Abstract
UvrD, a highly conserved helicase involved in mismatch repair, nucleotide excision repair (NER), and recombinational repair, plays a critical role in maintaining genomic stability and facilitating DNA lesion repair in many prokaryotic species. In this report, we focus on the UvrD homolog in Helicobacter pylori, a genetically diverse organism that lacks many known DNA repair proteins, including those involved in mismatch repair and recombinational repair, and that is noted for high levels of inter- and intragenomic recombination and mutation. H. pylori contains numerous DNA repeats in its compact genome and inhabits an environment rich in DNA-damaging agents that can lead to increased rearrangements between such repeats. We find that H. pylori UvrD functions to repair DNA damage and limit homologous recombination and DNA damage-induced genomic rearrangements between DNA repeats. Our results suggest that UvrD and other NER pathway proteins play a prominent role in maintaining genome integrity, especially after DNA damage; thus, NER may be especially critical in organisms such as H. pylori that face high-level genotoxic stress in vivo.
Genomic alterations due to recombination, deletions, and duplications are a major source of genetic plasticity in all prokaryotes (14, 15). To minimize such damage, bacteria have evolved mechanisms involving mismatch repair (MMR) proteins such as MutSLH, nucleotide excision repair (NER) proteins such as UvrABCD, and recombinational repair proteins such as RecBCD and RecFOR (32, 45, 55, 66). Helicobacter pylori, a gram-negative gastric bacterium found in humans, is unusual because it lacks many of the genes of major recognized DNA repair and recombination pathways, including mutSLH, recCD, and recFO, common to other bacteria (5, 60). H. pylori has a high rate of recombination and mutation, with H. pylori variants arising over relatively short time periods (months) in individual hosts (1, 17, 56). As essentially the only persistent bacterium in its niche, the high level of genetic diversity in H. pylori may facilitate adaptation to its environment through the continual generation of variants and selection for highest fitness (65).
H. pylori possesses a homolog of the DNA repair helicase, UvrD (annotated as HP1478 in sequenced strain 26695 and as JHP1371 in sequenced strain J99) (5, 60). In Escherichia coli, UvrD works in concert with MutSHL in the MMR pathway and with UvrABC in the NER system (Fig. 1). Disruption of MMR leads to increased spontaneous mutations and homologous recombination (22, 51), whereas disruption of NER increases sensitivity to UV and other agents of DNA damage (12, 33). UvrD also has an important role in restarting stalled replication forks and facilitates this process by displacing RecA protein from DNA (20) (Fig. 1).
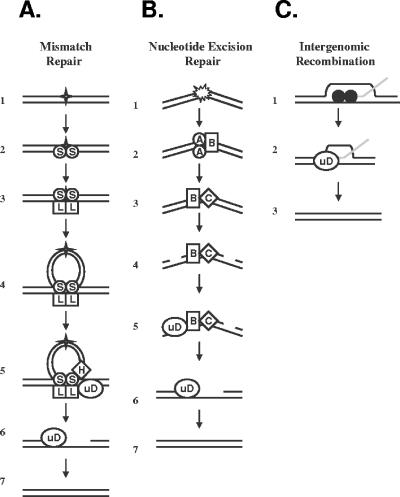
FIG. 1.
Proposed mechanisms for UvrD in mismatch repair, nucleotide excision repair, and intergenomic recombination. (A) In E. coli, the MMR pathway corrects mismatches, small insertions, and deletions (represented by the star) (row 1). The MutS1 dimer recognizes the mismatch (row 2) and recruits MutL (row 3). The MutS1-MutL complex translocates along the DNA, forming a loop (row 4). MutH endonuclease nicks the DNA (row 5). UvrD helicase, which interacts physically with MutL (23), is recruited to the lesion and removes it (row 5), leaving a gap (row 6) which is filled in by DNA polymerase III, restoring the template (row 7). (B) The NER pathway repairs a variety of bulky lesions, including pyrimidine dimers and alkylated base pairs, symbolized in row 1. The lesion is recognized by UvrA, which recruits UvrB helicase (row 2). UvrA subsequently detaches, and UvrC endonuclease is recruited to the lesion (row 3). The UvrBC complex nicks the DNA 5′ and 3′ of the lesion (row 4). UvrD helicase, which interacts physically with UvrB (2), is recruited to the site (row 5) and removes the lesion, leaving a gap (row 6) that subsequently is filled by DNA polymerase I and restored (row 7). (C) In intergenomic recombination, strand invasion is mediated by RecA (filled circles) (row 1). The UvrD helicase dismantles the RecA-DNA complex (row 2) (20, 44, 63), leading to restoration of the template (row 3).
We asked what role a uvrD homolog would play in an organism such as H. pylori, which is highly diverse and lacks known repair proteins, including MutSHL. Would H. pylori UvrD limit illegitimate recombination and mutation in the absence of MMR?
As in other bacteria with small genomes, the H. pylori chromosome possesses a large number of direct, nonrandomly distributed repeats throughout its genome that can promote genomic rearrangements (7). Previous studies have shown that a helicase, RecG, functions to limit deletions between such repeats (31). We asked whether the UvrD helicase would likewise influence deletions. H. pylori resides in the human gastric mucosa, where it is exposed to various sources of DNA damage, including oxidative radicals and nitrosative species (9, 13, 46). Therefore, we also explored the effect of uvrD mutation on inter- and intragenomic recombination after exposure to DNA damage.
Our studies demonstrate that H. pylori UvrD, like its E. coli homolog, plays an important role in repairing damaged DNA. We find that UvrD has no effect on spontaneous mutation but limits homologous intergenomic recombination in H. pylori despite the absence of MutSHL, indicating UvrD functional independence from MutSHL in this regard. In contrast to the RecG helicase, UvrD does not influence deletions between H. pylori DNA repeats under usual in vitro conditions, but with DNA damage, deletion frequency is significantly increased in the UvrD mutant. These findings indicate important roles for H. pylori UvrD in maintaining genomic integrity in the stressful environments in which it normally resides in these organisms.
MATERIALS AND METHODS
Statistical analyses.
Student's t test was used where appropriate, with a P value < 0.05 deemed significant.
Phylogeny and amino acid alignment of UvrD homologs.
GenBank (www.ncbi.nih.gov/GenBank/) was searched for homologs to H. pylori HP1478 by use of the BLAST-p algorithm (41). Alignments of amino acid sequences were constructed using the program ClustalX and the Gonnet 250 protein weight matrix (57) and visualized using the program Genedoc (www.psc.edu/biomed/genedoc) in three-shading mode. A phylogenetic tree was constructed using the program Mega 3 (34) and the neighbor-joining method (52) with 1,000 bootstrap replicates.
Bacterial strains and plasmids.
