Abstract
Multilocus sequence typing (MLST), a sequence-based method to characterize bacterial genomes, was used to examine the genetic structure in a large collection of Medicago-nodulating rhizobial strains. This is the first study where MLST has been applied in conjunction with eBURST analysis to determine the population genetic structure of nonpathogenic bacteria recovered from the soil environment. Sequence variation was determined in 10 chromosomal loci of 231 strains that predominantly originated from southwest Asia. Genetic diversity for each locus ranged from 0.351 to 0.819, and the strains examined were allocated to 91 different allelic profiles or sequence types (STs). The genus Medicago is nodulated by at least two groups of rhizobia with divergent chromosomes that have been classified as Sinorhizobium meliloti and Sinorhizobium medicae. Evidence was obtained that the degree of genetic exchange among the chromosomes across these groups is limited. The symbiosis with Medicago polymorpha of nine strains placed in one of these groups, previously identified as S. medicae, ranged from ineffective to fully effective, indicating that there was no strong relationship between symbiotic phenotype and chromosomal genotype.
Plants of the genus Medicago are legumes, which have the ability to form a symbiosis with soil bacteria commonly referred to as rhizobia. This symbiosis is mutually beneficial because these bacteria, when residing in root nodules, can fix atmospheric dinitrogen into a form (ammonium) that can be used by the plant for growth. Because of this property, many legumes are essential in low-input sustainable agriculture; alfalfa (Medicago sativa) and annual medics (Medicago spp.) have worldwide application as forage and green manure crops.
Over many decades there has been a concerted effort to collect and evaluate medics from various locations worldwide. Subsequent to their acquisition, plant introductions were usually evaluated for their potential application in agriculture. These programs were and are relatively easy since a plant's morphology, physiology, and performance are readily measured. By comparison, the collection of rhizobial microsymbionts, capable of nodulating these medics, has been more arbitrary since there is no efficient method to discriminate between genetically related clusters of these bacteria. Traditionally, a limited number of rhizobial cultures were isolated and then were tested on several varieties of a crop and those with the best overall performance were chosen for manufacture of inoculants. In the case of Medicago, USDA 1002 (strain 3Doa2 or ATCC 9930) was isolated in 1919 by USDA scientists in Virginia from a soil on the Arlington Farm (now the location of the Pentagon). This strain was subsequently chosen as the type for the species (then Rhizobium meliloti) based on its superior symbiotic performance.
With the development of techniques in molecular biology, it has become possible to examine newly isolated cultures for a much broader array of characters. These techniques have been used to investigate genetic diversity among strains, and the data have also been used to provide support for decisions in rhizobial taxonomic classification. One of the more popular techniques is DNA fingerprinting by PCR. This method depends on the use of primers for repeat sequences that reside throughout bacterial genomes (30). One such analysis was developed with rhizobia that form symbioses with Medicago (7). Other PCR-related techniques that have been applied to obtain DNA fingerprints of rhizobial genomes are random amplified polymorphic DNA (32) and amplified fragment length polymorphism (31). These tools have been used to examine genetic diversity among Medicago-nodulating rhizobia isolated in Croatia (3), Italy (5, 19, 20), Latin America (8), Tunisia (1, 33), and the Caucasian and Central Asian regions (2, 22). Even though with these methods the existence of genetic diversity among Medicago-nodulating rhizobia has been revealed, without sequence data it is not possible to obtain conclusive estimates of levels of their divergence. The data used to estimate divergence are inconclusive because PCR products of equal molecular size across lanes need not necessarily be identical. Besides the qualitative nature of fingerprint data, another limitation of the approach is that results are not portable and consequently numerous standard reference strains need to be included in every analysis.
DNA fingerprinting is one of a variety of methods, collectively referred to as a consensus or polyphasic approach, that are used in the characterization of rhizobia. However, there is lack of agreement among bacterial taxonomists and population geneticists on which of these methods are the most appropriate to define bacterial species limits (9, 25). As a consequence, rhizobial taxonomic decisions mostly have been subjective and arbitrary.
In two recent reviews, a theoretical basis for the use of a multilocus genotypic approach to define bacterial species limits was discussed (9, 25). The advantage of such an approach is that evidence can be provided to infer both founding genotypes and likely patterns of descent within clusters of closely related strains. The first attempt to apply a multilocus approach for genotyping Medicago-nodulating rhizobia was reported by Eardly et al. (10), who used multilocus enzyme electrophoresis (MLEE) to characterize 231 strains that originated predominantly from southwest Asia. Because results generated by MLEE also are not portable, comparisons of results with large numbers of strains in separate data sets are complex and require that multiple reference standards be included in each analysis. A significant improvement over MLEE is the multilocus sequence typing (MLST) method, where alleles of at least seven loci are identified by direct nucleotide sequence comparisons (16) rather than electrophoretic migration patterns of enzyme electromorphs. The combinations of alleles for the loci examined in each strain are then used to derive an allelic profile or sequence type (ST). The ST data are maintained in a database that can be manipulated to permit subsequent comparisons of large numbers of bacterial genotypes without the need to repeatedly analyze reference cultures. MLST results for numerous pathogenic bacterial species have already been reported (11), but to our knowledge this strategy has never been applied to rhizobia.
