Abstract
Detailed structural analysis of Lactococcus lactis peptidoglycan was achieved by identification of its constituent muropeptides separated by reverse phase high-performance liquid chromatography. Modification of the classical elution buffer allowed direct and sensitive analysis of the purified muropeptides by matrix-assisted laser desorption ionization-time of flight mass spectrometry. The structures of 45 muropeptides were assigned for L. lactis strain MG1363. Analysis of the muropeptide composition of an MG1363 dacB mutant showed that the dacB-encoded protein has l,d-carboxypeptidase activity and is involved in peptidoglycan maturation.
Peptidoglycan is the major component of the gram-positive bacterial cell wall and ensures its rigidity and stability. Although its basic structure is characteristic of a given bacterial species, peptidoglycan is in a dynamic state throughout the bacterial life span, and its structure is the result of complex biosynthetic, maturation, and degradation reactions (11). Structural analysis of the peptidoglycan constituent muropeptide is a powerful method that allowed elucidation of the roles of biosynthesis enzymes involved in the design of cell wall architecture (3) and to characterize changes in peptidoglycan structure leading to antibiotic resistance (1, 8, 16). Also, the technique allowed revelation of peptidoglycan covalent modifications, such as O-acetylation or de-N-acetylation, which could play essential roles in the control of the activities of exogenous (25) and endogenous (17) cell wall-degrading enzymes.
Lactococcus lactis is the model gram-positive lactic acid bacterium. Its peptidoglycan hydrolase complement was previously characterized (7, 12, 13, 22). Bacterial peptidoglycan hydrolases are involved in different cellular functions during growth, such as cell separation, cell wall turnover, and cell wall expansion (21). Their activities can also lead to bacterial autolysis by hydrolysis of the protective cell wall peptidoglycan. Since these potentially lethal enzymes are synthesized during bacterial growth, their activities should be controlled. As mentioned above, covalent structural modification of peptidoglycan is one of the proposed mechanisms that could control peptidoglycan hydrolase activity (17, 21). Thus, the analysis of the L. lactis peptidoglycan structure constitutes the basis for further studies of the mechanisms that regulate synthesis and degradation of the L. lactis cell wall. Earlier studies revealed that L. lactis (formerly Streptococccus lactis) has A4α-type peptidoglycan, with a monomer primary structure (GlcNAc-MurNAc-l-Ala-α-d-Glu-l-Lys-d-Ala) and a d-Asp in the interpeptide bridge, attached to the ɛ-amino group of Lys (19). In this study, we achieved detailed analysis of the muropeptide composition of Lactococcus lactis. Also, using the method developed, we identified an l,d-carboxypeptidase in L. lactis involved in peptidoglycan maturation.
Muropeptide composition of L. lactis MG1363.
Lactococcus lactis subsp. cremoris MG1363 was grown on M17 medium containing 0.5% glucose at 30°C. Peptidoglycan was extracted from early-exponential-phase cells by a method adapted from Billot-Klein et al. (5) for Enterococcus faecium and from Atrih et al. (3) for Bacillus subtilis. Briefly, a 500-ml culture (optical density at 600 nm, 0.3) was chilled in an ice bath, and bacteria were harvested by centrifugation (4,200 ×g; 10 min; 4°C). The cells were boiled in 4% sodium dodecyl sulfate and washed several times with distilled H2O. The cell walls were purified using pronase (90 min; 60°C), followed by trypsin (16 h; 37°C). They were then treated with 48% hydrofluoric acid at 4°C for 16 h. The insoluble material was washed several times with 0.25 M Tris-HCl, pH 8.0, and with distilled H2O until the pH reached 5. Purified peptidoglycan was digested with mutanolysin, a muramidase from Streptomyces globisporus (Sigma). The digestion yield, calculated as the percentage of solubilized muramic acid after mutanolysin treatment, reached more than 95%.
