Abstract
Sensory adaptation and chemotaxis by Escherichia coli require a specific pentapeptide at the chemoreceptor carboxyl terminus. This sequence binds the two enzymes of receptor adaptational modification, enhancing catalysis, but with different binding features and mechanisms. We investigated the relative importance of each pentapeptide side chain for the two enhancing interactions.
Adaptation in bacterial chemotaxis is mediated by covalent modification, CheR-catalyzed methylation of specific glutamates in the chemoreceptor cytoplasmic domain and CheB-catalyzed demethylation of those methyl glutamates (7, 11, 14). In Escherichia coli and related organisms efficient methylation and demethylation each require a pentapeptide interaction sequence, asparagine-tryptophan-glutamate-threonine-phenylalanine (NWETF) at the chemoreceptor carboxyl terminus (3, 17). The X-ray structure of CheR bound to an isolated pentapeptide identified side chains and other features that make specific contacts with the methyltransferase (8). Alterations of some of these side chains by site-directed mutagenesis reduced in vivo levels of steady-state methylation and efficiency of chemotaxis (15). Interaction of the pentapeptide and methylesterase CheB is distinctly different from the pentapeptide-CheR interaction in affinity, location of the binding site, and mechanism of enzymatic enhancement (1, 3, 4). For instance, the Kd of the pentapeptide-CheR complex is ∼2 μM but the interaction of CheR with substrate sites on the receptor is at least 50- to 100-fold weaker (3, 17). In contrast the Kd of the pentapeptide-CheB complex is ∼150 μM but effective demethylation at receptor concentrations of a few micromolar implies that interaction of the enzyme with substrate sites is significantly stronger (1, 3, 4). It had not been known which features of the pentapeptide were crucial for interaction with CheB. The three-dimensional structure of a CheB-pentapeptide complex has not been determined, and determination may not be feasible because the low affinity of CheB for the pentapeptide makes crystallization of the complex challenging.
Here, we compare effects of pentapeptide alterations on enhancement of CheR and CheB activities and use affinity chromatography combined with biochemical site-directed mutagenesis to assess relative effects of the alterations on enzyme binding.
Substitutions in the modification-enhancing pentapeptide.
We constructed plasmids carrying tar controlled by a modified lac promoter and coding for chemoreceptor Tar with an alanine substitution at each of the respective pentapeptide positions. We did this by introducing 1.4-kb XbaI and AvaI fragments from tar with the desired mutations (15) in place of the corresponding fragment of pNT201 (6). PCR-based mutagenesis of pNT201 created tar coding for Tar-E551R. These plasmids were introduced into CP362, a strain deleted of chromosomal copies of the methyl-accepting chemotaxis proteins but otherwise wild type for chemotaxis (13). Tar-mediated chemotaxis was assayed using formation of chemotactic rings on semisolid agar plates (12) containing a Tar-linked attractant, aspartate or maltose, or the complex amino acid mixture tryptone, which contains aspartate. On tryptone plates, we observed a substantial defect for Tar with alanine in place of tryptophan in the NWETF pentapeptide, which we designate NAETF, a modest defect for Tar-NWETA, and no significant defect for the other substituted receptors, confirming previous data (15). On maltose and aspartate plates, the Tar-NAETF defect remained substantial but the Tar-NWETA defect was diminished (data not shown).
Effects on CheR- and CheB-mediated receptor modifications.