MLST also can be applied in the efficient management of genetic resource collections. For example clones of the same strain can be easily identified and eliminated.
Our primary goal was to use MLST to identify distinct groups of Sinorhizobium spp. and to estimate the relationships between these groups with the analysis of 10 chromosomal loci in 231 diverse strains of Medicago-nodulating rhizobia that had previously been characterized by MLEE (10). These results were then used in a follow-up study with several representative strains to examine any possibility of a relationship between chromosomal genotype and symbiotic phenotype.
MATERIALS AND METHODS
Bacterial strains and genomic DNA extraction.
The Medicago-nodulating rhizobial strains included in this analysis were those previously described by Eardly et al. (10) with the exception that 102F84, M193b, M269, and M296 were omitted and the type strains for Sinorhizobium meliloti, USDA 1002, and Sinorhizobium medicae, A321, were included. Each culture was plated to check for purity and was subsequently grown in 10 ml modified arabinose-gluconate broth (24) for 3 days to prepare DNA by using the Qiamp DNAeasy minikit (QIAGEN, Valencia, CA). DNA samples were stored at −20°C, and the cultures were stored as glycerol suspensions at −70°C and as lyophilized samples.
Plant tests.
Seeds of Medicago polymorpha were surface sterilized with concentrated H2SO4 for 3 min and were washed five times with sterile distilled water. The treated seeds were germinated on sterile water agar, seedlings were sown in sterile 50:50 (wt/wt) sand-vermiculite-filled Leonard jars (15), and 2 ml of modified arabinose-gluconate-grown late-log-phase broth cultures was used to inoculate each jar. The cultures tested for symbiosis were the type strains for the species S. meliloti (USDA 1002) and S. medicae (A321) and the MLEE group B strains CC169, M3, M16, M58, M75, M161, M173, and M254. Each treatment was prepared in three replications, and three jars without inoculated bacteria served as controls. The plants were grown in a greenhouse without supplemental lighting in two duplicate experiments for 30 and 42 days. The plants were uprooted, and the tops were cut off to determine nitrogenase activities as described by van Berkum et al. (29). Determinations for the concentration of ethylene in each chamber were as described by van Berkum and Sloger (27). The plant tops were dried at 60°C for 2 days to determine dry matter (6).
PCR primer design and PCR amplification of chromosomal loci.
Loci for MLST analysis were chosen by referring to the complete genomic sequence of strain 1021 (4) to select 10 genes distributed across the chromosome (Table 1). The entire open reading frame of each locus was used to select primers that would amplify a portion of each gene between 200 and 500 bp in size by using the primer design software package Oligo Primer Analysis Software version 6.65 (Molecular Biology Insights, Inc., Cascade, CO). The oligonucleotides selected (Table 1) with Oligo were synthesized by Sigma-Genosys (The Woodlands, TX) and were received as dried preparations. Upon receipt the primers were dissolved with 10 mM Tris buffer, pH 8.0, to a final concentration of 1,000 pmol and were stored at −20°C. The PCRs for each locus were then optimized by using the FailSafe PCR PreMix selection kit (Epicentre, Madison, WI) and the thermal cycle protocol described by van Berkum and Fuhrmann (26) with an MJ Research PTC-225 Peltier thermal cycler (MJ Research Inc., Waltham, MA) using genomic DNAs of both USDA 1002 and A321 as templates. These 24 reactions were analyzed by horizontal agarose gel electrophoresis to select the FailSafe PCR system (Epicentre, Madison, WI) determined to be the most suitable for PCR amplification with the DNA preparations of all 231 strains used in this investigation. The presence of a single PCR product of the expected molecular size for each primer pair using each template was verified by horizontal gel electrophoresis. Each PCR product was then purified, especially to remove the PCR primers, by using the Ampure PCR purification system (Agincourt Bioscience Corporation, Beverly, MA).
TABLE 1.