The soluble muropeptides obtained from peptidoglycan digestion were reduced by sodium borohydride as described previously (3) and were separated by reverse phase high-performance liquid chromatography (RP-HPLC) using a Hypersil ODS column (C18; 250 by 4.6 mm; 5 μm; ThermoHypersil-Keystone) at 50°C. Muropeptides were eluted for 5 min with 10 mM ammonium phosphate buffer, pH 5.6 (buffer A), and then with a 270-min methanol (0 to 20%) linear gradient in buffer A at a flow rate of 0.5 ml min−1. Sodium azide was added (180 μg liter−1 of buffer A) to avoid baseline drift at 202 nm. The use of ammonium phosphate buffer for muropeptide separation provides two main improvements compared to the standard use of sodium phosphate buffer (9). First, muropeptides can be analyzed directly by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry without a time-consuming desalting step. Second, the sensitivity of muropeptide analysis by MALDI-TOF mass spectrometry is significantly improved, owing to the elimination of α-cyano-4-hydroxycinnamic acid matrix clusters, which interfere with muropeptide detection (20).
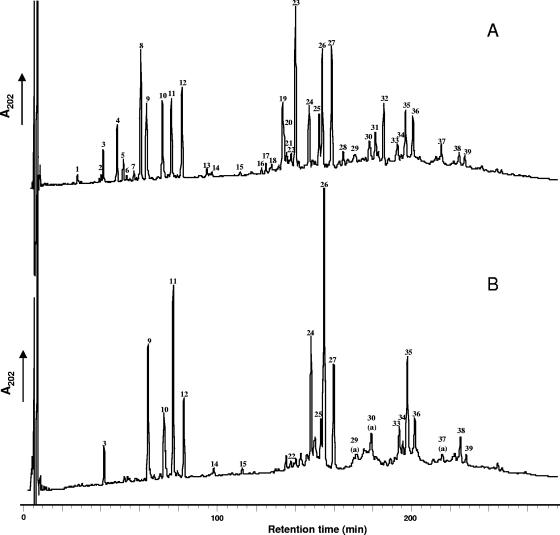
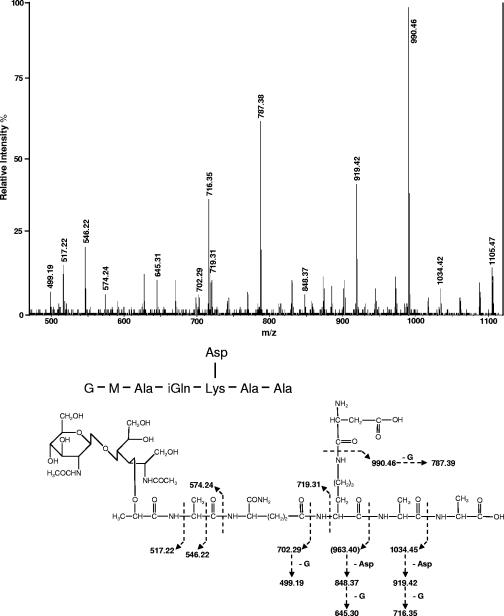
Under the conditions described above, 39 peaks were detected by RP-HPLC separation of the reduced muropeptides obtained from mutanolysin digestion of L. lactis MG1363 peptidoglycan (Fig. 1A). Amino acid and amino sugar composition analysis of all the peaks after acid hydrolysis, using the Waters Picotag method (4), indicated that they contained muropeptides (data not shown). The amino acids detected were Ala, Glu, Lys, and Asp, in agreement with the A4α structure found for L. lactis peptidoglycan (19). The peaks were analyzed by MALDI-TOF mass spectrometry with a Voyager DE STR mass spectrometer (Applied Biosystems, Framingham, MA), on a 1-μl aliquot for most of the peak fractions with an α-cyano-4-hydroxycinnamic acid matrix. For the less abundant fractions, 100 μl was first concentrated on a Zip-Tip C18 pipette tip (Millipore) and eluted with 1 μl of 50% acetonitrile, 0.15% trifluoroacetic acid before analysis. The structures of 45 muropeptides (Table 1) were tentatively assigned from the precise masses obtained by MALDI-TOF mass spectrometry and from amino acid and amino sugar analyses. Monomers represent about 35% of the total muropeptides, and the cross-linking index was calculated to be 