Abstract
σ28 RNA polymerase is an alternative RNA polymerase that has been postulated to have a role in developmental gene regulation in Chlamydia. Although a consensus bacterial σ28 promoter sequence has been proposed, it is based on a relatively small number of defined promoters, and the promoter structure has not been systematically analyzed. To evaluate the sequence of the σ28-dependent promoter, we performed a comprehensive mutational analysis of the Chlamydia trachomatis hctB promoter, testing the effect of point substitutions on promoter activity. We defined a −35 element recognized by chlamydial σ28 RNA polymerase that resembles the consensus −35 sequence. Within the −10 element, however, chlamydial σ28 RNA polymerase showed a striking preference for a CGA sequence at positions −12 to −10 rather than the longer consensus −10 sequence. We also observed a strong preference for this CGA sequence by Escherichia coli σ28 RNA polymerase, suggesting that this previously unrecognized motif is the critical component of the −10 promoter element recognized by σ28 RNA polymerase. Although the consensus spacer length is 11 nucleotides (nt), we found that σ28 RNA polymerase from both Chlamydia and E. coli transcribed a promoter with either an 11- or 12-nt spacer equally well. Altogether, we found very similar results for σ28 RNA polymerase from C. trachomatis and E. coli, suggesting that promoter recognition by this alternative RNA polymerase is well conserved among bacteria. The preferred σ28 promoter that we defined in the context of the hctB promoter is TAAAGwwy-n11/12-ryCGAwrn, where w is A or T, r is a purine, y is a pyrimidine, n is any nucleotide, and n11/12 is a spacer of 11 or 12 nt.
Bacteria use alternative forms of RNA polymerase to regulate the transcription of separate classes of genes through specific recognition of distinct promoter elements. In the human pathogen Chlamydia, an alternative RNA polymerase containing σ28 has been proposed to be a stage-specific regulator of gene expression (21) because it transcribes hctB, a gene that is only expressed at late time points in the chlamydial developmental cycle (1, 5). hctB is the only known σ28-regulated gene in Chlamydia, and we are interested in analyzing the promoter that is recognized by chlamydial σ28 RNA polymerase as an approach for identifying additional σ28-regulated genes.
Although a consensus bacterial σ28 promoter is available, there are limitations in utilizing this sequence to find chlamydial σ28-dependent promoters. This consensus sequence consists of two promoter elements spaced 11 nucleotides (nt) apart (TAAAnnnn-n11-GCCGATAA, where n is any nucleotide, and n11 is a spacer of 11 nt) (3, 11), and additional sequences in the upstream −35 element have been proposed to form an extended σ28 promoter (TAAAGTTT-n11-GCCGATAA) (12). However, these consensus promoter sequences were derived from the alignment of about a dozen σ28-dependent promoters compared to over 100 for the consensus σ70 promoter (8, 10, 15). This small sample population also limits the ability to determine which positions in the σ28 promoter are most important for transcriptional activity and the nucleotide preference at these positions. Additionally, the consensus sequence was mostly derived from promoters in Escherichia coli, Salmonella, and Bacillus, and it is not known if σ28 promoter structure is well conserved among other bacteria.
As an alternative to deriving a consensus from known promoter sequences, we can also define a promoter by determining which promoter sequences are most highly transcribed by RNA polymerase. We have previously used this approach to establish the promoter for σ66, the chlamydial homolog of σ70 (18, 19). We performed a comprehensive mutational analysis on a single σ66 promoter and determined the optimal promoter sequence transcribed by Chlamydia trachomatis σ66 RNA polymerase and E. coli σ70 RNA polymerase in vitro. The sequences determined for both polymerases closely resemble the E. coli consensus σ70 promoter, supporting the use of this approach for defining promoter structure.
In this study, we have used a similar comprehensive mutational analysis on the hctB promoter to determine the σ28 promoter sequence in Chlamydia. Point substitutions at many positions in both the −35 and −10 promoter elements caused large decreases in promoter activity with an in vitro transcription assay using chlamydial σ28 RNA polymerase. These results allowed us to determine the relative nucleotide preference at each position in the promoter. From this analysis, we propose a sequence for the chlamydial σ28-dependent promoter that resembles the consensus bacterial σ28 promoter but with greater prominence given to a distinct sequence motif (CGA) in the −10 element that appears to be critical for promoter activity. We have also determined the optimal spacer length between the two promoter elements. We obtained very similar results with E. coli σ28 RNA polymerase, suggesting that the promoter specificity of σ28 RNA polymerase is conserved between Chlamydia and E. coli.
MATERIALS AND METHODS
Cloning of the σ28 gene.
Cloning of the C. trachomatis serovar L2 σ28 gene into a His-tagged expression vector pRSET-C was previously described (21).
E. coli fliA, which encodes the gene for σ28, was cloned into a His-tagged expression vector pRSET-C to produce plasmid pMT1379. The insert (containing the entire σ28 gene with the exclusion of the start codon) was amplified by PCR from E. coli K-12 genomic DNA by Tgo DNA polymerase, using PCR primers T688 (5′-AATTCACTCTATACCGCTGAAGGT) and T628 (5′-CCCGGTACCTTATAACTTACCCAGTTTAGTGCGTA). The PCR product was digested with KpnI and cloned into pRSET-C between KpnI and blunted BamHI sites.
Overexpression and purification of σ28.
C. trachomatis serovar L2 His6-σ28 was overexpressed in E. coli BL21(DE3) and purified, as previously described (21), to a concentration of 35.7 μg/ml.
