Abstract
Posttranslational modification is an efficient mechanism for controlling the activity of structural proteins, gene expression regulators, and enzymes in response to rapidly changing physiological conditions. Here we report in vitro and in vivo evidence that the acuABC operon of the gram-positive soil bacterium Bacillus subtilis encodes a protein acetyltransferase (AcuA) and a protein deacetylase (AcuC), which may control the activity of acetyl-coenzyme A (CoA) synthetase (AMP-forming, AcsA) in this bacterium. Results from in vitro experiments using purified proteins show that AcsA is a substrate for the acetyl-CoA-dependent AcuA acetyltransferase. Mass spectrometry analysis of a tryptic digest of acetylated AcsA (AcsAAc) identified residue Lys549 as the sole modification site in the protein. Unlike sirtuins, the AcuC protein did not require NAD+ as cosubstrate to deacetylate AcsAAc. The function of the putative AcuB protein remains unknown.
Acetyl-coenzyme A (Ac-CoA) synthetase (Acs; EC 6.2.1.1; AMP-forming), a ubiquitous enzyme in nature, is responsible for the reversible conversion of acetate to Ac-CoA. In eukaryotes, Acs is the major source of Ac-CoA (28). In prokaryotes, acetate can be activated to Ac-CoA by Acs (high-affinity pathway) or by the ATP:acetate phosphotransferase (Ack, acetate kinase; EC 2.7.2.1)/Ac-CoA:orthophosphate acetyltransferase (Pta; EC 2.3.1.8) system (low-affinity pathway) (3). In Bacillus subtilis, a gram-positive soil bacterium, acetate catabolism requires AcsA, the product of the acsA gene, while the Ack-Pta pathway operates primarily in acetate excretion (7). A previous report by Grundy and coworkers (8) described in B. subtilis a three-gene operon proximal (161 bp apart), but divergently transcribed from acsA (see Fig. S1 in the supplemental material), whose functions were needed for utilization of acetoin and 2,3-butanediol as a carbon source. This operon was annotated as acuABC (acetoin utilization) to reflect its putative involvement in acetoin catabolism. Whether AcuABC functions were directly or indirectly involved in the use of acetoin as carbon and energy source remained an open question.
Growth analysis showed that acuABC mutant strains of B. subtilis grew poorly on acetoin and butanediol and were partially deficient in sporulation while utilizing these compounds. The observed residual growth suggested the existence of an additional acetoin catabolic pathway, which was subsequently identified by Huang and coworkers to be encoded by the acoABCL operon (12).
Bioinformatics analyses predicted that AcuA was a protein acetyltransferase of the GNAT family (30), AcuB was homologous to proteins with unknown function, and AcuC was homologous to class I histone deacetylases (HDACs) (31). Class I and class II HDACs are an ancient protein family, with class differentiation based on sequence similarity. A group of NAD+-dependent deacetylases related to Saccharomyces cerevisiae Sir2 (sirtuins) are referred to as class III HDACs (6).
The similarity of AcuA and AcuC to protein acetyltransferases and deacetylases suggested the possibility that the acuABC operon might encode a protein acetylation/deacetylation posttranslational modification system in B. subtilis (16).
In Salmonella enterica, Acs activity is modulated by acetylation/deacetylation of a single lysyl residue (Lys609), which is critical to the synthesis of the acetyl-AMP intermediate from acetate and ATP (10, 27). In S. enterica, two enzymes comprise the protein acetylation/deacetylation system that controls Acs activity. The Ac-CoA-dependent protein acetyltransferase (Pat) enzyme acetylates (deactivates) Acs, while the NAD+-dependent CobB sirtuin deacetylase enzyme removes acetyl groups from acetylated Acs (AcsAc), hence reactivating Acs (27, 29). Acetylation and deacetylation of Acs therefore controls acetate metabolism at a posttranslational level.
The effect of mutations in pat or cobB can be assessed in vivo. An S. enterica lacking CobB sirtuin deacetylase activity cannot grow on 10 mM acetate because Pat acetylates (inactivates) Acs. Inactivation of pat in a cobB strain blocks Acs acetylation, restoring growth on 10 mM acetate (29).