35.8% (Table 2). Most of the monomers contained d-Asp or d-Asn interpeptide bridge residues. Only muropeptides 1, 3, and 6 did not contain Asp in their amino acid compositions. The muropeptide composition of L. lactis has a high level of complexity due to several structural variations leading to high number of different muropeptide structures. First, contrary to what was found in several gram-positive bacteria, such as E. faecium, Lactobacillus casei, or B. subtilis, muropeptides with intact pentapeptide side chains were found in mature peptidoglycan, in addition to muropeptides with tri- and tetrapeptide side chains. Second, several muropeptides differed by 1 mass unit, suggesting a difference of one amidation. Analysis of monomers 4, 9, and 10 by MALDI tandem mass spectrometry (MS-MS) with a Q/Star XL mass spectrometer (Sciex, Toronto, Canada) indicated that they contained Asp side chains, and thus, according to their MALDI-TOF masses, they contained α-amidated-iGlu (iGln) residues. The MS-MS spectrum of monomer 10 is shown in Fig. 2. The fragment ion with 990.46 m/z corresponds to a loss of an Asp residue (a mass defect of 115.01 Da) from the molecular ion. The monomers 8, 11, and 12, which have a mass defect of 1 compared to monomers 4, 9, and 10, respectively, contain Asn and iGln according to their masses. The variability found in monomers was also found in the acceptor chains of dimers and trimers, leading to a high number of different muropeptide structures. Finally, a small proportion of muropeptides were modified by de-N-acetylation (5.5%) (peaks 7, 20a, 21, 31a, and 33a), leading to a mass defect of 42, or by O-acetylation (3.2%) (peaks 13, 14, 15, 28, 29a, 30a, and 37a), leading to an extra 42 Da. Anhydromuropeptides, which are usually found in gram-negative bacteria and are proposed to end glycan chains (10), were not detected in L. lactis peptidoglycan. Their absence may be due either to very small undetected amounts of these anhydromuropeptides, which would be indicative of long glycan strands, or to the presence of other muropeptides at the ends of glycan strands.
FIG. 1.
RP-HPLC separation of muropeptides from L. lactis MG1363 (A) and the MG1363 dacB mutant (B). Peak numbers refer to Table 1. A few peaks contain two muropeptides identified as a and b in Table 1. On the dacB mutant chromatogram, muropeptide a is indicated when muropeptide b is absent.
TABLE 1.
