Abstract
The RNA replicase extracted from Brome mosaic virus (BMV)-infected plants has been used to characterize the cis-acting elements for RNA synthesis and the mechanism of RNA synthesis. Minus-strand RNA synthesis in vitro requires a structure named stem-loop C (SLC) that contains a clamped adenine motif. In vitro, there are several specific requirements for SLC recognition. We examined whether these requirements also apply to BMV replication in barley protoplasts. BMV RNA3s with mutations in SLC were transfected into barley protoplasts, and the requirements for minus- and plus-strand replication were found to correlate well with the requirements in vitro. Furthermore, previous analysis of replicase recognition of the Cucumber mosaic virus (CMV) and BMV SLCs indicates that the requirements in the BMV SLC are highly specific. In protoplasts, we found that BMV RNA3s with their SLCs replaced with two different CMV SLCs were defective for replication. In vitro results generated with the BMV replicase and minimal-length RNAs generally agreed with those of in vivo BMV RNA replication. To extend this conclusion, we determined that, corresponding with the process of infection, the BMV replicases extracted from plants at different times after infection have different levels of recognition of the minimal promoters for plus- and minus-strand RNA syntheses.
Positive-strand RNA virus genomes are translated by the host ribosome soon after entry into the cell. These viral proteins, along with host factors, form a complex called the replicase that initiates RNA synthesis at the 3′ end of the genomic RNA to generate the complementary minus strand. This minus strand then acts as a template to produce multiple copies of the positive strand and, in some cases, the subgenomic RNA (3).
Structural and sequence requirements for viral RNA synthesis are just beginning to be elucidated. One approach to analyze viral RNA synthesis uses viral replicases extracted from infected plants (3, 12). The replicase preparation from Brome mosaic virus (BMV), a tripartite positive-strand RNA virus (15), has been used as a model system for in-depth studies of the cis-acting elements that direct RNA synthesis and the mechanism of RNA synthesis (1, 2, 4, 5, 8, 9, 19, 27, 28, 31). BMV is an excellent system for these studies, since a wealth of genetic evidence exists on the replication proteins and the viral RNAs. However, while results of BMV RNA synthesis in vitro have correlated with RNA replication in plant cells (5, 8, 9, 16, 20), some recent work concerning the mechanism of promoter recognition has advanced beyond the support of in vivo evidence (16, 17, 24). In addition, there are justifiable questions as to whether the results gleaned from studies of minimal-length RNAs reflect requirements in infected cells.
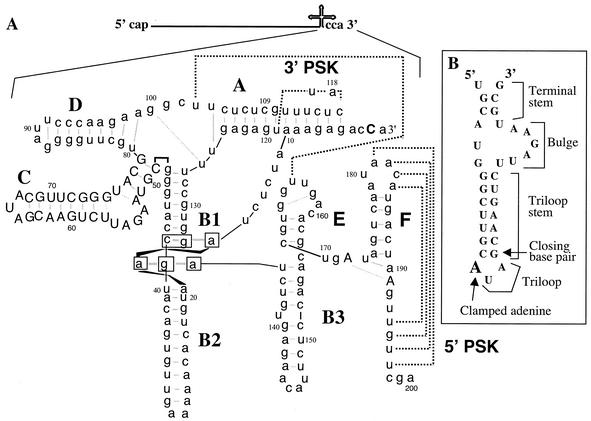
The best-characterized BMV core promoter is the one that directs the initiation of minus-strand RNA synthesis. BMV has three genomic RNAs that share a highly conserved sequence of ∼200 nucleotides (nt) at the 3′ end of each RNA (15). This sequence is required for minus-strand RNA synthesis and has been proposed to mimic a tRNA on the basis of computer modeling, chemical, probing, and phylogenetic analyses (10, 11, 14). A structure named stem-loop C (SLC) does not have a direct analog in canonical tRNA tertiary structure (Fig. 1A) and may be present, in part, to interact with the replicase. In the absence of the rest of the tRNA-like sequence, SLC can bind the replicase with a relative affinity that is only about threefold lower than that of the entire 3′ tRNA-like sequence (4). When the 8-nt sequence from the 3′ end of the BMV RNA is added to the 3′ end of SLC to generate RNA SLC+8, de novo initiation of viral RNA synthesis can take place efficiently from the terminal CCA3′ (preferred initiation nucleotide underlined) (5). Furthermore, some mutations in the SLC portion of BMV RNA3 that affected RNA synthesis in protoplasts also affected RNA synthesis in vitro from SLC+8 (reference 26 and reference within).
FIG. 1.
Structure of the BMV tRNA-like 3′ region and the nomenclature for the motifs in SLC examined in this study. (A) Proposed secondary structure of the BMV 3′-terminal 200 nt based on the structure described by Fechter et al. (10). The nucleotides within SLC are in capital letters. Individual RNA motifs are in bold letters A to F. PSK and dashed lines indicate the two pseudoknot motifs and base pairing that form the pseudoknot. The nucleotides that participate in an unusual 3-bp interaction at the junction of stems B1 and B2 are boxed. (B) Secondary structure of SLC with the names of the various motifs. The clamped adenine is represented by a larger boldface A.
Nuclear magnetic resonance spectroscopy was used to show that SLC contains two discrete domains, a large internal loop flanked on one side by a short stem—the terminal stem—followed by a longer stem called the triloop stem, which is capped with an AUA triloop (Fig. 1). The 5′ adenylate in the triloop interacts extensively with the terminal stem to form a clamped adenine motif (CAM) that is critical for recognition by the replicase (16). Based on the solution structure of SLC, predictions were made and tested in vitro. A change of the 3′ adenine in the triloop to a guanosine did not affect synthesis. This change actually retained a CAM, but it was at an altered angle relative to the stem (17). There are also sequences and/or structural requirements of the triloop stem and the closing base pair for RNA synthesis in vitro (Fig. 1B) (16, 24). We wanted to determine whether these in vitro observations hold true in the context of BMV RNA3 transfected into barley protoplasts along with wild-type RNA1 and RNA2. The results show that in vitro results do tend to predict defects in vivo. Further, we found that the ratio of replicase-initiated plus- to minus-strand BMV RNAs is not static but changes significantly during the course of BMV infection.