We used S. enterica strains to investigate whether the B. subtilis AcuA and AcuC proteins could substitute in vivo for Pat and CobB functions, respectively. We also determined in vitro whether isolated AcuA and AcuC had the predicted enzymatic activities. We show here that the B. subtilis AcuA and AcuC proteins comprise a protein acetylation/deacetylation system that uses AcsA as a substrate. The role of AcuB remains unclear. Unlike sirtuins, AcuC deacetylase activity did not require NAD+ and releases free acetate. We propose that, in B. subtilis, the AcuABC proteins comprise a regulatory system that modulates the activity of the AcsA enzyme as a function of a physiological signal other than NAD+.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
S. enterica serovar Typhimurium LT2 (hereafter referred to as S. enterica) and B. subtilis strains and their genotypes are listed in Table 1. S. enterica strains were grown on no-carbon medium (1), supplemented with MgSO4 (1 mM) and l-methionine (0.5 mM). Acetate was used as the sole carbon and energy source for cells grown in minimal medium. l-(+)-Arabinose (250 μM) or isopropyl-β-d-thiogalactopyranoside (IPTG; 400 μM) was used to induce expression of cloned genes. Luria-Bertani broth (LB) was used as rich medium. B. subtilis strains were grown Spizizen's minimal medium (26), supplemented with 20% casamino acids and 50 mg/ml glutamic acid. Acetate was used as the carbon and energy source for cells grown in minimal medium. Antibiotics were used as needed at the following concentrations: ampicillin, 100 (μg/ml); chloramphenicol, 25 (μg/ml); kanamycin, 50 (μg/ml), neomycin, 5 (μg/ml), spectinomycin, 100 (μg/ml). All chemicals were purchased from Sigma.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmida | Relevant genotype | Source or reference |
|---|---|---|
| B. subtilis strains | ||
| SMY | Prototroph | 6 |
| SMY derivatives | ||
| JE8609 | acuA::neo+ | 6 |
| JE8610 | acuC::spc+ | |
| S. enterica strains | ||
| LT2 | Wild type | Laboratory collection |
| ER2566 | Overexpression strain | New England Biolabs |
| LT2 derivative, TR6583 | metE205 ara-9 | K. Sanderson via J. Roth |
| TR6583 derivatives | ||
| JE4175 | pBAD30 | Laboratory collection |
| JE7246 | Δ1231 (acs) pta::cat+(pBAD30) | Laboratory collection |
| JE7724 | cobB1176::Tn10d(Tc) pta102::MudJ(pBAD30) | |
| JE7725 | cobB1176::Tn10d(Tc) pta102::MudJ(pCOBB8) | |
| JE7738 | Δ1231 (acs) pta::cat+(pACS7) | |
| JE7739 | Δ1231 (acs) pta::cat+(pACSA1) | |
| JE7801 | cobB1176::Tn10d(Tc)(pACUA1) | |
| JE7802 | pat1::Tn10d(Tc)(pACUA1) | |
| JE7803 | cobB1206::MudJ pat1::Tn10d(Tc)(pACUA1) | |
| JE7855 | cobB1176::Tn10d(Tc) pta102::MudJ(pACUA1) | |
| JE7856 | pat1::Tn10d(Tc) pta::cat+(pACUA1) | |
| JE8056 | cobB1176::Tn10d(Tc) pta102::MudJ(pACUC3) | |
| Plasmids | ||
| pBAD30 | Expression vector, ParaBAD bla+ | 11 |
| pACS7 | S. enterica acs+ cloned into pBAD30 | Laboratory collection |
| PACSA1 | B. subtilis acsA+ cloned into pBAD30 | |
| PCOBB8 | S. enterica cobB+ cloned into pBAD30 | Laboratory collection |
| pACUC3 | B. subtilis acuC+ cloned into pBAD30 | |
| pACUA1 | B. subtilis acuA+ cloned into pBAD30 | |
| PACSA2 | B. subtilis acsA+ cloned into pTYB12 bla+ | New England Biolabs |
| pACUA2 | B. subtilis acuA+ cloned into pTYB12 | |
| pACUC4 | B. subtilis acuC+ cloned into pTYB12 | |
| pACUA42 | B. subtilis acuA cloned into pET42a (kan+) | Novagen |
Unless otherwise stated, strains listed were constructed during the course of this work.
Genetic techniques. (i) Phage P22-mediated transduction.
Lysates of the high-transducing mutant HT105/1 int-201 (23, 24) of bacteriophage P22 grown on appropriate strains were used as donors to move chromosomal markers into selected S. enterica strains. Protocols for P22-mediated transductions reported elsewhere were used without modification (5).
(ii) Mobilization of plasmids.
Plasmids were introduced into S. enterica strains by transformation (21) or electroporation (20).
(iii) Construction of the B. subtilis acuC::spc+ strain.
A spectinomycin resistance cassette was introduced into B. subtilis strain SMY by the long-flanking homology PCR technique described previously (32). The spectinomycin cassette came from plasmid pDG1726 (9) by way of the Bacillus Genetic Stock Center (BGSC no. ECE101).
Recombinant DNA techniques. (i) PCR.
All amplifications used TripleMaster polymerase (Eppendorf) and were performed in an Eppendorf Mastercycler gradient PCR thermal cycler (Brinkmann Instruments). Primers were purchased from Integrated DNA Technologies.
(ii) Cloning and overexpression of B. subtilis genes.
Template DNA was from B. subtilis strain 1A775 (prototroph; Bacillus Genetic Stock Center, Ohio State University). Primers for amplification of the B. subtilis acsA, acuA, and acuC genes were designed from database sequence obtained from NCBI. For complementation studies in S. enterica, genes were cloned into vector pBAD30 (11). For protein overproduction, the acuA, acuC, and acsA genes were cloned into the pTYB12 (New England Biolabs) vector for N-terminal chitin purification. The acuA gene was also cloned into the pET42a (Novagen) vector for N-terminal glutathione transferase (GST) affinity chromatography purification.
Biochemical techniques. (i) Overproduction and isolation of proteins.
The B. subtilis acsA, acuA, and acuC genes were overexpressed in E. coli strain ER2566 (17). Strains carrying plasmids were grown overnight at 37°C and diluted 1:200 into 1 liter of LB broth supplemented with ampicillin. Cultures were shaken vigorously at room temperature until they reached an absorbance of 0.5 at 650 nm. Expression of the cloned genes was induced by the addition of IPTG (400 μM), followed by overnight incubation at room temperature with shaking (29). In all cases, cells were harvested by centrifugation and disrupted by passing the cell slurry twice through a French pressure cell (Spectronic Instruments) at 1.26 kPa. Cell debris was removed by centrifugation at 12,000 × g for 45 min at 4°C.
All proteins were purified at 4°C from cell extracts using chitin or GST affinity chromatography. AcsA, AcuA, and AcuC proteins were purified by chitin chromatography (pTYB12 constructs). In addition, GST-AcuA was purified by GST affinity chromatography (pET42a constructs). Purified proteins were dialyzed against 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer (HEPES; 0.05 M; pH 7.5) containing KCl (0.1 M) and glycerol (25%, vol/vol). Proteins were flash-frozen in liquid nitrogen and stored at −80°C until used.