Structures, molecular masses, and quantities of muropeptides from Lactococcus lactis MG1363 and MG1363 dacB mutant
| Peaka | Proposed structureb | Observed m/z | Calculatedd [M+Na]+ | % of all peakse
|
|
|---|---|---|---|---|---|
| MG1363 | dacB mutant | ||||
| Monomers | 35.4 | 33.7 | |||
| 1 | Tri | 848.49 | 848.39 | 0.4 | 0 |
| 2 | Tri-D missing GlcNAc | 760.34 | 760.41 | 0.3 | 0 |
| 3 | Tetra | 919.19 | 919.42 | 1.5 | 1.6 |
| 4 | Tri-D | 963.35 | 963.41 | 3.4 | 0 |
| 5 | Tri*-D with (NH2)f | 963.25 | 963.41 | 1.1 | 0 |
| 6 | Penta | 990.46 | 990.46 | 0.3 | 0.3 |
| 7 | Tri-N (deAC) | 920.38 | 920.42 | 0.6 | 0 |
| 8 | Tri-N | 962.40 | 962.43 | 6.6 | 0 |
| 9 | Tetra-D | 1,034.44 | 1,034.45 | 5.0 | 8.1 |
| 10 | Penta-D | 1,105.52 | 1,105.49 | 5.9 | 5.5 |
| 11 | Tetra-N | 1,033.40 | 1,033.47 | 4.1 | 11.5 |
| 12 | Penta-N | 1,104.56 | 1,104.50 | 5.0 | 5.3 |
| 13 | Tri-N (Ac) | 1,004.49 | 1,004.44 | 0.4 | 0 |
| 14 | Tetra-D (Ac) | 1,076.37 | 1,076.46 | 0.6 | 0.9 |
| 15 | Tetra-N (Ac) | 1,075.51 | 1,075.48 | 0.2 | 0.5 |
| Dimers | 44.6 | 44.3 | |||
| 16 | Tri-D-Tetra-D | 1,956.98 | 1,956.86 | 0.4 | 0 |
| 17 | Tri-N-Tetra | 1,841.07 | 1,840.85 | 0.6 | 0 |
| 18 | Tri-N-Tetra-Dc missing GlcNAc | 1,752.95 | 1,752.80 | 0.5 | 0 |
| 19 | Tri-N-Tetra-Dc | 1,956.01 | 1,955.88 | 4.4 | 0 |
| 20a | Tri-N-Tetra-Dc (deAc) | 1,914.01 | 1,913.87 | 1.2 | 0 |
| 20b | Tri-N-Tetra-N missing GlcNAc | 1,751.94 | 1,751.82 | 1.2 | 0 |
| 21 | Tri-N-Tetra-N (deAc) | 1,912.94 | 1,912.88 | 1.3 | 0 |
| 22 | Tetra-N-Tetra | 1,912.05 | 1,911.89 | 1.2 | 0.4 |
| 23 | Tri-N-Tetra-N | 1,954.91 | 1,954.90 | 9.1 | 0 |
| 24 | Tetra-N-Tetra-Dc | 2,027.05 | 2,026.91 | 4.0 | 8.9 |
| 25 | Penta-N-Tetra-Dc | 2,097.88 | 2,097.95 | 3.8 | 4.0 |
| 26 | Tetra-N-Tetra-N | 2,025.97 | 2,025.93 | 7.3 | 19.7 |
| 27 | Penta-N-Tetra-N | 2,096.97 | 2,096.97 | 7.3 | 8.1 |
| 28 | Tri-N-Tetra-N (Ac) | 1,996.95 | 1,996.90 | 0.9 | 0 |
| 29a | Tetra-N-Tetra-Dc (Ac) | 2,068.89 | 2,068.93 | 0.3 | 1.0 |
| 30a | Tetra-N-Tetra-N (Ac) | 2,067.95 | 2,067.94 | 1.1 | 2.2 |
| Trimers | 17.5 | 19.6 | |||
| 29b | Tri-N-Tetra-D-Tetra-Dc | 2,949.22 | 2,949.33 | 0.4 | 0 |
| 30b | Tri-N-Tetra-N-Tetra-Dc | 2,948.28 | 2,948.35 | 0.8 | 0 |
| 31a | Tri-N-Tetra-N-Tetra-N (deAC) | 2,905.42 | 2,905.32 | 1.8 | 0 |
| 31b | Tri-N-Tetra-N-Tetra-Dc | 2,948.39 | 2,948.35 | 0.8 | 0 |
| 32 | Tri-N-Tetra-N-Tetra-N | 2,947.29 | 2,947.36 | 3.9 | 0 |
| 33a | Tetra-N-Tetra-N-Tetra-N (deAC) | 2,976.26 | 2,976.38 | 0.6 | 1.3 |
