Abstract
The gastric pathogen Helicobacter pylori must adapt to fluctuating conditions in the harsh environment of the human stomach with the use of a minimal number of transcriptional regulators. We investigated whether H. pylori utilizes the stringent response, involving signaling through the alarmone (p)ppGpp, as a survival strategy during environmental stresses. We show that the H. pylori homologue of the bifunctional (p)ppGpp synthetase and hydrolase SpoT is responsible for all cellular (p)ppGpp production in response to starvation conditions. Furthermore, the H. pylori spoT gene complements the growth defect of Escherichia coli mutants lacking (p)ppGpp. An H. pylori spoT deletion mutant is impaired for stationary-phase survival and undergoes a premature transformation to a coccoid morphology. In addition, the spoT deletion mutant is unable to survive specific environmental stresses, including aerobic shock and acid exposure, which are likely to be encountered by this bacterium during infection and transmission.
Helicobacter pylori is an ɛ-proteobacterium that infects over half the world's population and is associated with a spectrum of gastric diseases (5). This bacterium, for which no environmental reservoir has been demonstrated, inhabits the harsh environment of human and primate stomachs. Within the environment of the stomach, as well as during the as-yet-ill-defined transmission process, H. pylori must endure rapid fluctuations in pH, oxygen tension, and chemical insults from the host immune system. The regulatory mechanisms of H. pylori's survival strategies in the onslaught of these environmental stresses are of interest, in part because the bacterium's small genome encodes few transcriptional regulators, including just three sigma factors (29). Insight into H. pylori's stress response programs will abet the design of better antibiotics to treat infections, since resistance to traditional therapies is increasing among H. pylori clinical isolates (19).
A well-characterized strategy used by Escherichia coli to survive stresses, such as starvation, is termed the stringent response (10). The stringent response involves the global regulation of transcription characterized by the repression of ribosomal genes and derepression of specific stress response genes (16). The molecular signal or “alarmone” for this response is the small hyperphosphorylated guanosine nucleotide ppGpp that binds directly to RNA polymerase and alters its promoter specificity and transcription rate (17). In E. coli, ppGpp and its precursor pppGpp are produced by the ribosome-associated synthetase, RelA, when it senses the presence of uncharged tRNAs. In addition, a bifunctional protein, SpoT, encodes both (p)ppGpp synthetase and hydrolase activities and is crucial for maintaining appropriate levels of (p)ppGpp in the cell. Mutants lacking both of these enzymes produce no detectable (p)ppGpp and exhibit a range of phenotypes, including an inability to survive in minimal media and during stationary phase.
Many other bacteria possess a single bifunctional (p)ppGpp synthetase and hydrolase (23). The distribution of these single bifunctional enzymes was previously thought to be restricted to gram-positive bacteria, but recent work has demonstrated that multiple gram-negative bacteria, particularly those of the α- and ɛ-proteobacterial families, also harbor single bifunctional synthetase/hydrolase enzymes (14, 18, 35). The ability to regulate transcription through production of the small signaling molecule (p)ppGpp is emerging as an important trait for multiple pathogens to survive environments specific to the infection and transmission processes (7). For example, a spoT mutant of the related ɛ-proteobacterium Campylobacter jejuni survives poorly in a low-CO2/high-O2 environment and is impaired in adhesion, invasion, and intracellular survival in cultured epithelial cells (14).
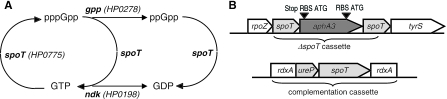
H. pylori has been reported to lack a stringent response (30); however, recent experiments have contradicted this claim and demonstrated that H. pylori produces (p)ppGpp under starvation and low-pH conditions (34). The H. pylori genome contains a homologue of the C. jejuni spoT gene, HP0775 or JHP0712 in the 26695 and J99 genomes, respectively (1, 32). Other H. pylori genes predicted to be involved in (p)ppGpp metabolism include gppA (HP0278, JHP0263), a guanosine-5-triphosphate-3-diphosphate pyrophosphatase, and ndk (HP0198, JHP0184), a nucleoside diphosphate kinase (Fig. 1A).
FIG. 1.
(A) Putative (p)ppGpp metabolic pathway of H. pylori according to Cashel et al. (10). (B) Constructs used for the construction of the ΔspoT and spoT* strains.
Here we report that the H. pylori spoT homologue encodes a functional (p)ppGpp synthetase that is responsible for (p)ppGpp production in H. pylori and can complement an E. coli mutant lacking relA and spoT. We further demonstrate that SpoT function is required for H. pylori to survive stationary phase, aerobiosis, and acid shock, all conditions that the bacterium is likely to experience during infection and transmission.
MATERIALS AND METHODS
Strains and growth conditions.