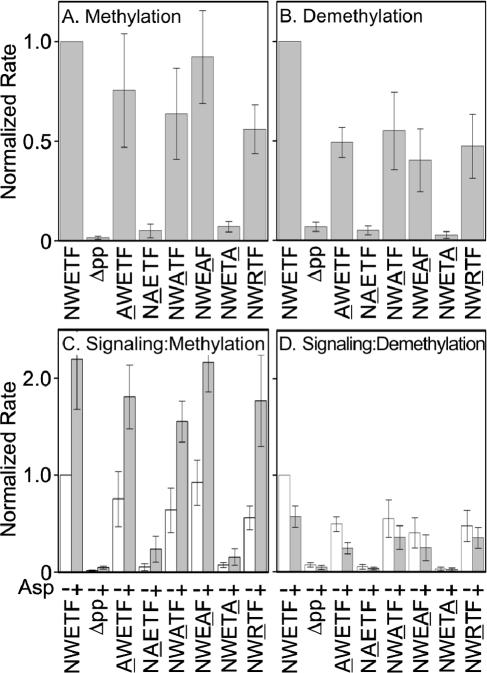
We determined initial rates in vitro of CheR-catalyzed methylation (Fig. 1A) and phospho-CheB-catalyzed demethylation (Fig. 1B). For reference, Fig. 1 shows rates of modification for Tar with the wild-type NWETF pentapeptide or deleted of this sequence (Δpp). Each substituted receptor was modified by the two enzymes, albeit in some cases at a rate no higher than for Tar lacking the pentapeptide entirely (Δpp). Substitutions replacing the aromatic side chains of tryptophan (NAETF) or phenylalanine (NWETA) drastically reduced rates of both CheR- and CheB-catalyzed reactions (Fig. 1A and B). Replacing glutamate with alanine (NWATF) or oppositely charged arginine (NWRTF) resulted in modest reductions for both reactions. Replacing the asparagine (AWETF) or the threonine (NWEAF) had little effect on methylation but some effect on demethylation. In several cases, the CheB-catalyzed reaction was more sensitive to changes in the pentapeptide than CheR-catalyzed methylation (Fig. 1A and B). We also tested the alanine-substituted forms of Tar for deamidation by phospho-CheB and observed effects similar to those for demethylation (data not shown).
FIG. 1.
In vitro methylation and demethylation. Bars are mean values of initial rates (three or more independent experiments), normalized to wild-type Tar, for methylation of membrane-embedded receptors catalyzed by CheR (A and C) and demethylation catalyzed by phospho-CheB (B and D) in conditions as described previously (12) except CheR was 0.1 μM. Phospho-CheB was generated by the presence of an excess of phosphoramidate (12). (A and B) Rates for membrane-embedded receptors in the absence of attractant. (C and D) Altered rates upon occupancy with a saturating concentration (1 mM) of the Tar-linked attractant aspartate. Initial rates were determined by linear fits of time courses (12). Error bars are standard deviations.
We tested transmembrane signaling by determining the effects of a saturating concentration of aspartate on initial rates of methylation and demethylation (Fig. 1C and D). For wild-type Tar, aspartate binding to the periplasmic domain alters the conformation of the cytoplasmic domain, resulting in an increased rate of methylation and a decreased rate of demethylation (see first pairs of bars in Fig. 1C and D). Similar changes were observed for Tar with each pentapeptide substitution. Thus alternations in the interaction site for the enzymes of adaptation did not have substantial effects on transmembrane signaling, consistent with the independence of the receptor functions of transmembrane signaling and adaptation (11).
Relative effects on enzyme binding investigated with affinity columns.
To what degree did effects of pentapeptide substitutions on receptor modification reflect reduced binding of the respective enzymes? We could not investigate potential reductions in binding using quantitative approaches like equilibrium dialysis or calorimetry because the low-affinity interaction of CheB and the wild-type pentapeptide was already at the limits of those techniques (1). Instead we utilized a qualitative approach that provided sensitivity to weaker interactions. This was affinity chromatography with isolated peptides coupled to a resin (3, 5). Peptides were synthesized and coupled by their sole amino group, at the amino terminus, to the resin in a 1-ml Hi-Trap N-hydroxysuccinimide (NHS)-activated column (Amersham Pharmacia). Since coupling to this standardized commercial resin was performed with excess peptide, each with the same reactive group, we assumed approximately equal efficiency of coupling. One set of peptides contained individual alanine substitutions. The other set had changes that affected charges, substitution of alanine or arginine for glutamate, displacement of the terminal carboxylate by extension with an alanine, or neutralization of the terminal carboxylate to an amide in its normal or displaced position. Extracts (2, 3) of RP3098 harboring pME43 (16) or pCW/cheB (9, 10) and thus containing enhanced amounts of CheR or CheB but no other chemotaxis proteins were dialyzed extensively against 50 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, and 2 mM dithiothreitol plus 10% glycerol (TEDG) and applied at ∼2 mg protein/ml to a 1-ml affinity column equilibrated with the same buffer. Columns were washed with three bed volumes of TEDG and then with two volumes containing 5 mg/ml of pentapeptide NWETF. Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Representative results are shown in Fig. 2. In the conditions used, resin-coupled NWETF bound all CheR or CheB in the sample and released it when treated with free pentapeptide (3, 5). The amount of enzyme bound and specifically eluted provided a qualitative indication of the relative binding of enzyme to the altered pentapeptides.