E. coli His6-σ28 was expressed in E. coli BL21(DE3) cells freshly transformed with pMT1379. A total of 250 ml of cells was grown at 37°C to an optical density at 600 nm of 0.5 and induced with 2 mM isopropyl-β-d-thiogalactosidase. After 3 h, cells were collected by centrifugation, resuspended in 10 ml of buffer N (10 mM Tris [pH 8.0], 0.3 M NaCl, 10 mM β-mercaptoethanol) containing 20 mM imidazole, and disrupted with a Branson Sonifier 450 (four times for 30 seconds each time). E. coli σ28 protein was then purified from the pellet under denaturing conditions. The protein pellet was solubilized with 5 ml of buffer B (20 mM Tris [pH 8.0], 500 mM NaCl) containing 6 M guanidine hydrochloride. Proteins were purified with a 1-ml nickel HiTrap chelating column (Amersham Bioscience, Piscataway, N.J.). Bound proteins were washed sequentially with 10 ml of buffer B containing imidazole at a concentration of 5 mM and then 30 mM. His-tagged σ28 protein was eluted with 5 ml of buffer B containing 250 mM imidazole. Purified σ28 protein was dialyzed overnight with two changes of 500 ml of storage buffer (50 mM Tris [pH 8.0], 200 mM KCl, 10 mM MgCl2, 10 μM ZnCl2, 1 mM EDTA, 5 mM 2-β-mercaptoethanol, 20% glycerol). The concentration of the purified E. coli σ28 protein was approximately 115.8 μg/ml.
Construction of the wild-type hctB transcription template.
The hctB promoter region (−39 to +6) from C. trachomatis serovar L2 was amplified from genomic DNA by PCR with primers T327 (5′-CCCGAATTCTTTATTAAAGTTTTTCATTCTCCTTGTC) and T335 (5′-ATTTATTTGATCTATCGACAAGGAGAAT). The promoter insert was cloned upstream of a promoterless G-less cassette transcription template in plasmid pMT1125 (20). Transcription of the plasmid by σ28 RNA polymerase produced a 130-nt transcript.
Construction of hctB transcription templates containing mutations.
Individual mutant promoters were produced by PCR, with the desired mutation introduced on an oligonucleotide primer. Each template contained the hctB promoter region from −39 to +6. A 5-bp substitution was introduced into the −35 element by altering the sequence from −32 to −28 (AAGTT to CCTGG) or into the −10 element by changing the sequence from −14 to −10 (GTCGA to TGATC). A total of 84 mutant hctB promoters (−39 to +6) with single base pair substitutions in the −35, −10, or flanking regions were produced, so that the effect of all possible single substitutions from −37 to −24 and −17 to −4 could be tested. Mutants with different spacing lengths of 9, 10, 11, and 13 nt were generated with insertions and deletions in the middle of the spacer. The mutant hctB promoters were cloned upstream of a promoterless G-less cassette transcription template in plasmid pMT1125 as previously described (20).
In vitro transcription.
Transcription reactions were performed as previously described (21). Each chlamydial σ28 transcription reaction was performed with C. trachomatis σ28 RNA polymerase that had been reconstituted by mixing 1 μl of C. trachomatis recombinant His6-σ28 with 1 μl of heparin-agarose-purified C. trachomatis RNA polymerase at 4°C for 15 min, immediately prior to the transcription reaction. For E. coli σ28, each transcription reaction was carried out with E. coli σ28 RNA polymerase reconstituted from 1 μl of E. coli recombinant His6-σ28 and 0.03 U of E. coli core enzyme (Epicenter, Madison, Wis.).
Calculation of promoter activity.
The relative promoter activity was determined by normalizing the promoter activity of each mutant promoter to that of the wild-type hctB promoter, which was defined as 100% activity. Three measurements of relative promoter activity were obtained for each promoter, and a mean and a standard deviation were calculated. Relative changes in promoter activity (n-fold) were obtained by comparing the relative promoter activity of each mutant promoter to that of the wild-type hctB promoter.
Generation of the sequence logo.
All sequence logos were derived using WebLogo, which is available online at http://weblogo.berkeley.edu. The format for data input into this site is a series of sequences. To adapt our data to this format, we first calculated the proportion of promoter activity attributable to each of the four possible nucleotides at each of the 28 positions that we tested. This 28-by-4 matrix of individual contributions was transformed into representative nucleotide sequences using a custom computer algorithm written by Dennis Kibler (University of California, Irvine), and entered into the online WebLogo form. The resulting sequence logo consists of stacks of letters at each position. The height of the stack indicates the importance of a particular position. The height of an individual letter within a stack indicates the relative preference for that nucleotide based on transcriptional activity (with a maximum height defined as 2 bits).
RESULTS
Substitution of the −35 and −10 regions of the hctB promoter decreased transcription by C. trachomatis σ28 RNA polymerase.
In previous studies, we have found that a comprehensive mutational approach for defining a promoter sequence works best with a strong promoter because it increases the likelihood of measuring the effect of an individual point substitution on transcriptional activity (18). The C. trachomatis hctB promoter was considered to be a good candidate because it strongly resembles the σ28 extended consensus promoter (Fig. 1) and is highly transcribed by both chlamydial and E. coli σ28 RNA polymerases (12, 21). To test this premise, we determined that 5-bp substitutions in either the −35 or the −10 promoter element of the hctB promoter caused an almost complete loss of promoter activity using an in vitro transcription assay with C. trachomatis σ28 RNA polymerase (data not shown). These findings confirm that each element contains sequences that are important for promoter activity.