H. pylori strain G27 (11) was used in these studies. E. coli wild-type MG1655 (CF1648) and isogenic mutant ΔrelA (CF1652) and ΔrelA ΔspoT (CF1693) strains (37) were obtained from Michael Cashel (National Institutes of Health). H. pylori was grown on blood agar (BA) plates consisting of Columbia agar (Difco) supplemented with 5% defibrinated horse blood (Hemostat), 0.02 mg of β-cyclodextrin (Sigma) ml−1, 20 μg of vancomycin (Sigma) ml−1, and 8 μg of amphotericin B (Sigma) ml−1. Selective plates were made with 10 μg of kanamycin (Fisher) ml−1 or 0.08 M sucrose. BA plates were grown in incubators at 37°C and 10% CO2. H. pylori cultures were passaged to fresh BA plates every 3 to 4 days and restarted from frozen stocks after three passages. Liquid media for H. pylori consisted of filter-sterilized brucella broth (Difco) supplemented with 10% fetal bovine serum (Gibco) and 20 μg of vancomycin ml−1. Liquid cultures were grown in 50-ml conical tubes with loosened lids (BD Falcon) and shaking at 37°C in anaerobic jars (Oxoid) with CampyGen microaerobic sachets (Oxoid).
Construction of the ΔspoT and spoT* strains.
The putative H. pylori spoT, HP0775, was PCR amplified from strain G27 genomic DNA using the primers SpoTXba1F forward (5′-GCTCTAGAGCTGAAGGGAAAATTGATATAGAC-3′) and SpoTKpn1R reverse (5′-GCGGTACCCAATCCGCCTTATCTGTGG-3′). The HP0775 PCR product and pBSSK+ cloning vector (Stratagene) were then digested and ligated to generate pK220-1. The primers SpoTEcoRI (5′-CGAATTCCAGGGCGTTTTCAATTTTG-3′) and SpoTBamHI (5′-GGGGATCCCCGTCGGTTTTAGCGGGTTTAT-3′) were then used to PCR amplify outward from HP0775 and around pK220-1. The pK220-1 PCR product and pUC18 K-2, which contains a kanamycin-resistant nonpolar cassette with two internal ribosomal binding sites (20), were digested and ligated to generate pK220-2. pK220-2 was constructed so that the downstream ribosomal binding site is in frame with the downstream HP0775 fragment. The HP0775 chromosomal locus was disrupted via homologous recombination by transforming strain G27 with purified pK220-2. The resulting ΔspoT strain was then verified by PCR and sequencing of the genomic locus.
The selective kan sacB cassette was cloned into the vector pRdxA (31) and transformed into wild-type H. pylori to generate a rdxA::kan sacB strain with a kanamycin resistance (Kanr) and sucrose sensitivity (Sucs) phenotype. The primers SpoTXba1F forward and SpoTKpn1R reverse were modified to contain BamHI and EcoRI restriction sites, respectively, and HP0775 was PCR amplified and cloned into pRdxA::ureP::gfp (obtained from Nina Salama, Fred Hutchison Cancer Research Center) to generate the construct pRdxA::ureP::HP0775 in which HP0775 expression is regulated by promoter sequences encoded in the 750-bp region directly upstream of the H. pylori ureA (HP0073) gene. This construct was then used to transform the rdxA::kan sacB strain, and replace the kan sacB cassette with ureP::HP0775 via homologous recombination. The appropriate rdxA::ureP::HP0775 strain was selected and screened based on its Sucr and Kans phenotype. The rdxA::ureP::HP0775 strain was then transformed with pK220-2 to disrupt the native HP0775 allele via homologous recombination. The genotype of the complemented spoT* (ΔspoT rdxA::ureP::HP0775) transformants was verified by PCR and sequencing of the genomic loci.
(p)ppGpp assays.
The production of (p)ppGpp in response to minimal media was assayed as previously described (14, 35). H. pylori strains were grown overnight in normal growth media to early exponential phase (optical density at 600 nm [OD600] of ∼0.4), diluted back to an OD600 of 0.2, and incubated in normal growth media for an additional 2 h, at which point all strains had reached an OD600 of ∼0.3. For the nutrient deprivation treatment 0.25 OD600 equivalents of each culture were removed, pelleted by centrifugation at 10,000 rpm for 5 min, and washed once with minimal medium consisting of 50 mM MOPS (morpholinepropanesulfonic acid; pH 7.4), 1 mM MgSO4, 0.25 mM CaCl2, 19 mM glutamic acid, and 0.004 mM biotin. Samples were resuspended in 250 μl of minimal medium, 32P (Perkin Elmer) was added at 100 μCi ml−1, and cultures were labeled for 1 h at 37°C. After labeling, 50-μl samples were removed and added to an equal volume of 2 M formic acid and placed on ice for at least 15 min. Samples were spun for 5 min at 16,000 × g in a microcentrifuge, and 3 μl of the supernatant was spotted directly onto polyethyleneimine (PEI) cellulose thin-layer chromatography plates (Sigma), dried, and developed in 1.5 M KH2PO4 for ∼2.5 h. Nucleotides were visualized by autoradiography.