Primer sequences for the 10 loci used in MLST analysis of the chromosomes of 231 Medicago-nodulating rhizobia
| Locus and primer sequencea | Product length (bp) | Locus location |
|---|---|---|
| asd | 534 | 3617313…3618347 |
| F 5<2w9>-CGG CCG GGA GAT GCT GAA CA-3′ | ||
| R 5′-ATG CGC TTG GTG AAC TTC TTG-3′ | ||
| edD | 552 | 766634…768454 |
| F 5′-GGC ATC ATC ACC TCC TAC AA-3′ | ||
| R 5′-CGG CGT GCC GGG ATT-3′ | ||
| gap | 486 | 2975447…2976457 |
| F 5′-CGG TCC GGT CGA GAC CAA-3′ | ||
| R 5′-CGG TAG AGA TCC TTG TGC AT-3′ | ||
| glnD | 343 | 429607…432456 |
| F 5′-GTG CGC TGC CAC ATG CAY TT-3′ | ||
| R 5′-CCG GRT CRC GCT TGA A-3′ | ||
| gnd | 400 | 2091007…2092437 |
| F 5′-GGG CCG GCT CAA CTC CTA-3′ | ||
| R 5′-CGG CAT CGG CAG GTT-3′ | ||
| nuoE1 | 293 | 1381334…1382161 |
| F 5′-GCG CGC KCA GGA GCA GGA-3′ | ||
| R 5′-CGC AGG CGC CCT GAC ATT-3′ | ||
| ordL2 | 456 | 770925…772211 |
| F 5′-GCG GCG CGG TCG TCA Tx-3′ | ||
| R 5′-CGC CAT GGC CGG AAT A-3′ | ||
| recA | 357 | 1948178…1949224 |
| F 5′-CCG GTT CGC TCG GCC TCG ATA-3′ | ||
| R 5′-CGC CCA TCT CGC CCT CGA TTT-3′ | ||
| sucA | 533 | 3313082…3316078 |
| F 5′-GCT CGG CCT CGA ATA-3′ | ||
| R 5′-CCG TCA GCG ACA GGT-3′ | ||
| zwf | 535 | 769253…770728 |
| F 5′-GGG GGC ACC GGC GAT CTT G-3′ | ||
| R 5′-AGC GCA GTG CCA TCA GAT TCT-3′ |
F, forward; R, reverse; asd, aspartate-semialdehyde dehydrogenase; edd, phosphogluconate dehydratase; gap, glyceraldehyde 3-phosphate dehydrogenase; glnD, protein-PII uridylyltransferase; gnd, 6-phosphogluconate dehydrogenase; nuoE1, NADH dehydrogenase I chain E protein; ordL2, putative oxidoreductase protein; recA, DNA strand exchange and recombination protein; sucA, 2-oxoglutarate dehydrogenase E1; zwf, glucose-6-phosphate 1-dehydrogenase.
Sequence analysis.
The purified PCR products were used in two sequencing reactions each with one of the original PCR primers. An Applied Biosystems 377 DNA sequencer or 3730 DNA analyzer in combination with a Dye Deoxy Terminator Cycle sequencing kit (Applied Biosystems, Foster City, CA) was used for sequencing the purified PCR products as described previously (28). The 16S rRNA gene sequences were determined as previously described (26) for strains 15B4, 74B15, 56A14, CC2013, S33, M7, and M58.
Data analysis.
A Microsoft Access database was created to compile the data collected with the 231 strains. The sequence length entered for each locus was identical, and the same alleles were identified using the software Sequence Comparator version 2.0.1, written by Keith Jolley. As additional alleles were identified they were assigned different allele numbers in the database. In each case, allelic variation was verified by confirming the sequence disparity using Genedoc (18) and then checking the electropherograms produced by the sequencing analysis to substantiate differences. In the case of ambiguities the sequencing analysis was repeated.
When the database was completed, the relationship function in Microsoft Access was used to create a “query” of the allelic allocation for each of the 10 loci across the 231 strains. The 10 entries across the strains were then listed according to an ascending value. The resulting tabulation of the data was then exported as a Microsoft Excel file to prepare data input files for the program Sequence Type Analysis and Recombinational Tests (START), written by Keith Jolley, University of Oxford (14); the statistics for population genetics computer program written by T. S. Whittam (23); and eBURST (12). START was used to determine both the allele and profile frequencies and to create an UPGMA (unweighted-pair group method average linkages) tree to portray the genetic relationships among the STs. Sequence types were classified as single-locus variants (SLVs), double-locus variants (DLVs), or singletons (STs differing at three or more loci). The computer program by T. S. Whittam was used to calculate the genetic distances among the alleles of each locus and to derive the mean value. The number of groups, the clonal complex of each group, and a population snapshot of the chromosomal variation were generated using eBURST. The null hypothesis of linkage equilibrium for the multilocus sequence data as defined by Maynard et al. (16) was evaluated using the program LIAN 3.0 (13).
Nucleotide sequence accession numbers.
Sequences of the alleles for each locus were deposited with GenBank under accession numbers DQ423249 through DQ423361. The 16S rRNA gene sequences of strains 15B4, 74B15, 56A14, CC2013, S33, M7, and M58 were deposited with GenBank under accession numbers DQ423242 through DQ423248, respectively.
RESULTS
MLST analysis of 231 Medicago-nodulating rhizobial strains yielded allele frequencies that ranged from 8 to 18 alleles per locus with a mean value of 11.3 (see Table 3); 91 different profiles or STs were identified. Multiple strains were placed in 22 of the 91 STs (Table 2). The type strain for S. meliloti was representative of only 26% of the chromosomal types identified with MLST. Genetic diversity across the 10 loci in the MLST analysis varied from 0.351 to 0.819 with a mean (H) of 0.593 (Table 3).