MATERIALS AND METHODS
Mutant construction and transcription.
The cDNA copies of wild-type BMV RNA1, RNA2, and RNA3 are contained in pB1TP3, pB2TP5, and pB3TP8, respectively, and are capable of producing infectious transcripts when the T7 RNA polymerase is used (13). Mutations within the SLC region of pB3TP8 were made by annealing oligonucleotides containing the desired mutations to a complementary oligonucleotide and by using Taq polymerase to complete the double-stranded sequence. The double-stranded DNA fragments were digested with restriction enzymes Bsu36I and BsmI and ligated into pB3TP8. The presence of the introduced mutations and the absence of spurious mutations were confirmed by sequencing the region encompassing the engineered mutation. Using a Message Machine kit as described by the manufacturer (Ambion, Austin, Texas), capped full-length transcripts of BMV RNA1, RNA2, and RNA3 and the mutant derivatives were generated from EcoRI-linearized plasmids.
Protoplast isolation, inoculation, and Northern blot analyses.
Protoplasts were generated from 5-day-old primary barley leaves as described by Kroner et al. (18) and inoculated with 0.5 μg each of capped full-length transcripts of RNA1, RNA2, and either RNA3 or the mutant derivatives of RNA3. Transfected protoplasts were incubated in 500 μl of incubation medium for 21 h, unless stated otherwise, under conditions of constant 23°C temperature and illumination. Following the incubation period, total RNA was extracted and Northern blot analysis was carried out using probes specific for the 3′-terminal 200 nt of the minus-strand BMV RNA3. Due to the conservation of the sequence at the 3′ terminus of BMV RNAs, this probe recognizes all three BMV RNAs. The blots were then stripped in a low-salt buffer at 95°C to remove the probe, checked by phosphorimage analysis to confirm that no radiolabel remained, and probed with an RNA that recognizes plus-strand BMV RNAs. Finally, blots were probed with an 80-nt sequence that specifically detects the 18S ribosomal RNAs. Hybridizations and washing of the membrane were performed under conditions that do not allow cross-recognition of the plus- and minus-strand RNAs. Autoradiograms for minus-strand RNAs were usually exposed for 2 to 3 h, while those for plus-strand BMV RNAs and the 18S rRNA were exposed for 15 to 30 min.
Extraction of the BMV replicase during a time course.
Preparations of enriched BMV replicase used 6- to 7-day-old barley. The primary leaves were inoculated with 0.1 or 0.3 mg of purified BMV/ml. The secondary leaves were harvested at specified times after inoculation. A total of 10 to 25 g of secondary leaves was stored at −50°C until use. The leaves were homogenized and prepared for gel filtration chromatography as described by Sun et al. (31).
RNA synthesis in vitro.
RNA-dependent RNA synthesis used the previously characterized minimal BMV promoter templates that can direct genomic minus- and subgenomic plus-strand RNA synthesis (SLdel+8 and −20/13, respectively), which were previously described by Kim et al. (16) and Siegel et al. (27), respectively. The assay and electrophoresis conditions were as described by Adkins et al. (2).
RESULTS
Effect of mutations in the SLC triloop on RNA synthesis.
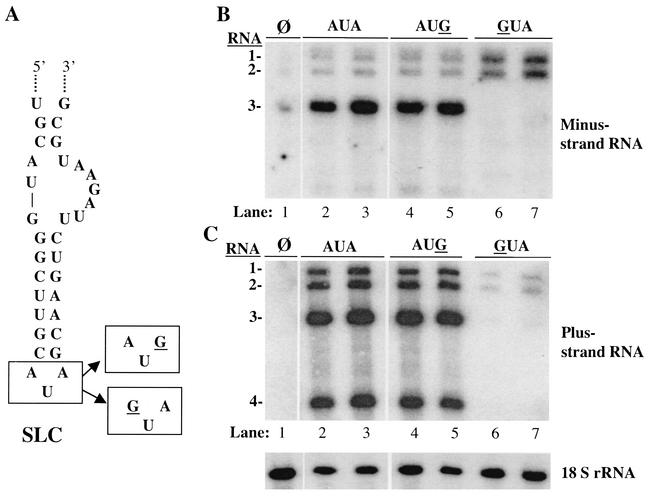
To examine whether the requirements for replicase-RNA interaction that had previously been determined from biochemical analysis applied in vivo, we first examined the change of the SLC triloop from 5′AUA3′ to 5′AUG3′ (mutation underlined). This change caused an altered CAM to form but was capable of directing at least wild-type levels of RNA synthesis by the BMV replicase (16, 17). BMV RNA3 with only the triloop changed from 5′AUA3′ to 5′AUG3′ or 5′GUA3′ was transfected along with BMV RNA1 and RNA2 (Fig. 2A). As positive controls, protoplasts were transfected with transcripts of wild-type BMV RNA1, RNA2, and RNA3 (Fig. 2B, lanes 2 and 3). Northern blot analysis probing for minus-strand and plus-strand BMV RNAs was performed. Significant levels of minus-strand RNA1, RNA2, and RNA3 and a lower amount of a molecule approximately the length of minus-strand RNA4 were observed (Fig. 2B, lanes 2 and 3). Robust amounts of plus-strand RNA1, RNA2, RNA3, and RNA4 were also observed (Fig. 2C, lanes 2 and 3). With 18S rRNA used as an internal loading control, slight variations in the RNAs were sometimes observed (Fig. 2). Therefore, to normalize for possible differences in the recovery of the RNA and/or transfection efficiency, we measured the RNA3 relative to the RNA2 within the same sample. The ratios of the minus- and plus-strand wild-type RNA3s relative to the comparable RNA2s were 9 and 4.3, respectively (Table 1). To ensure that we were able to detect differences in RNA accumulation, RNA3 with the clamped adenine changed to a guanine in a mutant named GUA (mutation underlined) was examined. Consistent with previous results on the effects of a mutation at this position (26), both minus- and plus-strand RNA3 accumulation was significantly decreased for the GUA mutation (Fig. 2B and C, lanes 6 and 7). The ratio of minus-strand RNA3 to RNA2 was 0.5, while the ratio of plus-strand RNA3 to RNA2 was 0.8 (Table 1). We noted that in the protoplasts in which RNA3 failed to replicate, there was a differential effect on the accumulation of minus- and plus-strand RNA1 and RNA2. In the absence of minus-strand RNA3 replication, minus-strand RNA1 and RNA2 increased slightly relative to transfections with RNA3 replication (Fig. 2B). Also, the absence of plus-strand RNA3 appears to cause a decrease in the amounts of plus-strand RNA1 and RNA2 (Fig. 2B and C, lanes 5 and 6). We speculate that the products from RNA3 or RNA3 itself is required for maximal plus-strand RNA1 and RNA2 replication. This effect is not the central theme of this work (since SLC is responsible for minus-strand RNA synthesis) and does not significantly affect the interpretation of the results. With regard to this work, the adenine that forms a CAM is critical for recognition by the BMV replicase in vivo, which is in agreement with previous in vitro observations (5, 16).