E. coli functional complementation assay.
E. coli relA and spoT were amplified from wild-type MG1655 genomic DNA using the primers RelABglIIF.2 forward (5′-GGAAGATCTGCTGGATATGTTCCCACACACG-3′), RelAXhoIR.2 reverse (5′-ATTCCGCTCGAGCCCTTTCCTCAAACCGCTAT-3′), SpoTBglIIF.2 forward (5′-GGAAGATCTAGGAAGCCGCTGAATTACAA-3′), and SpoTXhoIR.2 reverse (5′-ATTCCGCTCGAGATGAGGTTTGTGGACCTGCT-3′). H. pylori HP0775 was amplified from wild-type G27 genomic DNA with spoTBglIIF Forward (5′-GGAAGATCTCCGCTGAAGGGAAAATTGATATAGAC-3′) and spoTKpn1R reverse. The PCR products were cloned into the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible pQE-30 expression vector (QIAGEN) in frame and transformed into the various E. coli strains. Transformants were maintained on standard LB agar media supplemented with 50 μg of ampicillin ml−1. For the functional complementation assays, overnight cultures from LB plates were streaked onto M9 minimal medium plates supplemented with 50 μg of ampicillin ml−1, 100 μg ml−1 of each of the amino acids serine, methionine, and glycine (12), and ∼140 μg of IPTG ml−1. The plates were incubated overnight at 37°C and then photographed.
Growth curves and stress assays.
Prior to all experiments, H. pylori cultures from BA plates were used to inoculate 2 ml of liquid medium and incubated overnight as described above. To assay viability at each time point, 100-μl samples of culture were serially diluted in 1× phosphate-buffered saline, and various dilutions were plated in duplicate on BA plates. Plates were incubated for 3 to 4 days, and then the CFU were counted. Each assay was replicated at least three times. For the growth curves, the overnight wild-type, ΔspoT, and spoT* cultures were used to prepare 2-ml cultures with an OD600 of 0.05. The cultures were grown in the same anaerobic jar shaking horizontally at 150 rpm. The viability of the cultures was assayed at 0, 24, and 50 h. For the aerobic stress experiments, the overnight wild-type, ΔspoT, and spoT* cultures were used to prepare 2-ml cultures of 0.5 OD600. The cultures were incubated in lidless anaerobic jars at 37°C with shaking at 150 rpm under normal atmospheric conditions. The viability of the cultures was assayed every 2 h over a 6-h time course. For the pH stress experiments, pH 4 acidic medium supplemented with 0.5 mM urea, which promotes H. pylori survival in acid (27), was prepared by adding 4.8 M HCl to standard growth media and filter sterilizing. The addition of acid to the growth media caused a precipitate to form, which was lost during the filtering process. To determine whether the components of the precipitate were critical for H. pylori survival, we adjusted the filtered acidic medium to pH 7 and assayed the viability of our strains. Loss of the precipitate components did not impair growth of the cultures during the course of these experiments (data not shown). The acidity of the media was monitored at each time point sampled and found to be unchanged during the course of the experiment. To ensure that the sensitivity of the ΔspoT mutant to acid shock was not due to impaired urease activity, urease activity assays were performed on the wild-type, ΔspoT, and spoT* strains by growing them in Difco urea broth. All three strains exhibited the same amount of urease activity.
Transmission electron microscopy.
Wild-type, ΔspoT, and spoT* bacteria were grown for 48 h in liquid media from a starting OD600 of 0.05. Samples (10 μl) of each strain were obtained at 12, 24, and 48 h and applied to copper grids (400 mesh) with Butvar support film coated with carbon, rinsed twice, and negatively stained for 1 min with 1% phosphotungstate (pH 7). Grids were visualized and photographed in a Philips CM12 TEM at an acceleration voltage of 80 kV. Bacterial counts were conducted on three separate grids for each genotype at each time point with a total of more than 100 cells per genotype analyzed.
RESULTS
Construction of an HP0775 disruption mutation in H. pylori.
The H. pylori open reading frame (ORF) HP0775 is annotated as spoT and is 41% identical to the C. jejuni spoT gene. It is located in a putative operon with gene content similar to that of the C. jejuni gene, upstream of several genes that are likely to encode essential functions based on sequence and transposon mutagenesis studies (28). We engineered a nonpolar disruption of HP0775 by replacing nucleotides corresponding to amino acids 57 to 704 with a cassette encoding Kanr flanked by ribosome-binding sites, lacking a transcriptional termination region, and containing a start codon in frame with the 3′ end of the HP0775 ORF (Fig. 1B) (20). To verify that phenotypes observed with the HP0775::Kanr strain (hereafter referred to as ΔspoT) were due to loss of the HP0775 gene product, we engineered a complemented strain (hereafter referred to as spoT*) in which the HP0755 ORF was introduced into the rdxA locus of the ΔrdxA::kan sacB strain by selecting and screening for a Sucr and Kans phenotype. Because HP0755 lies in a large putative operon, the promoter sequence of which is not readily apparent, we expressed the copy of HP0755 inserted in the rdx gene with the urease promoter, which we had shown to be effective for transgene expression at this locus (6). The resultant ΔrdxA::ureP::HP0775 strain was transformed with the HP0775::Kanr construct to yield the spoT* strain.