FIG. 1.
The sequence of the hctB promoter aligned with bacterial core and extended σ28 consensus sequences. The −35 and −10 promoter elements are separated by a spacer length of 11 or 12 nt.
Many point substitutions in the −35 promoter region decreased transcription by C. trachomatis σ28 RNA polymerase.
We next tested whether single base substitutions had an effect on promoter activity with our chlamydial σ28 in vitro transcription assay. For both the −35 and −10 elements of the hctB promoter, we examined eight positions within the element and an additional three flanking positions on either side. At each position, we constructed transcription templates containing each of the three possible point substitutions. Altogether, we individually tested 84 mutant promoters and compared the level of transcription to the wild-type hctB promoter. A summary of the contribution of each mutant promoter to transcriptional activity is provided in Table 1. The majority of point substitutions produced a significant decrease in transcription, as will be discussed further.
TABLE 1.
Promoter activities of hctB promoter templates with single point substitutions
| Position | Wild-type residue | Relative promoter activity (± SD) with the indicated substitutiona
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
C. trachomatis RNA polymerase
|
Escherichia coli RNA polymerase
|
||||||||
| A | C | G | T | A | C | G | T | ||
| −37 | T | 21.5 ± 2.2 | 31.9 ± 3.9 | 27.4 ± 4.8 | 100 | 10.0 ± 2.1 | 18.3 ± 5.0 | 11.2 ± 3.1 | 100 |
| −36 | A | 100 | 16.2 ± 2.7 | 36.1 ± 6.9 | 46.4 ± 10.5 | 100 | 9.1 ± 3.3 | 18.7 ± 7.2 | 26.8 ± 12.5 |
| −35 | T | 101.8 ± 18.2 | 129.7 ± 21.5 | 53.3 ± 13.9 | 100 | 70.6 ± 35.1 | 109.7 ± 54.3 | 26.7 ± 11.4 | 100 |
| −34 | T | 0.0 ± 0.3 | 3.1 ± 0.7 | 1.4 ± 0.3 | 100 | 0.0 ± 0.3 | 1.5 ± 0.1 | 0.3 ± 0.1 | 100 |
| −33 | A | 100 | 11.2 ± 2.4 | 1.2 ± 0.2 | 13.3 ± 2.0 | 100 | 5.3 ± 0.9 | 1.5 ± 0.3 | 4.4 ± 0.5 |
| −32 | A | 100 | 5.7 ± 0.9 | 0.9 ± 0.2 | 6.5 ± 1.8 | 100 | 2.5 ± 0.2 | 0.5 ± 0.1 | 2.1 ± 0.2 |
| −31 | A | 100 | 4.6 ± 1.0 | 22.4 ± 5.2 | 7.6 ± 0.1 | 100 | 2.3 ± 0.3 | 19.9 ± 0.4 | 4.5 ± 0.3 |
| −30 | G | 1.1 ± 0.6 | 15.1 ± 1.2 | 100 | 7.4 ± 1.2 | 0.6 ± 0.1 | 7.4 ± 0.8 | 100 | 2.1 ± 0.4 |
| −29 | T | 119.6 ± 5.2 | 13.0 ± 0.7 | 44.1 ± 2.2 | 100 | 82.1 ± 3.9 | 6.1 ± 0.6 | 20.1 ± 0.6 | 100 |
| −28 | T | 130.2 ± 6.6 | 8.5 ± 1.1 | 22.1 ± 1.0 | 100 | 75.8 ± 2.4 | 4.8 ± 0.2 | 9.2 ± 0.5 | 100 |
| −27 | T | 7.1 ± 2.0 | 96.4 ± 4.1 | 7.7 ± 0.9 | 100 | 2.7 ± 0.4 | 76.8 ± 4.7 | 4.2 ± 0.2 | 100 |
| −26 | T | 15.5 ± 4.5 | 29.3 ± 8.9 | 32.2 ± 10.4 | 100 | 7.5 ± 2.1 | 13.2 ± 2.6 | 14.0 ± 3.8 | 100 |
| −25 | T | 84.5 ± 4.5 | 23.4 ± 3.7 | 22.5 ± 2.4 | 100 | 70.4 ± 10.5 | 14.8 ± 3.5 | 10.8 ± 3.0 | 100 |
| −24 | C | 28.5 ± 2.6 | 100 | 80.2 ± 7.9 | 26.7 ± 3.4 | 11.5 ± 2.0 | 100 | 61.9 ± 14.4 | 12.3 ± 2.3 |
| −17 | C | 35.4 ± 6.8 | 100 | 25.3 ± 4.6 | 34.8 ± 11.1 | 27.7 ± 1.9 | 100 | 14.5 ± 1.5 | 21.4 ± 1.7 |
| −16 | T | 141.0 ± 26.5 | 54.4 ± 10.6 | 54.8 ± 13.0 | 100 | 121.0 ± 15.1 | 30.8 ± 6.3 | 37.3 ± 4.3 | 100 |
| −15 | T | 31.5 ± 5.3 | 34.5 ± 6.0 | 59.2 ± 16.8 | 100 | 16.2 ± 4.5 | 12.3 ± 1.1 | 36.9 ± 11.2 | 100 |
| −14 | G | 33.4 ± 5.8 | 12.6 ± 3.5 | 100 | 16.4 ± 4.6 | 9.9 ± 1.7 | 2.4 ± 0.4 | 100 | 3.0 ± 0.4 |
| −13 | T | 42.6 ± 7.2 | 55.8 ± 7.7 | 0.9 ± 0.03 | 100 | 28.9 ± 0.9 | 53.5 ± 7.2 | 13.3 ± 0.6 | 100 |