(ii) Protein purity analysis.
Protein purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using Fotodyne's FOTO/Eclipse electronic documentation and analysis system, including software packages FOTO/Analyst PC Image version 5.0 and TotalLab 1D gel analysis, version 2003, from NonLinear Dynamics, Ltd.
Assays. (i) AcuA-dependent acetylation of AcsA.
To determine whether AcsA was a substrate for acetylation by AcuA, 3 μg of AcuA protein (94% homogeneous), and 5 μg of AcsA protein (99% homogeneous) (see Fig. S2 in the supplemental material) were added to a reaction mixture (total volume, 100 μl) containing HEPES buffer (0.05 M; pH 7.5), Tris(2-carboxyethyl)phosphine hydrochloride (TCEP; 200 μM), and [1-14C]acetyl-CoA (20 μM; specific activity, 47 mCi/mmol; Moravek Biochemicals). Reactions were incubated at 37°C for 1 h, and proteins were precipitated by the addition of trichloroacetic acid (TCA) to a final concentration of 5% (wt/vol). Precipitated proteins were incubated on ice for 30 min and were collected by centrifugation at 18,000 × g for 10 min in a Beckman Microfuge 18 centrifuge. The supernatant was discarded, and the precipitated protein was washed twice with 500 μl of ice-cold ethanol. Tubes containing protein were inverted and allowed to air dry overnight. The protein content of the samples was analyzed by SDS-PAGE (14) and Coomassie blue staining (22); radioactivity was monitored using a Packard Cylone phosphorimager.
(ii) AcuC-dependent deacetylation of acetylated AcsAAc.
The acetyltransferase reaction was performed under the conditions listed above, but untagged AcuA was replaced with GST-AcuA to facilitate its removal from the mixture prior to deacetylation. After a 2-h incubation period at 37°C, GST-AcuA protein was removed from the reaction using 20 μl of a 50% (vol/vol) slurry of GST-Mag beads (Novagen) equilibrated with the acetyltransferase reaction buffer (0.05 M HEPES buffer, pH 7.5, containing 200 μM TCEP). Incubation and removal of the magnetic beads were performed per the manufacturer's instructions. After removal of the beads, 5 μg of purified AcuC deacetylase (78% pure) was added to the sample. Samples were incubated at 37°C for 2 h, and the reaction was stopped by precipitation with TCA. Reaction products were analyzed by SDS-PAGE and phosphor imaging analysis.
The products of the AcuC reaction were also analyzed by thin-layer chromatography (TLC) using Whatman Silica Gel 60A with polyester support, developed with a methanol-chloroform (2:3) mobile phase. After development, TLC plates were air dried in a fume hood, and the amount of free acetate generated was calculated as a percentage of total radiolabel initially present in acetylated AcsA protein as determined by phosphor imaging analysis. The relative mobility (Rf) of acetate was 0.66; acetyl-CoA remained at the site of application. Authentic radiolabeled acetyl-CoA and acetate were used as controls.
(iii) Acetyl-CoA synthetase activity.
Conditions for the synthesis of acetyl-CoA from ATP, acetate, and CoA were as previously reported (33), with the following modifications. All reaction components except for potassium acetate were incubated for 3 min at 37°C prior to starting the reaction. Each reaction mixture contained AcsA protein (0.5 μg; 8 pmol), and the assay was started by the addition of potassium acetate (0.2 μmol). The specific activity of AcsA in crude cell extracts was measured using 120 μg of protein.
We measured AcsA activity in crude cell extracts of B. subtilis SMY (acuABC+), acuABC, and acuC strains. For this purpose, a 5-ml sample of an overnight culture was used to inoculate 1 liter of minimal medium supplemented with 50 mM acetate as the carbon and energy source. Cultures were grown at 37°C until they reached an optical density at 650 nm of 0.4 (∼8 h). Cells were harvested by centrifugation at room temperature at 10,543 × g using a Beckman Avanti J-25I centrifuge equipped with a JLA-16,250 rotor. Cells were resuspended in 10 ml of 50 mM HEPES buffer, pH 7.5; lysozyme was added to a final concentration of 1 mg/ml, and the culture was incubated without shaking for 1 h at 37°C. DNase I was added to the broken cell suspension to a final concentration of 0.5 mg/ml and incubated at 37°C without shaking for 10 min. Cell debris was removed by centrifugation as above. Crude cell extracts were filtered through a 0.45-μm-pore-size filter and dialyzed against 50 mM HEPES buffer, pH 7.5, containing 100 mM KCl and 25% (vol/vol) glycerol. Three 1-liter buffer exchanges were applied; total cell protein was determined as described below.
(iv) Protein measurements.
Protein concentration was determined with the Bradford protein assay protocol (2) using a commercially available kit from Bio-Rad Laboratories and bovine serum albumin as standard.
(v) Heat stability of AcuA.
AcuA protein samples (3 μg in 10 μl of 0.05 M HEPES buffer, pH 7.5, containing 0.2 mM TCEP-HCl) were incubated in PCR tubes (200 μl) in a sand-heating temperature block set at 100°C. Tubes were removed after 5, 30, and 60 min of heating, and samples were centrifuged using a Beckman-Coulter Microfuge 18 centrifuge at 18,000 × g at room temperature for one minute. AcuA protein acetyltransferase activity present in the sample supernatant was assayed as described above.
AcsA peptide fingerprinting by quadrupole-time of flight mass spectrometry.