The HP0775-deficient mutant fails to produce (p)ppGpp under nutrient deprivation conditions.
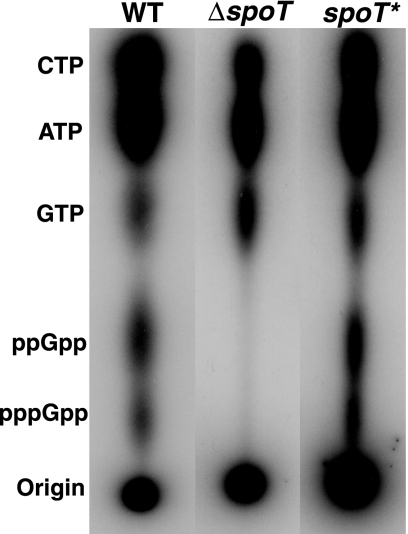
To test whether the H. pylori HP0775 gene is required for production of (p)ppGpp, we subjected the ΔspoT mutant to nutrient deprivation, a condition which has been shown to induce (p)ppGpp in H. pylori (34). Wild-type G27 (WT), ΔspoT, and spoT* strains were incubated in minimal media with 32P for 1 h. As expected, the WT and spoT* strains accumulated significant amounts of (p)ppGpp upon exposure to this stress. In contrast, (p)ppGpp was absent from the ΔspoT mutant extracts (Fig. 2). Plate counts of parallel bacterial cultures before and after 1 h of incubation in minimal medium showed that all three strains remained viable during the experiment; thus, the absence of (p)ppGpp production in the ΔspoT strain was not due to bacterial death. These data suggest that HP0775 encodes the enzymatic activity responsible for (p)ppGpp production in H. pylori.
FIG. 2.
The H. pylori ΔspoT mutant is deficient for (p)ppGpp production under nutrient deprivation conditions. WT, ΔspoT, and spoT* cultures were incubated for 1 h in minimal medium (MOPS-MGS without mannitol or phosphate) in the presence of 32P. Samples were resolved by thin-layer chromatography, and nucleotides were visualized with autoradiography. The WT and complemented strains produced abundant (p)ppGpp, whereas the spoT mutant was defective for (p)ppGpp production.
The H. pylori HP0775 gene functionally complements an E. coli (p)ppGpp-null mutant.
In E. coli, (p)ppGpp metabolism is regulated by the RelA synthetase and the bifunctional synthetase/phosphohydrolase SpoT. Null mutations in relA give rise to strains that produce only basal levels of (p)ppGpp and are unable to grow on minimal medium plates (10). In E. coli, the spoT gene is essential, but a ΔrelA ΔspoT double mutant is viable, indicating the importance of regulating (p)ppGpp levels in the cell (37). The double mutant produces no detectable (p)ppGpp and is unable to grow on minimal medium plates.
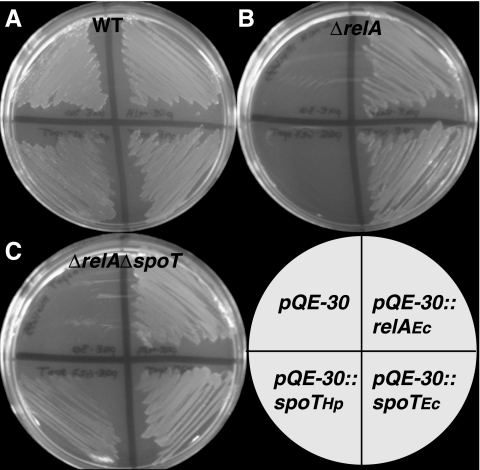
To test whether the H. pylori HP0775 gene could functionally complement either of the (p)ppGpp biosynthetic genes in E. coli, we transformed E. coli strain MG1655 (WT) and isogenic mutant strains ΔrelA and ΔrelA ΔspoT with IPTG-inducible expression constructs encoding E. coli relA, E. coli spoT, and H. pylori HP0775. The transformed E. coli strains were then plated on minimal medium plates in the presence of IPTG and assayed for growth. The growth of WT E. coli strains on minimal medium was not adversely affected by the presence of either H. pylori HP0775 or extra copies of E. coli relA or spoT (Fig. 3). The extrachromosomal copies of both E. coli genes were able to functionally complement both E. coli mutant strains, as was expected, except for the case of the complementation of the double mutant by relA (37), which we discuss below. Significantly, the H. pylori HP0775 gene was able to restore partial growth of the ΔrelA ΔspoT double mutant strain on minimal medium; however, it was unable to complement the ΔrelA single mutant. We interpret this to mean that the H. pylori HP0775 gene at least partially complements the E. coli relA/spoT genes in vivo. This result supports the idea that H. pylori SpoT functions in vivo in (p)ppGpp metabolism and validates the annotation of HP0755 as spoT.
FIG. 3.