The E. coli and H. pylori strains and plasmids used in this study are listed in Table 1. Wild-type E. coli strain JJC40 (10) and its isogenic uvrD mutant, JJC212, kindly provided by Benedicte Michel, were routinely grown in Luria-Bertani broth (LB) at 37°C. H. pylori strains were routinely grown at 37°C in 5% CO2 on Trypticase soy agar (TSA) plates or brucella agar (BA) with 10% newborn calf serum (NCS) plates supplemented with the appropriate antibiotic.
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Plasmids | ||
| pVacAKm | vacA::aphA in pGEMT-Easy | 31 |
| pRecAKm | recA::aphA in pGEMT-Easy | 31 |
| pUvrD | HP1478 in pGEMT-Easy | This work |
| pUvrDKm | HP1478::aphA in pGEMT-Easy | This work |
| pVacA0 | vacA with 0 bp deletion cassette | 7 |
| pVacA50 | vacA with 50 bp deletion cassette | 7 |
| pVacA100 | vacA with 100 bp deletion cassette | 7 |
| pUvrD-0 | HP1478 with 0 bp deletion cassette | This work |
| pUvrD-0 | HP1478 with 50 bp deletion cassette | This work |
| pUvrD-100 | HP1478 with 100 bp deletion cassette | This work |
| pAD1 | ureAB fragment in pUC18 | 6 |
| pHPuD | pAD1 with HP1478 and CAT cassette | This work |
| pECuD | pAD1 with E. coli uvrD and CAT cassette | This work |
| Strains | ||
| H. pylori | ||
| JP26 | Wild-type strain | 31 |
| JP26/vacA::aphA | vacA with aphA insertion | 31 |
| JP26/recA::aphA | recA with aphA insertion | 31 |
| JP26/uvrD::aphA | HP1478 with aphA insertion | This work |
| JP26 HPuDcomp | uvrD::aphA complemented with HP1478 in ureAB locus | This work |
| JP26 ECuDcomp | uvrD::aphA complemented with E. coli uvrD in ureAB locus | This work |
| JP26/vacA::0 | vacA interrupted with 0 bp deletion cassette | 7 |
| JP26/vacA::50 | vacA interrupted with 50 bp deletion cassette | 7 |
| JP26/vacA::100 | vacA interrupted with 100 bp deletion cassette | 7 |
| JP26/recA::100 | recA interrupted with 100 bp deletion cassette | 7 |
| JP26/uvrD::0 | HP1478 interrupted with 0 bp deletion cassette | This work |
| JP26/uvrD::50 | HP1478 interrupted with 50 bp deletion cassette | This work |
| JP26/uvrD::100 | HP1478 interrupted with 100 bp deletion cassette | This work |
| JP26/uvrB::0 | HP1114 interrupted with 0 bp deletion cassette | This work |
| JP26/uvrB::100 | HP1114 interrupted with 100 bp deletion cassette | This work |
| E. coli | ||
| JJC40 | Wild-type AB1157 hsdR | 10 |
| JJC212 | JJC40 uvrD::Tn5 (Kanr) | B. Michel |
Plasmid pECuD was constructed using primers ECUDXbaI and ECUDSmaI and plasmid pHPuD was constructed using primers HPUDXbaI and HPUDSmaI as described previously (30, 31) (see Table S2 in the supplemental material). The features of all recombinant plasmids were confirmed by PCR.
Construction of H. pylori mutants used to assess susceptibility to UV, intergenomic recombination frequencies, and spontaneous mutation frequencies.
Fragments of HP1478 (uvrD homolog), HP0887 (vacA), and HP0153 (recA) open reading frames (ORFs) were amplified by PCR using primers based on sequenced strain 26695 (see Table S2 in the supplemental material) and cloned into pGEMT-Easy (Promega, Wisconsin) to create pUvrD, pVacA, and pRecA, respectively (31). The aphA cassette, conferring kanamycin resistance (Kanr), was used to interrupt each ORF to create pUvrDKm, pVacAKm, and pRecAKm, respectively. The vacA locus was chosen as a control for the presence of the aphA cassette, since it is not involved in recombination and its construction has been documented previously (7, 31). The recA locus was also interrupted, since several phenotypes that are related to this study have been observed in mutants (7, 31, 54, 58). H. pylori strain JP26 was transformed to Kanr with pUvrDKm, pVacAKm, and pRecAKm to create JP26 uvrD::aphA, vacA::aphA, and recA::aphA, respectively, as described previously (64). Chromosomal DNA was isolated from transformants, and the correct insertion of the aphA cassette into the expected ORF was confirmed by PCR in each case.
Construction of H. pylori mutants used to assess deletion frequencies.
A unique BamHI site was created in pUvrD and pVacA by use of inverse PCR and primers UDinvR and UDinvF based on sequenced strain 26695 (see Table S2 in the supplemental material), and each ORF was subsequently interrupted with a deletion cassette (see Fig. 4B) containing either 0-, 50-, or 100-bp repeats to create pUvrD-0, pUvrD-50, and pUvrD-100 and pVacA-0, pVacA-50, and pVacA-100. The construction and use of the deletion cassettes and JP26 uvrB::100 and recA::100 strains has been previously described (7). H. pylori strain JP26 was subsequently transformed to chloramphenicol resistance (Catr) with these plasmids to create JP26 uvrD::0, uvrD::50, and uvrD::100 and JP26 vacA::0, vacA::50, and vacA::100. Chromosomal DNA was isolated from transformants, and the correct insertion of the deletion cassette into the expected ORF was confirmed by PCR in each case.
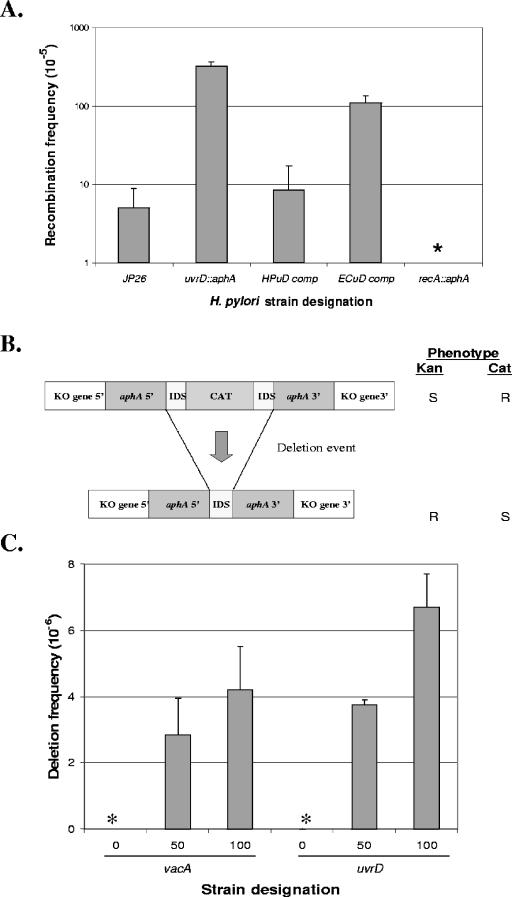
FIG. 4.
