Abstract
In this study, reverse transcriptase PCR was employed to construct a transcriptional profile of Mycoplasma pneumoniae lipoprotein genes contained in six multigene families. Most genes were found to be expressed. Many truncated lipoprotein genes were expressed, often polycistronically with other truncated genes, indicating that these genes may still be functional.
There are several mechanisms used by pathogenic bacteria to evade host defenses, including antigenic variation on their surfaces. Antigenic variation of cell surface lipoproteins has been reported in a number of Mycoplasma species (3).
Sequencing of the Mycoplasma pneumoniae genome has revealed that it harbors 46 lipoprotein genes in six multigene families, all of unknown function (4, 7). These multigene families include genes that encode proteins with sequence similarity to the lipoproteins but that lack the characteristic amino-terminal prolipoprotein signal sequence. In contrast, the closest relative of M. pneumoniae, M. genitalium, has only 21 putative lipoprotein genes, with 1 to 3 genes in each family.
No studies have focused specifically on the expression of the lipoprotein multigene families in M. pneumoniae. In this study, we employed reverse transcriptase PCR (RT-PCR) to investigate the expression of all the lipoprotein genes in the six lipoprotein multigene families in this organism. We also examined the transcriptional organization of lipoprotein genes to find possible operon structures. Furthermore, we analyzed the phylogenetics of the lipoprotein genes and explored the relationships between the genes within each of the gene families.
MATERIALS AND METHODS
M. pneumoniae strain M129 was cultured as described previously (16). RNA was isolated by applying methods described earlier (2, 13) and treated with DNase (Novagen) prior to RT-PCR. Oligonucleotide primer sequences used in this study and PCR cycling conditions are provided in the supplemental material (see Table S1). Primer pairs for RT-PCR were chosen to enable synthesis of a specific PCR product from each open reading frame (ORF) as well as regions that spanned selected ORFs to evaluate transcriptional linkage between lipoprotein genes. All primer pairs were first tested in PCRs using M129 genomic DNA as a template. All RT-PCR experiments were repeated at least twice using a RobusT I RT-PCR Kit (Finnzymes, Finland). Control reactions without reverse transcriptase or template were performed for each primer set. The RT-PCR products were analyzed by electrophoresis in 1.5% agarose gels. In some cases RT-PCR products were confirmed by DNA sequencing using an Applied Biosystems DNA sequencing system and Big Dye Terminator, version 3.1, chemistry (Perkin Elmer Applied Biosystems Division). The genome sequence of M. pneumoniae M129 was downloaded from http://www.zmbh.uni-heidelberg.de/M_pneumoniae/. Similarity searches were performed using BlastX and BlastN (http://www.ncbi.nlm.nih.gov/BLAST/); a cutoff value of 1e-15 was used unless otherwise stated, and dot plot analysis was performed with DNASTAR version 5.0 (www.DNAstar.com). Phylogenetic and molecular evolutionary analyses were conducted using the MEGA program, version 3.0 (10). Results of searches for lipoproteins in multigene families were viewed using BLink (http://www.ncbi.nlm.nih.gov), using a cutoff value of 1e-10.
RESULTS
Genomic distribution of lipoprotein genes.
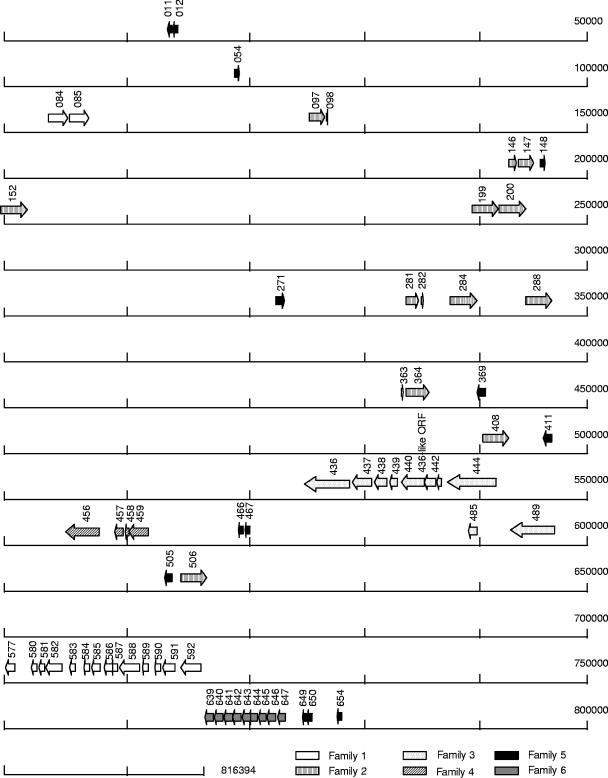
Himmelreich et al. (7) and Dandekar et al. (4) described 46 putative lipoprotein genes that could be grouped in six lipoprotein families. These families also included proteins with similarity to lipoproteins but without the lipoprotein signature sequence at the amino-terminal end. We examined the M. pneumoniae genome and found that a number of truncated ORFs could be linked together to form a sequence with high similarity to one or more full-length ORFs in the same family. We also added the MPN436-like ORF previously described by Jaffe et al. (9) to family 3 because of its high sequence similarity to ORFs in this family. Using similarity searches we found that the truncated ORF MPN467 belonged to family 5, not to family 2, as previously annotated. The resultant six lipoprotein multigene families with 67 members are shown in Table 1, and the genomic distribution of the family members is shown inFig. 1.