The H. pylori ΔspoT gene complements the E. coli (p)ppGpp-null mutant. MG1655 E. coli relA and spoT and H. pylori HP0775 were cloned into the IPTG-inducible expression vector pQE-30. WT (A), ΔrelA (B), and ΔrelA ΔspoT (C) E. coli strains were then transformed with the expression constructs shown in the key and plated onto M9 minimal medium plates supplemented with IPTG.
The H. pylori ΔspoT mutant has a stationary-phase survival defect.
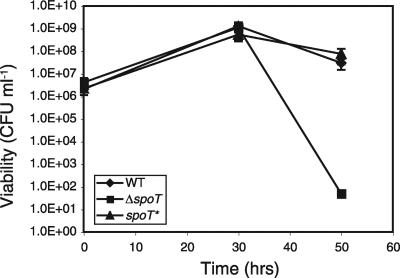
The importance of the stringent response in stationary-phase survival in E. coli and other gram-negative bacteria has been well documented. This phenomenon was thought to depend upon the alternative sigma factor rpoS; however, SpoT has recently been shown to be required for stationary-stage survival in C. jejuni, a bacterium that, similar to H. pylori, lacks rpoS (14). H. pylori strain G27 exhibits a characteristic growth curve in liquid culture with rapid growth over the first 24 h, followed by stationary phase and a gradual decline in viable counts over the following day (4). To test whether the spoT gene is required for any aspect of H. pylori growth, we assayed the viability of WT, ΔspoT, and spoT* liquid cultures over 50 h under standard laboratory growth conditions. All strains exhibited similar population dynamics during log and early stationary phases (Fig. 4 and data not shown). However, during late stationary phase, at approximately 50 h, the ΔspoT cultures exhibited significantly lower viability than the WT and spoT* cultures (Fig. 4).
FIG. 4.
The ΔspoT mutant has a stationary-phase survival defect. WT, ΔspoT, and spoT* H. pylori strains were grown by shaking under microaerophilic conditions at 37°C, and numbers of CFU ml−1 were determined at the time points indicated. A representative assay of at least three trials is shown. Error bars indicate the standard deviations of replicate plating. The limit of detection of the assay was 10 CFU ml−1.
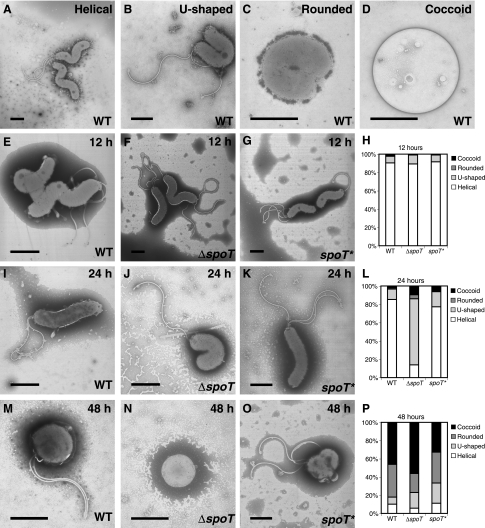
The H. pylori ΔspoT mutant initiates the coccoid transition prematurely.
To further characterize the loss of viability of the ΔspoT cultures, we examined the cellular morphology of cultures grown over 48 h. Viable H. pylori organisms have a characteristic helical shape. Older cultures have been described to adopt a different cellular morphology, termed the coccoid state, in which the cells become rounded up and can no longer be cultured (15). We documented this transformation to the coccoid state over the 48-h growth curve of a wild-type culture (Fig. 5). We were able to identify several distinct morphologies along this trajectory (Fig. 5A to D). Early in the transition cells adopted a U-shaped morphology. Subsequently, the inner and then the outer membranes fused the two arms of the U into a single rounded cell, which often still contained flagella. The final coccoid state was distinct in that it lacked flagella, and its membrane appeared to be less electron dense. We classified the morphologies of cell populations of WT, ΔspoT, and spoT* liquid cultures at 12, 24, and 48 h for more than 100 cells for each strain at each time points. At the 12-h time point the three strains appeared similar and were largely helical. At 24 h, a striking difference was seen in the ΔspoT mutant culture, in which the majority of the cells had initiated the coccoid transition and adopted the U-shaped morphology, in contrast to the wild-type and complemented populations that were still largely helical in morphology. By 48 h the majority of the cells in all three populations were nonhelical, but the ΔspoT culture contained more coccoid cells than the WT and spoT* cultures.
FIG. 5.
The ΔspoT mutant initiates the coccoid transformation prematurely. Transmission electron micrographs of WT (A-D, E, I, and M), ΔspoT (F, J, and N), and spoT* (G, K, and O) cells during growth in liquid culture over 48 h. Typical stages of the helical-to-coccoid transformation (A to D). Representative cells for each culture at 12 (E to G), 24 (I to K), and 48 (M to O) hours are shown. The distributions of morphologies of more than 100 cells of each genotype for each time point are shown (H, L, and P). Bars, 1 μm.
The H. pylori ΔspoT strain is more sensitive to the environmental stresses of anaerobiosis and acidity.