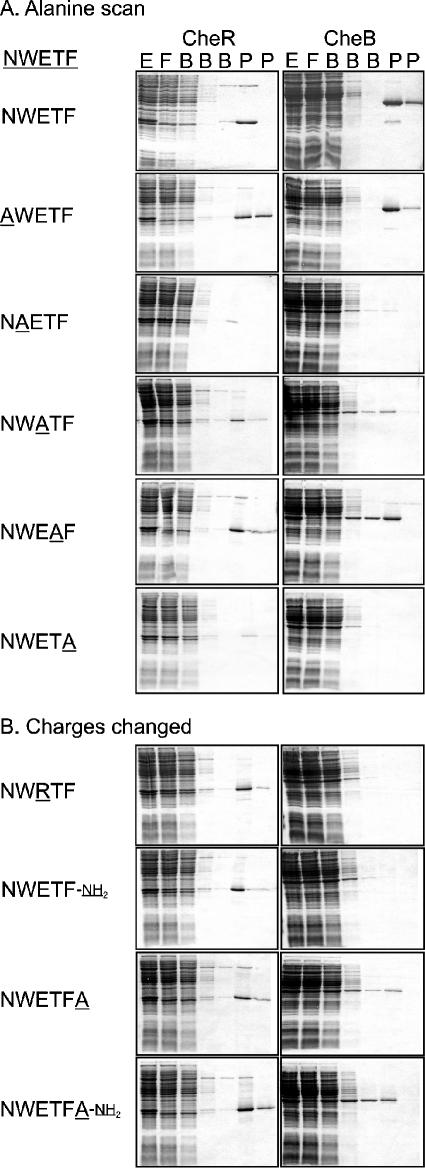
FIG. 2.
Retention and elution of CheR and CheB in affinity columns carrying altered chemoreceptor pentapeptides. Cell extracts (E) containing methyltransferase (CheR) or methylesterase (CheB) were applied to affinity columns carrying the indicated peptide. Fractions were collected during application of the sample (F) in a column volume, with washing by three column volumes of buffer (B) and two column volumes of buffer containing the NWETF pentapeptide (P). Protein content of the same volume of each fraction was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and staining with Coomassie brilliant blue. Representative results for the two families of altered peptides are shown in panels A and B, respectively.
For both CheR and CheB, substitution of alanine for the aromatic residues tryptophan and phenylalanine greatly reduced retention, and substitution of alanine for glutamate resulted in partial reductions (Fig. 2). In contrast, replacement of the asparagine by alanine did not significantly reduce retention of either protein. Replacement of threonine by alanine did not affect retention of CheR but reduced retention of CheB. The patterns of effects of alanine substitutions at the five positions were similar for CheR and CheB, but for every position, the effect was greater for CheB than CheR (Fig. 2).
Differences between effects on CheB and CheR binding were greater for alterations affecting the charged groups. Neutralization of the terminal carboxylate by an amide linkage or reversal of negative charge on the glutamate by a positive charge of an arginine eliminated detectable retention of CheB but caused only modest reduction in retention of CheR (Fig. 2). Extension of the carboxyl terminus by a single alanyl residue reduced retention of CheB substantially but retention of CheR only modestly. If the alanine extension ended in an amide instead of a carboxylate, there was a substantial reduction in retention of CheB but not CheR.