Inter- and intragenomic recombination in H. pylori wild-type and mutant strains. (A) Recombination frequencies. H. pylori strains were transformed to streptomycin resistance with an 800 bp A128G rpsL PCR product; bars represent means ± standard deviations of recombination frequency. The strains transformed were the JP26 wild type and its recA and uvrD mutants and the uvrD mutant complemented with either HP1478 or E. coli uvrD downstream of the ureA promoter to create JP26 HPuDcomp or ECuDcomp, respectively. The asterisk indicates that no recombinants were observed for the recA strain, as expected. The uvrD mutant showed a significant (P < 0.05) increase in recombination frequency compared to wild-type results. Both H. pylori (HPuDcomp) uvrD and E. coli (ECuDcomp) uvrD complement the uvrD mutant to a significant (P < 0.05) degree, with H. pylori uvrD complementing to a greater extent than E. coli uvrD. (B) Schematic of constructs used to assess deletion frequency in H. pylori. H. pylori mutants were created by inserting a deletion cassette construct with either 0 bp (control) or 100 bp flanking identical repeat segments (IDS) within vacA (control) or HP1478 (indicated as KO gene 5′ and KO gene 3′) (7). Insertion of the complete cassette into a host H. pylori strain confers resistance to chloramphenicol. The chloramphenicol cassette can subsequently be deleted by recombination between the two flanking IDS DNA repeats, resulting in resistance (R) to kanamycin and susceptibility (S) to chloramphenicol. (C) Deletion frequencies. The deletion cassette, as described above, was inserted into vacA (control) or HP1478, the insertion was confirmed, and deletion frequencies were calculated. Asterisks indicate that no deletions were detected (frequency < 10−8). As expected, H. pylori strains with the cassette in vacA or HP1478 showed progressively higher deletion frequencies with increasing IDS size. Strains with the deletion cassette in HP1478 show a small but not significant (P = 0.06) difference in deletion frequencies between flanking DNA repeats of 50 and 100 bp compared to control (vacA) mutants with comparable cassettes. Bars represent means ± standard deviations for four to six replicate experiments.
Spontaneous mutation rate assay to assess mismatch repair function.
Since a point mutation in rpoB confers resistance to rifampin in H. pylori (25) as well as in E. coli, rifampin resistance can be used as a phenotype to calculate spontaneous mutation rates. Nine colonies of each strain to be tested were picked and expanded to TSA plates. After 48 h growth at 37°C in 5% CO2, the bacteria were suspended in 1 ml Brucella broth and serially diluted onto TSA plates alone or onto rifampin plates. H. pylori strains were plated on BA plates supplemented with rifampin (7.5 ug/ml) and 10% NCS; E. coli strains were plated on LB plates supplemented with rifampin (50 ug/ml). Mutation rates were determined from frequencies based on the Lea-Coulson algorithm (35).
Recovery from DNA damage.
E. coli or H. pylori cells to be tested were grown on TSA plates for 24 h or 48 h, respectively, and suspended in 1 ml phosphate-buffered saline (PBS). Equal amounts of suspension were inoculated on TSA plates at dilutions that would produce 100 to 500 CFU per plate postexposure. Cells then were exposed to 0 to 1,400 kJ/m2 UV at 312 nm (Stratagene Transluminator, La Jolla, CA) and incubated at 37°C, and colonies were counted and survival rates calculated.
Streptomycin resistance frequency assay to assess intergenomic recombination.
H. pylori strains were grown on TSA plates for 48 h and harvested into 1 ml of PBS, and 25 μl was spotted onto a fresh TSA plate combined with 30 ng of donor DNA and incubated for 18 h at 37°C in 5% CO2. Donor DNA was an 800 bp PCR product of H. pylori rpsL from streptomycin-resistant (Str) strain JP26 with A128G (61). The transformation mixture was then harvested into 1 ml PBS, and 100 μl of the appropriate serial dilutions was plated onto either TSA or BA plates containing 10% NCS and 25 μg/μl streptomycin. The plates were incubated for 4 days at 37°C in 5% CO2, and the total recombination frequency was determined by calculating the number of Str colonies divided by the total number of CFU. As a negative control, H. pylori strains with no DNA added were also tested in parallel in each experiment; no colonies were seen in any case. To assess the influence of DNA damage on intergenomic recombination, H. pylori strains to be tested were incubated at 37°C in a 5% CO2 environment for 48 h and subsequently exposed to 0 to 1,400 kJ/m2 UV irradiation at 312 nm, and then assays were performed as described above.
Deletion frequency assays in H. pylori.
To assess recombination frequencies in the H. pylori strains containing the deletion cassette or control cassettes, the cells were grown on TSA plates for 48 h at 37°C (5% CO2), allowing for deletions to occur, and then harvested and washed twice in PBS, and 25, 100, and 200 μl aliquots were spread on BA plates supplemented with NCS and 25 μg/ml kanamycin. As further controls, 200 μl from each suspension was inoculated to BA plates containing NCS, kanamycin (25 μg/ml), and chloramphenicol (20 μg/ml); as expected, in no experiments were strains with double resistance identified, confirming the specificity of the deletion process (7). Total CFU and numbers of Kanr deletion mutants were determined by plating serial dilutions onto TSA plates and BA plates with kanamycin (25 μg/ml), respectively. Plates were incubated at 37°C in a 5% CO2 environment for 96 h, colonies were counted, and deletion frequencies were calculated.
To assess the influence of DNA damage on intragenomic recombination, H. pylori strains vacA100 and uvrD100 were passed to TSA plates and exposed to 700 kJ/m2 UV irradiation at 312 nm, and then assays performed as described above.
Complementation of the JP26 uvrD::aphA mutant.
Plasmid pHPuD, with ORF HP1478 placed downstream of the ureAB promoter, was constructed using primers HPUDXbaI and HPUDSmaI and used to introduce HP1478 in trans into the genome of mutant JP26 uvrD::aphA via natural transformation, as described previously (6, 64), to create JP26 HPuDcomp. Transformants were selected based on Catr, and the correct insertion of uvrD and flanking regions into ureA was confirmed by PCR of the chromosomal DNA. In parallel experiments, pECuD was used to construct JP26 ECuDcomp, exactly as described above, by use of primers ECUDXbaI and ECUDSmaI.
Complementation of E. coli uvrD mutant.
E. coli strain JJC212 (uvrD null mutant) was transformed with pHPuD, pECuD, and pAD1 (no insert) to create strains JJC212-HPuD, JJC212-ECuD, and JJC212-AD1, respectively. For the wild-type and transformed E. coli strains, recovery after exposure to DNA damage was assessed using overnight cultures that were serially diluted onto TSA plates and exposed to 0 to 1,400 kJ/m2 UV irradiation. After incubation of plates overnight at 37°C, total numbers of colonies were counted and survival rates were determined.