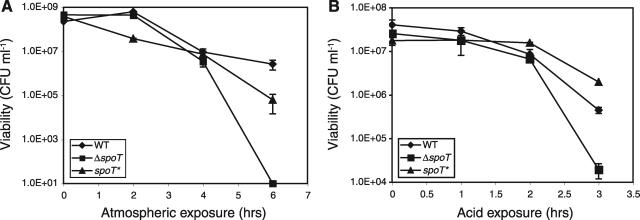
In numerous bacteria the stringent response has been identified as a key factor for survival during exposure to stressful environments. For H. pylori, the ability to survive stresses such as unfavorable atmospheric conditions and acidity are essential to its ability to infect and persist successfully within a human stomach. H. pylori is a microaerophilic and capnophilic and grows best at 3 to 7% O2 and 5 to 10% CO2, conditions similar to those in its gastric niche. A ΔspoT mutant of C. jejuni, which requires similar growth conditions, is extremely sensitive to ambient atmospheric conditions (14). We tested whether the stringent response might play a critical role in H. pylori survival during exposure to higher O2 and lower CO2 atmospheric conditions. We assayed viability while incubating WT, ΔspoT, and spoT* cultures in ambient atmospheric conditions for a 6-h time course. The WT and spoT* cultures exhibited similar patterns of viability and declined gradually over the 6-h period. In four independent trials, the WT and spoT* population sizes after 6 h of aerobic stress were within 2 orders of magnitude of each other, with one or the other showing a more rapid decline in different trials. In contrast, at between 4 and 6 h of exposure to ambient atmosphere, the ΔspoT cultures consistently exhibited a reduction in viability of 4 to 6 orders of magnitude relative to the WT and spoT* strains (Fig. 6A).
FIG. 6.
The ΔspoT mutant has impaired survival in aerobic and acidic stress. (A) WT, ΔspoT, and spoT* H. pylori strains were incubated under ambient atmospheric conditions at 37°C and shaken at 150 rpm. (B) WT, ΔspoT, and spoT* H. pylori strains were incubated by shaking in pH 4 media with 0.5 mM urea, at 37°C. Representative assays of at least three trials are shown. Error bars indicate the standard deviations of replicate plating. The limits of detection of the assays were 10 CFU ml−1 in panel A and 100 CFU ml−1 in panel B.
The ability of H. pylori to survive in an acidic environment is critical to its ability to chronically infect the human stomach. Acid exposure has been shown to elicit (p)ppGpp production in WT H. pylori cultures (34). To test the role of the H. pylori stringent response in acid stress survival, WT, ΔspoT, and spoT* strains were grown in pH 4 growth medium supplemented with 0. 5 mM urea for 3 h, during which time period the acidity of the media remained unchanged. During the first 2 h of acidic exposure all strains were relatively unaffected by the treatment. After 3 h, however, the ΔspoT strain consistently exhibited a significant decrease in viability relative to the WT and spoT* strains (Fig. 6B). In three independent trials the spoT* strain exhibited a slight but significant increase in viability relative to the WT strain at 3 h, which may have been due to the fact that the levels of the spoT gene in the spoT* strain, expressed from the acid-inducible urease promoter (25), may have exceeded those in the WT strain and conferred extra protection against the acid stress. These data indicate that the H. pylori stringent response is required for survival of specific environmental stresses.
DISCUSSION
H. pylori is one of the most widespread pathogens of humans, yet its survival strategies during infection and transmission between human hosts are not well understood. The compact H. pylori genome encodes few transcription factors, raising the question of how this organism adapts to the constantly changing environment of the human stomach. The experiments described here demonstrate that H. pylori has a functional homologue of the stringent response regulator SpoT responsible for (p)ppGpp synthesis. This small molecule alarmone is a global regulator of transcription in E. coli that functions through direct interaction with RNA polymerase (3). One consequence of ppGpp binding is to alter polymerase affinity for certain promoters, thus conferring a level of transcriptional regulation independent of the organism's complement of transcriptional activators and repressors (17). The global transcriptional regulation of the stringent response may be especially important for bacterial species such as H. pylori with limited repertoires of transcriptional regulators.
SpoT function is conserved between H. pylori and E. coli.
Our data indicate that the H. pylori spoT gene not only is required for (p)ppGpp synthesis in H. pylori but that it can also functionally complement the minimal medium growth defect of an E. coli relA and spoT double mutant lacking all (p)ppGpp synthetase and hydrolase functions. Interestingly, the H. pylori spoT gene was not able to rescue an E. coli relA single mutant, possibly because its (p)ppGpp hydrolase activity prevents accumulation of sufficiently high (p)ppGpp levels for cell survival under starvation conditions. Unexpectedly, despite the fact that an E. coli spoT-null allele is lethal in the presence of a wild-type copy of relA (37), we were able to generate the ΔrelA ΔspoT double mutant with an extrachromosomal copy of the E. coli relA gene. The viability of this strain is likely due to the fact that under the conditions of the strain construction (abundant nutrients and no IPTG), the relA transgene was not induced, thus mimicking the situation of a strong loss-of-function relA allele (relA1) in a spoT-null mutant background, which has been shown to be viable (37). The fact that induction of relA under starvation conditions was not toxic to the double mutant and could promote its growth on minimal media (lacking nutrients and containing IPTG) leads us to hypothesize that (p)ppGpp synthetase activity in the absence of hydrolase activity (the situation in a relA+ spoT cell) is toxic to the cell only under conditions when levels of (p)ppGpp are normally low. Under nutrient starvation conditions, however, when (p)ppGpp is normally abundant, we speculate that cells can tolerate RelA activity that is not counteracted by SpoT activity. Our (p)ppGpp labeling and complementation data, together with conservation of key residues in the hydrolytic domain (23) and the absence of another putative (p)ppGpp hydrolase in the H. pylori genome, indicate that the H. pylori spoT gene encodes a bifunctional enzyme that is essential for the production and hydrolysis of the stringent response alarmone (p)ppGpp.