TABLE 3.
Numbers of alleles and genetic diversity across chromosomes of 231 Medicago-nodulating rhizobia
| Locus | No. of alleles | H |
|---|---|---|
| asd | 16 | 0.571 |
| edD | 18 | 0.819 |
| zwf | 10 | 0.697 |
| gap | 9 | 0.572 |
| glnD | 10 | 0.731 |
| gnd | 8 | 0.351 |
| nuoE | 11 | 0.450 |
| ordL | 9 | 0.633 |
| recA | 10 | 0.498 |
| sucA | 12 | 0.604 |
| Mean value for total | 11.3 | 0.593 |
TABLE 2.
STs with multiple strains
| ST | Representative straina | Strain(s) with the same ST |
|---|---|---|
| 1 | USDA 1002 | A145, ATCC 9930, N6B5, 128A9, 3-3, 128A13, 5C6, 17C3, 5A13, M91, M99, M100, 5A10, 5A12, 5B19, 15B65, 15B69, 27-2, 128A8, 54032, LS2A, M68, U45, M94 |
| 4 | 17B6 | 1322, M248, N6B9, M275, M287, M163, M164, M182, M288, M291, 15A5, M47, M52, M266, M276, M35 |
| 5 | 56A14 | USDA 1948, 128A10, 128A14, 15B66, 56A11, 56A13, 128A12, 15B61, 56A4, 56A18, 15B1, 56A2, 56A17 |
| 6 | 74B3 | M289, N6B4, 128A6, M101 |
| 8 | 74B12 | 17C1, N6B6 |
| 10 | 102F85 | S14 |
| 11 | A321 | 12, CC 169 |
| 12 | 1021 | M250, M255, M267, M199, M200, M224, M240, 5B17, M183, M192, 5C15, 19A19, 74B6, 74B14, 311, CC 2003, M11, M5, RCR2011, RF22, S26, M252, M32 |
| 13 | M7 | M204 |
| 14 | M56 | M281 |
| 15 | M58 | M104, M173, M16, M53, M9 |
| 30 | 19A4 | 19A11, 74B20 |
| 31 | 19A8 | 17C5, 19A20, N4A2, N4A5, N4A6, N4A8, N4A10, N4A11, 19A16, 74B5, 17A6, 17A7, 19A6, 74B9, B294, L5-30, N4A3, U54 |
| 33 | M207 | M176, M180, M184, M191, M197 |
| 34 | M210 | M202 |
| 36 | M256 | M258, M253, M260, M20, M265 |
| 40 | Sa-10 | V7 |
| 44 | 102F82 | M264 |
| 46 | N4A7 | N4A9, N6B3 |
| 51 | 1944 | 17A8, 17B7, 17C2 |
| 60 | M76 | M105, M165, M292, M6, M44, M49, M50, M201, M21 |
| 65 | M181 | M51 |
USDA 1002 is the type strain for Sinorhizobium meliloti; A321 is the type strain for Sinorhizobium medicae. All other STs consisted of a single strain.
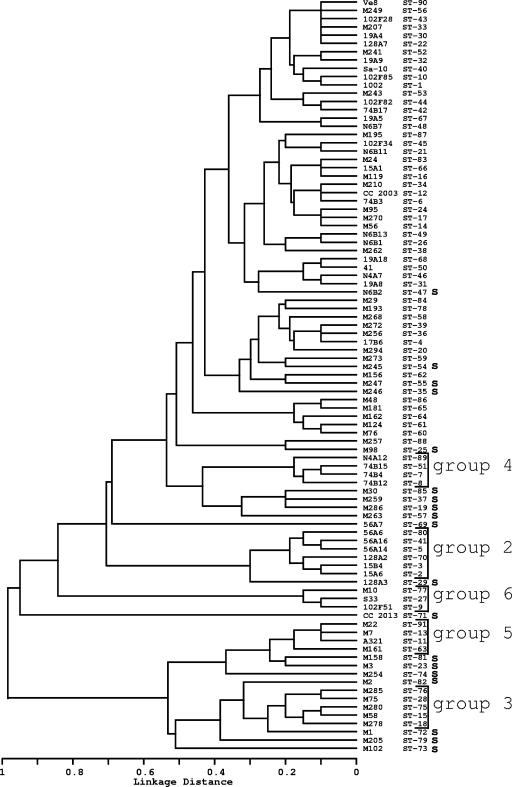
An UPGMA tree was constructed with the MLST data set of 10 loci and 91 STs using the software program START. The STs of rhizobia that have been defined as S. medicae and S. meliloti (21) were separated at a linkage distance of 0.98 (Fig. 1). Bootstrap support for the separation of clusters was 98%. The only other significant bootstrap value obtained was in support of the STs of group 6. Alleles of the S. medicae cluster were distinctive at 9 of the 10 loci examined; the gnd locus was the exception. Only a single allele of the gnd locus was found in all 16 STs placed in the S. medicae cluster. This allele was also shared with an additional 57 STs not placed in this cluster. In strain CC2013 (ST-71) alleles were also distinctive at 9 of the 10 loci examined. The nuoE allele found in CC2013 also was present in strains 102F51 (ST-9), S33 (ST-27), M10 (ST-77), and M30 (ST-85).