| −12 | C | 2.0 ± 0.2 | 100 | 0.7 ± 0.3 | 6.1 ± 1.1 | 0.7 ± 0.03 | 100 | 0.3 ± 0.1 | 0.9 ± 0.2 |
| −11 | G | 2.7 ± 0.5 | 3.6 ± 0.4 | 100 | 1.0 ± 0.3 | 0.9 ± 1.3 | 1.3 ± 0.4 | 100 | 0.2 ± 0.1 |
| −10 | A | 100 | 0 ± 0.5 | 0.1 ± 0.1 | 1.9 ± 0.5 | 100 | 2.9 ± 0.8 | 1.4 ± 0.5 | 13.3 ± 1.3 |
| −9 | T | 124.8 ± 6.8 | 39.4 ± 5.5 | 10.9 ± 0.5 | 100 | 119.3 ± 19.6 | 48.3 ± 6.3 | 6.7 ± 1.5 | 100 |
| −8 | A | 100 | 4.8 ± 0.9 | 39.0 ± 6.3 | 30.2 ± 4.7 | 100 | 2.6 ± 0.9 | 19.1 ± 2.6 | 19.0 ± 4.2 |
| −7 | G | 55.3 ± 9.2 | 38.4 ± 8.2 | 100 | 45.9 ± 6.6 | 37.7 ± 3.3 | 20.3 ± 1.5 | 100 | 34.5 ± 5.2 |
| −6 | A | 100 | 30.0 ± 7.6 | 71.9 ± 22.0 | 36.4 ± 8.9 | 100 | 8.0 ± 1.5 | 44.1 ± 13.2 | 13.5 ± 3.5 |
| −5 | T | 183.9 ± 24.3 | 39.3 ± 6.3 | 34.8 ± 3.0 | 100 | 153.5 ± 31.0 | 13.1 ± 0.6 | 12.0 ± 1.1 | 100 |
| −4 | C | 51.8 ± 7.7 | 100 | 160.2 ± 36.0 | 47.0 ± 12.4 | 33.1 ± 7.6 | 100 | 174.9 ± 24.6 | 32.7 ± 6.5 |
The relative promoter activity was determined by normalization to the activity of the wild-type hctB promoter, which was defined as 100%. Each value represents the mean of three independent experiments and their standard deviation.
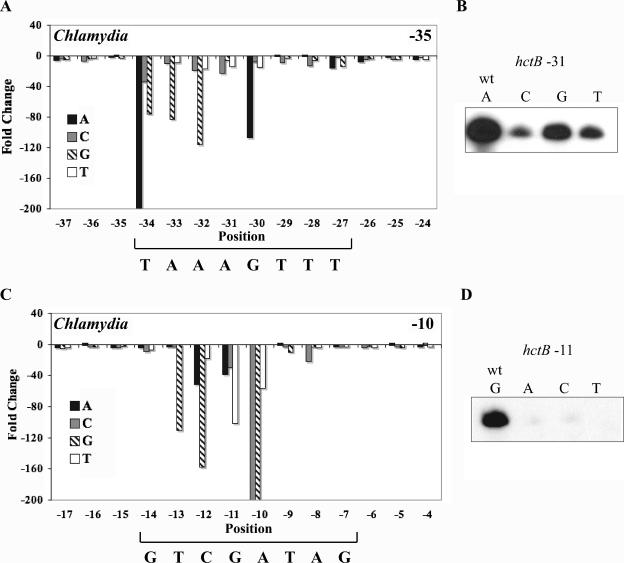
Most substitutions within the core −35 promoter element (TAAA) produced large decreases in promoter activity, with a greater than 10-fold reduction in transcription. For example, at position −34, substitution of the T with an A completely abrogated transcription. Changing the T to a C or G decreased transcription by 33-fold and 75-fold, respectively (Fig. 2A). From these results, it is clear that a T is the preferred nucleotide at position −34 for transcriptional activity within this promoter context. The least effect in the core −35 promoter element was at −31, but even here, alteration of the wild-type A decreased transcription from 4.7-fold to 22-fold, depending on the substitution (Fig. 2B).
FIG. 2.
Effect of point substitutions within the hctB promoter on in vitro transcription by C. trachomatis σ28 RNA polymerase. All three possible point substitutions were tested at each position in the −35 element from −37 to −24 (A) and in the −10 element from −17 to −4 (C). The wild-type sequence of the predicted element is shown below each graph. Changes in promoter activity are depicted as the decrease (n-fold) relative to wild-type promoter activity. Decreases greater than 200-fold are not shown as extending below the bottom axis. Each bar represents the mean of three independent experiments. Sample transcription of DNA templates containing the wild-type (wt) hctB promoter and point substitutions of positions −31 (B) and −11 (D) are shown.
We also tested the GTTT sequence directly downstream of the TAAA core −35 element that has been proposed to be part of an extended σ28 promoter (12). Point substitutions in each of these four positions from −30 to −27 decreased transcription, although to different degrees. For instance, the greatest effect was at position −30, where the wild-type G was preferred and a C, T, or A substitution produced a 7-, 14-, and 106-fold decrease in promoter activity, respectively. There was a lesser effect at positions −29 and −28, where T and A were equally favored, and at −27, where the pyrimidines (T or C) were preferred.