Loss of stationary-phase viability is preceded by a premature coccoid transition in the ΔspoT mutant.
In several bacteria, including E. coli (37) and C. jejuni (14), mutants lacking (p)ppGpp production exhibit aberrant elongated morphologies. H. pylori ΔspoT mutants undergo a precipitous decline in viability by 50 h of growth in liquid culture. This decline in viability is preceded a day earlier by a striking change in cell morphology. Unlike E. coli and C. jejuni, however, the H. pylori ΔspoT mutant appears to enter prematurely into U-shaped and coccoid morphologies similar to those normally observed during later growth stages in wild-type H. pylori strains. The biological significance of the coccoid state has been greatly debated. Some have proposed that it represents a dormant, spore-like stage and is possibly the transmissible form of the organism (24), although analysis of the proteome content argues against this model (8). Our observation that more of the ΔspoT mutant cells appeared coccoid at 24 and 48 h than the wild-type population is consistent with a role for spoT in protection against stationary-phase stresses, but it cannot in itself account for the dramatic difference in viability between these populations at 50 h. The fact that H. pylori precociously adopts the coccoid form in the absence of spoT argues against the coccoid morphology being part of a developmental program of stress resistance that is regulated by the stringent response, as in Bacillus subtilis (26).
SpoT confers protection against specific environmental stresses.
Besides being impaired in stationary-phase survival, the ΔspoT mutant is susceptible to specific environmental stresses, including atmospheric exposure (containing higher oxygen and lower carbon dioxide levels than are optimal for H. pylori growth) and growth in acidified media. Both of these stresses are likely encountered by H. pylori during transmission between hosts and transit through the gastric lumen before the bacteria can situate themselves in the neutral pH environment of the gastric mucosa (27). Acid exposure has been shown to elicit numerous transcriptional changes in H. pylori with a great deal of variation depending on the specific acid exposure regiment (2, 9, 22, 36). In one study, the gppA component of the (p)ppGpp metabolic pathway was found to be upregulated in acidic media (22). Specific transcriptional regulators, including the metal-responsive transcription factors NikR and Fur (9, 13, 33) and the two-component ArsRS system (25), have been shown to control gene expression in response to acid. Acid shock was also recently found to induce (p)ppGpp production in H. pylori under nonstarvation (i.e., rich medium) conditions (34). Together, these data are consistent with a model in which global transcriptional regulation by (p)ppGpp signaling provides an additional level of gene expression control that protects H. pylori from the specific stresses of its environment.
Acknowledgments
We thank Jeannie Selker and Kurt Langworthy for assistance with electron microscopy, Alice Barkan for use of her radioactive room, David Baltrus and Kevin Bourzac for help with genetic manipulations of H. pylori, and Julie Toplin for technical assistance. We are grateful to Benjamin Shepherd and Nina Salama for providing the rdx::urease gene construct and Michael Cashel for the ΔrelA and ΔrelA ΔspoT mutant E. coli strains.