FIG. 1.
Linkage distance among 231 Medicago-nodulating rhizobia derived from allelic variation among 10 chromosomal loci. A matrix of the strain identification and the ST followed by the allele labels for each was used in the START (Sequence Type Analysis and Recombinational Tests, version 1.05) program to generate the UPGMA dendrogram. The program was written by Keith Jolley, University of Oxford (14). MLST group affiliation is indicated. Singletons are identified by an “s”; ST numbers followed by a blank belong to group 1.
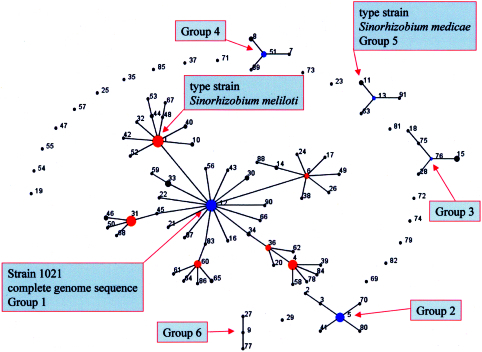
The likely patterns of chromosomal evolutionary descent and the identity of the probable founding genotypes were inferred from an eBURST analysis (12) of the MLST data. In this analysis the number of resamplings made for bootstrapping was 1,000 and the number of identical loci for group definition was nine. Using these parameters six groups were identified among the 91 STs. The largest of these groups (group 1) contained 50 STs, which represented 164 of the 231 strains (Fig. 2). Within group 1 the predicted founder was ST-12 with a bootstrap confidence value of 81%, while ST-1 was a distant second with 55%. Four other STs within this group (ST-1, ST-4, ST-60, and ST-31) were identified as cofounders (Fig. 2). The strain 1021 (for which a genome sequence has been determined) belonged to ST-12, while the type strain for S. meliloti, USDA 1002 (also ATCC 9930), was placed in ST-1. The remaining groups had fewer STs and were numbered in order of descending size (groups 2 through 6). There were six STs representing 19 of the 231 strains in group 2 with ST-5 as the predicted founder (74%). There were five, four, four, and three STs in groups 3, 4, 5, and 6, respectively. The type strain for S. medicae (A321) was placed in group 5, which consisted of four STs representing seven strains. Two of these strains had the same ST as A321, but the probability that this ST was the predicted founder was low (4%). ST-13 and ST-91 had a greater probability of representing the predicted founder of group 5 (60% and 10%, respectively). Some of the strains that Rome et al. (21) identified as S. medicae were placed in two groups (groups 3 and 5) while the remaining seven strains were associated singletons (Fig. 1 and 2).
FIG. 2.
Population snapshot of 231 Medicago-nodulating rhizobia derived from the allelic variation of 10 chromosomal loci. A matrix of the ST followed by the allele labels for each was used in eBURST (11) to generate a diagram of the evolutionary patterns among the strains. The snapshot was produced by setting the group definition to 0/10 genes. The sizes of the circles are related to the numbers of strains within each ST. The founder and cofounder genotypes are colored blue and red, respectively. Distances between STs indicated by connecting lines are arbitrary.
Significant linkage disequilibrium was observed among the 91 STs (Table 4). Within the S. medicae cluster (groups 5 and 3 and their associated singletons in Fig. 1) linkage disequilibrium was not evident. Linkage disequilibrium also was not evident among the STs of group 1 (Table 4). Similar results were previously reported among the multilocus enzyme genotypes of the same strains (10, 16).
TABLE 4.
Test of the null hypothesis of linkage equilibrium among 91 multilocus STs in a global collection of 231 Medicago-nodulating rhizobial strainsa
An eBURST population snapshot of the six groups and 19 singletons was used to establish possible patterns of evolutionary descent of the STs of the groups (Fig. 2). The most extensive clonal complex was that of group 1, with ST-12 representing the founder genotype. Members of the ST-12 founder complex (ST-12 with SLVs ST-16, ST-21, ST-22, ST-30, ST-33, ST-34, ST-43, ST-45, ST-56, ST-66, ST-83, ST-87, and ST-90 and DLV ST-59) were isolated from nodules of both M. sativa and the annual medics in approximately equal numbers (Table 5). Approximately half of these strains originated from Pakistan and Syria, while the remainder were from Turkey, Jordan, Nepal, and France and from countries outside the native range of Medicago. Cofounders ST-1 and ST-31 with their SLVs and DLVs were predominantly isolated from root nodules of M. sativa in Pakistan and Nepal (Table 5). Cofounders ST-6, ST-36, and ST-60 with their SLVs and DLVs were predominantly isolated from annual medics in Jordan and Syria (Table 5).
