Google Scholar]
- 12.Christensen, S. K., and K. Gerdes. 2003. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 48:1389-1400. [DOI] [PubMed] [Google Scholar]
- 13.Christensen, S. K., and K. Gerdes. 2004. Delayed-relaxed response explained by hyperactivation of RelE. Mol. Microbiol. 53:587-597. [DOI] [PubMed] [Google Scholar]
- 14.Clowes, R. C., and W. Hayes. 1968. Experiments in microbial genetics. Wiley and Sons, New York, N.Y.
- 15.Cole, S. T., K. Eiglmeier, S. Ahmed, N. Honore, L. Elmes, W. F. Anderson, and J. H. Weiner. 1988. Nucleotide sequence and gene-polypeptide relationships of the GlpABC operon encoding the anaerobic sn-glycerol-3-phosphate dehydrogenase of Escherichia coli K-12. J. Bacteriol. 170:2448-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 17.Cozzarelli, N. R., J. P. Koch, S. Hayashi, and E. C. Lin. 1965. Growth stasis by accumulated l-α-glycerophosphate in Escherichia coli. J. Bacteriol. 90:1325-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donahue, J. L., J. L. Bownas, W. G. Niehaus, and T. J. Larson. 2000. Purification and characterization of glpX-encoded fructose 1,6-bisphosphatase, a new enzyme of the glycerol-3-phosphate regulon of Escherichia coli. J. Bacteriol. 182:5624-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eagle, H., R. Fleischman, and A. D. Musselman. 1950. The effective concentrations of penicillin in vitro and in vivo for streptococci, pneumococci, and Treponema pallidum. J. Bacteriol. 59:625-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falla, T. J., and I. Chopra. 1998. Joint tolerance to beta-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob. Agents Chemother. 42:3282-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freedberg, W. B., W. S. Kistler, and E. C. Lin. 1971. Lethal synthesis of methylglyoxal by Escherichia coli during unregulated glycerol metabolism. J. Bacteriol. 108:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 24.Heath, R. J., and C. O. Rock. 1999. A missense mutation accounts for the defect in the glycerol-3-phosphate acyltransferase expressed in the plsB26 mutant. J. Bacteriol. 181:1944-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hohorst, H. 1963. l-(−)-Glycerol-1-phosphate determination with glycerol-1-phosphate dehydrogenase, p. 215-219. In H. Bergmeyer (ed.), Methods of enyzymatic analysis. Academic Press, Inc., New York, N.Y.
- 26.Kanehisa, M., S. Goto, M. Hattori, K. F. Aoki-Kinoshita, M. Itoh, S. Kawashima, T. Katayama, M. Araki, and M. Hirakawa. 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34:D354-D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy, E. P., M. K. Rumley, H. Schulman, and L. M. Van Golde. 1976. Identification of sn-glycero-1-phosphate and phosphoethanolamine residues linked to the membrane-derived oligosaccharides of Escherichia coli. J. Biol. Chem. 251:4208-4213. [PubMed] [Google Scholar]
- 28.Keren, I., N. Kaldalu, A. Spoering, Y. Wang, and K. Lewis. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13-18. [DOI] [PubMed] [Google Scholar]
- 29.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korch, S. B., T. A. Henderson, and T. M. Hill. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50:1199-1213. [DOI] [PubMed] [Google Scholar]
- 31.Larson, T. J., and A. T. van Loo-Bhattacharya. 1988. Purification and characterization of glpQ-encoded glycerophosphodiester phosphodiesterase from Escherichia coli K-12. Arch. Biochem. Biophys. 260:577-584. [DOI] [PubMed] [Google Scholar]
- 32.Lewis, K. 2001. The riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Licking, E. 1999. Getting a grip on bacterial slime. Business Week 13 September 1999, p. 98-100.
- 34.Lin, E. C., J. P. Koch, T. M. Chused, and S. E. Jorgensen. 1962. Utilization of l-α-glycerophosphate by Escherichia coli without hydrolysis. Proc. Natl. Acad. Sci. USA 48:2145-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, E. C. C. 1976. Glycerol dissimilation and its regulation in bacteria. Annu. Rev. Microbiol. 30:535-578. [DOI] [PubMed] [Google Scholar]
- 36.MacLean, M. J., L. S. Ness, G. P. Ferguson, and I. R. Booth. 1998. The role of glyoxalase I in the detoxification of methylglyoxal and in the activation of the KefB K+ efflux system in Escherichia coli. Mol. Microbiol. 27:563-571. [DOI] [PubMed] [Google Scholar]
- 37.Moyed, H. S., and K. P. Bertrand. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moyed, H. S., and S. H. Broderick. 1986. Molecular cloning and expression of hipA, a gene of Escherichia coli K- 12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 166:399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oram, M., and L. M. Fisher. 1991. 4-Quinolone resistance mutations in the DNA gyrase of Escherichia coli clinical isolates identified by using the polymerase chain reaction. Antimicrob. Agents Chemother. 35:387-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pichersky, E., L. D. Gottlieb, and J. F. Hess. 1984. Nucleotide sequence of the triose phosphate isomerase gene of Escherichia coli. Mol. Gen. Genet. 195:314-320. [DOI] [PubMed] [Google Scholar]
- 41.Pilch, D. S., M. Kaul, and C. M. Barbieri. 2005. Ribosomal RNA recognition by aminoglycoside antibiotics, p. 179-204. In M. J. Waring, J. B. Chaires, and B. A. Armitage (ed.), DNA binders and related subjects. Springer, New York, N.Y.
- 42.Raj, V. S., H. Tomitori, M. Yoshida, A. Apirakaramwong, K. Kashiwagi, K. Takio, A. Ishihama, and K. Igarashi. 2001. Properties of a revertant of Escherichia coli viable in the presence of spermidine accumulation: increase in l-glycerol-3-phosphate. J. Bacteriol. 183:4493-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scherrer, R., and H. S. Moyed. 1988. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J. Bacteriol. 170:3321-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 46.Tomasz, A. 1979. Mechanism of the irreversible anti-microbial effects of penicillins—how the beta-lactam antibiotics kill and lyse bacteria. Annu. Rev. Microbiol. 33:113-137. [DOI] [PubMed] [Google Scholar]
- 47.Tomasz, A., A. Albino, and E. Zanati. 1970. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227:138-140. [DOI] [PubMed] [Google Scholar]
- 48.Wiuff, C., R. M. Zappala, R. R. Regoes, K. N. Garner, F. Baquero, and B. R. Levin. 2005. Phenotypic tolerance: antibiotic enrichment of noninherited resistance in bacterial populations. Antimicrob. Agents Chemother. 49:1483-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfson, J. S., D. C. Hooper, G. L. McHugh, M. A. Bozza, and M. N. Swartz. 1990. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and beta-lactam antimicrobial agents. Antimicrob. Agents Chemother. 34:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamagishi, M., H. Matsushima, A. Wada, M. Sakagami, N. Fujita, and A. Ishihama. 1993. Regulation of the Escherichia coli Rmf gene encoding the ribosome modulation factor. Growth phase-dependent and growth rate-dependent control. EMBO J. 12:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, B., and T. J. Larson. 1996. Action at a distance for negative control of transcription of the glpD gene encoding sn-glycerol 3-phosphate dehydrogenase of Escherichia coli K-12. J. Bacteriol. 178:7090-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]