TABLE 1.
Members of lipoprotein multigene families in M. pneumoniae, their closest M. pneumoniae and M. genitalium homologs, and the expression of lipoprotein genes as detected by RT-PCR and proteomic methods
| Familya | Gene annotation (NCBI) | M. pneumoniae gene | Lipoprotein signal sequenceb | Position (NCBI)c | Closest M. pneumoniae homolog (% identity and E value)d | Closest M. genitalium homolog (% identity and E value)d | Gene or protein detected bye:
|
||
|---|---|---|---|---|---|---|---|---|---|
| RT-PCR | MS | 2D | |||||||
| 1 | MPN084 | R02_orf524 | − | 105592..107166 | MPN588 (41%, 4e-93) | MG068 (49%, 3e-126) | + | + | |
| MPN083 | R02_orf533 | + | 103941..105542 | MPN588 (39%, 1e-83) | MG067 (55%, 3e-166) | + | + | + | |
| MPN592 | D02_orf521 | + | c(714431..715996) | MPN588 (40%, 7e-96) | MG395 (43%, 5e-96) | + | + | ||
| MPN591 | D02_orf353V | − | c(712847..713908)f | MPN592 (44%, 1e-103) | MG395 (38%, 6e-75) | +g | +j | + | |
| MPN590 | D02_orf217L | + | c(712037..712690) | MPN592 (39%, 2e-40) | MG395 (36%, 1e-28) | − | + | ||
| MPN589 | D02_orf157L | − | c(711423..711896) | MPN591 (64%, 7e-42) | MG395 (37%, 4e-06) | − | − | ||
| MPN588 | D02_orf531 | + | c(709615..711210) | MPN592 (41%, 2e-98) | MG395 (45%, 9e-115) | + | + | ||
| MPN587 | D02_orf150 | − | c(709105..709557) | MPN585 (58%, 2e-39) | MG068 (40%, 2e-15) | − | + | ||
| MPN586 | D02_orf347 | − | c(708186..709229) | MPN582 (56%, 3e-72) | MG068 (39%, 7e-43) | + | + | ||
| MPN585 | D02_orf302 | + | c(707204..708112) | MPN582 (53%, 2e-77) | MG068 (41%, 1e-46) | + | + | ||
| MPN584 | D02_orf135L | − | c(706735..707142) | MPN586 (67%, 3e-35) | MG395 (37%, 2e-14) | +h | − | ||
| MPN583 | D02_orf225L | − | c(705554..706231) | MPN586 (48%, 3e-45) | MG068 (33%, 9e-20) | +g,i | − | ||
| MPN582 | D02_orf439 | + | c(703797..705116) | MPN586 (56%, 2e-84) | MG068 (36%, 2e-54) | + | + | ||
| MPN581 | D02_orf265V | − | c(702824..703621) | MPN084 (43%, 8e-58) | MG068 (45%, 8e-53) | + | + | ||
| MPN580 | D02_orf140 | − | c(702334..702756) | MPN586 (48%, 4e-22) | MG395 (40%, 3e-20) | + | − | ||
| MPN577 | D02_orf346 | − | c(700067..701107) | MPN582 (38%, 1e-61) | MG068 (35%, 1e-53) | + | − | ||
| 2 | MPN152 | E07_orf794 | + | 199823..202207 | MPN284 (62%, 0.0) | MG260 (47%, 4e-178) | + | + | |
| MPN147 | E07_orf485 | − | 193875..195332 | MPN288 (53%, 7e-141) | MG260 (47%, 4e-125) | + | + | ||
| MPN146 | E07_orf265 | − | 193071..193868 | MPN506 (60%, 7e-86) | MG260 (52%, 5e-72) | +k | + | ||
| MPN098 | R02_orf147 | − | 126947..127390 | MPN364 (57%, 2e-42) | MG260 (49%, 4e-37) | + | − | ||
| MPN097 | R02_orf541 | + | 125024..126649 | MPN152 (62%, 2e-170) | MG260 (51%, 4e-127) | +l | + | ||
| MPN506 | P02_orf793 | + | 614865..617246 | MPN288 (72%, 0.0) | MG260 (50%, 0.0) | − | + | + | |
