Abstract
Fluorescent retroviral envelope (Env) proteins were developed for direct visualization of viral particles. By fusing the enhanced green fluorescent protein (eGFP) to the N terminus of the amphotropic 4070A envelope protein, extracellular presentation of eGFP was achieved. Viruses incorporated the modified Env protein and efficiently infected cells. We used the GFP-tagged viruses for staining retrovirus receptor-positive cells, thereby circumventing indirect labeling techniques. By generating cells which conditionally expressed the GFP-tagged Env protein, we could confirm an inverse correlation between retroviral Env expression and infectivity (superinfection). eGFP-tagged virus particles are suitable for monitoring the dynamics of virus-cell interactions.
The early steps of retroviral infection are under investigation in order to elucidate the underlying mechanisms and to find ways for inhibiting virus infection. The key proteins of the infection process are the retroviral surface protein (SU) and a transmembrane protein (TM), which are cleaved from the precursor protein Env by the cellular processing machinery. TM is integrated into the cellular membrane. SU is located extracellularly and linked to TM by noncovalent interactions. Virus budding results in the release of viruses enveloped with the cellular membrane and spiked with trimers of SU-TM heterodimers. Because of the weak association of SU and TM, SU is frequently shed from the surface of cells and virus particles.
Until 1999, electron microscopy was the only method for visualization of retrovirus particles. Electron microscopy can provide detailed information about the morphology of viral particles. It has been also used to study virus-cell interactions in the case of human immunodeficiency virus (9). Recently, confocal laser scanning microscopy (CLSM) was used for visualization of immunostained retroviral particles (8), opening new perspectives for the analysis of retroviruses. However, both electron microscopy and the confocal analysis of immunostained particles are restricted to the visualization of a fixed state. Thus, these methods do not allow monitoring of dynamic processes such as the early phase of retroviral infection.
In recent years, fluorescent proteins and in particular variants of the green fluorescent protein (GFP) have become an important tool for imaging cellular proteins and structures in living cells as well as for the monitoring of cellular processes. Here, we report the tagging of retroviral particles with a fluorescent protein by N-terminal fusion of enhanced GFP (eGFP) to the amphotropic SU. The fusion protein is efficiently incorporated into cellular and viral membranes, resulting in green-fluorescent virus particles capable of transducing cells. GFP-tagged virus particles thus should allow direct monitoring of virus-cell interactions by CLSM.
N-terminal fusion of eGFP to Env4070A maintains infection potential.
For direct visualization of retroviral particles, we developed a fusion gene comprising the amphotropic retroviral 4070A envelope gene and the gene encoding the enhanced GFP (eGFP). eGFP was fused to the N terminus of the retroviral SU protein, resulting in an eGFP-Env fusion protein designated G-FLAEP (enhanced green fluorescent amphotropic envelope protein; see Fig. 1 for details). We assessed the ability of G-FLAEP to assemble into virus particles that were able to interact with the retroviral receptor Pit-2 and efficiently transduce cells. With DNA-calcium phosphate coprecipitation (10), NIH 3T3 cells stably expressing gag-pol (Env− cells [10]) were cotransfected with both a G-FLAEP-encoding plasmid (G-FLAEP-C, Fig. 1) and a packageable retroviral vector plasmid encoding the neomycin phosphotransferase gene (10 and 1 μg, respectively). As a control, a wild-type Env4070A expression plasmid together with the same retroviral vector was transfected. G418-resistant cell pools of approximately 500 clones each were established. Subsequently, virus particles were collected from G-FLAEP and wild-type Env producer cells by ultracentrifugation with a Beckman SW40 rotor (Beckman, Munich, Germany) at 30,000 rpm for 1.5 h at 4°C. Virus pellets resuspended in phosphate-buffered saline were excited with blue light.
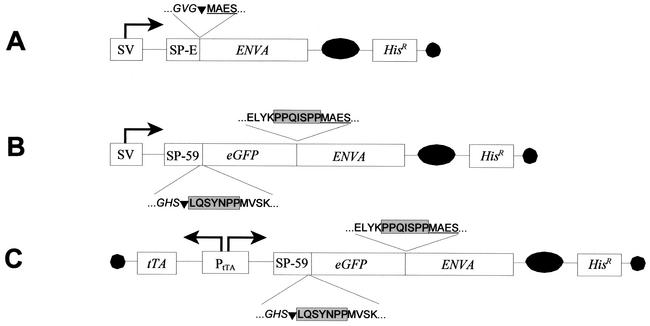
FIG. 1.