FIG. 2.
Role of the SLC triloop nucleotides in RNA synthesis in transfected protoplasts. Where there is a mutation, the mutated nucleotide is underlined. (A) Structure of SLC and the two mutations in the triloop that were examined. Only the affected portions of RNA3 in the three RNAs are shown. (B) Autoradiogram showing the effect of mutations on genomic minus-strand accumulation at 21 h posttransfection. The names of the RNAs used in the transfected protoplasts are shown above the autoradiogram, and the identities of the various RNAs in the autoradiogram are indicated to the left of the autoradiogram. (C) Autoradiogram showing the plus-strand RNAs that accumulated and the amount of 18S rRNA that was present in each sample. Results in the autoradiograms of minus-, plus-, and 18S ribosomal RNAs were all derived from one Northern blot that was probed and stripped sequentially with probes for the specific RNAs. Some samples unrelated to the results in this experiment were removed. The symbol “φ” indicates a reaction that did not result in transfection with BMV RNAs. Contamination in the loading of the lanes labeled with this symbol is evident but does not affect the interpretation of the results.
TABLE 1.
Effect of mutations in SLC on RNA synthesis in vitro and in protoplasts
| Nucleotide change or RNA3 derivative | Percent RNA synthesis in vitroa | Negative-strand synthesis ratio (RNA3/RNA2) | Positive-strand synthesis ratio (RNA3/RNA2) |
|---|---|---|---|
| Triloop | |||
| AUA (wt)b | 100 | 9.0 | 4.3 |
| AUGc | 132 ± 7 | 8.5 | 3.7 |
| GUA | 8 ± 1 | 0.5 | 0.8 |
| Triloop stem | |||
| G-C | 33 ± 7 | 8.7 | 5.2 |
| A-G | 9 ± 1 | 3.1 | 1.2 |
| Δ1bp | 95 ± 12 | 9.0 | 4.3 |
| In3 | 35 ± 3 | 0.2 | 0.5 |
| Bulge | |||
| mBulge (AAGA/UUCU) | 56 ± 4 | 9.0 | 3.5 |
| CMV/BMV chimera | |||
| C-SLC3 | 21 ± 1 | 0.3 | 0.4 |
| C-SLC5 | 23 ± 2 | 0.2 | 0.3 |
For the AUG mutant, minus- and plus-strand RNAs accumulated to levels similar to those of the wild-type transcripts. We used the ratios of RNA3 to RNA2 to normalize for slight differences in the overall levels of RNA accumulated. The ratio of minus-strand RNA in transfections with the AUG mutant was 8.5 (compared to a ratio of 9.0 for wild-type RNA3; Table 1). For positive-strand RNAs, the RNA3 to RNA2 ratio for the AUG transcripts was 3.7 (relative to 4.3 for the wild type; Table 1). These results demonstrate that the change of the triloop from AUA to AUG had no major effect on minus-strand RNA synthesis, consistent with the results from RNA synthesis by the replicase in vitro.
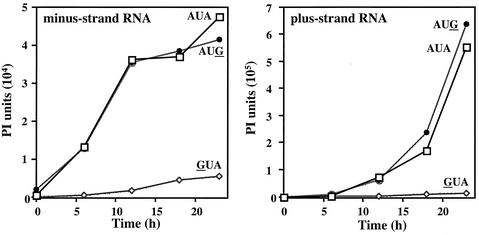
To examine further whether the AUG mutation has an effect on RNA replication, we analyzed its replication in barley protoplasts over time in direct comparison to the replication of wild-type BMV RNA. RNAs were extracted at 6, 12, 18, and 23 h after infection (Fig. 3). Quantification of the minus-strand RNAs revealed that the rate of accumulation of RNA with an AUG triloop was similar to that of the wild type. In contrast, an RNA3 with a change of the triloop to GUA accumulated to less than 10% of the level of either the wild-type or the AUG mutant. Similar accumulations were observed for the plus-strand RNAs (Fig. 3). Since the kinetics of RNA accumulation of the wild-type and the AUG versions of RNA3 are virtually identical, this argues against the accumulation of a reversion or a suppressing mutation that allowed the AUG mutant to gain the ability to replicate.