Pentapeptide interaction with CheR.
The X-ray structure of pentapeptide bound to CheR showed that all five peptide side chains and the terminal carboxylate interacted with the protein (8). However, structural information does not directly identify contributions of individual interactions to binding or activity. Our assays indicate that the only crucial residues for binding CheR and for enhancement of CheR-catalyzed methylation were tryptophan and phenylalanine and that glutamate made a detectable contribution. Previous work had characterized the effects of alanine substitutions in the pentapeptide on net methylation levels in cells containing both CheR and CheB and identified the importance of the aromatic residues as well as the lack of importance of the other residues for normal levels of methylation in these conditions (15). However, those data could not reveal whether the lowered or normal levels of methylation in the presence of both enzymes were the result of altered activity of only CheR, only CheB, or both. Our in vitro assays in the presence of only a single enzyme (Fig. 1) clearly establish the pattern of the effects of pentapeptide substitutions on the activity of the respective enzymes.
Synthesized peptides allowed us to test the effect of neutralizing the carboxyl-terminal charge. In the CheR-pentapeptide cocrystal, the terminal carboxylate forms a hydrogen bond with a CheR side chain and thus was a candidate contributor to the binding energy of the interaction (8). In addition, the structure of the cocrystal suggested that, if the peptide were longer, overall interaction would be perturbed (8). We found that neutralizing the carboxyl-terminal charge or extending the pentapeptide by a single alanine hardly perturbed the CheR interaction. Tolerance for an alanine extension is consistent with the modest effect of other chemoreceptor extensions beyond the pentapeptide sequence on CheR activity (12). Furthermore, combining extension of the peptide by an alanine and charge neutralization by an amide resulted in no detectable perturbation of CheR binding, perhaps because a crucial perturbing feature of the mispositioned negative charge was eliminated by neutralization.
Pentapeptide interaction with CheB.
Interaction of the pentapeptide with CheB is less well characterized than interaction with CheR but is quite interesting because it enhances enzyme activity allosterically (1) and occurs in the linker region between the two enzyme domains (4), both features that distinguish it from CheR-pentapeptide binding. Thus it is notable that the relative importance of the five pentapeptide side chains for binding CheB was very similar to the relative importance for binding CheR. The pentapeptide binds to a subdomain of CheR by forming a fourth strand of a β sheet with the tryptophan and phenylalanine side chains making substantial interactions with CheR (8), providing important contributions to activity and binding (Fig. 1 and 2). The central importance of the two aromatic side chains in pentapeptide binding to CheB could indicate a related mode of binding. However, the region of CheB that interacts with the pentapeptide is not a β sheet but an extended linker that connects the regulatory and catalytic domains. Perhaps binding to the linker involves β sheet hydrogen bonding and intercalation of the aromatic groups into hydrophobic regions of the protein surface.
For all substitutions that affected the binding of either enzyme, the effect on CheB was greater than on CheR (Fig. 2). A simple explanation would be the lower affinity of the NWETF-CheB interaction (Kd ∼ 150 μM [1]), versus the NWETF-CheR interaction (Kd ∼ 2 μM [17]). For instance, if the functional pentapeptide concentration in the affinity column was near the weaker dissociation constant, then a substitution that caused a 10-fold reduction in affinity for both proteins would greatly reduce retention of CheB and hardly affect CheR retention. In any case, the consistently greater effect of pentapeptide alterations on CheB in comparison to CheR expands the list of features that distinguishes the interaction of the reaction-enhancing pentapeptide with the two enzymes of adaptational modification.
A biochemical approach for identifying features important for protein-protein interactions.
The combination of peptide synthesis and peptide-bearing affinity columns was effective in investigating specific contributions to protein-protein interaction, even relatively weak ones, in which one of the partners interacted via a short, contiguous sequence of residues. The approach provided a biochemical version of site-specific mutagenesis. In addition, it allowed investigation of features, such as a terminal charge, which cannot be altered by DNA manipulation. Many protein-protein interactions, particularly those involved in signaling, involve recognition of short, contiguous sequences. Thus, the approach we have utilized could be useful for the study of other systems.