Schematic representation of amphotropic envelope constructs. All constitutive expressing plasmids are based on pSBC-1 (2) and pSBC-2-his (D. Spitzer, unpublished data) and express the respective env genes in a bicistronic message with the histidinol dehydrogenase gene (Hisr). (A) pEnvA, the wild-type 4070A envelope expression plasmid. env4070A was PCR amplified from an amphotropic murine leukemia virus genome. pEnvA comprises the endogenous murine leukemia virus-derived signal peptide (SP-E). (B) pG-FLAEP-C, the constitutive G-FLAEP expression plasmid. This plasmid encodes the fusion protein G-FLAEP, comprising the CD59 signal peptide sequence SP-59, eGFP, and the signal peptide-deleted env4070A gene linked through a spacer. eGFP was derived from pEGFP-1 (Clontech) by PCR, and the spacer was designed on the basis of CD59. (C) pG-FLAEP-AR, the autoregulated plasmid comprising the bidirectional tTA-dependent promoter (1) driving both the transactivator tTA and G-FLAEP. The bicistronic message comprising the G-FLAEP gene and Hisr gene was integrated into pTHTG (11), replacing the expressed gene in this vector. SV, simian virus 40 early promoter/enhancer; PtTA, bidirectional tTA-dependent promoter; SP, signal sequence; EnvA, mature amphotropic Env4070A; eGFP, coding sequence of the enhanced green fluorescent protein; tTA, tetracycline-dependent transactivator; black circle, simian virus 40 polyadenylation signal; black oval, poliovirus internal ribosomal entry site. The amino acid sequence of fusion sites is indicated: eGFP sequence (regular) and EnvA sequence (underlined) as well as spacer sequences (boxed), and signal peptide sequences (italics), including the cleavage sites (inverted triangles) are depicted.
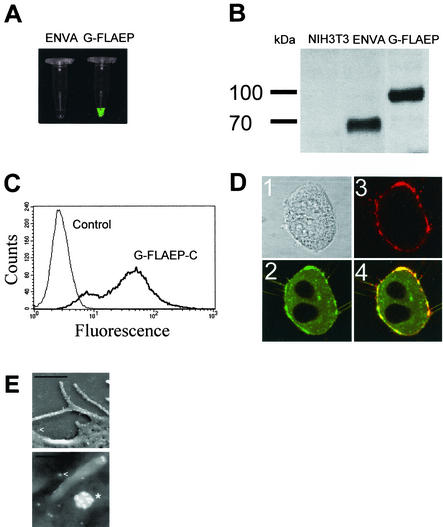
Concentrated G-FLAEP virus preparations but not control viruses exhibited significant green fluorescence (Fig. 2A). Virus particles generated from these cells were subjected to Western blot analysis under reducing conditions (Fig. 2B). With a polyclonal goat-derived antiserum directed against SU and a horseradish peroxidase-coupled anti-goat immunoglobulin antibody for detection (ECL System; Amersham, Braunschweig, Germany), the G-FLAEP fusion protein appeared as a signal of approximately 100 kDa, compared to approximately 70 kDa for the unmodified SU protein. No degradation products and no reversion to wild-type SU was observed (Fig. 2B). To exclude formation of intra- or intermolecular protein bonds due to the presence of the additional eGFP ligand in G-FLAEP, we performed a Western blot under nonreducing conditions with the same anti-SU antiserum (not shown). With respect to their molecular weights, we found essentially the same banding pattern for the EnvA and G-FLAEP supernatants as described for the reducing gel conditions shown in Fig. 2B. This finding proves the absence of any irregular bond formations in G-FLAEP.
FIG. 2.