FIG. 3.
Time course of minus-strand (left panel) and plus-strand (right panel) RNA accumulation in protoplasts transfected with RNA3 containing a wild-type AUA triloop or mutant AUG (mutation underlined) or GUA triloops. Transfected cells were lysed, and RNA was extracted at the times indicated on the horizontal axes. Where there was a mutation in RNA3, the mutated nucleotide is underlined in the name of the RNA. The results of a Northern blot were quantified using a PhosphorImager and expressed in phosphorimage (PI) units.
Additional requirements for features in SLC.
A number of requirements in the triloop stem were previously determined from biochemical analyses. These include a proper length of the triloop stem and specific requirement for a closing base pair of the triloop (16, 24). More specifically, a transversion of the closing base pair from 5′CG3′ to 5′GC3′ (mutation underlined) in the context of the wild-type triloop decreased RNA synthesis of SLC+8 to one-third that of the wild type (16). The requirement for the closing base pair is not absolute, however, since a change from the normal 5′CG3′ to a 5′GC3′ in the context of the AUG triloop did not affect RNA synthesis (17). These results indicate that unlike that for the clamped adenine, there is some flexibility in the requirement for the closing base pair. A similar situation exists for the triloop stem: a deletion of 1 bp did not significantly affect RNA synthesis, while an insertion or deletion of 3 bp prevented RNA synthesis (24). These observations are consistent with an induced fit recognition mechanism wherein the CAM acts as a primary determinant for recognition by the replicase and subsequent interactions between the replicase and the RNA can be adjusted by their mutual interactions (17).
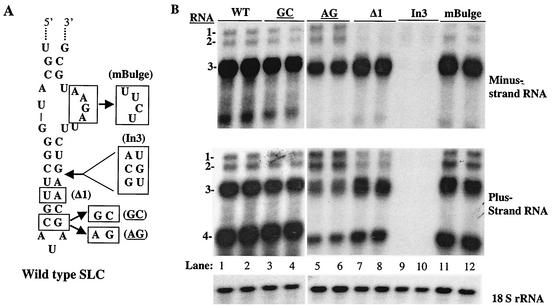
An RNA3 containing a closing base pair of 5′GC3′ was examined in protoplasts. The levels of minus- and plus-strand RNA synthesized from this mutant RNA were comparable to those of wild-type RNA3 (Table 1). Hence, the results in vivo did not agree with those in vitro for SLC with an AUA triloop. However, the results were consistent with an SLC with an AUG triloop, which no longer requires the closing base pair in vitro (16, 17). The closing base pair must remain stably base paired, since a change to 5′AG3′ resulted in an RNA in which the ratio of RNA3 to RNA2 was reduced to 3.1 and 1.2, approximately threefold below that of transfections with wild-type RNAs (Table 1).
The effects of changing the triloop stem on RNA3 replication in protoplasts were examined. A deletion of 1 bp in an RNA3 derivative named Δ1 in the SLC triloop stem had only a minor effect on RNA synthesis (Fig. 4B, lanes 7 and 8) (Table 1). However, a 3-bp insertion in SLC, resulting in an RNA3 named In3, reduced minus- and plus-strand RNA accumulation to 3% and 11% of the amounts seen with wild-type RNA3 (Fig. 4B, lanes 9 and 10) (Table 1). These results are consistent with the effects of the presence of BMV replicase on RNA synthesis in vitro, for which a specific length of the closing stem is required (24).
FIG. 4.
Effect of mutations in the triloop stem and the closing base pair on BMV RNA3 replication. (The names of mutated nucleotides are underlined.) (A) Schematic of SLC, with the insertion, deletion, and point mutations shown in boxes. Only the affected regions of RNA3 are shown. The names of the mutant RNAs are in parentheses. (B) Autoradiogram showing effects of mutations on genomic minus- and plus-strand (as indicated to the right of the autoradiograms) accumulation. Total RNA was extracted from transfected protoplasts, and Northern analysis was carried out using probes specific for plus-strand, minus-strand, and 18S RNAs. The names of the mutant RNA3s used in the transfections are shown above the autoradiograms, and the identities of the RNAs in each image are indicated to the left of the autoradiograms.
Lastly, based on observations of a highly flexible bulge sequence detected in nuclear magnetic resonance analysis of SLC, biochemical analysis of mutant RNAs, and a comparison of bromoviral sequences (16), we proposed that the bulge acts as a flexible region rather than a specific sequence (16). Consistent with this hypothesis, versions of SLC in which deletions that result in a highly flexible sequence which replaces the terminal stem remain capable of efficient RNA synthesis (16, 24) and the removal of the bulge that should result in a more stable stem debilitated RNA synthesis (8). To examine the sequence requirements of the bulge, we made RNA mBulge, in which the bulge was changed from the normal purine-rich sequence of 5AGAA3′ to a pyrimidine-rich 5′UCUU3′. In vitro, mBulge directed RNA synthesis at 56% of that of SLC+8. In the context of RNA3 in protoplasts, mBulge was found to produce minus- and plus-strand RNAs at ratios of 9 and 3.5, which was similar to those for wild-type RNA3 (Fig. 4B, lanes 11 and 12) (Table 1). Hence, a change of the bulge sequence had only minor effects in vitro and no detectable effect in vivo.
A cellular tRNA cannot replace the BMV tRNA-like sequence.