RESULTS
Analyses of UvrD family homologs.
The H. pylori UvrD homolog from sequenced strain 26695 (60), annotated as HP1478 for that strain, has 35.4% identity and 57.1% similarity to E. coli UvrD, which has been extensively characterized for its roles in NER, MMR, and recombinational repair (for a review, see references 28 and 53). Alignment of UvrD amino acid sequences from H. pylori with the closely related Campylobacter jejuni, as well as Pseudomonas aeruginosa and E. coli, showed that all five sequences possess the conserved helicase domains, suggesting probable conservation of function (see Fig. S1A in the supplemental material).
To understand ancestral relationships, a phylogeny was constructed using UvrD sequences from 15 representative bacterial species. As expected, the H. pylori UvrD homologs were most closely related with each other and cluster with that from the related C. jejuni (see Fig. S1B in the supplemental material); interestingly, ancestral relationships were closer with the distantly related Thermotoga and Aquifex hyperthermophile species than with Enterobacteriaceae and related organisms.
Role of H. pylori UvrD in MMR.
In organisms with intact MMR, including E. coli, P. aeruginosa, and Deinococcus radiodurans, the absence of UvrD function results in higher spontaneous mutation frequencies (42, 48, 49) resulting from failure to unwind the mismatched strand after it has been recognized (see Fig. 1). H. pylori lacks homologs of MutS1, MutL, and MutH, the mismatch recognition and excision proteins (5, 30, 50, 60), and therefore the uvrD mutant was not expected to display a mutation rate significantly different from that of the wild type unless it participated in an unrecognized mismatch repair pathway. We found that the absence of UvrD function in H. pylori does not lead to a change in mutation rates, as expected (Fig. 2A). These results also exclude the possibility of an alternative mismatch repair pathway involving UvrD.
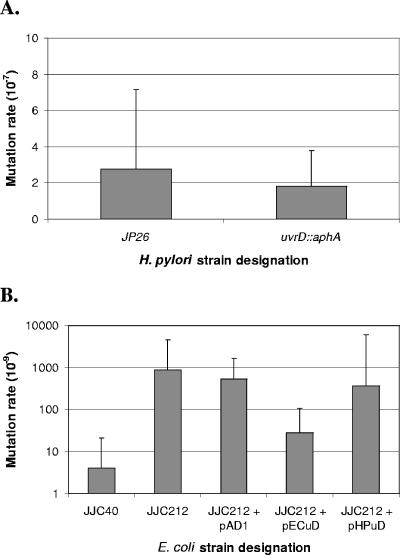
FIG. 2.
Spontaneous mutation rates of H. pylori and E. coli wild-type, mutant, and complemented strains. (A) Spontaneous mutation rates of the H. pylori wild type (JP26) and the uvrD mutant were determined based on resistance to rifampin and calculated using the method of Lea-Coulson (35). Differences were not significant (P > 0.05). (B) Spontaneous mutation rates of E. coli wild-type (JJC40), uvrD mutant (JJC212), and uvrD mutant strains transformed with vector pECuD (E. coli uvrD), pHPuD (H. pylori uvrD), or pAD1 (no insert) by use of the method of Lea-Coulson. The E. coli uvrD mutant was partially complemented (P < 0.05) by E. coli UvrD expression but not by H. pylori UvrD expression (P > 0.05).
E. coli uvrD mutants display elevated mutation rates.
We asked whether H. pylori UvrD could complement this phenotype in E. coli uvrD mutants to determine the extent of functional similarity between H. pylori and E. coli UvrD. To address this question, JJC212, the E. coli uvrD mutant, was transformed with pAD1 (no insert control), pECuD (E. coli uvrD), or pHPuD (H. pylori uvrD), and mutation rates were calculated (Fig. 2B). As expected, transformation of JJC212 with pAD1 resulted in no change, whereas transformation with pECuD resulted in a partial but significant (P < 0.01) decrease in mutation rate. Transformation of JJC212 with pHPuD did not result in a significant degree of complementation (Fig. 2B), demonstrating that H. pylori UvrD was unable to complement the MMR defect in E. coli.
Role of H. pylori UvrD in nucleotide excision repair.
Next, we asked whether H. pylori UvrD could complement the UV sensitivity phenotype of E. coli uvrD mutants. E. coli strains JJC40 and JJC212 were assayed for sensitivity to UV exposure (312 nm), and JJC212 was complemented in trans with pECuD or pHPuD or with pAD1 alone. As expected, JJC212 showed markedly increased susceptibility to UV damage compared to wild-type results (47), and transformation of JJC212 with pAD1 had no effect on survival (Fig. 3A). Transformation of JJC212 with either pECuD or pHPuD resulted in partial but significant complementation (P < 0.05 for both 700 and 1,400 kJ/m2 exposures). That the H. pylori UvrD homolog could complement the UV sensitivity of E. coli uvrD mutants indicated that HP1478 encodes a UvrD homolog and that this protein can interact with other NER proteins in E. coli (Fig. 3A).
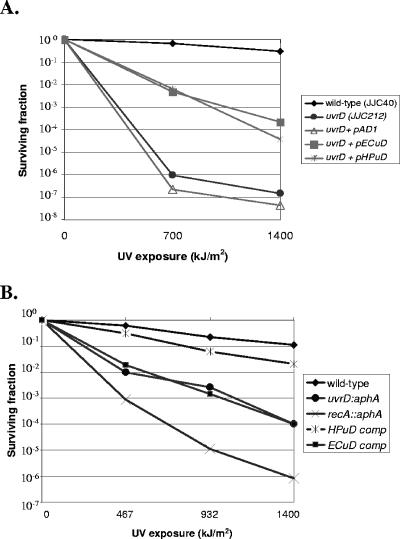
FIG. 3.
Susceptibility to UV of E. coli and H. pylori wild-type, mutant, and complemented strains. (A) E. coli wild-type strain JJC40, uvrD mutant strain JJC212, and JJC212 complemented with vector pAD1 alone (no insert) or with E. coli uvrD in pAD1 (pECuD) or H. pylori HP1478 in pAD1 (pHPuD) were examined for susceptibility to UV (0 to 1,400 kJ/m2). Results shown are representative of five experiments. Both pHPuD and pECuD complemented the E. coli uvrD mutant to a significant (P < 0.05) degree at 700 and 1,400 kJ/m2, whereas pAD1 did not (P > 0.05), as expected. (B) H. pylori wild-type strain JP26, mutant strains JP26 uvrD::aphA and JP26 recA::aphA, and strain JP26 uvrD::aphA complemented in trans with an H. pylori uvrD homolog (HPuDcomp) or an E. coli uvrD homolog (ECuDcomp) were assayed for survival after exposure to UV (0 to 1,400 kJ/m2). Results shown are representative of five experiments. As expected, the recA::aphA and uvrD::aphA strains were UV sensitive. Both H. pylori and E. coli uvrD complemented the uvrD::aphA mutant to a significant (P < 0.05) degree at the UV exposures tested, with the H. pylori uvrD complementing to a greater degree than E. coli uvrD.