TABLE 5.
Host plant and geographic origin of Medicago-nodulating rhizobia of the group 1 clonal complex
| Founder and cofounder | STs included | % of rhizobia from host plant of origin
|
Geographic locations of origin (% of rhizobia) | |
|---|---|---|---|---|
| M. sativa | Annual medics | |||
| ST-1 | ST-10, ST-32, ST-40, ST-42, ST-44, ST-48, ST-52, ST-53, ST-67 | 73 | 27 | France (5), Jordan (8), Pakistan (38), Syria (16), Nepal (5), exteriora (27) |
| ST-6 | ST-14, ST-17, ST-24, ST-26,ST-38, ST-49, ST-88 | 23 | 77 | Jordan (38), Pakistan (15), Syria (23), Nepal (23) |
| ST-31 | ST-46, ST-50, ST-68 | 92 | 8 | Pakistan (38), Jordan (4), Nepal (42), exterior (16) |
| ST-36 | ST-4, ST-20, ST-39, ST-58, ST-62, ST-78, ST-84 | 12 | 88 | Jordan (50), Pakistan (8), Syria (31), Nepal (4), Turkey (4), exterior (4) |
| ST-60 | ST-61, ST-64, ST-65, ST-86 | 100 | Jordan (7), Syria (86), Turkey (7) | |
| ST-12 (founder) | ST-16, ST-21, ST-22, ST-30, ST-33, ST-34, ST-43, ST-45, ST-56, ST-59, ST-66, ST-83, ST-87, ST-90 | 43 | 57 | France (2), Jordan (11), Pakistan (23), Syria (26), Nepal (4), Turkey (17), exterior (17) |
Locations exterior to the native range of Medicago include Australia, Canada, Poland, South America, Sweden, and New Zealand.
Groups 2 through 6 each contained fewer STs than group 1. The group 2 strains all originated from M. sativa in Pakistan. The group 3 strains all were isolated from annual medics and originated from Syria (60%) and Jordan (40%). The group 4 strains all originated from M. sativa and Pakistan with the exception of one annual medic strain from Nepal. All group 5 strains originated from annual medics and were collected in six different countries. Group 6 had two strains isolated from M. sativa growing in the United States and one strain from an annual medic growing in Syria. Of the 19 singletons (STs that differed at three or more loci), only three originated from M. sativa.
In addition to sequencing of the 10 loci for MLST, the 16S rRNA gene sequences were determined for a select set of strains. The 16S rRNA gene sequences of strains 56A14 (ST-5) and 74B15 (ST-51), representing groups 2 and 4, respectively, and of USDA 1002 (S. meliloti) were identical. The 16S rRNA gene sequences of group 6 strain S33 (ST-27) and of the singleton CC2013 (ST-71) were identical to each other, and both had a single nucleotide difference compared with the USDA 1002 allele. Strains M58 (ST-15) and M7 (ST-13) represent groups 3 and 5, respectively, and possessed a 16S rRNA gene sequence identical with that reported for strain A321 (21). The 16S rRNA gene sequence of these three strains had four base pair differences from the corresponding sequence of USDA 1002.
The symbiotic reactions of several strains that were placed in the cluster defined as S. medicae (21) were tested with M. polymorpha as the plant host. This was done because it has been reported that only S. medicae forms an effective symbiosis with this host legume species. If substantiated, this would represent a phenotypic trait that could be used to distinguish strains that were defined as S. medicae. The type strains for S. meliloti (USDA 1002) and S. medicae (A321) could be distinguished by the plant test since they produced an ineffective and an effective symbiotic response on M. polymorpha, respectively (Table 6 ). However, a mixed response was obtained with eight of the other strains placed in the S. medicae cluster: four were ineffective, one was intermediate in effectiveness, and the three remaining strains were fully effective, similar to strain A321 (Table 6). In a repeat test, M16, M161, M173, and M58 were again observed to nodulate and to establish an ineffective response with M. polymorpha while strains M254 and M3 were effective.
TABLE 6.
Symbiotic reaction of several MLEE group B strains, identified as genospecies 2 or Sinorhizobium medicae (21), on Medicago polymorphaa
| Strainb | MLEE group | Host origin | Nodule typec | Nitrogenase activityd | Above-ground plant dry mattere |
|---|---|---|---|---|---|
| USDA 1002T (ST-1) | A | M. sativa | W | 0.08 | 20 |
| A321T (ST-11) | B | Medicago truncatula | P | 1.09 | 91 |
| CC169 (ST-11) | B | Medicago rugosa | P | 1.04 | 86 |
| M3 (ST-23) | B | Medicago orbicularis | P | 1.23 | 75 |
| M16 (ST-15) | B | M. orbicularis | W/P | 0.25 | 37 |
| M58 (ST-15) | B | Medicago rotata | W | 0.01 | 24 |
| M75 (ST-28) | B | Medicago radiata | W | 0.01 | 12 |
| M161 (ST-63) | B | Medicago noeana | W | 0.01 | 10 |
| M173 (ST-15) | B | Unspecified | W | 0.01 | 16 |
| M254 (ST-74) | B | M. rotata | P | 0.91 | 86 |
| Uninoculated | None | 0.00 | 13 |
Results are the means of three replicate jars containing two plants grown for 30 days.