| MPN408 | F11_orf760 | + | 490375..492657 | MPN506 (48%, 0.0) | MG260 (44%, 0.0) | + | + | + | |
| MPN364 | H91_orf677 | − | 433547..435580 | MPN288 (60%, 0.0) | MG260 (46%, 3e-159) | + | + | ||
| MPN363 | H91_orf102 | + | 433200..433508 | MPN284 (81%, 2e-41) | MG260 (59%, 1e-26) | +m | + | ||
| MPN288 | A65_orf787 | + | 344716..347079 | MPN506 (71%, 0.0) | MG260 (50%, 0.0) | + | + | + | |
| MPN284 | A65_orf794 | + | 337770..340154 | MPN288 (64%, 0.0) | MG260 (49%, 0.0) | + | + | + | |
| MPN282 | A65_orf166 | − | 334768..335268 | MPN200 (55%, 9e-44) | MG260 (50%, 6e-38) | + | + | ||
| MPN281 | A65_orf377 | + | 333280..334413 | MPN097 (66%, 5e-129) | MG260 (57%, 1e-103) | + | + | ||
| MPN200 | GT9_orf798 | + | 241983..244379 | MPN506 (58%, 0.0) | MG260 (45%, 0.0) | + | + | + | |
| MPN199 | GT9_orf760 | + | 239417..241699 | MPN506 (39%, 1e-136) | MG185 (51%, 9e-175) | + | + | + | |
| 3 | MPN489 | P02-orf1300 | + | c(592398..596300) | MPN485 (93%, 4e-121) | MG338 (46%, 0.0) | − | + | + |
| MPN485 | P02_orf316 | − | c(589030..589980)n | MPN489 (95%, 0.0) | MG338 (30%, 6e-47) | * | + | ||
| MPN444 | H08_orf1325 | + | c(537762..541739) | MPN436 (24%, 9e-54) | MG309 (58%, 0.0) | + | + | + | |
| MPN442 | H08_orf150 | + | c(536089..536541) | MPN436 (77%, 4e-59) | MG307 (60%, 8e-41) | + | − | ||
| MPN436-like ORFo
|
− | c(535005..536168) | MPN436 (57%, 4e-105) | MG307 (44%, 1e-74) | + | + | |||
| MPN440 | H08_orf726 | − | c(532818..534998) | MPN436 (69%, 0.0) | MG307 (52%, 7e-172) | +p | + | ||
| MPN439 | H08_orf237 | + | c(531949..532662) | MPN436 (74%, 1e-95) | MG307 (57%, 4e-74) | * | + | ||
| MPN438 | H08_orf345 | − | c(530856..531893) | MPN436 (44%, 6e-57) | MG307 (39%, 8e-41) | * | − | ||
| MPN437 | H08_orf572o | − | c(528920..530638) | MPN440 (74%, 0.0) | MG307 (52%, 1e-155) | * | + | ||
| MPN436 | A05_orf1244 | + | c(524877..528611) | MPN440 (69%, 0.0) | MG307 (53%, 0.0) | + | + | + | |
| 4 | MPN459 | H08_orf591 | − | c(560123..561898) | MPN456 (82%, 0.0) | MG321 (64%, 0.0) | + | + | |
| MPN458 | H08_orf157b | − | c(559729..560202) | MPN456 (73%, 8e-44) | MG321 (55%, 1e-32) | + | + | ||
| MPN457 | H08_orf329V | − | c(558798..559787) | MPN456 (74%, 1e-125) | MG321 (64%, 2e-103) | +q | + | ||
| MPN456 | H08_orf1005 | + | c(555398..558415) | MPN459 (82%, 0.0) | MG321 (63%, 0.0) | + | + | + | |
| 5 | MPN148 | E07_orf140 | − | 195875..196297r | MPN505 (77%, 2e-90) | +g | +j | ||
| MPN054 | D09_orf123 | + | 68627..68998 | MPN012 (65%, 3e-36) | +s | − | |||
| MPN012 | D12_orf235 | − | c(13558..14265) | MPN505 (57%, 2e-57) | + | − | |||
| MPN011 | D12_orf231 | + | c(12838..13533) | MPN411 (44%, 8e-43) | + | − | |||
| MPN654 | E09_orf129 | + | c(779342..779731) | MPN271 (80%, 3e-44) | + | − | |||