The observation that SLC+8 can direct RNA synthesis in vitro demonstrates that this element is sufficient for recognition by the BMV replicase and to direct proper placement of the initiation site in the replicase. To examine whether these requirements can be fulfilled by a more minimal tRNA, we replaced the 3′ 200 nt of BMV RNA3 with the sequence of the barley tyrosine tRNA, tRNATyr (24), and assessed whether the chimeric RNA was able to replicate in barley protoplasts cotransfected with BMV RNA1 and RNA2. In addition, we added a fused version of the tRNATyr in which the tψc stem-loop was replaced with SLC. None of these chimeras were able to direct minus-strand synthesis in protoplasts (data not shown). Therefore, in addition to providing the elements for replicase recognition and initiation, the BMV 3′ tRNA-like sequence must fulfill additional roles in BMV infection process.
CMV and BMV chimeras.
The Cucumber mosaic virus (CMV) SLC differs from the one in BMV in that a highly conserved dinucleotide in the loop, 5′CA3′, replaces the CAM in acting as a specificity element for RNA synthesis (29). Sivakumaran et al. have previously determined that a minimal-length RNA consisting of the CMV SLC attached to an initiation sequence was an inefficient template for RNA synthesis by the BMV replicase (29). RNA synthesis in vitro was improved approximately twofold when the RNA contained the entire CMV tRNA-like sequence, indicating that elements within the tRNA-like structure in addition to SLC can help the BMV replicase recognize this chimeric RNA. Rao and Grantham (25) determined that replacement of the 3′ tRNA region of the BMV RNA3 with the CMV tRNA-like sequence allowed viral RNA replication in protoplasts, although the results were primarily qualitative. These results support our claim of the importance of the CAM for the BMV replicase and indicate that proper interaction between the SLC and other portions of the tRNA-like sequence contributes to RNA synthesis. In vitro, Chapman and Kao had previously found that the tRNA-like portion of the BMV 3′ region can bind the replicase about threefold better than SLC (5).
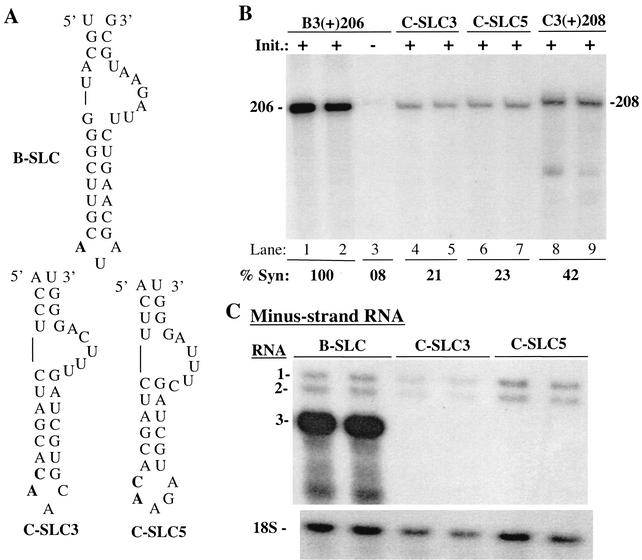
We first examined RNA synthesis in vitro from a ∼206-nt sequence containing the tRNA-like portion of the BMV RNA. Two constructs were made in which the BMV SLC was replaced with comparable sequences from CMV strains from Fny (group 1A) (containing a 3-nt loop of 5′AAC3′) and from Ixora (group II) (containing a predicted 5-nt loop [5′CAAGA3′]) (Fig. 5A). In addition to differences in the triloop region, nucleotides in the bulge of the two CMV strains were different from those of the BMV SLC (Fig. 5A). The amount of products from the 206-nt 3′ sequence of BMV RNA3, B3(+)206, was assigned a value of 100%. A change of the initiation CCA site to GGA reduced synthesis to 8% (Fig. 5B, lane 3). The CMV 3′ 208 nt from strain Fny was found to give 42% synthesis, while chimeric RNAs B3(+)cSLC3 and B3(+)cSLC5 produced 21 and 23%, respectively, of the products of B3(+)206 (Fig. 5B). These results confirm and extend previous findings that the BMV replicase does not efficiently recognize the CMV SLC sequence in vitro. However, other portions of the CMV tRNA-like sequence may induce the BMV replicase to increase recognition of the mutant SLC about twofold.
FIG. 5.
Effects of replacing the BMV SLC with CMV SLC motifs on RNA synthesis in vitro and in transfected protoplasts. (A) Relevant features in the chimeric BMV and CMV RNA3s under analysis. The sequence of SLC from BMV RNA3 is indicated as B-SLC. SLCs from strain Fny and strain Ixora are shown as C-SLC3 and C-SLC5, respectively. The clamped adenine and the highly conserved CA residues in the CMV SLCs are in bold letters. Other portions of the BMV RNA3 are not shown. (B) Autoradiogram showing the effects of chimeric RNA3 on RNA synthesis by the BMV RNA replicase in vitro. Templates for these assays consist of the 3′ 206 or 208 nt of the BMV RNA or chimeric RNAs. Quantifications of the results from this experiment and at least two additional independent experiments are shown below the autoradiogram. The symbol “+” indicates that the RNA is capable of initiation of RNA synthesis, while a “−” indicates that the RNA has a change of the initiation CCA sequence to a GGA. (C) Results of Northern blot analysis comparing RNA synthesis from BMV RNA and the two chimeric RNA3s. Only the minus-strand and 18S RNAs from this blot are shown. All three combinations of RNAs were tested in the same set of protoplasts. The identities of the RNA3 used in the transfections along with BMV RNA1 and RNA2 are shown above the autoradiogram.
Next, we examined whether BMV RNA3 with SLC replaced by the comparable sequence from Fny (C-SLC3) or Ixora (C-SLC5) can replicate in protoplasts. Both chimeric RNA3s were poor templates for RNA synthesis in protoplasts, with minus-strand RNA3 synthesis at a ratio of 0.3 to that of RNA2 (Table 1). The amount of plus-strand RNA synthesis was comparably reduced, with ratios to RNA2 of 0.4 and 0.3 for C-SLC3 and C-SLC5, respectively. Similar to the in vitro results, the CMV SLC is not efficiently recognized by the BMV replicase in vivo, even in the context of the BMV tRNA-like sequence.