Acknowledgments
We thank Ikuro Kawagishi (Nagoya University) for the plasmids encoding forms of Tar with substitutions in the pentapeptide, Angela Lilly for construction of the tar-bearing plasmids used in our studies, and Gerhard Munske (Washington State University) for synthesis of peptides.
This work was supported in part by grant GM29963 to G.L.H. from the National Institute of General Medical Sciences.
REFERENCES
- 1.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 2002. Allosteric enhancement of adaptational demethylation by a carboxyl-terminal sequence on chemoreceptors. J. Biol. Chem. 277:42151-42156. [DOI] [PubMed] [Google Scholar]
- 2.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 1998. Comparison in vitro of a high- and a low-abundance chemoreceptor of Escherichia coli: similar kinase activation but different methyl-accepting activities. J. Bacteriol. 180:6713-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 1999. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc. Natl. Acad. Sci. USA 96:10667-10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 2001. Location of the receptor-interaction site on CheB, the methylesterase response regulator of bacterial chemotaxis. J. Biol. Chem. 276:32984-32989. [DOI] [PubMed] [Google Scholar]
- 5.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 2000. Using HiTrap NHS-activated columns for identification, purification and analysis of an enzyme that recognizes a linear docking site on its protein substrate. Life Sci. News 2000:10-11. [Google Scholar]
- 6.Borkovich, K. A., N. Kaplan, J. F. Hess, and M. I. Simon. 1989. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. USA 86:1208-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourret, R. B., and A. M. Stock. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625-9628. [DOI] [PubMed] [Google Scholar]
- 8.Djordjevic, S., and A. M. Stock. 1998. Chemotaxis receptor recognition by protein methyltransferase CheR. Nat. Struct. Biol. 5:446-450. [DOI] [PubMed] [Google Scholar]
- 9.Gegner, J. A., and F. W. Dahlquist. 1991. Signal transduction in bacteria: CheW forms a reversible complex with the protein kinase CheA. Proc. Natl. Acad. Sci. USA 88:750-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gegner, J. A., D. R. Graham, A. F. Roth, and F. W. Dahlquist. 1992. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell 70:975-982. [DOI] [PubMed] [Google Scholar]
- 11.Hazelbauer, G. L. 2004. Bacterial chemotaxis: a model for sensory receptor systems. In G. Adelman and B. H. Smith (ed.), Encyclopedia of neuroscience, 3rd ed. Elsevier, Amsterdam, The Netherlands.
- 12.Lai, W.-C., and G. L. Hazelbauer. 2005. Carboxyl-terminal extensions beyond the conserved pentapeptide reduce rates of chemoreceptor adaptational modification. J. Bacteriol. 187:5115-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park, C., D. P. Dutton, and G. L. Hazelbauer. 1990. Effects of glutamines and glutamates at sites of covalent modification of a methyl-accepting transducer. J. Bacteriol. 172:7179-7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkinson, J. S., P. Ames, and C. A. Studdert. 2005. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 8:116-121. [DOI] [PubMed] [Google Scholar]
- 15.Shiomi, D., H. Okumura, M. Homma, and I. Kawagishi. 2000. The aspartate chemoreceptor Tar is effectively methylated by binding to the methyltransferase mainly through hydrophobic interaction. Mol. Microbiol. 36:132-140. [DOI] [PubMed] [Google Scholar]
- 16.Simms, S. A., E. W. Cornman, J. Mottonen, and J. Stock. 1987. Active site of the enzyme which demethylates receptors during bacterial chemotaxis. J. Biol. Chem. 262:29-31. [PubMed] [Google Scholar]
- 17.Wu, J., J. Li, G. Li, D. G. Long, and R. M. Weis. 1996. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry 35:4984-4993. [DOI] [PubMed] [Google Scholar]