| MPN650 | E09_orf101 | + | c(775034..775339) | MPN012 (57%, 3e-17) | + | + | |||
| MPN649 | E09_orf136L | − | c(774639..775049) | MPN011 (48%, 2e-26) | +t | − | |||
| MPN505 | P02_orf253 | − | c(613188..613949) | MPN369 (84%, 2e-114) | + | + | |||
| MPN467 | P01_orf101 | + | c(569740..570045) | MPN271 (94%, 4e-39) | + | − | |||
| MPN466 | P01_orf140 | − | c(569288..569710) | MPN369 (81%, 5e-59) | +u | + | |||
| MPN411 | A05_orf252 | + | c(495282..496040) | MPN271 (63%, 8e-84) | + | + | |||
| MPN369 | H91_orf253 | + | c(439894..440655) | MPN505 (84%, 4e-121) | + | − | |||
| MPN271 | A65_orf251a | + | 321736..322491 | MPN369 (77%, 1e-98) | + | + | |||
| 6 | MPN647 | E09_orf290 | + | c(772359..773231) | MPN644 (64%, 3e-98) | MG439 (53%, 4e-76) | + | + | |
| MPN646 | E09_orf277 | + | c(771503..772336) | MPN643 (62%, 2e-97) | MG440 (56%, 2e-78) | + | + | ||
| MPN645 | E09_orf283a | + | c(770652..771503) | MPN644 (63%, 1e-84) | MG439 (53%, 7e-71) | + | + | ||
| MPN644 | E09_orf283b | + | c(769798..770649) | MPN647 (65%, 6e-95) | MG439 (54%, 2e-71) | + | + | ||
| MPN643 | E09_orf302 | − | c(768969..769877) | MPN646 (62%, 1e-92) | MG440 (59%, 4e-77) | + | + | ||
| MPN642 | E09_orf279 | + | c(768130..768969) | MPN640 (73%, 1e-103) | MG439 (50%, 6e-67) | + | + | ||
| MPN641 | E09_orf276 | + | c(767297..768127) | MPN646 (62%, 2e-96) | MG440 (57%, 5e-80) | + | − | ||
| MPN640 | E09_orf300 | + | c(766395..767297) | MPN642 (73%, 1e-99) | MG439 (47%, 1e-60) | + | + | ||
| MPN639 | E09_orf287o | − | c(765524..766387) | MPN643 (33%, 6e-36) | MG440 (32%, 2e-39) | + | + | ||
Reclassifications are given for each family (plus sign indicates pairings). For family 1 (16 ORFs; 11 members), reclassifications are as follows: MPN587+MPN586, MPN590+MPN589, MPN585+MPN584, and MPN581+MPN580. For family 2 (15 ORFs; 11 members), reclassifications are as follows: MPN281+MPN282, MPN146+MPN147, MPN364+MPN363, and MPN097+MPN098. For family 3 (10 ORFs; 5 members), reclassifications are as follows: MPN439+MPN438+MPN437 and MPN442+MPN436 like ORF+MPN440. For family 4 (4 ORFs; 2 members), the only reclassification is MPN459+MPN458+MPN457. For family 5 (13 ORFs; 11 members), reclassifications are as follows: MPN650+MPN649 and MPN467+MPN466. For family 6 (9 ORFs; 9 members), there are no reclassifications.
As predicted by PROSITE.
c, complementary strand.
Identity is percent amino acid similarity; E values are from BlastX searches.
For RT-PCR, results are from this study. MS, protein detected by mass spectrometry (9); 2D, protein detected by two-dimensional gel electrophoresis and mass spectrometry (12, 14). An asterisk indicates an inconsistent result.
N-terminal extension c(713908..714283), 125 amino acids.
Amino-terminal extension detected by RT-PCR.
MPN585+MPN584 expressed as a single transcript.