Differential RNA synthesis by the BMV replicase in vitro.
The general agreement between RNA synthesis by the BMV replicase in vitro and BMV replication in protoplasts gives credence to the use of BMV replicase and minimal-length RNAs to examine the requirements of viral RNA synthesis in vitro. We sought to examine whether in vitro replicase-dependent RNA synthesis can be used to examine temporal changes in the modes of RNA synthesis. In a typical infection with wild-type BMV RNA, minus-RNAs accumulated prior to the detection of plus-strand genomic and subgenomic RNAs (Fig. 6A; compare minus- and plus-strand RNAs at 6 h and 12 h posttransfection).
FIG. 6.
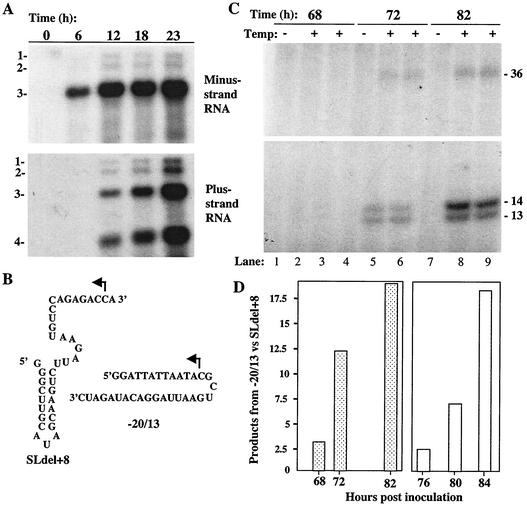
BMV RNA replication and RNA synthesis by the BMV replicase as a function of time. (A) Results of a time course experiment with BMV RNAs synthesized in transfected protoplasts. The times at which the protoplasts were harvested after transfection are indicated above the autoradiogram. Identities of the RNAs are listed to the left of the autoradiograms. (B) Sequences of the minimal BMV promoter templates used to assess minus-strand RNA and subgenomic RNA initiation in vitro. The arrows identify the initiation cytidylate. RNA SLdel+8 produces a 36-nt RNA product (16), and Proscript −20/13 should produce a 13-nt runoff transcript but frequently produces a transcript that contains one nontemplated nucleotide (27). (C) Qualitative analysis of the RNA synthesized from the three promoters by the replicases prepared at three different times. The upper panel shows the genomic minus-strand product from SLdel + 8, while the lower panel shows the subgenomic initiation products from −20/13. The presence of the two bands is indicative of nontemplated nucleotide addition, a feature previously described for the BMV replicase (27). (D) Quantification of the ratio of the BMV genomic minus-strand RNA synthesis to subgenomic plus-strand synthesis from replicases harvested and purified in two independent time course experiments. The products of RNA synthesis were normalized for the potential number of radiolabeled CMPs incorporated.
The template for characterization of genomic minus-strand RNA synthesis used SLdel+8, which produces a 36-nt product in a manner dependent on the CAM (16). Subgenomic RNA synthesis used Proscript −20/13, which produces a 13-nt RNA (27) (Fig. 6B). Should RNA synthesis by the BMV replicase mimic the accumulation of the different classes of BMV RNAs in vivo, we predict that replicase preparations harvested over a time course would direct differing levels of RNA synthesis from the two classes of minimal promoters in vitro. These experiments are complicated, however, by the replicases from the different time points being purified separately. To remove this complication, comparisons were based on the ratio of the RNAs synthesized from the different promoter templates by the same fraction after normalization for radiolabel incorporation.
BMV-infected barley was harvested at different times after inoculation, and the BMV replicase was enriched as described in Materials and Methods. This procedure removes most RNase activity and allows efficient RNA synthesis from exogenously provided RNAs. Fractions harvested from barley infected for less than 60 h had robust RNA synthesis from a number of templates that are not specific to the BMV RNAs (data not shown). Identical activity was observed from mock-inoculated leaves, indicating that cellular RNA polymerase(s) is likely the source of this polymerase activity (data not shown). Usually by 70 h after inoculation with BMV, the purported cellular polymerase activity decreased significantly and BMV promoter-specific synthesis predominated (Fig. 6C and B) and BMV core promoter-dependent synthesis was observed. Our normal replicase preparations were usually harvested 85 to 90 h after inoculation, when the cellular RNA polymerase activity did not interfere with interpretation of BMV replicase activity.
Barley inoculated with 0.1 mg of BMV/ml had peak replicase activity at 76, 80, and 84 h postinoculation. The initial screen for replicase activity was identified by RNA synthesis from the BMV subgenomic Proscript −20/13. The fractions were then tested for the ability to synthesize products from −20/13 and the genomic minus-strand promoter SLdel + 8 (Fig. 6C). The relative amounts of product were normalized for CMP (cytidine 5′-monophosphate) incorporation. The ratios of subgenomic plus-strand RNA to genomic minus-strand RNA were 2, 7, and 18 for the replicase preparations at 76, 80, and 84 h, respectively. In a second set of barley samples (inoculated with BMV at 0.3 mg/ml), three preparations harvested at 68, 72, and 82 h postinoculation were found to have replicase activity. In these three preparations, the ratios of subgenomic to genomic minus-strand RNAs were 3, 13, and 19, respectively. The ratios of genomic plus-strand RNA to genomic minus-strand RNA increased from 0.08 to 0.14 to 0.3 for the replicase preparation at 76, 80, and 84 h, respectively. The increase in plus-strand RNA relative to minus-strand RNA over time indicates that the replicase can change its preferred mode of RNA synthesis in a manner paralleling the situation in vivo.