Mutations in the three positions upstream and downstream of the predicted −35 promoter element had much less of an effect on transcriptional activity. The largest effect was a sevenfold decrease for a T to A substitution at position −26. Only 3 of the 18 point substitutions in these flanking positions produced a decrease in transcription of at least fivefold compared to the wild-type hctB promoter. In contrast, 18 of the 24 substitutions in the extended −35 element reduced transcription by fivefold or greater. These results demonstrate that the flanking sequences contribute little to overall promoter activity compared to the extended version of the −35 element.
The results for the entire −35 region can be visualized in a graphical format shown in Fig. 2A, which displays relative decreases in promoter activity (n-fold) for each point substitution and position compared to the wild-type hctB promoter. The most important positions were −34 to −30 (TAAAG) with a smaller contribution at −28 (A or T) and −27 (a pyrimidine). The sequence at position −29 within the extended −35 region and the flanking positions from −37 to −35 and −26 to −24 had minimal effects on transcription.
Point substitutions in the −10 promoter region that decreased transcription by C. trachomatis σ28 RNA polymerase are clustered.
Although many point substitutions in the −10 region affected the activity of the hctB promoter, the ones with the largest effect were physically clustered. Fourteen of the 24 point substitutions caused greater than a fivefold decrease in transcription, and the greatest effects were at positions −12 to −10 (Fig. 2C). At −12, a substitution of C to G decreased transcription 157-fold while A and T substitutions resulted in decreases of 50-fold and 17-fold, respectively. Substitutions at −11 (Fig. 2D) and −10 caused more than a 20-fold reduction in promoter activity. In contrast, the effects of substitutions at the other positions (−14, −13, −9, and −8) were relatively modest, and there was no nucleotide preference at −7. The sequences flanking the predicted −10 promoter had little or no effect on transcription. From the graphical representation of the results shown in Fig. 3A, it is apparent that the CGA sequence at positions −12 to −10 in the −10 element was most important for promoter activity.
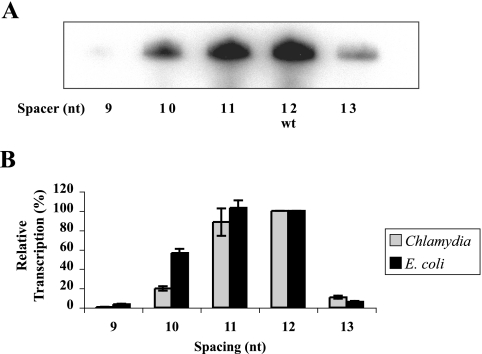
FIG. 3.
Effect of the spacer length on transcription. (A) In vitro transcription by C. trachomatis σ28 RNA polymerase of hctB promoter templates containing a spacer of 9 to 13 nt as indicated. The 12-nt wild-type (wt) spacer is marked for reference. (B) Graph showing quantification of the transcription results for C. trachomatis and E. coli RNA polymerases. Reactions were performed in triplicate, and standard deviations are marked by error bars. Results for each RNA polymerase were normalized to a promoter activity of 100% for the wild-type hctB promoter.
Spacer length affected promoter activity.
We also tested the effect of altering the length of the spacer between the −35 and −10 elements of the hctB promoter. This chlamydial promoter is unusual in having a 12-nt spacer, unlike the known σ28 promoters in E. coli, Salmonella, and Bacillus, which have a spacer of 11 nt (11). We tested mutant hctB promoters with a spacer of 9, 10, 11, or 13 nt and compared transcription by C. trachomatis σ28 RNA polymerase to the wild-type promoter (Fig. 3A). Our results show that the promoter activity was similar for a spacer of 11 and 12 nt, indicating that these spacer lengths are equally acceptable (Fig. 3B). In contrast, the other spacer lengths caused large decreases in transcription by C. trachomatis σ28 RNA polymerase. A 1-nt change in the spacer length to 10 or 13 nt decreased activity by four- and eightfold, respectively, while a 9-nt spacer produced a large decrease of 140-fold.
Transcription with E. coli σ28 RNA polymerase produced similar results.
Although a consensus σ28 promoter has been derived from an alignment of known E. coli σ28 promoters (12, 14, 16), the relative importance of each position has not been defined. We decided to take advantage of our panel of mutant hctB promoters and repeated our analysis with E. coli σ28 RNA polymerase to define the E. coli σ28 promoter.
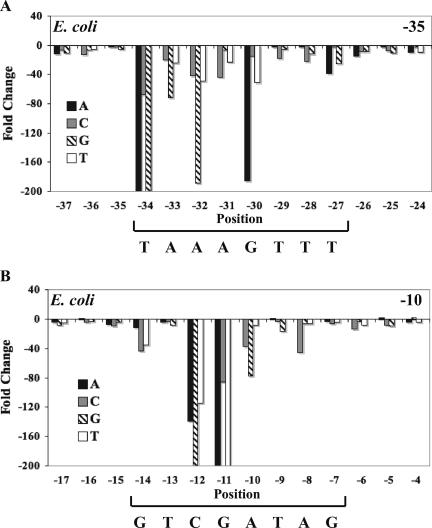
We found that the effect of individual point substitutions on transcription by E. coli σ28 RNA polymerase was very similar to the results obtained with chlamydial σ28 RNA polymerase. In some instances, the magnitude of the effect was different, but the overall pattern in the −35 and −10 elements was the same with respect to both the positions of importance for promoter activity and the preferred nucleotides at those positions. In general, we measured larger decreases in transcription with E. coli σ28 RNA polymerase than with the chlamydial enzyme (compare Fig. 4 with Fig. 2A and C). One notable difference was at position −14 in the −10 element, where substitution of the wild-type G produced large decreases in transcription by E. coli σ28 RNA polymerase, whereas it had little effect on chlamydial σ28 RNA polymerase. Changing the G to a C, A, or T at this position decreased E. coli transcription by 43-, 10-, and 34-fold, respectively (Fig. 4B). Point substitutions in the flanking positions also had a somewhat larger effect on transcription by E. coli σ28 RNA polymerase. For example, altering the T at −15 to a C decreased transcription by eightfold, compared to a threefold decrease with chlamydial σ28 RNA polymerase. The effect of altering the spacer length was similar for the two σ28 RNA polymerases. Like the chlamydial enzyme, E. coli σ28 RNA polymerase tolerated a spacer length of 11 and 12 nt equally well (Fig. 3B). There was a slight difference with a 10-nt spacer, which only caused a twofold reduction in transcription by E. coli σ28 RNA polymerase compared to a fourfold decrease with chlamydial σ28 RNA polymerase (Fig. 3B). These results demonstrate that the basic promoter specificity of σ28 RNA polymerase in C. trachomatis and E. coli is generally well-conserved.