Carboxyl-terminal extension not detected by RT-PCR.
Amino-terminal extension detected in mass spectrometry.
MPN147+MPN146 expressed as a single transcript.
MPN098+MPN097 expressed as a single transcript.
MPN364+MPN363 expressed as a separate transcript.
Extension c(589030..590475), 482 amino acids.
Reference 9.
MPN442+MPN436 likeORF+MPN440 expressed as a single transcript.
MPN458+MPN457 expressed as a single transcript.
Amino-terminal extension 195539..196297, 253 amino acids.
Carboxyl-terminal extension detected in RT-PCR.
MPN650+MPN649 expressed as a single transcript.
MPN467+MPN466 expressed as a single transcript.
FIG. 1.
Genomic location of the six lipoprotein multigene families. The truncated ORFs in family 1 could be grouped together with other truncated ORFs to form several full-length genes: MPN587 with MPN586, MPN590 with MPN589, MPN585 with MPN584, and MPN581 with MPN580. MPN591 and MPN583 lacked amino- or carboxyl-terminal sequences that were encoded by adjacent sequences that had been classified as noncoding sequences in the gene map. Each of the truncated ORFs in family 2 could be linked with an adjacent truncated ORF, MPN281 with MPN282, MPN146 with MPN147, MPN364 with MPN363, and MPN097 with MPN098. The full-length sequences generated from each of these pairs had a significant amino acid sequence identity (61%, 55%, 61%, and 56%, respectively) with full-length gene products. In family 3 truncated ORFs MPN439, MPN438, and MPN437 could be linked together, as could MPN442, the MPN436-like ORF, and MPN440, and they shared high sequence identity (65% and 70%, respectively) with the full-length ORF MPN436. There was also sequence similarity with MPN436 in the noncoding regions between the truncated ORFs, indicating that these truncated ORFs probably resulted from several mutations in two originally full-length ORFs. The three overlapping truncated ORFs in family 4, MPN459, MPN458, and MPN457, could be linked together to form a sequence with high similarity (82%) to the single full-length ORF. In family 5 the truncated ORFs MPN650 and MPN649 could be linked, as well as MPN467 and MPN466, and these pairs had high levels of identity with full-length genes (48% and 75%, respectively). Family 6 consisted of nine full-length ORFs. No truncated ORFs were found.
Associations between members of lipoprotein gene families and other M. pneumoniae genes.
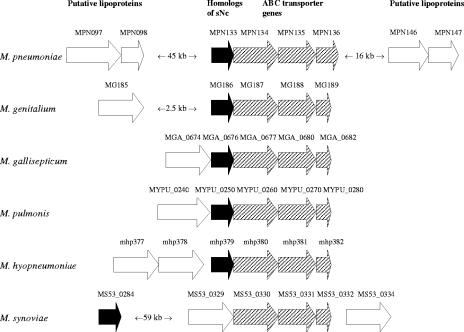
We found that four members of family 2, MPN097, MPN098, MPN146, and MPN147, were positioned on either side, although not immediately adjacent to, a putative operon of ABC transporter genes that also contained a homolog of the staphylococcal nuclease gene. Homologous lipoprotein genes are located immediately adjacent to homologous putative ABC transporter operons in the genomes of M. hyopneumoniae, M. pulmonis, M. gallisepticum, M. genitalium, and M. synoviae (Fig. 2).
FIG. 2.
Genomic locations of lipoprotein genes homologous to members of family 2 and of homologous putative ABC transporter operons in M. pneumoniae M129, M. genitalium G37, M. gallisepticum R, M. pulmonis UAB CTIP, M. hyopneumoniae 232, and M. synoviae 53. Open arrows, lipoprotein genes; solid arrows, staphylococcal nuclease (sNc) gene homologs; crosshatched arrows, ABC transporter genes.
BlastX analysis of family 3 ORF MPN436 found 29% (E value, 8e-09) identity to a predicted homolog of the permease component of an antimicrobial peptide ABC transporter system (COG0577) in M. genitalium G-37. In family 4, MPN459 and MPN456 had 63% (2e-99) and 61% (8e-97) identity, respectively, with a homolog of a predicted component of a predicted oligopeptide ABC transport system (COG4166) in M. genitalium G-37.
BLink was used to determine which Mycoplasma species possessed homologs of the M. pneumoniae lipoprotein families. Family 1 and family 2 homologs were common in many Mycoplasma species, whereas family 3 and 4 homologs were found only in M. genitalium, M. penetrans, and M. gallisepticum. Homologs of family 6 were found only in M. genitalium. Family 5 genes were found to be unique to M. pneumoniae.