AcsA protein (30 μg) was digested with trypsin, and the resulting peptides were extracted using C18 ZipTips (Varian Inc.) and analyzed by micro-liquid chromatography tandem mass spectrometry (MS/MS) using a Micromass Q-TOF2 hybrid quadrupole/orthogonal time of flight mass spectrometer (Waters Corp) to locate protein acetylation sites. Chromatography of peptides prior to mass spectral analysis was accomplished using C18 reverse-phase high-pressure liquid chromatography (HPLC) columns made in-house from which eluted species are directly micro-electrosprayed. Columns were made using lengths of fused silica tubing (outside diameter, 365 μm; internal diameter, 100 μm) with pulled tips (1-μm orifice) that were packed to 12-cm with Zorbax Eclipse XDB-C18 (Agilent, Palo Alto, CA), 5-μm 300-Å-pore-size media. An Agilent 1100 series HPLC instrument delivered solvents A (0.1% [vol/vol] formic acid in water) and B (95% [vol/vol] acetonitrile, 0.1% [vol/vol] formic acid) at 1 μl/min to load sample or 150 to 200 nl/min to elute peptides over a 180-min 10% (vol/vol) B to 70% (vol/vol) B gradient. Voltage was applied upstream of the column through a platinum wire electrode inserted into the fluid path via a PEEK T-junction. As peptides eluted from the HPLC column/electrospray source, MS/MS spectra were collected from 400 to 2,200 m/z; redundancy was limited by dynamic exclusion. Collision energy profiles were empirically predetermined for different peptide charge states. MS/MS data were converted to pkl file format using Micromass Protein Lynx Global Server, version 2.1.5 (Waters Corp). Resulting pkl files were used to search a nonredundant (nrNCBI) B. subtilis amino acid sequence database using Mascot Search Engine (Matrix Science, London, United Kingdom) with methionine oxidation, glutamic and aspartic acid deamidation, and lysine acetylation as variable modifications. Putative modifications identified by Mascot were confirmed using manual assignments of MS/MS spectra.
RESULTS
B. subtilis genes acsA and acuC complement S. enterica mutants defective in Acs and CobB, respectively.
We used strains of S. enterica lacking acs or cobB functions to investigate whether the acsA and acuC genes of B. subtilis could replace the S. enterica functions during growth on 10 mM acetate. The pta gene was inactivated in all the S. enterica strains to prevent acetate utilization through the low-affinity pathway (3).
AcsA.
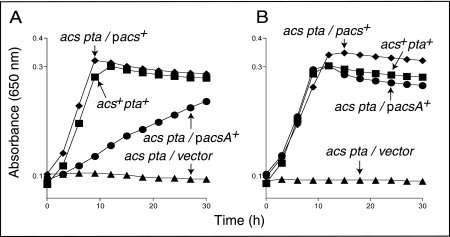
The lack of growth of an S. enterica acs pta strain on acetate (Fig. 1A, triangles) was corrected when the wild-type allele of the S. enterica acs gene was expressed in trans (Fig. 1A, diamonds). Expression of the B. subtilis acsA+ gene also restored growth of the S. enterica acs pta strain on acetate, but the level of enzyme needed to be increased (Fig. 1A versus B, circles). These results indicated that the B. subtilis AcsA enzyme has the function of the S. enterica Acs enzyme.
FIG. 1.
B. subtilis AcsA compensates for the lack of Acs protein during growth of an S. enterica acs mutant on low acetate. Growth kinetics on low acetate (10 mM) in the absence (A) or the presence (B) of l-(+)-arabinose. Strain JE4175 (acs+), squares; strain JE7246 (acs pta/empty cloning vector), triangles; strain JE7738 (acs pta/pACS7 ParaBAD-acs+), inverted triangles; strain JE7739 (acs pta/pACSA1 ParaBAD-acsA), circles. On the graph, plasmid pacs+ is pACS7 and pacsA+ is pACSA1.
AcuC.
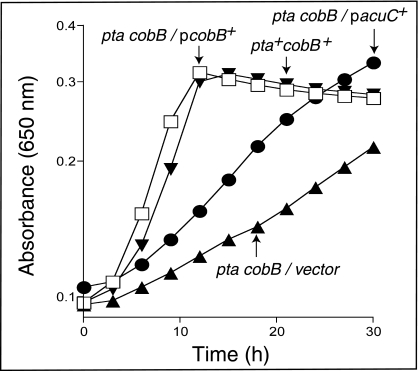
An S. enterica cobB mutant, which cannot deacetylate AcsAc (27), exhibited a partial defect in growth on acetate (Fig. 2, triangles). Introduction of plasmid pCOBB8, in which the S. enterica cobB gene is expressed under control of the ParaBAD promoter, restored growth on acetate in the presence or absence of arabinose (Fig. 2, squares). Expression of the B. subtilis acuC+ gene partially restored growth of the S. enterica cobB mutant, but in this case addition of arabinose did not improve the efficiency of complementation (Fig. 2, circles).
FIG. 2.
AcuC partially compensates for the lack of CobB sirtuin deacetylase during growth of an S. enterica cobB mutant strain on low acetate. (A) Growth kinetics on 10 mM acetate without l-(+)-arabinose. (B) Growth kinetics on low acetate (10 mM) with l-(+)-arabinose (250 μM). Strain JE4175 (acs+), diamonds; strain JE7724 (cobB pta/empty cloning vector), triangles; strain JE7725 (cobB pta/pCOBBB8 ParaBAD-cobB+), squares; strain JE8056 (cobB pta/pACUC3 ParaBAD− acuC+), circles. On the graph, plasmid pcobB+ is pCOBB8 and pacuC+ is pACUC3.
High levels of AcuA acetyltransferase are toxic to S. enterica.