FIG. 4.
Effect of point substitutions within the hctB promoter on in vitro transcription by E. coli σ28 RNA polymerase. All three possible point substitutions were tested at each position from −37 to −24 (A) and from −17 to −4 (B). The wild-type sequence of each predicted promoter element is shown below the respective graph. Changes in promoter activity are shown as the decrease (n-fold) relative to wild-type promoter activity. Decreases greater than 200-fold are not shown as extending below the bottom axis. Each bar represents the mean of three independent experiments.
Derivation of an optimal σ28 promoter sequence.
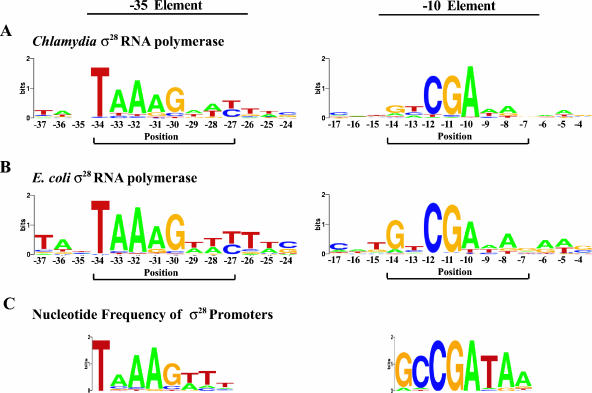
A convenient way of representing the contribution of the four possible nucleotides at each position in the promoter is with a sequence logo (http://weblogo.berkeley.edu) where the height of each letter (A, C, G, or T) is a measure of the relative importance of that nucleotide to transcription (Fig. 5). At positions where the nucleotide choice had no effect on promoter activity, such as at flanking position −35, the sequence logo shows the height of the stack of letters as zero. In contrast, if a particular nucleotide at a given position is absolutely required for promoter activity, it is assigned the maximum height, which is defined as 2 bits (4), and each of the other three nucleotides has a height of zero. If there is a relative preference for one nucleotide, it is shown as the tallest letter at that position, with its height representing the extent of the preference over the other nucleotides. This representation makes it easy to see both the positions where the sequence has the greatest effect on promoter activity and the preferred nucleotide at these positions. For example, with chlamydial σ28 RNA polymerase (Fig. 5A), the most important sequences in the −35 element appear to be TAAAG at positions −34 to −30, which correspond to the core consensus σ28 promoter plus the first position of the extended region. The remaining three positions of the extended promoter had a more modest effect on promoter activity.
FIG. 5.
Sequence logos for the −35 and −10 elements of the σ28-dependent promoter. (A) Sequence recognized by C. trachomatis σ28 RNA polymerase in the context of the C. trachomatis hctB promoter. (B) Sequence recognized by E. coli σ28 RNA polymerase in the same promoter context. (C) The sequence logo based on the nucleotide frequencies of known bacterial σ28 promoters. Details of the sequence logo format are presented in the Materials and Methods and Results sections. All sequence logos were derived using WebLogo, which is available online at http://weblogo.berkeley.edu.
In the −10 element, our analysis shows that a CGA motif at positions −12 to −10 was most important for transcription by chlamydial σ28 RNA polymerase, while other sequences in the consensus −10 promoter element had a smaller role. Overall, the promoter sequences recognized by C. trachomatis σ28 RNA polymerase are consistent with the consensus bacterial σ28 promoter, but our analysis provides additional information about the relative importance of individual positions in the promoter.
When we display the results of the analysis with E. coli σ28 RNA polymerase in the sequence logo format (Fig. 5B), we can see the overall similarity to the chlamydial results. The nucleotide preferred at each position is similar, although the E. coli results show a slightly greater sequence preference as a consequence of the larger effects of point substitutions on transcription by E. coli σ28 RNA polymerase. For instance, the extended portion of the −35 element from −30 to −27 seems to be more important for E. coli σ28 RNA polymerase. In the −10 element, there were two positions in which there was a notable difference in the degree to which a particular nucleotide was preferred over all others: at −14, G had greater importance while at −10, A was less important for promoter activity compared to C. trachomatis σ28 RNA polymerase. Overall, our comparative analysis demonstrates that promoter recognition by σ28 RNA polymerase from the two bacterial genera appears to be well conserved and supports the published consensus and extended σ28 promoter sequences. From the sequence logo analysis, we can derive a σ28 promoter with the sequence TAAAGwwy-n11/12-ryCGAwrn, where w is A or T, y is a pyrimidine, r is a purine, and n11/12 is a spacer of 11 or 12 nt.
