Abstract
As one of the final steps in the bacterial growth cycle, daughter cells must be released from one another by cutting the shared peptidoglycan wall that separates them. In Escherichia coli, this delicate operation is performed by several peptidoglycan hydrolases, consisting of multiple amidases, lytic transglycosylases, and endopeptidases. The interactions among these enzymes and the molecular mechanics of how separation occurs without lysis are unknown. We show here that deleting the endopeptidase PBP 4 from strains lacking AmiC produces long chains of unseparated cells, indicating that PBP 4 collaborates with the major peptidoglycan amidases during cell separation. Another endopeptidase, PBP 7, fulfills a secondary role. These functions may be responsible for the contributions of PBPs 4 and 7 to the generation of regular cell shape and the production of normal biofilms. In addition, we find that the E. coli peptidoglycan amidases may have different substrate preferences. When the dd-carboxypeptidase PBP 5 was deleted, thereby producing cells with higher levels of pentapeptides, mutants carrying only AmiC produced a higher percentage of cells in chains, while mutants with active AmiA or AmiB were unaffected. The results suggest that AmiC prefers to remove tetrapeptides from peptidoglycan and that AmiA and AmiB either have no preference or prefer pentapeptides. Muropeptide compositions of the mutants corroborated this latter conclusion. Unexpectedly, amidase mutants lacking PBP 5 grew in long twisted chains instead of straight filaments, indicating that overall septal morphology was also defective in these strains.
Most eubacteria owe their shape and mechanical stability to the peptidoglycan sacculus, a semirigid structure composed of multiple carbohydrate chains of alternating N-acetylglucosamine and N-acetylmuramic acid that are interconnected by covalent cross-links between short peptides (14). Far from static, this structure exists in a dynamic state throughout the life of a cell. In Escherichia coli, the peptidoglycan is in constant turnover (34, 35), and the cutting of specific peptide and carbohydrate bonds is important for cell growth, division, separation of daughter cells, and lysis (10, 11, 13-15, 39).
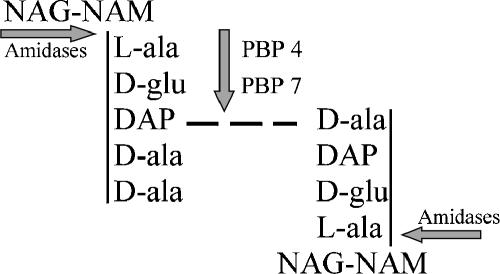
Although enzymes capable of cleaving bonds in the murein sacculus are ubiquitous among the bacteria (39), surprisingly few of these proteins fit into a well-defined role in normal peptidoglycan biochemistry. One reason for this ambiguity is an apparent redundancy, so that the loss of any single enzyme may be masked by the presence of others with equivalent or compensatory activities (13, 39). The situation is exemplified by the widely distributed family of N-acetylmuramyl-l-alanine amidases, represented in E. coli by three closely related members: AmiA, AmiB, and AmiC (10, 11, 14). These periplasmic amidases remove side chains from peptidoglycan by cleaving the bond between the peptide and N-acetylmuramic acid (Fig. 1).
FIG. 1.
Activities of endopeptidases and amidases on peptidoglycan subunits. Endopeptidases PBP 4 and PBP 7 cleave cross-linked peptide side chains, while amidases cut the bond connecting the peptide to the NAM component of the polymerized glycan backbone. The figure shows a pentapeptide monomer (upper left) cross-linked (dashed line) to a tetrapeptide monomer (lower right). NAG, N-acetylglucosamine; NAM, N-acetylmuramic acid; l-ala, l-alanine; d-glu, d-glutamic acid; DAP, diaminopimelic acid; d-ala, d-alanine.
The peptidoglycan-specific amidases play an important role in cleaving the septum to release daughter cells after cell division in E. coli (10). Mutants lacking the AmiC protein separate poorly, in that ∼30% of the population exists as chains of three to six unseparated cells (10). If amiA alone is deleted, only 5 to 10% of the population is in chains of three to four cells, and deleting amiB by itself produces no chaining effect at all. Thus, AmiC appears to be the principle septum-cleaving enzyme, and because the enzyme localizes specifically to the septal ring, it is ideally positioned for cleaving the divisional septum (1). But even given the preeminent role of AmiC, cell chaining is exacerbated in mutants lacking all three amidases; in these strains, more than 90% of the cells exist as unseparated chains from 6 to 24 cells long (10). Thus, the three amidases have overlapping or redundant functions with regard to cell separation. Yet the continued existence of some short chains indicates that residual septum cleavage occurs, suggesting that at least one other enzyme can separate daughter cells, albeit with reduced efficiency.