K.G. is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences and was supported by Research Scholar grant RSG-03-101-01-MBC from the American Cancer Society. E.C.G. is a recipient of a Burroughs Wellcome Fund Career Development Award and a Canada Research Chairs Award and is supported by Canadian Institutes for Health Research grant MOP-68981.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Ang, S., C. Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J. T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Artsimovitch, I., V. Patlan, S. Sekine, M. N. Vassylyeva, T. Hosaka, K. Ochi, S. Yokoyama, and D. G. Vassylyev. 2004. Structural basis for transcription regulation by alarmone ppGpp. Cell 117:299-310. [DOI] [PubMed] [Google Scholar]
- 4.Baltrus, D. A., and K. Guillemin. 2006. Multiple phases of competence occur during the Helicobacter pylori growth cycle. FEMS Microbiol. Lett. 255:148-155. [DOI] [PubMed] [Google Scholar]
- 5.Blaser, M. J., and J. C. Atherton. 2004. Helicobacter pylori persistence: biology and disease. J. Clin. Investig. 113:321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourzac, K. M., L. A. Satkamp, and K. Guillemin. 2006. The Helicobacter pylori cag pathogenicity island protein CagN is a bacterial membrane-associated protein that is processed at its C terminus. Infect. Immun. 74:2537-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braeken, K., M. Moris, R. Daniels, J. Vanderleyden, and J. Michiels. 2006. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 14:45-54. [DOI] [PubMed] [Google Scholar]
- 8.Bumann, D., H. Habibi, B. Kan, M. Schmid, C. Goosmann, V. Brinkmann, T. F. Meyer, and P. R. Jungblut. 2004. Lack of stage-specific proteins in coccoid Helicobacter pylori cells. Infect. Immun. 72:6738-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bury-Mone, S., J. M. Thiberge, M. Contreras, A. Maitournam, A. Labigne, and H. De Reuse. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:623-638. [DOI] [PubMed] [Google Scholar]
- 10.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 11.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, N. Figura, et al. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 90:5791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danchin, A. 1977. A new technique for selection of sensitive and auxotrophic mutants of Escherichia coli: isolation of a strain sensitive to an excess of one-carbon metabolites. Mol. Gen. Genet. 150:293-299. [DOI] [PubMed] [Google Scholar]
- 13.Gancz, H., S. Censini, and D. S. Merrell. 2006. Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect. Immun. 74:602-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaynor, E. C., D. H. Wells, J. K. MacKichan, and S. Falkow. 2005. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol. Microbiol. 56:8-27. [DOI] [PubMed] [Google Scholar]
- 15.Kusters, J. G., M. M. Gerrits, J. A. Van Strijp, and C. M. Vandenbroucke-Grauls. 1997. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect. Immun. 65:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kvint, K., C. Hosbond, A. Farewell, O. Nybroe, and T. Nystrom. 2000. Emergency derepression: stringency allows RNA polymerase to override negative control by an active repressor. Mol. Microbiol. 35:435-443. [DOI] [PubMed] [Google Scholar]
- 17.Magnusson, L. U., A. Farewell, and T. Nystrom. 2005. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13:236-242. [DOI] [PubMed] [Google Scholar]
- 18.Masuda, S., and C. E. Bauer. 2004. Null mutation of HvrA compensates for loss of an essential relA/spoT-like gene in Rhodobacter capsulatus. J. Bacteriol. 186:235-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Megraud, F. 2004. Helicobacter pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut 53:1374-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendrygal, K. E., and J. E. Gonzalez. 2000. Environmental regulation of exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 182:599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merrell, D. S., M. L. Goodrich, G. Otto, L. S. Tompkins, and S. Falkow. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 71:3529-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittenhuber, G. 2001. Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J. Mol. Microbiol. Biotechnol. 3:585-600. [PubMed] [Google Scholar]
- 24.Nilsson, H. O., J. Blom, W. Abu-Al-Soud, A. A. Ljungh, L. P. Andersen, and T. Wadstrom. 2002. Effect of cold starvation, acid stress, and nutrients on metabolic activity of Helicobacter pylori. Appl. Environ. Microbiol. 68:11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pflock, M., S. Kennard, I. Delany, V. Scarlato, and D. Beier. 2005. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect. Immun. 73:6437-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhaese, H. J., J. A. Hoch, and R. Groscurth. 1977. Studies on the control of development: isolation of Bacillus subtilis mutants blocked early in sporulation and defective in synthesis of highly phosphorylated nucleotides. Proc. Natl. Acad. Sci. USA 74:1125-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sachs, G., D. L. Weeks, K. Melchers, and D. R. Scott. 2003. The gastric biology of Helicobacter pylori. Annu. Rev. Physiol. 65:349-369. [DOI] [PubMed] [Google Scholar]
- 28.Salama, N. R., B. Shepherd, and S. Falkow. 2004. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J. Bacteriol. 186:7926-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scarlato, V., I. Delany, G. Spohn, and D. Beier. 2001. Regulation of transcription in Helicobacter pylori: simple systems or complex circuits? Int. J. Med. Microbiol. 291:107-117. [DOI] [PubMed] [Google Scholar]
- 30.Scoarughi, G. L., C. Cimmino, and P. Donini. 1999. Helicobacter pylori: a eubacterium lacking the stringent response. J. Bacteriol. 181:552-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smeets, L. C., J. J. Bijlsma, S. Y. Boomkens, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 182:3948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 33.van Vliet, A. H., F. D. Ernst, and J. G. Kusters. 2004. NikR-mediated regulation of Helicobacter pylori acid adaptation. Trends Microbiol. 12:489-494. [DOI] [PubMed] [Google Scholar]
- 34.Wells, D. H., and E. C. Gaynor. 2006. Helicobacter pylori initiates the stringent response upon nutrient and pH downshift. J. Bacteriol. 188:3726-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wells, D. H., and S. R. Long. 2002. The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol. Microbiol. 43:1115-1127. [DOI] [PubMed] [Google Scholar]
- 36.Wen, Y., E. A. Marcus, U. Matrubutham, M. A. Gleeson, D. R. Scott, and G. Sachs. 2003. Acid-adaptive genes of Helicobacter pylori. Infect. Immun. 71:5921-5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA-null mutants can be eliminated by spoT-null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]