Characterization of G-FLAEP. (A) Concentrated virus preparations from EnvA and G-FLAEP producer cells were excited with blue light and analyzed with a charge-coupled device camera. (B) Western blot analysis of Env proteins of recombinant virus with particles from producer cells expressing EnvA and G-FLAEP. A polyclonal goat-derived anti-Env antibody directed against SU was used (kindly provided by R. Friedrich, Giessen, Germany). This antibody detects amphotropic and ecotropic murine leukemia virus. Supernatant of NIH 3T3 parental cells was used as a control. See text for details. (C) Fluorescence activity of G-FLAEP. The fluorescence signal of G-FLAEP-C-expressing cells was analyzed by flow cytometry with a FACScan flow cytometer (Becton Dickinson, Bedford, Mass.). NIH 3T3 cells were used as a control. (D) Colocalization of eGFP and SU domains. Fixed G-FLAEP-expressing NIH 3T3 cells were immunostained with a polyclonal anti-SU antibody and analyzed by confocal microscopy with a laser scanning confocal imaging system (MRC-1024 UV; Bio-Rad, Munich, Germany) equipped with a 5-mW krypton-argon laser (488 nm, 568 nm, and 647 nm). Fixed cells were subsequently incubated with the polyclonal goat serum and indocarbocyanine-labeled anti-goat IgG F(ab′)2 fragments. Images: 1, transmission image; 2, eGFP detection with a fluorescein isothiocyanate filter; 3, SU detection with indocarbocyanine-conjugated secondary antibodies; 4, merged pictures 2 and 3. (E) Detection of G-FLAEP after staining with immunogold particles by indirect immunoscanning electron microscopy. Bright dots, 10-nm gold particles bound to G-FLAEP on cells or on virions or to shed protein; star, viral particle; arrowheads, shed G-FLAEP protein complexes. Bars in the upper and lower images represent 1,000 nm and 200 nm, respectively.
We next determined the virus titer of G-FLAEP producer cells. For this purpose, we collected the supernatants of the above-described cells for transduction of NIH 3T3 cells in the presence of Polybrene (4 μg/ml). The supernatant of G-FLAEP-expressing producer cells efficiently transduced NIH 3T3 cells with a titer of 4.1 × 105 G418-resistant CFU/ml. This is comparable to that of the above-described control cells expressing Env4070A, which give rise to 4.3 × 105 CFU/ml. This implies, as a prerequisite for further studies, that in G-FLAEP, the eGFP domain perturbs neither the interaction of SU and TM nor the formation of Env trimers with respect to binding and postbinding events within the retrovirus infection process. Thus, the functional eGFP domain of G-FLAEP does not seem to interfere with any stage of the retroviral replication cycle.
Characterization of G-FLAEP viruses and cells.
G-FLAEP-expressing cells which showed strong fluorescence upon excitation with blue light were analyzed by flow cytometry. Figure 2C shows the fluorescence profile of G-FLAEP virus producer cells expressing G-FLAEP and nontransfected NIH 3T3 cells as a control. The fluorescence shift indicates that the fluorescent properties of eGFP were maintained in the fusion protein.
It was expected that eGFP, as part of a type I membrane protein, would be located both intra- and extracellularly. The distribution of G-FLAEP within the producer cells was studied by CLSM (Fig. 2D). Indeed, G-FLAEP fluorescence was detected in the cytoplasm, in intracellular vesicles, and at the plasma membrane, while the nuclei did not stain and appeared black (Fig. 2D). The cytoplasm exhibited high fluorescent signal intensities, which reflect synthesis and transport of retroviral Env proteins to the plasma membrane. To demonstrate that the eGFP domain in G-FLAEP was located at the extracellular side of the plasma membrane, we performed indirect immunostaining and CLSM. G-FLAEP producer cells were immunostained with a polyclonal goat-derived anti-SU antibody and an indocarbocyanine-labeled secondary antibody. Image 3 in Fig. 2D shows that red indocarbocyanine fluorescence was detected exclusively at the outer cell surface. Merging of images resulted in a yellow-stained plasma membrane, which proves that the eGFP domain of G-FLAEP is extracellularly colocalized with the retroviral SU domain (image 4 in Fig. 2D). These results demonstrate that the eGFP-SU fusion protein G-FLAEP is transported to the cell surface and is properly presented at its outer side.
Immunoscanning electron microscopy was performed to localize G-FLAEP on cells at higher resolution. Cells tagged with the anti-SU antibody were incubated with protein A-coated colloidal gold particles, fixed (3% formaldehyde-2% glutaraldehyde), and incubated in an aqueous 2% osmium tetroxide solution. Dehydrated samples were subjected to critical-point drying with liquid CO2 and coated with a thin carbon layer (1 carbon string) with a Balzers MED 020. Samples were examined with a Zeiss DSM982 Gemini field emission scanning electron microscope at an acceleration voltage of 5 kV. The study confirmed that G-FLAEP protein was evenly distributed on the surface of the plasma membrane. In addition, these proteins were also located on distinct budded retroviral particles (Fig. 2E).