The additional septum-cleaving enzymes belong to two other classes of peptidoglycan hydrolases: lytic transglycosylases, which cut the glycan chain, and endopeptidases, which cut peptide cross-links at a different site than that cleaved by amidases (Fig. 1). When the sltY gene, encoding lytic transglycosylase Slt70, was deleted from a triple amidase mutant, the chain length increased to 8 to 40 cells, indicating that this enzyme also plays a role in cell separation (10). Still, the existence of eight-cell chains suggests that cell separation occurs via another route, possibly due to the action of endopeptidases. An E. coli mutant lacking all three murein endopeptidases (PBP 4, PBP 7, and MepA) continues to separate normally and grows as single cells (11), indicating that these enzymes are not primary septum-cleaving factors. However, a mutant lacking the three endopeptidases and all three amidases grows as chains from 6 to 80 cells long (11), indicating that the chaining phenotype of the amidase mutant is exacerbated by removing the endopeptidases. The cumulative effect of deleting all amidases, all endopeptidases, and Slt70 is even more severe (11). Therefore, although AmiC appears to be the major cell separation factor in E. coli, it is aided or can be replaced by other peptidoglycan hydrolases.
E. coli contains two low-molecular-weight (LMW) penicillin-binding protein (PBP) endopeptidases, PBP 4 and 7, which could play a role in septum cleavage either by directly cutting peptide side chains (Fig. 1) or by modifying the peptidoglycan to provide alternate substrates for the amidases. Because the individual endopeptidases were not examined for their contributions to cell separation (11), we deleted individual PBPs and amidases to see which enzymes facilitated cell separation. Amidase mutants lacking PBP 4 grew as longer chains, identifying a specific biological role for this endopeptidase in cell separation. PBP 7 appeared to play a secondary role. In addition, we found that amidases apparently prefer different peptidoglycan substrates: AmiC is more likely to remove tetrapeptides from the glycan chains, while AmiA and AmiB act on either tetrapeptides or pentapeptides. Finally, particular combinations of PBPs and amidases affected the overall septum geometry in that certain mutants formed highly abnormal cell chains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phage.
Bacterial strains and plasmids are listed in Table 1. All bacteria were grown in Luria-Bertani medium with appropriate antibiotics where required: ampicillin (50 μg/ml), kanamycin (50 μg/ml), chloramphenicol (50 μg/ml), or spectinomycin (50 μg/ml). Yeast extract and tryptone were from Difco (Detroit, Mich.). All other chemicals and antibiotics were from Sigma Chemical Co. (St. Louis, Mo.) or Fisher Scientific (Pittsburgh, Pa.). Transduction via bacteriophage P1 was performed as previously described (27).