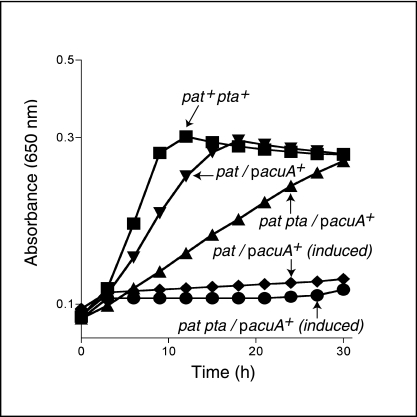
High levels of expression of the B. subtilis acuA+ gene arrested growth of the S. enterica pat strain on acetate (Fig. 3, inverted triangles versus diamonds). A simple explanation for this result is that excess AcuA results in Acs protein that is largely acetylated (inactivate), hence blocking the synthesis of Ac-CoA.
FIG. 3.
Deleterious effect of high levels of AcuA protein. Growth kinetics on acetate (10 mM). When added as inducer, l-(+)-arabinose was present in the medium at 250 μM. The growth behavior of the wild-type strain did not change with or without inducer. The growth behavior of negative control strains containing empty vector was like that of the uninduced strains (data not shown). Strain JE4175 (WT), squares; strain JE7802 (pat/pACUA1), triangles; strain JE7856 (pat pta/pACUA1), circles. On the graph, plasmid pacuA+ is pACUA1 (B. subtilis acuA+).
Also shown in Fig. 3 are results from experiments with strains lacking phosphotransacetylase (Pta) function. We learned from these experiments that the Pta enzyme of S. enterica makes an important contribution to the rate of growth of the pat pta+/pacuA+ strain (Fig. 3, triangles versus inverted triangles). This was an unexpected result given that the pat pta+/pacuA+ strain was grown on 10 mM acetate, a concentration below the one needed for Pta function (≥25 mM). This result suggested that even though AcuA levels were not high (no inducer added to the medium), the pool of Acs protein may be substantially acetylated, and the cell may respond by making more Pta enzyme. It would be of interest to determine the level of pta transcription in the pat pta+/pacuA+ strain.
Activity of AcsA in B. subtilis CE of wild-type and mutant strains.
To address the challenge of the inability to see a phenotype of acsA and acuABC mutants in liquid medium (6), we performed specific activity assays on wild-type and acuABC and acuC mutant strains. The wild-type cell extracts (CE) had no activity above background. The specific activity of AcsA in the acuABC mutant CE was 0.021 ± 0.002 μmol of AMP released min−1 mg of protein−1. No AcsA activity was detected in acuC mutant CE.
AcuA uses acetyl-CoA to acetylate AcsA.
Given the precedent in S. enterica for the control of Acs activity by acetylation, we hypothesized that in B. subtilis the AcuABC functions comprised a protein acetylation/deacetylation system. To determine whether AcuA had acetyltransferase activity we isolated the protein and performed acetyltransferase activity assays as described (29).
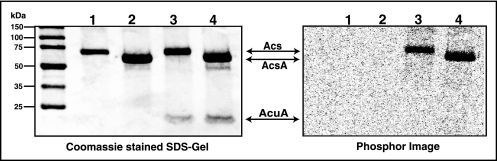
The B. subtilis AcuA protein and its putative AcsA substrate were purified using chitin or GST affinity chromatography. As shown in Fig. 4 (lanes 3 and 4), AcuA acetylated S. enterica Acs and B. subtilis AcsA proteins. The amount of acetyl-CoA used in the assay, 20 μM, is in the physiologically relevant range (4, 34).
FIG. 4.
AcuA has protein acetyltransferase activity. (Left) Coomassie blue-stained SDS-polyacrylamide gel. Lane 1, S. enterica Acs; lane 2, B. subtilis AcsA; lane 3, S. enterica Acs plus AcuA; lane 4, B. subtilis AcsA plus AcuA. (Right) Phosphor image of the left panel. Initial amounts of protein used in all cases: Acs, 3 μg; AcuA, 3 μg. Other components of the AcuA reaction mixture are described in Materials and Methods. Molecular markers are shown at left.
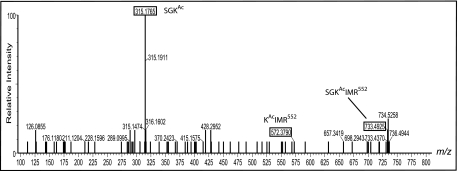
Matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis showed that B. subtilis AcsA was acetylated only once on residue lysine 549 (Lys549). Peptide fingerprinting identified the diagnostic peptide SGKIMR552 with an m/z 690.5 ion in the unacetylated state and an m/z 733.5 ion for the acetylated peptide; a difference of 42 atomic mass units was consistent with the presence of one acetyl group (Fig. 5) (27).
FIG. 5.
Site of acetylation in AcsA. B. subtilis AcsA protein peptide fingerprint showing 315.18 SGKAc, 572.38 KAcIMRR552, and 733.49 SGKAcIMR552 ions. Masses represent at least 65% protein coverage.
The activity of the AcuA protein was remarkably stable in respect to heat, considering that B. subtilis is a mesophilic bacterium. AcuA retained 93, 78, and 15% of its protein acetyltransferase activity after the protein was heated to 100°C for 5, 30, and 60 min, respectively (Fig. 6).
FIG. 6.