It has been shown that confocal microscopy allows visualization of single retrovirus particles upon indirect immunostaining (7, 8). The eGFP-tagged envelope protein G-FLAEP should further extend this method because it should allow the following of the dynamics of virus-cell interactions under physiological conditions. We could easily track G-FLAEP virus particles by real-time CLSM, thereby discriminating short, nonspecific interactions and extended binding resulting in virus uptake (data not shown). Further investigations with G-FLAEP should allow the elucidation of the exact mechanism of virus uptake. G-FLAEP should allow us to obtain more detailed insights into the retrovirus infection cycle both in the producer cell (synthesis, translocation of Env and budding) and in the attachment and entry events in cells being infected. Recently, the fluorescent tagging of a retroviral receptor was reported (3). Combining the G-FLAEP retroviral envelope protein with alternatively fluorescence-tagged retroviral receptors should enable investigation of the fate of envelope-receptor complexes during infection of living cells by confocal microscopy.
G-FLAEP viruses for direct cell surface staining of virus receptors.
A common technique to determine the presence and expression level of retroviral cell surface receptors relies on indirect detection of virus particles bound to the plasma membrane of target cells (4). Usually, target cells are incubated with the culture supernatant of producer cells. Virus particles and shed SU bind to the virus receptor and are then detected with antibodies directed against the retroviral SU. We investigated if this method could be improved by making use of eGFP-tagged viruses. For this purpose, we compared the fluorescence pattern of NIH 3T3 cells either treated for 1 h with the supernatant of G-FLAEP-expressing producer cells (4 μg of Polybrene per ml) or treated with amphotropic virus supernatant under identical conditions followed by a classical immunostaining procedure.
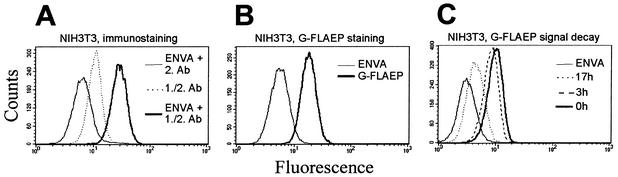
Figure 3A shows that indirect immunostaining was accompanied by a significant nonspecific fluorescence signal. This is attributed to nonspecific binding of antibodies in the absence of viral particles or SU proteins (Fig. 3A; compare the thin and dotted lines with mean fluorescence peaks at 6.76 and 10.85; specific ratio of fluorescence enhancement was 1.6). After treatment of the cells with amphotropic virus, the cells shifted to 28.21 arbitrary units of relative fluorescence, resulting in a 2.6-fold-enhanced signal. Interestingly, upon incubation of cells with G-FLAEP virus, we observed a 3.1-fold-enhanced specific fluorescence (Fig. 3B; compare mean fluorescence peaks at 5.86 and 18.02). Thus, the use of eGFP-tagged virus is at least as sensitive as the standard protocol for detection of retrovirus receptor levels.
FIG. 3.
Characterization of murine cells treated with G-FLAEP virus supernatant. Flow cytometry analysis of NIH 3T3 cells treated with virus supernatant in the presence of Polybrene. (A) Cells incubated with wild-type EnvA virus supernatant (EnvA) were subjected to indirect immunofluorescence staining with a polyclonal anti-SU primary antibody (1.Ab) and fluorescein isothiocyanate-conjugated secondary antibody (2.Ab). (B) Cells incubated with G-FLAEP virus supernatant were directly subjected to flow cytometry. (C) eGFP signal decay was monitored by flow cytometry analysis with NIH 3T3 cells stained with G-FLAEP virus supernatant (0 h represents the initial fluorescence signal).
The sensitivity of the eGFP virus assay can be further improved by immunological detection of G-FLAEP on NIH 3T3 target cells by immunostaining of SU with fluorescein isothiocyanate-labeled secondary antibodies. Since both eGFP and fluorescein isothiocyanate can be excited with 488-nm laser light and since the emitted fluorescent light is detectable with the same filter, the relative fluorescence intensity is further increased (ratio of 3.4; data not shown).
Importantly, the G-FLAEP-associated fluorescence of G-FLAEP virus-infected NIH 3T3 cells declined to background values within 20 h. Thus, this staining does not interfere with the virus titration method, which is based on flow cytometric analysis of eGFP-transduced cells 24 to 72 h postinfection (Fig. 3C).
This method will be of benefit for detection of low levels of receptors, at least on cells that have a certain unspecific antibody binding affinity. The better signal-to-noise ratio is of special advantage on cells that show enhanced background fluorescence due to unspecific antibody binding. In particular, it allows detection of receptor levels on amphotropic packaging cells which are not accessible for the classical receptor staining method because of their high endogenous Env expression, which is the readout parameter in this antibody-mediated assay.