DISCUSSION
In this study, we have used a mutational approach to determine the optimal promoter sequence recognized by σ28 RNA polymerase in the context of the C. trachomatis hctB promoter. Prior to this work, there has been little experimental data analyzing the determinants of promoter strength for σ28 RNA polymerase. Our approach was based on the observation that substitution of a single nucleotide in the promoter region can have a dramatic effect on the transcriptional activity of a strong promoter. By comparing each of the four possible nucleotides one position at a time, while keeping other sequences unchanged, we were able to identify the positions where the sequence was important for transcription and the relative preference for each nucleotide at that position. We then compiled the results to generate a composite view of the preferred nucleotide sequence for the σ28-dependent promoter. The promoter sequences we derived for σ28 RNA polymerase from C. trachomatis and E. coli were similar (Fig. 5) and also resembled the consensus σ28 promoter (3, 11). This conservation of σ28 promoter specificity is consistent with conservation at the level of the σ28 protein between C. trachomatis and E. coli (34% amino acid identity and an additional 56% similarity). In addition, the regions involved in promoter recognition, such as region 2.4 for −10 recognition (50% identity plus 25% similarity) and region 4.2 for −35 recognition (62% identity plus 17% similarity) show a higher level of conservation. There is also in vivo functional evidence of σ28 conservation as Chlamydia σ28 protein can complement a Salmonella enterica serovar Typhimurium σ28 mutant in motility studies (13).
In the −35 promoter element, our findings confirm the importance of the core −35 consensus sequence (TAAA) and provide experimental support for the extended −35 promoter that has been predicted from alignment of strongly transcribed σ28 promoters from E. coli and Salmonella (12). Our promoter analysis suggests that the most important position in the extended portion (GTTT) of the −35 element is the initial G, which is immediately downstream of the core −35 sequence. Our results are based on the sequence recognized by σ28 RNA polymerase from Chlamydia and E. coli in functional studies, but they are recognizably similar to the sequence depicting the frequency of each nucleotide in a compilation of known bacterial σ28 promoters (compare Fig. 5A and B with C). For example, the preferred −35 sequence determined with these two very different methods of promoter definition is essentially the same, although there are differences in the degree of nucleotide preference at some of the positions.
In the −10 promoter element, we have identified a CGA motif from −12 to −10 that was important for transcription by both chlamydial and E. coli σ28 RNA polymerases. This motif is not apparent in the −10 sequence derived from the nucleotide frequency of known bacterial σ28 promoters (Fig. 5C). We believe that we were able to identify this previously unrecognized promoter motif because our point substitution analysis allowed us to determine the relative importance of each position in the promoter, providing a higher-resolution view of the promoter sequence. Since this motif was important for σ28 RNA polymerase from two highly divergent bacteria, we propose that it is the hallmark of the −10 element and a critical determinant of σ28 promoter activity.
Our analysis also indicates that σ28 RNA polymerase is able to recognize promoters with a spacer length of 11 or 12 nt and, to a lesser extent, 10 nt. All functionally studied σ28 promoters in E. coli, Salmonella, and Bacillus have had an 11-nt spacer, although the list is relatively small (3, 11, 12). The Caulobacter flbF promoter was reported to have a 10-nt spacer (17), but the strength of this promoter compared to the highly transcribed E. coli promoters has not been determined. σ28 promoters with different spacer lengths have been predicted but not validated in Agrobacterium (2). When first characterized, the 12-nt spacer of the chlamydial hctB promoter was considered unusual compared to the canonical 11-nt spacer length (21). However, our study demonstrates that a 12-nt spacer length is well tolerated and explains why the hctB promoter was highly transcribed. We do not know whether a 12-nt spacer length is more common in Chlamydia or whether σ28 promoters with a 12-nt spacer remain to be identified in other bacteria.
The consensus σ70 promoter has been defined from a compilation of known σ70 promoters, but this approach has limitations for determining the promoter recognized by an alternative σ factor. The σ70 consensus promoter is based on a large cohort of individually defined E. coli promoters (8, 10, 15). For most alternative σ factors, however, far fewer promoters are available for comparison, and there is a greater likelihood of sample bias with this smaller pool. For σ28, a total of about 50 promoters in a range of bacteria have been predicted based on sequence similarity (6, 7, 11), but only about half have been functionally studied. Furthermore, this small number is overrepresented by promoters for a few class 3 flagellar genes from different bacteria. We propose that this sample bias accounts for the homogeneous nature of the −10 sequence that has been derived from an alignment of known σ28 promoters (Fig. 5C) and masked the importance of the CGA motif.
This comprehensive mutational analysis for defining a promoter has advantages and disadvantages compared to the derivation of a consensus sequence from known promoters. It is attractive because it can be performed on a single promoter, although we have found that this approach works best on a promoter that is highly transcribed so that the effect of a single point substitution can be measured. When we used this approach to determine the chlamydial σ66 promoter, we had better success defining the −35 element with the strong promoter for the dnaK operon than with the C. trachomatis rRNA promoter, whose core promoter sequence is not as strongly transcribed or conserved (19). The use of a single promoter, however, is an important caveat, as the sequence is defined within this promoter context. We have previously shown proof of principle by demonstrating that a mutational analysis of the C. trachomatis dnaK promoter identified −35 sequences recognized by both C. trachomatis σ66 and E. coli σ70 RNA polymerases that were identical to the σ70 consensus promoter (18).