Since H. pylori possesses homologs to all four NER proteins (UvrABCD) (5, 60) and since a role of H. pylori UvrB in UV repair has been shown previously (59), to understand UvrD NER function in an H. pylori background we next performed UV susceptibility assays using wild-type H. pylori strain JP26 and JP26 uvrD::aphA, an isogenic mutant (Fig. 3B). JP26 recA::aphA was used as a control, since recA mutants are highly susceptible to all types of DNA damage involving recombinational repair (39). As expected, the recA mutant showed extreme sensitivity to UV exposure (54, 58), and there was marked (>5 log10) reduction in survival of JP26 uvrD::aphA with 1,400 kJ/m2 UV exposure (Fig. 3B). Complementation of JP26 uvrD::aphA by expression of uvrD (HP1478) downstream of a strong (ureA) promoter restored the phenotype to nearly wild-type levels (Fig. 3B), demonstrating that the UV susceptibility phenotype was specific to the uvrD mutation and not due to polar effects on downstream genes. The H. pylori uvrD mutant also was complemented in trans through the expression of E. coli uvrD in the same ureA locus (Fig. 3B). There was partial (>2 log10) yet significant (P < 0.05) complementation by E. coli uvrD. This result obtained using H. pylori cells contrasts with the findings in the E. coli background, where the heterologous homolog was functionally equivalent to the native product (Fig. 3A). Thus, more-stringent H. pylori-specific requirements for UvrD structure and NER function exist in H. pylori than in E. coli.
Intergenomic recombination in mutant and complemented H. pylori strains.
In E. coli, uvrD mutants are hyperrecombinant (8, 18, 38, 67), but numerous factors, including disruption of MMR, may contribute to this phenotype. We studied H. pylori uvrD mutants, since H. pylori lacks MMR. Recombination frequency assays of H. pylori wild-type and uvrD mutants (Fig. 4A) were conducted by transformation of these cells with a linear 800 bp rpsL PCR product that confers streptomycin resistance upon recombination (29). For the control cells (JP26 recA::aphA), which are unable to undergo recombination, no transformants were detected, as expected. JP26 uvrD::aphA showed a significant (66-fold) increase in intergenomic recombination frequencies compared to wild-type results (P < 0.05). As observed for UV susceptibility, in trans complementation with H. pylori uvrD essentially completely restored the wild-type phenotype, whereas complementation with the E. coli uvrD only partially restored the wild-type phenotype (P < 0.05 compared with uvrD::aphA). Thus, H. pylori UvrD limits recombination, possibly in conjunction with other NER proteins such as UvrB, with which it physically interacts in E. coli (2), or independently of the NER pathway, possibly through repair of stalled replication forks (19), preventing genome rearrangements.
To distinguish between these possibilities, we also examined the recombination frequencies of H. pylori JP26 uvrB and uvrC mutants, which are defective in NER. In contrast to the uvrD mutant, the uvrB and uvrC mutants do not show recombination frequencies significantly different from those seen with wild-type H. pylori (see Fig. S2 in the supplemental material), indicating that defective NER will not necessarily lead to hyperrecombination. In total, our results indicate that the increased recombination in the H. pylori uvrD mutant (P < 0.05) results from independent UvrD helicase activity and does not require interaction with MMR and NER proteins. In E. coli, the UvrD helicase dismantles RecA-mediated strand invasion events (63), and it is possible that UvrD has a similar role in H. pylori, thereby limiting genetic diversification.
Effect of uvrD on deletions between flanking DNA repeats.
Since prokaryotic NER mutants have increased numbers of deletions between tandem repeats (11, 21), we next examined the role of H. pylori UvrD in deletions involving such direct DNA repeats. We employed a constructed chloramphenicol resistance cassette with flanking identical repeats of 50 or 100 bp (7, 31) inserted into uvrD (to produce a null mutation) or into vacA as a control (no known effect of its product on recombination) (7). There were small but not significant (P > 0.05) increases in deletion frequency for the uvrD mutants possessing the 50 and 100 bp identical repeats compared to the results seen with the wild-type (vacA) mutants with the parallel constructions (Fig. 4C); thus, UvrD did not significantly influence deletion frequencies under the basal growth conditions.
DNA damage-induced recombination in wild-type strains and uvrD mutants.
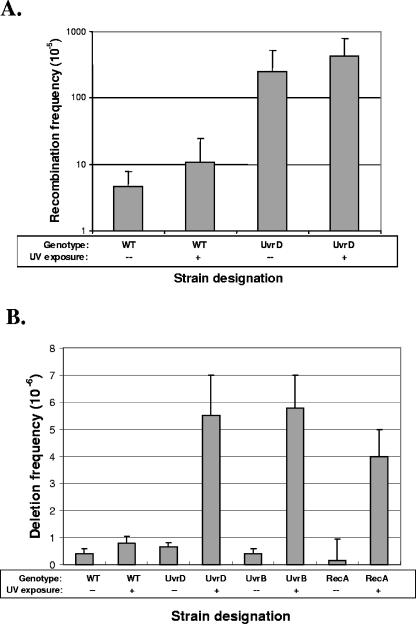
In NER-deficient eukaryotes, UV-induced damage increases chromosomal rearrangements, translocations, and recombination (3, 4, 21, 62). The role of NER in preventing recombination events in prokaryotes has not been characterized as completely, although UvrA and UvrB can suppress certain UV-induced recombination events in E. coli (24). Thus, we explored the role of H. pylori UvrD in DNA damage-induced rearrangements. First, wild-type and uvrD mutant H. pylori strains were exposed to UV (312 nm); by use of the 800 bp transforming DNA fragment (Fig. 4A), intergenomic recombination frequencies were assayed. The marginal increases in recombination frequency in both the wild-type and uvrD strains after UV exposure (Fig. 5A) indicated that UV-induced DNA damage has little effect on the already high rates of intergenomic recombination in H. pylori and that the DNA damage created by UV exposure does not lead to an increased propensity for recombination.
FIG. 5.