USDA 1002 and A321 are the type strains for S. meliloti and S. medicae, respectively.
W, small and white; P, large and pink; W/P, mixture of the two types.
Acetylene reduction rate is in μmol C2H4/plant/h.
Dry weight is in mg/plant.
DISCUSSION
In this study the allelic profiles of 231 rhizobial strains of the legume genus Medicago were examined. These strains, previously characterized by MLEE analysis (10), originated predominantly from southwest Asia, which in part is the native range of the host legume genus Medicago. The MLST data supported the conclusions made from the MLEE study that this plant genus is nodulated by at least two highly divergent groups of rhizobia. The increased resolving power of MLST made it possible to identify 91 STs separated by a maximum linkage distance of 0.98 (Fig. 1) compared with 50 electrophoretic types detected by MLEE separated by a maximum linkage distance of 0.75 (9). With eBURST analysis of the MLST data six groups and 19 singletons were identified (Fig. 2) compared with five groups and seven singletons established with the MLEE data (9). Also, by MLST analysis six cofounders with the founder ST-12 were identified compared with three cofounders using eBURST analysis of the MLEE data (9).
The presence of 91 STs among the 231 strains would indicate that there is extensive diversity in natural populations of Medicago-nodulating rhizobia. It seems likely that in this study only a small fraction of this diversity was revealed. It is anticipated that a much broader range of genotypes will be identified with an analysis of strains from other geographic regions.
In addition to its value for characterizing genetic diversity in bacterial populations, this method has potential practical applications as well. For example MLST data could be used for choosing unique strains for preservation in culture collections or for the selection of strains that might be useful in low-input sustainable agriculture.
One of the goals in this study was to determine if symbiotic host range and effectiveness might be associated with specific chromosomal genotypes. This question was based on the report of Rome et al. (21) and their conclusion that, while strains of S. medicae form effective symbioses with M. polymorpha, those of S. meliloti are ineffective with this legume species. The MLST analysis confirmed the separation of a cluster of strains that had previously been identified (by MLEE and other criteria) as belonging to the species S. medicae. However, from the subsequent plant tests it was apparent that there is no strict correlation between the chromosomal genotype of a strain and its symbiotic effectiveness on M. polymorpha. An explanation for this conclusion may be that the symbiotic determinants in S. meliloti and S. medicae reside on large extrachromosomal elements (e.g., pSymA) that may be subject to lateral transfer and recombination. The possibility that this may be the explanation for the results with the plant tests could be investigated with an MLST analysis of the extrachromosomal elements in the same strains.
Acknowledgments
We thank K. Lee Nash for excellent technical assistance.
REFERENCES
- 1.Badri, Y., K. Zribi, M. Badri, T. Huguet, and M. E. Aouani. 2003. Sinorhizobium meliloti nodulates Medicago laciniata in Tunisian soils. Czech J. Genet. Plant Breed. 39(Special Issue):178-183. [Google Scholar]
- 2.Biondi, E. G., E. Pilli, E. Giuntini, M. L. Roumiantseva, E. E. Andronov, O. P. Onichtchouk, O. N. Kurchak, B. V. Simarov, N. I. Dzyubenko, A. Mengoni, and M. Bazzicalupo. 2003. Genetic relationship of Sinorhizobium meliloti and Sinorhizobium medicae strains isolated from Caucasian region. FEMS Microbiol. Lett. 220:207-213. [DOI] [PubMed] [Google Scholar]
- 3.Bradić, M., S. Sikora, S. Redžepović, and Z. Štafa. 2003. Genetic identification and symbiotic efficiency of an indigenous Sinorhizobium meliloti field population. Food Technol. Biotechnol. 41:69-75. [Google Scholar]
- 4.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Pühler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carelli, M., S. Gnocchi, S. Fancelli, A. Mengoni, D. Paffetti, C. Scotti, and M. Bazzicalupo. 2000. Genetic diversity and dynamics of Sinorhizobium meliloti populations nodulating different alfalfa cultivars in Italian soils. Appl. Environ. Microbiol. 66:4785-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cousin, C., J. Grant, F. Dixon, D. Beyene, and P. van Berkum. 2002. Influence of biosolids compost on the bradyrhizobial genotypes recovered from cowpea and soybean nodules. Arch. Microbiol. 177:427-430. [DOI] [PubMed] [Google Scholar]