| 33b | Tetra-N-Tetra-N-Tetra-Dc | 3,019.25 | 3,019.38 | 1.2 | 2.5 |
| 34 | Tetra-N-Tetra-N-Tetra-N missing GlcNAc | 2,815.38 | 2,815.27 | 0.5 | 1.6 |
| 35a | Tetra-N-Tetra-N-Tetra-N | 3,018.64 | 3,018.40 | 3.8 | 8.5 |
| 35b | Penta-N-Tetra-N-Tetra-Dc | 3,090.63 | 3,090.42 | 0.2 | 0.5 |
| 36 | Penta-N-Tetra-N-Tetra-N | 3,089.34 | 3,089.44 | 3.4 | 4.2 |
| 37a | Tetra-N-Tetra-N-Tetra-N (AC) | 3,060.51 | 3,060.41 | 0.1 | 1.0 |
| Tetramers | 2.5 | 2.4 | |||
| 37b | Tri-N-Tetra-N-Tetra-N-Tetra-N | 3,939.86 | 3,939.83 | 1.1 | 0 |
| 38 | Tetra-N-Tetra-N-Tetra-N-Tetra-N | 4,010.74 | 4,010.87 | 0.8 | 1.8 |
| 39 | Penta-N-Tetra-N-Tetra-N-Tetra-N | 4,081.88 | 4,081.90 | 0.6 | 0.6 |
Peak numbers refer to Fig. 1. A few peaks contain two muropeptides, named a and b.
Tri, disaccharide tripeptide (l-Ala-d-iGln-l-Lys); Tetra, disaccharide tetrapeptide (l-Ala-d-iGln-l-Lys-d-Ala); Penta, disaccharide pentapeptide (l-Ala-d-iGln-l-Lys-d-Ala-d-Ala); Disaccharide, GlcNAc-MurNAc; D, d-Asp; N, d-Asn; Ac, acetylation; deAc, deacetylation; (NH2), amidation; iGln, α-amidated isoGlu (isoGln).
Positions of N and D in either peptide chain are arbitrary.
Sodiated molecular ions were the most abundant on MALDI-TOF mass spectra for all muropeptides. m/z values correspond to monoisotopic masses.
Percentage of each peak was calculated as the ratio of the peak area over the sum of areas of all the peaks identified in the table.
Position of amidation was not identified. Tri*, disaccharide tripeptide (l-Ala-d-iGlu-l-Lys).
TABLE 2.
Percentages of muropeptides carrying the given peptide chain as a monomer or as an acceptor peptide chain in dimers, trimers, and tetramersa
| Muropeptide | MG1363 | dacB mutant |
|---|---|---|
| Monomers | 35.4 | 33.7 |
| Tripeptide | 12.8 | 0 |
| Tetrapeptide | 11.4 | 22.6 |
| Pentapeptide | 11.2 | 11.1 |
| Dimers | 44.6 | 44.3 |
| Tripeptide | 19.9 | 0 |
| Tetrapeptide | 13.6 | 32.2 |
| Pentapeptide | 11.1 | 12.1 |
| Trimers | 17.5 | 19.6 |
| Tripeptide | 7.7 | 0 |
| Tetrapeptide | 6.2 | 14.9 |
| Pentapeptide | 3.6 | 4.7 |
| Tetramers | 2.5 | 2.4 |
| Tripeptide | 1.1 | 0 |
| Tetrapeptide | 0.8 | 1.8 |
| Pentapeptide | 0.6 | 0.6 |
| Cross-linking indexb | 35.8 | 37.0 |
FIG. 2.
MS-MS analysis of muropeptide 10. The muropeptide was desalted and analyzed by MS-MS with a MALDI source. The [M+Na]+ parental ion (m/z 1105.47) was selected for fragmentation. The fragments were also detected as sodiated ions. G, GlcNAc; M, MurNAc.
Identification of an l,d-carboxypeptidase in L. lactis.