Influence of DNA damage on H. pylori genome rearrangements. (A) Influence of UV exposure on intergenomic recombination. Recombination frequencies were measured using the 800 bp rpsL PCR product for H. pylori strains JP26 and JP26 uvrD::aphA, with 0 (control) or 700 kJ/m2 exposure to UV. As expected, with no UV exposure, the intergenomic recombination frequency of JP26 uvrD::aphA was higher (62-fold) and significantly (P < 0.05) different from the frequency seen with wild-type JP26. With 700 kJ/m2 of UV exposure, the intergenomic recombination frequencies for both wild-type JP26 and JP26/uvrD::aphA did not change significantly (P > 0.05) from the results for the unexposed control. (B) Influence of UV exposure on deletion frequency. Deletion frequencies were measured as indicated for H. pylori strains JP26 vacA::100, uvrD::100, uvrB::100, and recA::100 with no (control) or 700 kJ/m2 exposure to UV. With no UV exposure, there was no significant difference between the vacA::100 and uvrD::100 deletion strain results, as expected, or between uvrB::100 or recA::100 results. After 700 kJ/m2 of UV exposure, JP26 vacA::100 displayed a twofold increase in deletion frequency (P < 0.05) compared to the results seen with unexposed JP26 vacA::100. With the UV exposure, the JP26 uvrD::100 strain displayed a 9-fold increase (P < 0.05) in deletion frequency compared to unexposed JP26 uvrD::100, the JP26 uvrB::100 strain displayed a 14-fold increase (P < 0.05) in deletion frequency compared to unexposed JP26 uvrB::100, and the JP26 recA::100 strain showed a 27-fold increase (P < 0.05) in deletion frequency compared to unexposed JP26 recA::100.
We also examined the role of UvrD in DNA damage-induced intragenomic recombination, using the constructed H. pylori strains in which the deletion cassette was inserted into uvrD, uvrB, recA, or vacA (as a control). In the absence of UV, the wild type and the uvrD mutant had similarly low deletion frequencies (Fig. 5B), confirming our prior findings (Fig. 4C). Results were parallel for uvrB and recA, as anticipated by prior results (7). However, after UV exposure, the log10 increase of approximately 1 in deletion frequency in the uvrD mutant was significantly (P < 0.05) greater than in the wild-type background (Fig. 5B). The recA mutant also displayed an increase in deletion frequencies, consistent with the fact that intragenomic recombination between homologous sequences can be RecA independent (37). Parallel studies were undertaken of the uvrB mutant, which acts upstream of uvrD, to determine whether other components of the NER pathway would display a similar phenotype. In both the absence and presence of UV exposure, results for the uvrB and uvrD mutants were nearly identical, providing evidence that DNA damage induces intragenomic recombination in H. pylori, especially in NER-deficient strains.
DISCUSSION
Does HP1478 encode a UvrD protein?
The UV sensitivity of the HP1478 mutant, the partial cross-complementation by E. coli UvrD, and the partial complementation of the E. coli uvrD mutant by HP1478 provide phylogenetic and functional evidence that HP1478 is indeed a UvrD homolog. The partial complementation for both E. coli and H. pylori likely reflects primary and secondary structure differences between the two species, which may limit proper interaction between UvrD and other proteins of the NER pathway. That H. pylori UvrD can complement E. coli in the NER but not the MMR pathway suggests UvrD functional divergence from its E. coli counterpart consistent with the absence of H. pylori MMR. The closely related Wolinella succinogenes, Campylobacter, and Helicobacter species all lack MMR pathway homologs (16, 30), suggesting that these organisms may have lost MMR function during evolution. Interestingly, homologs of UvrD remain present in these species, suggesting that our findings for H. pylori may be applicable to other MMR-deficient organisms as well and that UvrD function, possibly due to its involvement in NER as well as its independent functions, has been retained.
As with other species, H. pylori uvrD mutants display sensitivity to UV exposure and increased frequency of intergenomic recombination (33, 47, 48). Increased sensitivity to UV likely results from inability of the cell to remove excised damaged DNA strands, one of the final steps in the NER pathway (Fig. 1), in the absence of a functional UvrD helicase. Increased recombination in uvrD mutants can arise through several possible mechanisms—absence of the appropriate helicase may create a stall in the NER pathways (or in MMR in organisms with intact pathways), with resultant generation of loose, recombinogenic DNA strands. It is also possible that UvrD acts independently as a helicase at fork-like DNA substrates, to unwind invading strands; as a 3′→5′ DNA helicase (26, 27, 53), UvrD may limit recombination by unwinding strand invasion events (D loops) and aborting potential recombination events (Fig. 1), consistent with UvrD disruption of RecA-mediated strand invasion in E. coli (44). Our experiments show that for H. pylori, the latter possibility most likely causes the increased recombination frequencies in uvrD mutants, since H. pylori lacks MMR. That exposure to UV, which increases substrates for NER, did not lead to increased recombination in uvrD mutants compared to wild-type results suggests that a stall in NER is not necessarily responsible for hyperrecombination in uvrD mutants and again points to an independent UvrD activity in limiting H. pylori genome rearrangements. That uvrB and uvrC mutants do not display increased recombination is consistent with this hypothesis (see Fig. S2 in the supplemental material).
In contrast, under basal conditions for intragenomic recombination, the UvrD mutant and the wild type showed similar phenotypes, but after DNA damage, there was significant elevation in intragenomic recombination frequencies of the uvrD mutants only. These findings indicate a different mechanism for generation of deletions between direct repeats compared to intergenomic recombination, which is consistent with deletions occurring in a RecA-independent process (7), whereas intergenomic recombination requires RecA function (54, 58). We propose the following explanations for these observations. First, the identical repeats are within 1.3 kb of one another (7); processes leading to strand breakage increase chances of slipped-strand mispairing and subsequent deletion of genetic material. DNA damage, especially in NER-deficient strains, results in increased strand breakage potential, potentially leading to increased deletions. Consistent with this explanation, the H. pylori uvrB mutant, which is as sensitive to UV damage as the uvrD strain (59), shows a DNA damage-induced deletion phenotype similar to that seen with the uvrD strains. A second possibility is that in the absence of adequate NER of UV-induced lesions, an alternative pathway for repair, such as recombinational repair, becomes predominant. RecA-mediated recombination also can lead to deletions between identical repeats in E. coli (36), and it is possible that when repair pathways become saturated due to defective NER, recombinational repair increases deletion rates. However, recombinational repair requires RecA function; that RecA mutants also exhibit increased deletion frequencies upon DNA damage (Fig. 5B) suggests that strand breakage and subsequent slipped-strand deletions may be a possible mechanism behind DNA damage-induced deletions in uvrD, uvrB, and recA mutants.
Thus, H. pylori UvrD has a major influence in limiting intergenomic recombination and deletions, particularly in response to genomic damage. Our studies suggest that this role of UvrD in maintaining genomic fidelity might be critical in vivo, since H. pylori lives in an environment that generates substantial genotoxic stress (9, 13, 46).