σ28 RNA polymerase has been proposed as a late regulator of gene expression in Chlamydia because its target gene, hctB, is only transcribed late in the chlamydial developmental cycle (1, 5, 9). However, hctB is the only σ28-regulated gene that has been identified in Chlamydia to date, and it is not known if σ28 RNA polymerase transcribes other late genes or whether σ28-dependent regulation is limited to late developmental expression. With the results of our promoter analysis, we are developing a computer algorithm to identify other candidate σ28-dependent promoters in the chlamydial genome. The relative promoter activities that we have measured for the four possible nucleotides at each promoter position allow us to construct a probability weight matrix for this algorithm. In particular, our results suggest that a CGA motif is the hallmark of the σ28 −10 promoter element and that the spacer length can be either 11 or 12 nt. In certain ways, our approach for studying σ28-dependent promoters in Chlamydia is the reverse of the historical process taken for σ70 promoters in E. coli. Individual σ70 promoters were first identified, and from there, a consensus promoter sequence was determined. In Chlamydia, where the total number of known promoters is small, we have instead used mutational analysis to determine the promoter sequences important for transcriptional activity as a means of identifying additional promoters. This approach is applicable to other bacteria and is particularly suited for alternative σ factors where few promoters are known.
Acknowledgments
We thank Pete Chandrangsu for technical support and Chris Schaumburg, Eike Niehus, Johnny Akers, Allan Chen, and Narae Park for their valuable suggestions. We are very grateful to Dennis Kibler for his help writing computer algorithms for data analysis.
This work was supported by a grant from the NIH (AI 44198). M.T. is supported by an NIH Independent Scientist Award (AI 057563) and H.H.Y.Y. is supported by a predoctoral training grant from the NIH (National Research Service Award 5 T15 LM007443 from the National Library of Medicine).
REFERENCES
- 1.Brickman, T. J., C. E. Barry III, and T. Hackstadt. 1993. Molecular cloning and expression of hctB encoding a strain-variant chlamydial histone-like protein with DNA-binding activity. J. Bacteriol. 175:4274-4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chesnokova, O., J. B. Coutinho, I. H. Khan, M. S. Mikhail, and C. I. Kado. 1997. Characterization of flagella genes of Agrobacterium tumefaciens, and the effect of a bald strain on virulence. Mol. Microbiol. 23:579-590. [DOI] [PubMed] [Google Scholar]
- 3.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahr, M. J., A. L. Douglas, W. Xia, and T. P. Hatch. 1995. Characterization of late gene promoters of Chlamydia trachomatis. J. Bacteriol. 177:4252-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilman, M. Z., J. S. Glenn, V. L. Singer, and M. J. Chamberlin. 1984. Isolation of sigma-28-specific promoters from Bacillus subtilis DNA. Gene 32:11-20. [DOI] [PubMed] [Google Scholar]
- 7.Gilman, M. Z., J. L. Wiggs, and M. J. Chamberlin. 1981. Nucleotide sequences of two Bacillus subtilis promoters used by Bacillus subtilis sigma-28 RNA polymerase. Nucleic Acids Res. 9:5991-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harley, C. B., and R. P. Reynolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res. 15:2343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatch, T. P. 1999. Developmental Biology, p. 29-68. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, D.C.
- 10.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmann, J. D. 1991. Alternative sigma factors and the regulation of flagellar gene expression. Mol. Microbiol. 5:2875-2882. [DOI] [PubMed] [Google Scholar]
- 12.Ide, N., T. Ikebe, and K. Kutsukake. 1999. Reevaluation of the promoter structure of the class 3 flagellar operons of Escherichia coli and Salmonella. Genes Genet. Syst. 74:113-116. [DOI] [PubMed] [Google Scholar]
- 13.Karlinsey, J. E., and K. T. Hughes. 2006. Genetic transplantation: Salmonella enterica serovar Typhimurium as a host to study sigma factor and anti-sigma factor interactions in genetically intractable systems. J. Bacteriol. 188:103-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kutsukake, K., Y. Ohya, and T. Iino. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lisser, S., and H. Margalit. 1993. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 21:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park, K., S. Choi, M. Ko, and C. Park. 2001. Novel sigmaF-dependent genes of Escherichia coli found using a specified promoter consensus. FEMS Microbiol. Lett. 202:243-250. [DOI] [PubMed] [Google Scholar]
- 17.Sanders, L. A., S. Van Way, and D. A. Mullin. 1992. Characterization of the Caulobacter crescentus flbF promoter and identification of the inferred FlbF product as a homolog of the LcrD protein from a Yersinia enterocolitica virulence plasmid. J. Bacteriol. 174:857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaumburg, C. S., and M. Tan. 2003. Mutational analysis of the Chlamydia trachomatis dnaK promoter defines the optimal −35 promoter element. Nucleic Acids Res. 31:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan, M., T. Gaal, R. L. Gourse, and J. N. Engel. 1998. Mutational analysis of the Chlamydia trachomatis rRNA P1 promoter defines four regions important for transcription in vitro. J. Bacteriol. 180:2359-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson, A. C., and M. Tan. 2002. Functional analysis of the heat shock regulator HrcA of Chlamydia trachomatis. J. Bacteriol. 184:6566-6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu, H. H. Y., and M. Tan. 2003. Sigma 28 RNA polymerase regulates hctB, a late developmental gene in Chlamydia. Mol. Microbiol. 50:577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]