DISCUSSION
Biochemical approaches are increasingly being used to examine the requirements for viral replication (for examples, see references 6, 7, 21, 22, 23, 29, 30, and 32). This reductionist approach is useful in asking highly focused questions uncomplicated by possibilities that must be taken into account during an infection. Attempts to correlate the requirements of the BMV replicase and RNA replication in protoplasts have been performed previously (8, 9). However, thorough analyses of the requirements in vitro have been correlated with those in vivo for relatively few systems.
We demonstrated in this work that the requirements for the BMV replicase found using minimal length RNAs generally parallel the requirements in vivo. For example, we determined that the requirement for the CAM in the promoter for minus-strand RNA synthesis applies to the situation in vivo. A change in the triloop that reduced the formation of the CAM also severely affected BMV RNA3 replication, in agreement with the results of Rao et al. (26). A mutation in the triloop of SLC that resulted in sterically altered CAM retained RNA synthesis in vitro and in vivo (17). Furthermore, previous claims that the closing base pair for the CAM and the length of the triloop stem are important in vitro were also found to hold true in vivo. Lastly, our hypothesis that the bulge of SLC is required to be flexible but that a specific sequence is not required is found to be supported in vivo.
We note, however, that while the in vitro results do predict defects in vivo, the absolute correlation of the amounts of RNA synthesis in vivo and in vitro is not linear. For example, an insertion of 3 bp in the minimal SLC reduced RNA synthesis to one-third in vitro, while the effect was more severe in the context of RNA3 in protoplasts. In addition, a change of the closing base pair from the wild-type CG to an AG reduced RNA synthesis to 9% of that of the control in the context of the minimal SLC but decreased RNA synthesis to ∼30% in protoplasts. Lastly, while a closing base pair for the stem-triloop is required (as we had observed in vitro), a change in the identity of the bases was tolerated better in vivo. It is likely that some mutations are modulated by other RNA elements in vivo which might ameliorate or exaggerate the effects observed in vitro. For example, increasing the length of the triloop stem might clash with neighboring structures within RNA3 while the change of the detrimental effects of the closing base pair might be partially alleviated by the contributions of other structures in the tRNA-like domain. The minimal-length RNAs are thus useful for identifying the essential requirements but cannot take into account additional contributions from other factors. This situation is similar to those seen in studies of core promoters in DNA that can mediate responses to other regulatory signals. In addition, while biochemical assays with the replicase allow examination of the requirements for promoter recognition and RNA synthesis, the intention is not to duplicate all of the activities associated with BMV replication in the infected cell. Consistent with this claim, a tRNATyr cannot replace the BMV 3′ 200 nt for replication in barley protoplasts, even when the tRNA contains an SLC (data not shown). We note that a tRNATyr fused to an SLC at multiple positions can direct RNA synthesis in vitro (24), suggesting that other interactions between the BMV tRNA-like sequence and cellular and/or viral factors can constrain interactions with the replicase. Despite the limitation of the biochemical system, it is useful first to understand the essential requirements and then to address the factors that modulate them.
Other features within the BMV RNA3 may modulate the activity of SLC. We observed that the tRNA-like region of the CMV RNA3 had a less severe effect on RNA synthesis than did the minimal length CMV SLC attached to an initiation sequence in vitro. Rao and Grantham (25) previously demonstrated this heterologous recognition of RNA3 in protoplasts, while our lab demonstrated that RNA synthesis of the CMV SLC attached to a BMV initiation sequence was inefficient. Second, while SLC is necessary and sufficient for RNA-dependent RNA synthesis, Chapman and Kao have demonstrated that the replicase binds to the tRNA-like sequence approximately threefold better than to SLC alone (5). There may even be influences from sequences beyond the tRNA-like sequence. The BMV sequence 5′ of the tRNA-like sequence, but not the CMV sequence, can induce initiation to occur from a mutated BMV tRNA-like 3′ end (4), indicating that initiation can be stimulated by sequence 5′ of the tRNA-like region. The fact that these effects are seen in vitro and in vivo indicates that many additional interactions required for BMV RNA replication can be dissected by a combination of in vitro and in vivo approaches.
RNA synthesis by the BMV replicase can also be used to examine changes that take place during an infection. Replicases isolated early in the infection process preferentially used the promoter for minus-strand RNA synthesis, while the replicases isolated later increased the ability for genomic and subgenomic plus-strand RNA synthesis. These shifts in the recognition of the minimal promoters parallel the production of the different classes of RNAs produced in vivo. This observation allows a proteomic approach for the examination of changes in the composition of the replicase that are correlated with promoter-specific RNA synthesis.
Acknowledgments
This work would not have been possible without the encouragement of Tim Hall at numerous ASV meetings.
Funding was provided by the National Science Foundation.