H. pylori has a high number of repetitive DNA elements that cluster around loci that can potentially influence immunogenicity and host response (7), suggesting that deletion and expansion may be adaptive strategies to target variation to specific locations; in specific contexts, particular phenotypes may have a selective advantage. Our results indicate that DNA damage accelerates the generation of variants in such loci. Thus, UvrD, along with other NER pathway products (e.g., UvrB), is crucial in modulating the rate of H. pylori variation by limiting DNA damage-induced rearrangements. The unexpected relatively high homology of UvrD from H. pylori with those of the extremophiles T. maritima and A. aeolicus, which have specialized DNA repair systems (40), may reflect that all of these organisms share environments characterized by high levels of genotoxic stress. The genome of T. maritima also possesses a high density of repetitive elements, with evidence of frequent lateral transfer and of genetic insertions and deletions (43), in similarity to that of H. pylori (7, 17, 56). These similarities suggest that inter- and intragenomic recombination provides a mechanism for adaptation of residents in such extreme environments, and we speculate that UvrD in these organisms has convergently evolved to limit these processes.
Supplementary Material
Acknowledgments
We thank Benedicte Michel for providing E. coli strains JJC40 and JJC212 and plasmids pGB2 and pGBuvrD, Alanna E. Gregory for assistance in conducting assays, Megan Fannon for assistance with computations, and Hannah Klein for assistance with the Lea-Coulson computations.
This work was supported in part by grants RO1GM62370 and 5T32 GM07308 from the National Institutes of Health, by the Medical Research Service of the Department of Veterans Affairs, and by the Diane Belfer Program in Microbial Ecology.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, B. 2000. A physical interaction of UvrD with nucleotide excision repair protein UvrB. Mol. Cell 10:592-597. [DOI] [PubMed] [Google Scholar]
- 3.Aledo, R., M. F. Avril, B. Dutrillaux, and A. Aurias. 1988. Spontaneous chromosomal anomalies in lymphocytes from Xeroderma pigmentosum. A study of ten patients. Ann. Genet. 31:211-215. [PubMed] [Google Scholar]
- 4.Aledo, R., G. Renault, M. Prieur, M. F. Avril, B. Chretien, B. Dutrillaux, and A. Aurias. 1989. Increase of sister chromatid exchanges in excision repair deficient Xeroderma pigmentosum. Hum. Genet. 81:221-225. [DOI] [PubMed] [Google Scholar]
- 5.Alm, R. A., and T. J. Trust. 1999. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J. Mol. Med. 77:834-846. [DOI] [PubMed] [Google Scholar]
- 6.Ando, T., D. A. Israel, K. Kusugami, and M. J. Blaser. 1999. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J. Bacteriol. 181:5572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aras, R. A., J. Kang, A. I. Tschumi, Y. Harasaki, and M. J. Blaser. 2003. Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc. Natl. Acad. Sci. USA 100:13579-13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arthur, H. M., and R. G. Lloyd. 1980. Hyper-recombination in uvrD mutants of Escherichia coli K-12. Mol. Gen. Genet. 180:185-191. [DOI] [PubMed] [Google Scholar]
- 9.Bagchi, D., T. R. McGinn, X. Ye, M. Bagchi, R. L. Krohn, A. Chatterjee, and S. J. Stohs. 2002. Helicobacter pylori-induced oxidative stress and DNA damage in a primary culture of human gastric mucosal cells. Dig. Dis. Sci. 47:1405-1412. [DOI] [PubMed] [Google Scholar]
- 10.Bierne, H., S. D. Ehrlich, and B. Michel. 1991. The replication termination signal terB of the Escherichia coli chromosome is a deletion hot spot. EMBO J. 10:2699-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bierne, H., M. Seigneur, S. D. Ehrlich, and B. Michel. 1997. uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol. Microbiol. 26:557-567. [DOI] [PubMed] [Google Scholar]
- 12.Caron, P. R., S. R. Kushner, and L. Grossman. 1985. Involvement of helicase II (uvrD gene product) and DNA polymerase I in excision mediated by the uvrABC protein complex. Proc. Natl. Acad. Sci. USA 82:4925-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, C. S., W. N. Chen, H. H. Lin, C. C. Wu, and C. J. Wang. 2004. Increased oxidative DNA damage, inducible nitric oxide synthase, nuclear factor κB expression and enhanced antiapoptosis-related proteins in Helicobacter pylori-infected non-cardiac gastric adenocarcinoma. World J. Gastroenterol. 10:2232-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devriendt, K., and J. R. Vermeesch. 2004. Chromosomal phenotypes and submicroscopic abnormalities. Hum. Genomics 1:126-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobrindt, U., and J. Hacker. 2001. Whole genome plasticity in pathogenic bacteria. Curr. Opin. Microbiol. 4:550-557. [DOI] [PubMed] [Google Scholar]
- 16.Eppinger, M., C. Baar, G. Raddatz, D. H. Huson, and S. C. Schuster. 2004. Comparative analysis of four Campylobacterales. Nat. Rev. Microbiol. 2:872-885. [DOI] [PubMed] [Google Scholar]
- 17.Falush, D., C. Kraft, N. S. Taylor, P. Correa, J. G. Fox, M. Achtman, and S. Suerbaum. 2001. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl. Acad. Sci. USA 98:15056-15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feinstein, S. I., and K. B. Low. 1986. Hyper-recombining recipient strains in bacterial conjugation. Genetics 113:13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flores, M. J., V. Bidnenko, and B. Michel. 2004. The DNA repair helicase UvrD is essential for replication fork reversal in replication mutants. EMBO Rep. 5:983-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flores, M. J., N. Sanchez, and B. Michel. 2005. A fork-clearing role for UvrD. Mol. Microbiol. 57:1664-1675. [DOI] [PubMed] [Google Scholar]
- 21.Garfinkel, D. J., and A. M. Bailis. 2002. Nucleotide excision repair, genome stability, and human disease: new insight from model systems. J. Biomed. Biotechnol. 2:55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glickman, B. W., and M. Radman. 1980. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc. Natl. Acad. Sci. USA 77:1063-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall, M. C., J. R. Jordan, and S. W. Matson. 1998. Evidence for a physical interaction between the Escherichia coli methyl-directed mismatch repair proteins MutL and UvrD. EMBO J. 17:1535-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanada, K., M. Iwasaki, S. Ihashi, and H. Ikeda. 2000. UvrA and UvrB suppress illegitimate recombination: synergistic action with RecQ helicase. Proc. Natl. Acad. Sci. USA 97:5989-5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heep, M., S. Odenbreit, D. Beck, J. Decker, E. Prohaska, U. Rieger, and N. Lehn. 2000. Mutations at four distinct regions of the rpoB gene can reduce the susceptibility of Helicobacter pylori to rifamycins. Antimicrob. Agents Chemother. 44:1713-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoeijmakers, J. H. 1993. Nucleotide excision repair I: from E. coli to yeast. Trends Genet. 9:173-177. [DOI] [PubMed] [Google Scholar]