Transcripts for the majority of ORFs in lipoprotein multigene families were detectable by RT-PCR.
We used RT-PCR to comprehensively study the transcription of all 67 putative lipoprotein genes contained in the six multigene families. Every gene was tested at least twice with an independently isolated RNA template. We also looked for any operon structure within the lipoprotein genes by using primers targeted to bridge the sequence between truncated ORFs. In total, 58 genes were detected by RT-PCR (Table 1).
Most of the ORFs in family 1 were expressed, with the exception of MPN590, MPN589, and MPN587. We also found that the truncated ORFs MPN581 and MPN580 were located on a single mRNA, while MPN585 and MPN584 were expressed as separate mRNAs. Two transcripts, MPN591 and MPN583, had noncoding regions at their 5′ ends, which, based on similarity searches, corresponded to the missing sections of the full-length ORFs.
In family 2, all full-length ORFs were detected by RT-PCR, with the exception of MPN506. Most of the paired truncated ORFs were transcribed on the same mRNAs, with the exception of MPN364 and MPN363, which were detected only on separate transcripts.
Of the three full-length ORFs in family 3, MPN444 and MPN436 were transcribed, while MPN489 was not detected by RT-PCR. Truncated ORFs MPN442, MPN436-like ORF, and MPN440 were transcribed polycistronically, whereas truncated ORFs MPN439, MPN438, MPN437, and MPN485 did not yield consistent products in RT-PCR, possibly as a consequence of low amounts of the respective mRNAs.
All four ORFs in family 4 were transcribed. Truncated ORFs MPN458 and MPN457 were located on a single transcript, while MPN459 was located on a separate transcript.
In family 5 all 13 ORFs were detected in RT-PCR. The paired truncated ORFs MPN650 and MPN649 were detected on a single transcript, as were MPN467 and MPN466. MPN148 and MPN054 were transcribed with predicted noncoding regions, containing amino-terminal and carboxyl-terminal sequences, respectively. Thus, these ORFs appear to have once been full-length ORFs that have since been truncated by a mutation in a start codon and by a frameshifting mutation, respectively.
In family 6, transcripts of all nine ORFs were detectable. The mRNAs for each gene were transcribed separately.
Phylogenetic relationships between lipoproteins within each family.
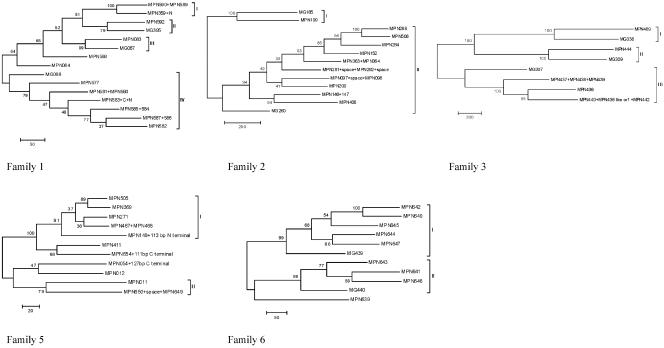
To investigate orthologous and paralogous relationships of putative lipoprotein genes within a family, we conducted phylogenetic analyses of members of families 1, 2, 3, 5, and 6 and the corresponding M. genitalium homologs. Phylogenetic trees (Fig. 3) were constructed using the maximum parsimony method. For the analysis, truncated ORFs were linked together to form a full-length sequence.
FIG. 3.