REFERENCES
- 1.Adkins, S., R. W. Siegel, J. H. Sun, and C. C. Kao. 1997. Minimal templates directing accurate initiation of subgenomic RNA synthesis in vitro by the brome mosaic virus RNA-dependent RNA polymerase. RNA 3:634-647. [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins, S., S. Stawicki, G. Faurote, R. Siegel, and C. Kao. 1998. Mechanistic analysis of RNA synthesis RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA 4:455-470. [PMC free article] [PubMed] [Google Scholar]
- 3.Buck, K. W. 1996. Comparison of the replication of positive-strand RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman, M., A. L. N. Rao, and C. Kao. 1998. The 5′ end of BMV RNAs promotes initiation of (−)-strand RNA synthesis in vitro and in vivo. Virology 252:458-467. [DOI] [PubMed] [Google Scholar]
- 5.Chapman, M. R., and C. Kao. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286:709-720. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, J. H., M. P. Ding, Y. H. Hsu, and C. H. Tsai. 2001. The partial purified RNA-dependent RNA polymerases from bamboo mosaic potexvirus and potato virus X infected plants containing the template-dependent activities. Virus Res. 80:41-52. [DOI] [PubMed] [Google Scholar]
- 7.Deiman, B. A. L. M., A. K. Konen, P. W. G. Verlaan, and C. W. A. Pleij. 1998. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J. Virol. 72:3965-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dreher, T. W., and T. C. Hall. 1988. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus-strand promoter activity. J. Mol. Biol. 201:31-40. [DOI] [PubMed] [Google Scholar]
- 9.Dreher, T. W., and T. C. Hall. 1988. Mutational analysis of the tRNA mimicry of brome mosaic virus RNA. J. Mol. Biol. 201:41-55. [DOI] [PubMed] [Google Scholar]
- 10.Fechter, P., R. Giege, and J. Rudinger-Thirion. 2001. Specific tyrosylation of the bulky tRNA-like structure of brome mosaic virus RNA relies solely on identity nucleotides present in its amino acid-accepting domain. J. Mol. Biol. 309:387-399. [DOI] [PubMed] [Google Scholar]
- 11.Felden, B., C. Florentz, R. Geige, and E. Westhof. 1994. Solution structure of the 3′ end of brome mosaic virus genomic RNAs. Conformational mimicry with canonical tRNAs. J. Mol. Biol. 235:508-531. [DOI] [PubMed] [Google Scholar]
- 12.Hardy, S. F., T. L. German, S. Loesch-Fries, and T. C. Hall. 1979. Highly active template-specific RNA-dependent RNA polymerase from barley leaves infected with Brome mosaic virus. Proc. Natl. Acad. Sci. USA 76:4956-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janda, M., R. French, and P. Ahlquist. 1987. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions on transcript infectivity. Virology 158:259-262. [DOI] [PubMed] [Google Scholar]
- 14.Kao, C. 2002. Lessons learned from the core RNA promoters of Brome mosaic virus and Cucumber mosaic virus. Mol. Plant Pathol. 3:53-59. [DOI] [PubMed] [Google Scholar]
- 15.Kao, C., and K. Sivakumaran. 2000. Brome mosaic virus, good for an RNA virologist's basic needs. Mol. Plant Pathol. 1:91-98. [DOI] [PubMed] [Google Scholar]
- 16.Kim, C.-H., C. Kao, and I. Tinoco. 2000. RNA motifs that determine specificity between a viral replicase and its promoter. Nat. Struct. Biol. 7:415-423. [DOI] [PubMed] [Google Scholar]
- 17.Kim, C.-H., and C. Kao. 2001. A mutant viral RNA promoter with an altered conformation retains efficient recognition by a viral RNA replicase through a solution-exposed adenine. RNA 7:1476-1485. [PMC free article] [PubMed] [Google Scholar]
- 18.Kroner, P. A., B. M. Young, and P. Ahlquist. 1990. Analysis of the role of brome mosaic virus 1a protein domains in RNA replication, using linker insertion mutagenesis. J. Virol. 64:6110-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh, L. E., T. W. Dreher, and T. C. Hall. 1988. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 16:981-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, W. A., T. W. Dreher, and T. C. Hall. 1985. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature 313:68-70. [DOI] [PubMed] [Google Scholar]
- 21.Osman, T. A. M., and K. W. Buck. 1996. Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J. Virol. 70:6227-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plante, C. K., H. Kim, N. Pillai-Nair, T. A. Osman, K. W. Buck, and C. L. Hemenway. 2000. Soluble template-dependent extracts from Nicotiana benthamiana plants infected with potato virus X transcribe both plus- and minus-strand RNA templates. Virology 275:444-451. [DOI] [PubMed] [Google Scholar]
- 23.Quadt, R., H. J. M. Rosdorff, T. W. Hunt, and E. M. J. Jaspars. 1992. Analysis of the protein composition of the alfalfa mosaic virus RNA-dependent RNA polymerase. Virology 182:309-315. [DOI] [PubMed] [Google Scholar]
- 24.Ranjith-Kumar, R., X. Zhang, and C. Kao. 2003. Enhancer-like activity of a brome mosaic virus RNA promoter. J. Virol. 77:1830-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao, A. L. N., and G. L. Grantham. 1994. Amplification in vivo of brome mosaic virus RNAs bearing 3′ noncoding region from cucumber mosaic virus. Virology 204:478-481. [DOI] [PubMed] [Google Scholar]
- 26.Rao, A. L. N., T. W. Dreher, L. E. Marsh, and T. C. Hall. 1989. Telomeric function of the transfer RNA-like structure of brome mosaic virus RNA. Proc. Natl. Acad. Sci. USA 86:5335-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegel, R. W., S. Adkins, and C. Kao. 1997. Sequence-specific recognition of a subgenomic promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. USA 94:11238-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sivakumaran, K., C. H. Kim, R. Tayon, Jr., and C. Kao. 1999. RNA sequence and secondary structural determinants in a minimal viral promoter that directs replicase recognition and initiation of genomic plus-strand RNA synthesis. J. Mol. Biol. 294:667-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sivakumaran, K., Y. Bao, M. J. Roossinck, and C. Kao. 2000. Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicase of Brome mosaic virus and Cucumber mosaic virus. J. Virol. 74:10323-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song, C., and A. E. Simon. 1995. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 254:6-14. [DOI] [PubMed] [Google Scholar]
- 31.Sun, J. H., S. Adkins, G. Faurote, and C. Kao. 1996. Initiation of (−)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology 226:1-12. [DOI] [PubMed] [Google Scholar]
- 32.Yoshinari, S., P. D. Nagy, A. E. Simon, and T. W. Dreher. 2000. CCA initiation boxes without unique promoter elements support in vitro transcription by three RNA-dependent RNA polymerases. RNA 6:698-707. [DOI] [PMC free article] [PubMed] [Google Scholar]