- 27.Hoeijmakers, J. H. 1993. Nucleotide excision repair. II: From yeast to mammals. Trends Genet. 9:211-217. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh, P. 2001. Molecular mechanisms of DNA mismatch repair. Mutat. Res. 486:71-87. [DOI] [PubMed] [Google Scholar]
- 29.Israel, D. A., A. S. Lou, and M. J. Blaser. 2000. Characteristics of Helicobacter pylori natural transformation. FEMS Microbiol. Lett. 186:275-280. [DOI] [PubMed] [Google Scholar]
- 30.Kang, J., S. Huang, and M. J. Blaser. 2005. Structural and functional divergence of MutS2 from bacterial MutS1 and eukaryotic MSH4-5 homologs. J. Bacteriol. 187:3528-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang, J., D. Tavakoli, A. Tschumi, R. A. Aras, and M. J. Blaser. 2004. Effect of host species on recG phenotypes in Helicobacter pylori and Escherichia coli. J. Bacteriol. 186:7704-7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 33.Kuemmerle, N. B., and W. E. Masker. 1980. Effect of the uvrD mutation on excision repair. J. Bacteriol. 142:535-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 35.Lea, E. A., and D. E. Coulson. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 36.Lovett, S. T. 2004. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 52:1243-1253. [DOI] [PubMed] [Google Scholar]
- 37.Lovett, S. T., P. T. Drapkin, V. A. Sutera, Jr., and T. J. Gluckman-Peskind. 1993. A sister-strand exchange mechanism for recA-independent deletion of repeated DNA sequences in Escherichia coli. Genetics 135:631-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundblad, V., and N. Kleckner. 1985. Mismatch repair mutations of Escherichia coli K12 enhance transposon excision. Genetics 109:3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lusetti, S. L., and M. M. Cox. 2002. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 71:71-100. [DOI] [PubMed] [Google Scholar]
- 40.Makarova, K. S., L. Aravind, N. V. Grishin, I. B. Rogozin, and E. V. Koonin. 2002. A DNA repair system specific for thermophilic archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 30:482-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGinnis, S., and T. L. Madden. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32:W20-W25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mennecier, S., G. Coste, P. Servant, A. Bailone, and S. Sommer. 2004. Mismatch repair ensures fidelity of replication and recombination in the radioresistant organism Deinococcus radiodurans. Mol. Genet. Genomics 272:460-469. [DOI] [PubMed] [Google Scholar]
- 43.Mongodin, E. F., I. R. Hance, R. T. Deboy, S. R. Gill, S. Daugherty, R. Huber, C. M. Fraser, K. Stetter, and K. E. Nelson. 2005. Gene transfer and genome plasticity in Thermotoga maritima, a model hyperthermophilic species. J. Bacteriol. 187:4935-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morel, P., J. A. Hejna, S. D. Ehrlich, and E. Cassuto. 1993. Antipairing and strand transferase activities of E. coli helicase II (UvrD). Nucleic Acids Res. 21:3205-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morimatsu, K., and S. C. Kowalczykowski. 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11:1337-1347. [DOI] [PubMed] [Google Scholar]
- 46.Normark, S., C. Nilsson, B. H. Normark, and M. W. Hornef. 2003. Persistent infection with Helicobacter pylori and the development of gastric cancer. Adv. Cancer Res. 90:63-89. [DOI] [PubMed] [Google Scholar]
- 47.Oeda, K., T. Horiuchi, and M. Sekiguchi. 1981. Molecular cloning of the uvrD gene of Escherichia coli that controls ultraviolet sensitivity and spontaneous mutation frequency. Mol. Gen. Genet. 184:191-199. [DOI] [PubMed] [Google Scholar]
- 48.Oliver, A., F. Baquero, and J. Blazquez. 2002. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 43:1641-1650. [DOI] [PubMed] [Google Scholar]
- 49.Ossanna, N., and D. W. Mount. 1989. Mutations in uvrD induce the SOS response in Escherichia coli. J. Bacteriol. 171:303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinto, V. A., A. Mathieu, S. Marsin, X. Veaute, E. J. O'Rourke, A. Labigne, and P. Radicella. 2004. Suppression of homologous and homeologous recombination by bacterial MutS2 protein. Mol. Cell 17:113-120. [DOI] [PubMed] [Google Scholar]
- 51.Rayssiguier, C., D. S. Thaler, and M. Radman. 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342:396-401. [DOI] [PubMed] [Google Scholar]
- 52.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 53.Sancar, A. 1996. DNA excision repair. Annu. Rev. Biochem. 65:43-81. [DOI] [PubMed] [Google Scholar]
- 54.Schmitt, W., S. Odenbreit, D. Heuermann, and R. Haas. 1995. Cloning of the Helicobacter pylori recA gene and functional characterization of its product. Mol. Gen. Genet. 248:563-572. [DOI] [PubMed] [Google Scholar]
- 55.Schofield, M. J., and P. Hsieh. 2003. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 57:579-608. [DOI] [PubMed] [Google Scholar]
- 56.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson, S. A., and M. J. Blaser. 1995. Isolation of the Helicobacter pylori recA gene and involvement of the recA region in resistance to low pH. Infect. Immun. 63:2185-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson, S. A., R. L. Latch, and J. M. Blaser. 1998. Molecular characterization of the Helicobacter pylori uvr B gene. Gene 209:113-122. [DOI] [PubMed] [Google Scholar]
- 60.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 61.Torii, N., T. Nozaki, M. Masutani, H. Nakagama, T. Sugiyama, D. Saito, M. Asaka, T. Sugimura, and K. Miki. 2003. Spontaneous mutations in the Helicobacter pylori rpsL gene. Mutat. Res. 535:141-145. [DOI] [PubMed] [Google Scholar]
- 62.Tsujimura, T., V. M. Maher, A. R. Godwin, R. M. Liskay, and J. J. McCormick. 1990. Frequency of intrachromosomal homologous recombination induced by UV radiation in normally repairing and excision repair-deficient human cells. Proc. Natl. Acad. Sci. USA 87:1566-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Veaute, X., S. Delmas, M. Selva, J. Jeusset, E. Le Cam, I. Matic, F. Fabre, and M. A. Petit. 2005. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 24:180-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, Y., K. P. Roos, and D. E. Taylor. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J. Gen. Microbiol. 139(Pt. 10):2485-2493. [DOI] [PubMed] [Google Scholar]
- 65.Webb, G. F., and M. J. Blaser. 2002. Dynamics of bacterial phenotype selection in a colonized host. Proc. Natl. Acad. Sci. USA 99:3135-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yasui, A., and S. J. McCready. 1998. Alternative repair pathways for UV-induced DNA damage. Bioessays 20:291-297. [DOI] [PubMed] [Google Scholar]
- 67.Zieg, J., V. F. Maples, and S. R. Kushner. 1978. Recombinant levels of Escherichia coli K-12 mutants deficient in various replication, recombination, or repair genes. J. Bacteriol. 134:958-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.