Phylogenetic trees of families 1, 2, 3, 5, and 6. Trees were generated using the maximum parsimony method (MEGA 3.0). The nucleotide substitution rate is shown as a bar under the tree, and bootstrap values (1,000 replicates) are given for each branch. Phylogenetic analysis of the family 1 members and family 1 homologs in M. genitalium placed them in at least four separate groups. Groups II, III, and IV had an orthologous association with different M. genitalium genes. Phylogenetic divergence between the genes in group IV mostly corresponded to their order in the genome. Thus, an ORF was more closely related to the ORFs closer to it on the gene map than to the ORFs farther away from it. The genes in group I were closely related and also lay together on the gene map. Further evidence supporting the clusters of the tree was found in the different gene lengths between the groups. MG068, containing 474 amino acids, is similar in size to the other genes in group IV, which is shorter than MG067 and MG395, which are 516 amino acids and 524 amino acids, respectively, and are in groups II and III. There were at least two orthologous groups in family 2. Group I only had two members, MG185 and MPN199, while the other group contained MG260 and the remaining 10 family 2 members. Family 3 formed three orthologous groups with an M. genitalium homolog in each group. Although there were more genes in group III, only one ORF was full-length. Family 5 members formed two groups with significant bootstrap values (>70). As there were no homologs in other species, it was not possible to further explore the origins of these two groups. Truncated ORFs were usually closely related to an intact member (e.g., MPN411 to MPN654 plus 111 bp of C-terminal sequence, MPN012 to MPN054 plus 127 bp of C-terminal sequence, MPN011 to MPN650 plus MPN649, and MPN271 to MPN467 plus MPN466).Family 6 ORFs formed at least two orthologous groups in the phylogenetic tree, and both groups had an M. genitalium homolog.
In phylogenetic analysis of families 1, 2, 3, and 6, all groups except for group I in family 1 had an orthologous association with a M. genitalium gene. As there were no homologs of family 5 genes in other species, it was not possible to further explore the origins of the two groups in this family.
DISCUSSION
In many Mycoplasma species only one or few lipoproteins within a multigene family are expressed at any one time (6). However, in M. pneumoniae we found that most of the genes were expressed, suggesting that the lipoprotein genes in multigene families may not be variably expressed. This is supported by data previously obtained using a proteomics approach for M. pneumoniae FH strain (9). However, we cannot exclude the possibility that different cells in the population may have been expressing different lipoprotein genes due to the absence of any selection pressure. The RT-PCR method used in this investigation is sensitive but qualitative. Hence, in some cases we might have detected low mRNA levels that may not have a biological significance.
Interestingly, many truncated lipoprotein genes were transcribed, often polycistronically, with other truncated ORFs. This was observed in all lipoprotein gene families that contained truncated genes. This indicates that these genes may not be pseudogenes and, furthermore, that the truncated gene products may retain function(s). The M. pneumoniae genome is reported to have few known transcriptional stop signals (5, 15). Therefore, even functionally unrelated genes could be transcribed polycistronically, as suggested by Benders et al. (1), although M. pneumoniae could employ mechanisms other than hairpin termination for transcriptional termination (15).
In studies of gene expression at the protein level, Regula et al. (12) and Ueberle et al. (14) were able to identify 305 proteins of the predicted 668 ORFs in M. pneumoniae using one-dimensional and two-dimensional gel electrophoresis and mass spectrometry, although they were unable to resolve alkaline proteins and membrane proteins using these methods. Jaffe et al. (9) used shotgun mass spectrometry combined with novel computational methods to build a proteogenomic map of M. pneumoniae strain FH based on the detection of expressed proteins alone. We detected transcription of 87% of the genomically predicted lipoprotein ORFs contained in multigene families in M. pneumoniae strain M129, compared to detection of 76% of the ORF products by Jaffe et al. in strain FH (9). The mass spectrometry and RT-PCR findings were in agreement for 45 of 67 ORFs. At the RNA level gene expression has been previously studied in microarrays (17). Weiner et al. (17) found that 606 of 688 ORFs were transcribed at 37°C, in agreement with our observation that most of the lipoprotein genes were transcribed.
Currently, the functions of most M. pneumoniae lipoproteins are not known, the exception being the lipoprotein that is a subunit of the FoF1-type ATPase (11), which is not a member of any of the six lipoprotein families. We observed that some lipoproteins were associated with ABC transporter genes (Fig. 2) or shared sequence similarity with ABC transporter genes (MPN436 and MPN459), suggesting that they may play a role in the transport of nutrients into the cell.
Our phylogenetic analyses suggest that gene duplication events have occurred frequently in all lipoprotein multigene families and that this duplication occurred prior to and after separation from M. genitalium. This is consistent with the suggestion by Himmelreich et al. (8) that the increased number of lipoprotein genes in the M. pneumoniae genome compared to that of M. genitalium was due to gene amplification. However, concurrent gene degradation has also occurred, as indicated by the many truncated ORFs in all of the families apart from family 6. Thus, the genomic repertoire for each group has resulted from an interaction between gene duplication and degradation. Each orthologous group in these families may be exposed to independent selective pressures, possibly related to their function, as indicated by the common retention of at least one full-length ORF in each group. Family 6 evolution differs from the other families. No truncated ORFs were found in this family, suggesting that the gene duplication events might have occurred recently and that the family may still be expanding.