TABLE 1.
E. coli strains
| Strain (reference)a | Genotypeb | PBP(s) deletedc | Amidase(s) deleted |
|---|---|---|---|
| CS109 | W1485 rph rpoS | ||
| APCS-1 (25) | CS109 yhbE-3650::res-npt-res (Kanr cassette insertion near dacB+) | ||
| APCS-3 (25) | CS109 yeh-7310::res-npt-res (Kanr cassette insertion near pbpG+) | ||
| BMCS04-1K (26) | CS109 mrcA::res-npt-res | 1a | |
| BMCS16-1K (25) | CS109 ybe-9207::res-npt-res (Kanr cassette insertion near dacA+) | ||
| BUDDY (16) | MC4100 tatC::spc | ||
| KM-32 (28) | argE3 his-4 leuB6 proA2 thr-1 ara-14 galK2 lacY1 mlt-1 xyl-5 thi-1 rpsL31 tsx-33 supE44 Δ(recC ptr recB recD)::Plac-bet exo cmr | ||
| KMRP-1K | KM-32 yje25::res-npt-res (Kanr cassette insertion near amiB+) | ||
| MC1061 (3) | araD139 Δ(ara- leu)7697 ΔlacX74 galU galK hsdR2 rpsL | ||
| MHD8 (10) | MC1061 amiC::Kan | AmiC | |
| MHD9 (10) | MC1061 amiA::Cam | AmiA | |
| MHD41 (10) | MC1061 amiB | AmiB | |
| MHD41-12K | MC1061 amiB yje25::res-npt-res | AmiB | |
| RPKM179-1K | KM-32 amiC::res-npt-res | AmiC | |
| RP1 | CS109 amiC::Kan | AmiC | |
| RP3 | CS203-1B amiC::Kan | 4, 7 | AmiC |
| RP4 | CS204-1 amiC::Kan | 5, 7 | AmiC |
| RP8 | CS219 amiC::Kan | 4, 5 | AmiC |
| RP9 | CS220-1 amiC::Kan | 4, 6 | AmiC |
| RP12 | CS322-1 amiC::Kan | 4, 5, 6 | AmiC |
| RP14 | CS446-1 amiC::Kan | 4, 5, 6, 7 | AmiC |
| RP19 | CS703-1 amiC::Kan | 1a, 4, 5, 6, 7, C, H | AmiC |
| RP20 | CS702-1 amiC::Kan | 1b, 4, 5, 6, 7, C, H | AmiC |
| RP21 | CS219 amiA::Cam | 4, 5 | AmiA |
| RP22-1K | CS219 amiB yje25::res-npt-res | 4, 5 | AmiB |
| RP24-1K | CS204 amiB yje25::res-npt-res | 5, 7 | AmiB |
| RP25-1K | CS11-2 amiB yje25::res-npt-res | 4 | AmiB |
| RP26 | CS11-2 amiA::Cam | 4 | AmiA |
| RP27-1K | CS9-19 amiB yje25::res-npt-res | 7 | AmiB |
| RP28 | CS9-19 amiA::Cam | 7 | AmiA |
| RP29-1K | CS12-7 amiB yje25::res-npt-res | 5 | AmiB |
| RP30 | CS12-7 amiA::Cam | 5 | AmiA |
| RP31 | CS12-7 amiC::Kan | 5 | AmiC |
| RP32 | CS11-2 amiC::Kan | 4 | AmiC |
| RP33 | CS9-19 amiC::Kan | 7 | AmiC |
| RP37 | CS109 amiA::Cam | AmiA | |
| RP40 | RP24-1 amiA::Cam | 5, 7 | AmiA, AmiB |
| RP41 | RP22-1 amiA::Cam | 4, 5 | AmiA, AmiB |
| RP42 | RP29-1 amiA::Cam | 5 | AmiA, AmiB |
| RP43 | RP25-1 amiA::Cam | 4 | AmiA, AmiB |
| RP44 | RP27-1 amiA::Cam | 7 | AmiA, AmiB |
| RP45 | CS14-2 amiC::Kan | C | AmiC |
| RP46 | CS15-3 amiC::Kan | H | AmiC |
| RP48 | CS17-1 amiC::Kan | 6 | AmiC |
| RP49 | CS109 amiB yje25::res | AmiB | |
| RP50 | RP30 ybe-9207::res-npt-res | AmiA | |
| RP52 | RP21 ybe-9207::res-npt-res | 4 | AmiA |
| RP53 | RP21 yhbE-3650::res-npt-res | 5 | AmiA |
| RP60-1K | CS16-1 amiB yje25::res-npt-res | 1b | AmiB |
| RP61-1K | CS17-1 amiB yje25::res-npt-res | 6 | AmiB |
| RP64-1K | CS703-1 amiB yje25::res-npt-res | 1a, 4, 5, 6, 7, C, H | AmiB |
| RP65-1K | BMCS04 amiB yje25::res-npt-res | 1a | AmiB |
| RP70 | CS331-1 amiC::Kan | 5, 6, 7 | AmiC |
| RP75 | RP49 amiA::Cam | AmiA, AmiB | |