A gene named dacB encoding a protein with 30% sequence identity to Enterococcus faecalis VanY d-Ala-d-Ala-carboxypeptidase (2) was identified in the complete genome sequence of L. lactis IL-1403 (6). The encoded protein, DacB, is a 249-residue protein with a putative N-terminal signal sequence. The dacB gene was disrupted in strain MG1363 by single-crossover integration of the pRV300 vector (15) containing a 460-bp internal fragment of the gene amplified with primers 5′-ACCTAAAGATGAAATGAACCC-3′ and 5′-AGCTTCATAACCAATTCCTG-3′. The muropeptide HPLC profile of the resulting dacB mutant was markedly different from that of the MG1363 parent strain (Fig. 1B). It was characterized by the complete disappearance of all muropeptides with tripeptide chains, whereas muropeptides with tetrapeptide or pentapeptide chains remained (Table 1). In the dacB mutant, the percentage of tetrapeptide side chain monomers or multimers increased significantly and was equal to the sum of the percentages of tripeptide and tetrapeptide side chain muropeptides found in MG1363, whereas the percentage of muropeptides with pentapeptide side chains was similar in the dacB mutant and MG1363 (Table 2). From these results, we conclude that DacB has l,d-carboxypeptidase activity and cleaves the peptide side chain of peptidoglycan between l-Lys and d-Ala. It is worth noting that another putative d-Ala-d-Ala carboxypeptidase (DacA) encoded in the IL-1403 genome sequence likely converts a pentapeptide side chain to a tetrapeptide side chain. The major modification of peptidoglycan in the dacB mutant does not significantly affect the peptidoglycan cross-linking index (Table 2).
Only a few bacterial l,d-carboxypeptidases were previously characterized at the molecular level (14, 23, 24). This study allowed us to identify L. lactis DacB as the first l-Lys-d-Ala-carboxypeptidase belonging to the M15 peptidase family in the MEROPS Database (18), which contains metallopeptidases involved in bacterial cell wall biosynthesis and metabolism with different specificities. Despite the described differences in peptidoglycan structure, the dacB mutant does not show growth defects (data not shown), indicating that l,d-carboxypeptidase activity is dispensable for L. lactis growth under laboratory conditions.
The technique developed for L. lactis muropeptide analysis will allow us to investigate the roles of other enzymes involved in peptidoglycan biosynthesis or covalent modifications, such as acetylation or deacetylation.
Acknowledgments
We warmly thank J. Van Heijenoort, S. Foster, J.-L Mainardi, and L. Gutman for helpful discussions at the starting point of this work. We are grateful to D. Mollé (UMR 1253 STLO, INRA-Agrocampus, 65 rue de Saint-Brieuc, 35042 Rennes Cedex, France) for MS-MS analysis, C. Besset for technical help, and V. Monnet and M. Yvon for critical reading of the manuscript.
REFERENCES
- 1.Antignac, A., I. G. Boneca, J. C. Rousselle, A. Namane, J. P. Carlier, J. A. Vazquez, A. Fox, J. M. Alonso, and M. K. Taha. 2003. Correlation between alterations of the penicillin-binding protein 2 and modifications of the peptidoglycan structure in Neisseria meningitidis with reduced susceptibility to penicillin G. J. Biol. Chem. 278:31529-31535. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, M., F. Depardieu, L. Cabanie, P. Reynolds, and P. Courvalin. 1998. Requirement of the VanY and VanX d,d-peptidases for glycopeptide resistance in enterococci. Mol. Microbiol. 30:819-830. [DOI] [PubMed] [Google Scholar]
- 3.Atrih, A., G. Bacher, G. Allmaier, M. P. Williamson, and S. J. Foster. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bidlingmeyer, B. A., S. A. Cohen, and T. L. Tarvin. 1984. Rapid analysis of amino acids using pre-column derivatization. J. Chromatogr. 336:93-104. [DOI] [PubMed] [Google Scholar]