Heat stability of the AcuA acetyltransferase activity. The acetyltransferase reaction was performed as described using AcuA protein heated to 100°C for various lengths of time prior to adding it to the reaction. (Left) SDS-polyacrylamide gel stained with Coomassie blue. Lane 1, AcsA; lane 2, AcsA plus AcuA; lane 3, AscA plus AcuA boiled for 5 min; lane 4, AcsA plus AcuA boiled for 30 min; lane 5, AcsA plus AcuA boiled for 60 min. (Right) Phosphor image of the left panel. Initial amounts of protein used in all cases: Acs, 3 μg; AcuA, 3 μg.
Inactivation and reactivation of AcsA by AcuA and AcuC.
Results from in vivo and in vitro experiments predicted that acetylation of AcsA would inactivate the enzyme. We tested this idea in vitro. While unacetylated AcsA enzyme had a specific activity of 3.3 ± 0.41 μmol of AMP released min−1 mg of protein−1, AcsAAc had no detectable activity under the same assay conditions. This result was similar to results obtained with the S. enterica enzyme (29). The specific activity of AcsAAc after incubation with AcuC was 2.8 ± 0.16 μmol of AMP released min−1 mg of protein−1, which corresponded to 84% of the activity of unmodified AcsA.
B. subtilis AcuC protein has NAD+-independent protein deacetylase activity.
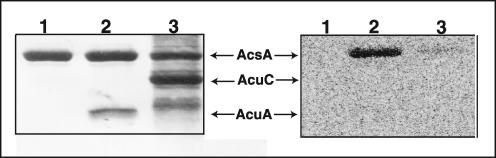
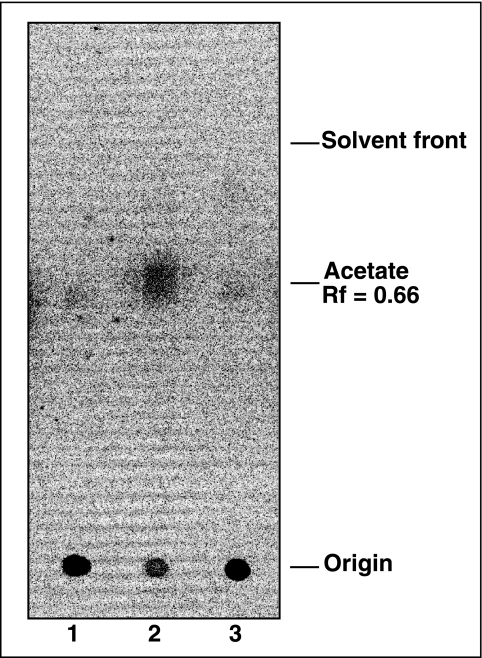
Results from complementation studies shown in Fig. 2 prompted us to determine whether the AcuC protein had protein deacetylase activity in vitro. AcsAAc protein was generated by incubation with AcuA and radiolabeled [1-14C]Ac-CoA. After removal of the AcuA acetyltransferase, AcuC removed approximately 55% of the radioactive acetyl moiety from AcsAAc after a 2-h incubation period at 37°C (Fig. 7). AcuC-dependent deacetylation of AcsAAc removed 80% of the label in the absence of any cofactor, releasing free acetate (Fig. 8, lane 2). The stoichiometry of acetate released to AcsAAc was 1:1, a result that was consistent with mass spectrometry data indicating the modification of a single residue of AcsA. Results presented in Fig. 8 show that AcuC does not use NAD+ as a substrate, as sirtuins do, including S. enterica CobB (13, 15, 25).
FIG. 7.
Protein deacetylase activity of AcuC. The left panel is a Coomassie blue-stained SDS-polyacrylamide gel. Lane 1, AcsA only; lane 2, AcsA and AcuA-GST; lane 3, Acs and AcuC. The right panel shows the phosphor image of the left panel. Protein amounts for reactions are as follows: Acs, 3 μg; AcuA-GST, 3 μg; and AcuC, 4 μg. Other conditions for the assay were as described in Materials and Methods.
FIG. 8.
Free acetate is the product of the AcuC deacetylation reaction. The acetylation reaction was performed as described. After 2 h proteins were salted out with 5.25 M ammonium sulfate (final concentration) and incubated at room temperature for 10 min. Proteins were removed by centrifugation for 10 min at 13,000 × g. Supernatants were removed and mixed with 100 μl of 50 mM HEPES (pH 7.5) containing 200 μM TCEP. Reaction mixtures were incubated at 37°C for 2 h and filtered through a 0.45-micron-pore-size Spin-X filter, and 20 ml of each mixture was spotted on a silica gel TLC that was developed with a chloroform:methanol (3:2) mobile phase. Lane 1, 14C-labeled AcsAAc, with no AcuC added; lane 2, 14C-labeled AcsAAc plus 5 μg of AcuC added; lane 3, 14C-labeled AcsAAc plus heat-inactivated AcuC (10 min at 100°C). The amount of free acetate was the percentage of total radiolabel initially associated with 14C-labeled AcsAAc protein.
Lack of Acu functions does not impair growth of B. subtilis on acetate.
We assessed the effect on the absence of Acu functions on growth of B. subtilis on 50 mM acetate. The doubling time of a culture of the wild-type SMY strain was 2 h, and the cell density of cultures of strains JE8609 (acuA::neo+) and JE8610 (acuC::spc+) doubled after 3 and 2.1 h, respectively. It appears that only when all Acu functions are absent a subtle but reproducible effect on acetate growth is observed. Further work is needed to understand the extent of the involvement of Acu functions in acetate catabolism in B. subtilis.