- 7.de Bruijn, F. J. 1992. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ. Microbiol. 58:2180-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Papa, M. F., L. J. Balagué, S. C. Sowinski, C. Wegener, E. Segundo, F. M. Abarca, N. Toro, K. Niehaus, A. Pühler, O. M. Aguilar, G. Martínez-Drets, and A. Lagares. 1999. Isolation and characterization of alfalfa-nodulating rhizobia present in acidic soils of central Argentina and Uruguay. Appl. Environ. Microbiol. 65:1420-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eardly, B. D., and P. van Berkum. 2005. Use of population genetic structure to define species limits in Rhizobiaceae. Symbiosis 38:109-122. [Google Scholar]
- 10.Eardly, B. D., L. Materon, N. H. Smith, D. A. Johnson, M. D. Rumbaugh, and R. K. Selander. 1990. Genetic structure of natural populations of the nitrogen-fixing bacterium Rhizobium meliloti. Appl. Environ. Microbiol. 56:187-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feil, E. J. 2004. How stable are the core genes of bacterial pathogens? ASM News 6:234-238. [Google Scholar]
- 12.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequencing typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847-848. [DOI] [PubMed] [Google Scholar]
- 14.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 15.Leonard, L. T. 1943. A simple assembly for use in the testing for cultures of rhizobia. J. Bacteriol. 45:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maynard Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholas, K. B., and H. B. Nicholas. 1997. Genedoc: a tool for editing and annotating multiple sequence alignments. Multiple Sequence Alignment Editor and Shading Utility, 2.6.001. http://www.psc.edu/biomed/genedoc.
- 19.Paffetti, D., C. Scotti, S. Gnocchi, S. Fancelli, and M. Bazzicalupo. 1996. Genetic diversity of an Italian Rhizobium meliloti population from different Medicago sativa varieties. Appl. Environ. Microbiol. 62:2279-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paffetti, D., F. Daguin, S. Fancelli, C. Scotti, S. Gnocchi, F. Lippi, and M. Bazzicalupo. 1998. Influence of plant genotype on the selection of nodulating Sinorhizobium meliloti strains by Medicago sativa. Antonie Leeuwenhoek 73:3-8. [DOI] [PubMed] [Google Scholar]
- 21.Rome, S., M. P. Fernandez, B. Brunel, P. Normand, and J.-C. Cleyet-Marel. 1996. Sinorhizobium medicae sp. nov., isolated from annual Medicago spp. Int. J. Syst. Microbiol. 46:972-980. [DOI] [PubMed] [Google Scholar]
- 22.Roumiantseva, M. L., E. E. Andronov, L. A. Sharypova, T. Dammann-Kalinowski, M. Keller, J. P. W. Young, and B. V. Simarov. 2002. Diversity of Sinorhizobium meliloti from the Central Asian alfalfa gene center. Appl. Environ. Microbiol. 68:4694-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Berkum, P. 1990. Evidence for a third uptake hydrogenase phenotype among the soybean bradyrhizobia. Appl. Environ. Microbiol. 56:3835-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Berkum, P., and B. D. Eardly. 2005. Impact of genomics on the reconstruction of evolutionary relationships of nitrogen-fixing bacteria and implications for taxonomy, p. 201-219. In R. Palacios and W. E. Newton (ed.), Genomes and genomics of nitrogen-fixing organisms. Springer-Verlag, Dordrecht, The Netherlands.
- 26.van Berkum, P., and J. J. Fuhrmann. 2000. Evolutionary relationships among the soybean bradyrhizobia reconstructed from 16S rRNA gene and internally transcribed spacer region sequence divergence. Int. J. Syst. Evol. Microbiol. 50:2165-2172. [DOI] [PubMed] [Google Scholar]
- 27.van Berkum, P., and C. Sloger. 1979. Immediate acetylene reduction by excised grass roots not previously preincubated at low oxygen tensions. Plant Physiol. 64:739-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Berkum, P., D. Beyene, and B. D. Eardly. 1996. Phylogenetic relationships among Rhizobium species nodulating the common bean Phaseolus vulgaris L. Int. J. Bacteriol. 46:240-244. [DOI] [PubMed] [Google Scholar]
- 29.van Berkum, P., R. E. Tully, and D. L. Keister. 1995. Nonpigmented and bacteriochlorophyll-containing bradyrhizobia isolated from Aeschynomene indica. Appl. Environ. Microbiol. 61:623-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Versalovic, J. M., M. Schneider, F. J. de Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 31.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams, J. G. K., A. R. Kubelik, K. J. Livak, J. A. Rafalski, and S. V. Tingey. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zribi, K., R. Mhamdi, T. Huguet, and M. E. Aouani. 2004. Distribution and genetic diversity of rhizobia nodulating natural populations of Medicago truncatula in Tunisian soils. Soil Biol. Biochem. 36:903-908. [Google Scholar]