In summary, we have performed a comprehensive analysis of lipoprotein multigene families in M. pneumoniae and investigated the transcription of all the genes in these families using RT-PCR. We found that most of the lipoprotein genes were transcribed in M. pneumoniae, in contrast to the limited numbers of lipoprotein genes expressed in many other mycoplasmas. Our examination of the transcription data and the genome sequence did not detect any evidence of phase variation. There is no evidence of local polynucleotide sequences upstream of the truncated ORFs that might be generating phase variable frameshift mutations, as has been seen in lipoproteins of some other mycoplasmas. However, we did detect some interesting associations between known genes, particularly ABC transporter genes, and the lipoprotein genes. The gene expression data and lipoprotein gene analysis presented in this paper lay a foundation for functional studies of lipoprotein multigene families in M. pneumoniae.
Supplementary Material
Acknowledgments
This work was supported by an NHMRC project grant.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Benders, G. A., B. C. Powell, and C. A. Hutchison, 3rd. 2005. Transcriptional analysis of the conserved ftsZ gene cluster in Mycoplasma genitalium and Mycoplasma pneumoniae. J. Bacteriol. 187:4542-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 3.Citti, C., G. F. Browning, and R. Rosengarten. 2005. Phenotypic diversity and cell invasion in host subversion by pathogenic mycoplasmas, p. 439-483. In A. Blanchard and G. Browning (ed.), Mycoplasmas: molecular biology, pathogenicity and strategies for control. Horizon Bioscience, Norfolk, United Kingdom.
- 4.Dandekar, T., M. Huynen, J. T. Regula, B. Ueberle, C. U. Zimmermann, M. A. Andrade, T. Doerks, L. Sánchez-Pulido, B. Snel, M. Suyama, Y. P. Yuan, R. Herrmann, and P. Bork. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ermolaeva, M. D., H. G. Khalak, O. White, H. O. Smith, and S. L. Salzberg. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 301:27-33. [DOI] [PubMed] [Google Scholar]
- 6.Glew, M. D., P. F. Markham, G. F. Browning, and I. D. Walker. 1995. Expression studies on four members of the pMGA multigene family in Mycoplasma gallisepticum S6. Microbiology 141:3005-3014. [DOI] [PubMed] [Google Scholar]
- 7.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Himmelreich, R., H. Plagens, H. Hilbert, B. Reiner, and R. Herrmann. 1997. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 25:701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaffe, J. D., H. C. Berg, and G. M. Church. 2004. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4:59-77. [DOI] [PubMed] [Google Scholar]
- 10.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 11.Pyrowolakis, G., D. Hofmann, and R. Herrmann. 1998. The subunit b of the FoF1-type ATPase of the bacterium Mycoplasma pneumoniae is a lipoprotein. J. Biol. Chem. 273:24792-24796. [DOI] [PubMed] [Google Scholar]
- 12.Regula, J. T., B. Ueberle, G. Boguth, A. Görg, M. Schnölzer, R. Herrmann, and R. Frank. 2000. Towards a two-dimensional proteome map of Mycoplasma pneumoniae. Electrophoresis 21:3765-3780. [DOI] [PubMed] [Google Scholar]
- 13.Tang, S. L., S. Nuttall, K. Ngui, C. Fisher, P. Lopez, and M. Dyall-Smith. 2002. HF2: a double-stranded DNA tailed haloarchaeal virus with a mosaic genome. Mol. Microbiol. 44:283-296. [DOI] [PubMed] [Google Scholar]
- 14.Ueberle, B., R. Frank, and R. Herrmann. 2002. The proteome of the bacterium Mycoplasma pneumoniae: comparing predicted open reading frames to identified gene products. Proteomics 2:754-764. [DOI] [PubMed] [Google Scholar]
- 15.Washio, T., J. Sasayama, and M. Tomita. 1998. Analysis of complete genomes suggests that many prokaryotes do not rely on hairpin formation in transcription termination. Nucleic Acids Res. 26:5456-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiner, J., III, R. Herrmann, and G. F. Browning. 2000. Transcription in Mycoplasma pneumoniae. Nucleic Acids Res. 28:4488-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiner, J., III, C. U. Zimmerman, H. W. Göhlmann, and R. Herrmann. 2003. Transcription profiles of the bacterium Mycoplasma pneumoniae grown at different temperatures. Nucleic Acids Res. 31:6306-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.