- 5.Billot-Klein, D., D. Shlaes, D. Bryant, D. Bell, J. van Heijenoort, and L. Gutmann. 1996. Peptidoglycan structure of Enterococcus faecium expressing vancomycin resistance of the VanB type. Biochem. J. 313:711-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis subsp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buist, G., J. Kok, K. J. Leenhouts, M. Dabrowska, G. Venema, and A. J. Haandrikman. 1995. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J. Bacteriol. 177:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Bustos, J., and A. Tomasz. 1990. A biological price of antibiotic resistance: major changes in the peptidoglycan structure of penicillin-resistant pneumococci. Proc. Natl. Acad. Sci. USA 87:5415-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glauner, B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172:451-464. [DOI] [PubMed] [Google Scholar]
- 10.Glauner, B., J. V. Holtje, and U. Schwarz. 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 263:10088-10095. [PubMed] [Google Scholar]
- 11.Holtje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huard, C., G. Miranda, Y. Redko, F. Wessner, S. J. Foster, and M. P. Chapot-Chartier. 2004. Analysis of the peptidoglycan hydrolase complement of Lactococcus lactis: identification of a third N-acetylglucosaminidase, AcmC. Appl. Environ. Microbiol. 70:3493-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huard, C., G. Miranda, F. Wessner, A. Bolotin, J. Hansen, S. J. Foster, and M. P. Chapot-Chartier. 2003. Characterization of AcmB, an N-acetylglucosaminidase autolysin from Lactococcus lactis. Microbiology 149:695-705. [DOI] [PubMed] [Google Scholar]
- 14.Korza, H. J., and M. Bochtler. 2005. Pseudomonas aeruginosa ld-carboxypeptidase, a serine peptidase with a Ser-His-Glu triad and a nucleophilic elbow. J. Biol. Chem. 280:40802-40812. [DOI] [PubMed] [Google Scholar]
- 15.Leloup, L., S. D. Ehrlich, M. Zagorec, and F. Morel-Deville. 1997. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl. Environ. Microbiol. 63:2117-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mainardi, J. L., M. Fourgeaud, J. E. Hugonnet, L. Dubost, J. P. Brouard, J. Ouazzani, L. B. Rice, L. Gutmann, and M. Arthur. 2005. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J. Biol. Chem. 280:38146-38152. [DOI] [PubMed] [Google Scholar]
- 17.Pfeffer, J. M., H. Strating, J. T. Weadge, and A. J. Clarke. 2006. Peptidoglycan O-acetylation and autolysin profile of Enterococcus faecalis in the viable but nonculturable state. J. Bacteriol. 188:902-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawlings, N. D., F. R. Morton, and A. J. Barrett. 2006. MEROPS: the peptidase database. Nucleic Acids Res. 34:D270-D272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smirnov, I. P., X. Zhu, T. Taylor, Y. Huang, P. Ross, I. A. Papayanopoulos, S. A. Martin, and D. J. Pappin. 2004. Suppression of alpha-cyano-4-hydroxycinnamic acid matrix clusters and reduction of chemical noise in MALDI-TOF mass spectrometry. Anal. Chem. 76:2958-2965. [DOI] [PubMed] [Google Scholar]
- 21.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249-262. [DOI] [PubMed] [Google Scholar]
- 22.Steen, A., G. Buist, K. J. Leenhouts, M. El Khattabi, F. Grijpstra, A. L. Zomer, G. Venema, O. P. Kuipers, and J. Kok. 2003. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J. Biol. Chem. 278:23874-23881. [DOI] [PubMed] [Google Scholar]
- 23.Templin, M. F., A. Ursinus, and J. V. Holtje. 1999. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 18:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tremplin, M. F., and J.-V. Höltje. 1998. Murein tetrapeptide l,d-carboxypeptidases, p. 1574-1576. In A. J. Barrett, N. D. Rawlings and J. F. Woessner (ed.), Handbook of proteolytic enzymes. Academic Press, New York, N.Y.
- 25.Vollmer, W., and A. Tomasz. 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275:20496-20501. [DOI] [PubMed] [Google Scholar]