Correlation between staining efficiency and resistance to superinfection.
We applied the direct staining method for the detection of Pit-2 receptor levels to a conditionally Env-expressing producer cell. Amphotropic producer cells generally exhibit low numbers of retroviral receptors on their plasma membrane due to intracellular Pit-2 receptor retention by newly synthesized Env proteins. As a consequence, viral envelope protein-expressing cells lack accessible retroviral receptors on their surface, resulting in resistance to superinfection (3).
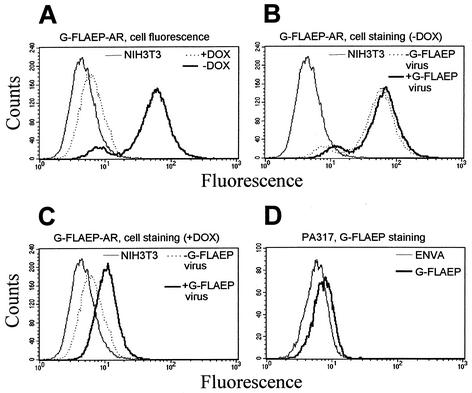
G-FLAEP was conditionally expressed in NIH 3T3 cells with an autoregulated expression system as described earlier (11). A bidirectional promoter (1) ensured the simultaneous expression of tTA (tet-off) and G-FLAEP which is coupled to a resistance marker in a bicistronic configuration (Fig. 1C). A total of 10 μg of the linearized plasmid was electroporated into 3 × 106 Env− cells and selected for resistance against 5 mM histidinol. The fluorescence patterns of a cell clone cultivated under inducing and repressing conditions is shown in Fig. 4A. The induced cells were characterized by a significant fluorescence. In the presence of doxycycline, G-FLAEP expression was downregulated close to the autofluorescence level of untransfected NIH 3T3 cells. To check for virus production in the repressed and induced state, we transfected these cells with and without doxycycline, with an eGFP and neomycin resistance transducing retroviral vector, pM5EGFP (A. Otto and D. Wirth, unpublished data). Only induced transfectants produced GFP-transducing virus particles, as determined on NIH 3T3 cells (data not shown).
FIG. 4.
Characterization of virus-producing cells stably expressing the autoregulated eGFP-EnvA fusion protein (G-FLAEP-AR). (A) Flow cytometry analysis of cells cultivated under repressed (with doxycycline [+DOX]) and induced (without doxycycline[−DOX]) conditions. The cells cultivated under the induced (B) and repressed (C) conditions were incubated with G-FLAEP virus and subjected to flow cytometry analysis. The overlay with the respective nontreated cells and noninfected NIH 3T3 control cells is shown. (D) The amphotropic packaging cell line PA317 was stained with G-FLAEP. Flow cytometry analysis of untreated cells is presented as a control. Results of representative experiments are shown.
An inverse correlation of G-FLAEP expression and the ability of these cells to become infected would be expected. This property was investigated by cultivating the cells under induced and repressed conditions. These cells were incubated with G-FLAEP virus particles from a different source. In parallel, the cells were infected with amphotropic retrovirus transducing a retroviral eGFP-carrying vector. Cells with low G-FLAEP expression (with doxycycline) showed strong staining with the labeled virus as well as high transduceability (Table 1). In contrast, cells cultured under inducing conditions (in the absence of doxycycline) were only weakly stained with G-FLAEP virus supernatant, accompanied by moderate infectivity.
TABLE 1.
Correlation between staining and transduction efficiency depending on amphotropic envelope protein expression level
| Cells | Envelope expressiona | Staining with G-FLAEP virusb | Transduction with EnvA virus (%)c |
|---|---|---|---|
| NIH 3T3 | No | 3.08 | 100 |
| PA317 | High | 1.25 | 2.6 |
| G-FLAEP-AR | |||
| Repressed | Low | 1.61 | 42.9 |
| Induced | Moderate/high | 1.14 | 9.6 |
NIH 3T3 cells do not exhibit Env expression, whereas PA317 have high levels of Env (per definition). Compare the data from Fig. 4 under repressed and induced conditions.
Ratios calculated from mean fluorescence peak shifts after treatment with G-FLAEP virus supernatant analysed by flow cytometry. Results from representative experiments are shown.
Transduction efficiency was determined by flow cytometry. The ratios of eGFP-positive cells were calculated by setting that for NIH 3T3 cells to 100%.