DISCUSSION
We have identified biochemical activities associated with two proteins encoded by the acuABC operon of B. subtilis. The newly identified activities are not directly involved in acetoin catabolism, supporting the conclusions reached by Huang et al. (12). The data reported here indicate that the acuABC operon of B. subtilis encodes enzymes that may comprise a posttranslational modification system previously unknown to exist in this bacterium. Our in vitro data show that the AcuA and AcuC activities modulate the activity of the acetyl-CoA synthetase (AcsA) enzyme as a function of its acetylation state without the involvement of NAD+. The in vitro and in vivo data presented here are consistent with the hypothesis that the AcuABC functions may control AcsA activity in vivo in B. subtilis.
Differences between Ac-CoA synthetase posttranslational control in B. subtilis and S. enterica.
There are important differences between the systems used by S. enterica and B. subtilis to acetylate/deacetylate Acs. First, the acetyltransferases are considerably different in size. The B. subtilis AcuA enzyme is 676 amino acids shorter than the S. enterica Pat enzyme. The fact that AcuA is an effective acetyltransferase leaves open the question of the function of the role of the large N-terminal domain of the S. enterica Pat enzyme. In addition, the S. enterica pat gene is transcribed alone, while acuA is the first of a three-gene operon. Secondly, unlike the S. enterica CobB sirtuin deacetylase, the B. subtilis AcuC deacetylase enzyme does not consume NAD+ and releases acetate as its by-product. Interestingly, the B. subtilis genome contains a gene that encodes a functional homolog of CobB (i.e., a sirtuin-like deacetylase). The activity of the B. subtilis CobB homolog is also NAD+ dependent and deacetylates AcsAAc protein in vitro and in vivo (J. Gardner and J. Escalante-Semerena, unpublished results). These results suggest that there maybe multiple systems controlling AcsA activity in B. subtilis. The possibility for interplay between the sirtuin-dependent and the sirtuin-independent systems for the control of AcsA or other proteins in B. subtilis may reflect on the complexity of the regulation these activities as a function of specific, as-yet-unidentified physiological conditions.
The function of the acuB gene has not been identified. It is noteworthy, however, that Noirot and coworkers identified AcuB as part of the protein interaction network involved in DNA replication in B. subtilis (18). Results from two-hybrid screens suggested that AcuB might interact with YabA, a protein that may be involved in DNA replication initiation control (19). The relevance of this interaction is unclear. We performed acetylation/deacetylation experiments using purified AcuB protein with either AcuA or AcuC; however, AcuB did not affect the ability of either protein to perform its function (data not shown). We cannot rule out, however, that AcuB may bind a signal molecule before eliciting an effect on either AcuA or AcuC. Other scenarios where AcuB serves as a coupling protein to allow AcuA or AcuC to interact with their targets are possible and should be explored.
Protein acetyltransferase activity may be regulated in vivo.
It is likely that in the wild-type strain of B. subtilis, the protein acetyltransferase enzyme is carefully maintained at a level that allows sufficient AcsA to remain unacetylated. This hypothesis is supported by the pronounced negative effect that high levels of acuA expression had on growth of S. enterica on low acetate. As mentioned in the Results section, one explanation for such an effect would be that excess AcuA results in AcsA protein that is largely acetylated (inactivate), effectively blocking the synthesis of Ac-CoA. The idea of tight regulation of AcuABC levels already exists at the transcriptional level, where acuABC expression is under control of the global regulatory protein CcpA (7).
The fact that CcpA controls acsA and acuABC expression (24) reflects on the importance of modulating AcsA activity both at the transcriptional and posttranslational level to maintain a pool of free CoA that can satisfy the requirement of other processes for this important coenzyme.
What is the connection between the AcuABC system and acetoin catabolism?
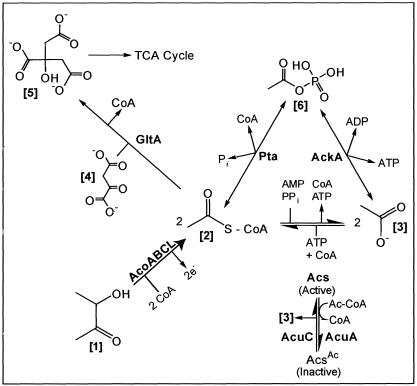
The work by Huang and coworkers clearly showed that AcuABC is not directly involved in acetoin catabolism (12). On the basis of our work, we speculate that in wild-type B. subtilis, AcuA acetyltransferase and AcuC deacetylase activities are needed to maintain a balance between free CoA and Ac-CoA during acetoin catabolism (Fig. 9).
FIG. 9.
Hypothetical role of AcuA and AcuC enzymes on acetoin catabolism. AcoABC, acetoin dehydrogenase enzyme system; AcoA, a subunit of the TPP-dependent acetoin dehydrogenase; AcoB, b subunit of the TPP-dependent acetoin dehydrogenase; AcoC, dihydrolipoamide dehydrogenase; AcuA, protein acetyltransferase; AcuC, protein deacetylase; GltA, citrate synthase. The bracketed numbers are as follows: 1, acetoin; 2, acetyl-CoA; 3, acetate; 4, oxaloacetate; 5, citrate. 2e−, two reducing equivalents.
The absence of the glyoxylate bypass in B. subtilis renders acetate and acetoin poor sources of carbon and energy when fed to this bacterium without a cosubstrate (e.g., glutamate) for the TCA cycle. We can use the scheme shown in Fig. 9 to analyze the behavior of an acuA strain during growth on acetoin. Inactivation of acuA would block AcsA acetylation (i.e., AcsA would remain active), resulting in an uncontrolled build up of Ac-CoA and depletion of the pool of free CoA. This imbalance in the CoA pool could be corrected by the hydrolysis of the acetyl moiety from Ac-CoA, recycling free CoA and probably excreting acetate into the medium. Under these conditions, we would expect only limited amounts (if any) of the exogenous acetate to be recaptured, since the cell would be starved for ATP.
Dealing with poor carbon and energy sources while controlling the pool of CoA.
The proposed acetylation/deacetylation system in B. subtilis suggests that gram-positive and gram-negative bacteria alike control the activity of the CoA-consuming Acs enzyme. This control may reflect changes in the pool of CoA and the need to maintain a balance between acylated and free coenzyme. Also keeping AcsA and other proteins or enzymes in the cell long after they have been made and controlling them by posttranslational modifications are ways for a cell to conserve protein synthesis resources when they are scarce. The work reported here hypothesizes that in B. subtilis the putative posttranslational modification AcuABC system may be responsive to as-yet-unidentified metabolic signals.
Supplementary Material
Acknowledgments
This research was supported by grant R01 DE10510 (NIDCR), and M.E.D. is the recipient of the Hein Fellowship at Forsyth.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Berkowitz, D., J. M. Hushon, H. J. Whitfield, J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-255. [DOI] [PubMed] [Google Scholar]
- 3.Brown, T. D., M. C. Jones-Mortimer, and H. L. Kornberg. 1977. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J. Gen. Microbiol. 102:327-336. [DOI] [PubMed] [Google Scholar]
- 4.Chohnan, S., H. Furukawa, T. Fujio, H. Nishihara, and Y. Takamura. 1997. Changes in the size and composition of intracellular pools of nonesterified coenzyme A and coenzyme A thioesters in aerobic and facultatively anaerobic bacteria. Appl. Environ. Microbiol. 63:553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis, R. W., D. Botstein, and J. R. Roth. 1980. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 6.Gregoretti, I. V., Y. M. Lee, and H. V. Goodson. 2004. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338:17-31. [DOI] [PubMed] [Google Scholar]
- 7.Grundy, F. J., A. J. Turinsky, and T. M. Henkin. 1994. Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA. J. Bacteriol. 176:4527-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundy, F. J., D. A. Waters, T. Y. Takova, and T. M. Henkin. 1993. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol. Microbiol. 10:259-271. [DOI] [PubMed] [Google Scholar]
- 9.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 10.Gulick, A. M., V. J. Starai, A. R. Horswill, K. M. Homick, and J. C. Escalante-Semerena. 2003. The 1.75Å crystal structure of acetyl-CoA synthetase bound to adenosine-5′-propylphosphate and coenzyme A. Biochemistry 42:2866-2873. [DOI] [PubMed] [Google Scholar]
- 11.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, M., F. B. Oppermann-Sanio, and A. Steinbuchel. 1999. Biochemical and molecular characterization of the Bacillus subtilis acetoin catabolic pathway. J. Bacteriol. 181:3837-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage and structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, and R. Sternglanz. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leipe, D. D., and D. Landsman. 1997. Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res. 25:3693-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 18.Noirot-Gros, M. F., E. Dervyn, L. J. Wu, P. Mervelet, J. Errington, S. D. Ehrlich, and P. Noirot. 2002. An expanded view of bacterial DNA replication. Proc. Natl. Acad. Sci. USA 99:8342-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noirot-Gros, M. F., M. Velten, M. Yoshimura, S. McGovern, T. Morimoto, S. D. Ehrlich, N. Ogasawara, P. Polard, and P. Noirot. 2006. Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 103:2368-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Toole, G. A., J. R. Trzebiatowski, and J. C. Escalante-Semerena. 1994. The cobC gene of Salmonella typhimurium codes for a novel phosphatase involved in the assembly of the nucleotide loop of cobalamin. J. Biol. Chem. 269:26503-26511. [PubMed] [Google Scholar]
- 21.Ryu, J.-I., and R. J. Hartin. 1990. Quick transformation of Salmonella typhimurium LT2. BioTechniques 8:43-45. [PubMed] [Google Scholar]
- 22.Sasse, J. 1991. Detection of proteins, p. 10.6.1-10.6.8. In F. A. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. Wiley Interscience, New York, N.Y. [Google Scholar]
- 23.Schmieger, H. 1971. A method for detection of phage mutants with altered transduction ability. Mol. Gen. Genet. 100:378-381. [DOI] [PubMed] [Google Scholar]
- 24.Schmieger, H., and H. Bakhaus. 1973. The origin of DNA in transducing particles of P22 mutants with increased transduction frequencies (HT-mutants). Mol. Gen. Genet. 120:181-190. [DOI] [PubMed] [Google Scholar]
- 25.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starai, V. J., I. Celic, R. N. Cole, J. D. Boeke, and J. C. Escalante-Semerena. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298:2390-2392. [DOI] [PubMed] [Google Scholar]
- 28.Starai, V. J., and J. C. Escalante-Semerena. 2004. Acetyl-coenzyme A synthetase (AMP forming). Cell Mol. Life Sci. 61:2020-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Starai, V. J., and J. C. Escalante-Semerena. 2004. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340:1005-1012. [DOI] [PubMed] [Google Scholar]
- 30.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiagalingam, S., K. H. Cheng, H. J. Lee, N. Mineva, A. Thiagalingam, and J. F. Ponte. 2003. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann. N. Y. Acad. Sci. 983:84-100. [DOI] [PubMed] [Google Scholar]
- 32.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 33.Williamson, J. R., and B. E. Corkey. 1969. Assays of intermediates of the citric acid cycle and related compounds by fluorometric enzyme methods, p. 494-497. In J. M. Lowenstein (ed.), Methods in enzymology, vol. XIII. Academic Press, New York, N.Y. [Google Scholar]
- 34.Wolfe, A. J. 2005. The acetate switch. Microbiol. Mol. Biol Rev. 69:12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.