To verify the results obtained from the artificial autoregulated expression system, we used the amphotropic packaging cell line PA317, which is known to be largely resistant to superinfection with amphotropic retrovirus due to expression of the amphotropic envelope gene. We determined the Pit-2 receptor expression level on PA317 cells via staining with G-FLAEP virus supernatant and correlated these data with their transduction capacity with an eGFP-encoding retroviral vector as described above. Parental NIH 3T3 cells served as a control. As expected, PA317 cells were only weakly stained with G-FLAEP virus supernatant (Fig. 4D), and their transduction frequency was below 3% (Table 1). The G-FLAEP-mediated staining and transduction experiments further demonstrated that the expression level of retrovirus receptors is a critical parameter for the infectability of a given virus-cell combination.
In summary, the data presented here show that the N-terminal extension of the retroviral Env protein provides virus particles that are detectable by CLSM and can be used to detect virus receptor levels on target cells. N-terminal extension of retroviral SU proteins has also been followed in order to alter or specify the host range of retroviruses. Consequently, these studies were designed to impair the intrinsic receptor binding activity of the SU moiety (6). Only a limited number of examples show the successful generation of modified SU proteins which bear a new (enzymatic) activity and simultaneously retain the function of SU (10). Interestingly, eGFP fusions to the retroviral 10A1 envelope protein resulted in virus with only 20% of the infectivity of wild-type 10A1 virus (data not shown), although Env4070A and Env10A1 are highly homologous. Recently, it was reported that fusing GFP to the ecotropic env gene resulted in noninfectious virus but that infectivity was achieved upon implementation of two point mutations within the env gene (5). Together, these observations illustrate that fusing eGFP to the SU protein is rather sensitive to the amino acid sequence of SU. Thus, for other SU proteins, tagging might have to be adapted.
REFERENCES
- 1.Baron, U., S. Freundlieb, M. Gossen, and H. Bujard. 1995. Co-regulation of two gene activities by tetracycline via a bidirectional promoter. Nucleic Acids Res. 23:3605-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dirks, W., M. Wirth, and H. Hauser. 1993. Dicistronic transcription units for gene expression in mammalian cells. Gene 128:247-249. [DOI] [PubMed] [Google Scholar]
- 3.Jobbagy, Z., S. Garfield, L. Baptiste, M. V. Eiden, and W. B. Anderson. 2000. Subcellular redistribution of Pit-2 P-i transporter/amphotropic leukemia virus (A-MuLV) receptor in A-MuLV-infected NIH 3T3 fibroblasts: involvement in superinfection interference. J. Virol. 74:2847-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kadan, M. J., S. Sturm, W. F. Anderson, and M. A. Eglitis. 1992. Detection of receptor-specific murine leukemia virus binding to cells by immunofluorescence analysis. J. Virol. 66:2281-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kizhatil, K., A. Gromley, and L. M. Albritton. 2001. Two point mutations produce infectious retrovirus bearing a green fluorescent protein-SU fusion protein. J. Virol. 23:11881-11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavillette, D., S. J. Russell, and F. L. Cosset. 2001. Retargeting gene delivery with surface-engineered retroviral vector particles. Curr. Opin. Biotechnol. 12:461-466. [DOI] [PubMed] [Google Scholar]
- 7.Pizzato, M., E. D. Blair, M. Fling, J. Kopf, A. Tomassetti, R. A. Weiss, and Y. Takeuchi. 2001. Evidence for nonspecific adsorption of targeted retrovirus vector particles to cells. Gene Ther. 8:1088-1096. [DOI] [PubMed] [Google Scholar]
- 8.Pizzato, M., S. A. Marlow, E. D. Blair, and Y. Takeuchi. 1999. Initial binding of murine leukemia virus particles to cells does not require specific Env-receptor interaction. J. Virol. 73:8599-8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pudney, J., and M. J. Song. 1994. Electron microscopic analysis of HIV-host cell interactions. Tissue Cell 26:539-550. [DOI] [PubMed] [Google Scholar]
- 10.Spitzer, D., H. Hauser, and D. Wirth. 1999. Complement-protected amphotropic retroviruses from murine packaging cells. Hum. Gene Ther. 10:1893-1902. [DOI] [PubMed] [Google Scholar]
- 11.Unsinger, J., A. Kroger, H. Hauser, and D. Wirth. 2001. Retroviral vectors for the transduction of autoregulated, bidirectional expression cassettes. Mol. Ther. 4:484-489. [DOI] [PubMed] [Google Scholar]