| RP77 | RP75 amiC::Kan | AmiA, AmiB, AmiC | |
| RP79 | CS17-1 amiA::Cam | 6 | AmiA |
| RP80 | CS18-3K amiA::Cam | DacD | AmiA |
| RP102 | RP19 amiA::Cam | 1a, 4, 5, 6, 7, C, H | AmiA, AmiC |
| RP103 | RP30 amiC::Kan | 5 | AmiA, AmiC |
| RP104 | RP32 amiA::Cam | 4 | AmiA, AmiC |
| RP106 | RP43 amiC::Kan | 4 | AmiA, AmiB, AmiC |
| RP107 | RP44 amiC::Kan | 7 | AmiA, AmiB, AmiC |
| RP108 | RP42 amiC::Kan | 5 | AmiA, AmiB, AmiC |
| RP109 | RP40 amiC::Kan | 5, 7 | AmiA, AmiB, AmiC |
| RP110 | RP41 amiC::Kan | 4, 5 | AmiA, AmiB, AmiC |
| RP113 | RP4 amiA::Cam | 5, 7 | AmiA, AmiC |
| RP114 | RP8 amiA::Cam | 4, 5 | AmiA, AmiC |
| RP117 | CS315-1 amiA::Cam | 4, 5, 7 | AmiA |
| RP118-1K | CS315-1 amiB yje25::res-npt-res | 4, 5, 7 | AmiB |
| RP120 | RP117 amiC::Kan | 4, 5, 7 | AmiA, AmiC |
| RP130 | RP3 amiA::Cam | 4, 7 | AmiA, AmiC |
| RP139 | RP14 amiA::Cam | 4, 5, 6, 7 | AmiA, AmiC |
| RP144 | RP118-1 amiC::Kan | 4, 5, 7 | AmiB, AmiC |
| RP145 | RP144 amiA::Cam | 4, 5, 7 | AmiA, AmiB, AmiC |
| RP149 | CS609-1 amiC::Kan | 1b, 4, 5, 7, C, H | AmiC |
| RP152 | RP149 amiA::Cam | 1b, 4, 5, 7, C, H | AmiA, AmiC |
| RP154 | CS610-1 amiA::Cam | 1b, 4, 6, 7, C, H | AmiA |
| RP156 | CS203-1B amiB yje25::res | 4, 7 | AmiB |
| RP157 | RP156 amiA::Cam | 4, 7 | AmiA, AmiB |
| RP158 | RP157 amiC::Kan | 4, 7 | AmiA, AmiB, AmiC |
| RP160 | CS447-1 amiC::Kan | 4, 6, 7, C | AmiC |
| RP164 | RP160 amiA::Cam | 4, 6, 7, C | AmiA, AmiC |
| RP167 | CS316-1 amiC::Kan | 4, 6, 7 | AmiC |
| RP168-1K | CS316-1 amiB yje25::res-npt-res | 4, 6, 7 | AmiB |
| RP169 | CS703-1 amiA::Cam | 1a, 4, 5, 6, 7, C, H | AmiA |
| RP172 | RP168-1 amiA::Cam | 4, 6, 7 | AmiA, AmiB |
| RP173 | RP172 amiC::Kan | 4, 6, 7 | AmiA, AmiB, AmiC |
| RP175 | CS703-1 tatC::Spc | 1a, 4, 5, 6, 7, C, H | |
| RP178 | RP154 amiC::Kan | 1b, 4, 6, 7, C, H | AmiA, AmiC |
| RP184-1K | CS446-1 amiC::res-npt-res | 4, 5, 6, 7 | AmiC |
| RP185-1K | CS331-1 amiC::res-npt-res | 5, 6, 7 | AmiC |
| RP188 | RP184-1 yhbE-3650::res-npt-res | 5, 6, 7 | AmiC |
| RP189 | RP184-1 ybe-9207::res-npt-res | 4, 6, 7 | AmiC |
| RP191 | RP184-1 yeh-7310::res-npt-res | 4, 5, 6 | AmiC |
Strains having prefixes CS or RP were derived from E. coli CS109. CS strains have been described previously (4, 25, 26), and RP strains were created for this work.
Strains were created by moving genes by P1 transduction from the following source strains: amiC::Kan (from MHD8), amiA::Cam (from MHD9), amiB yje25::res-npt-res (from MHD41-12K), amiC::res-npt-res (from RPKM179-1K), and tatC::spc (from BUDDY). Strains carrying the res-npt-res allele have a suffix K. After removal of the kanamycin gene from these strains by using the ParA resolvase (21, 28), the suffix K was removed from the strain name.
Numbers refer to the respective PBPs: 1a, PBP 1a; 1b, PBP 1b; 4, PBP 4; 5, PBP 5; 6, PBP 6; 7, PBP 7; C, AmpC; H, AmpH.
Molecular techniques.
Plasmids were isolated using QIAprep spin miniprep kits (QIAGEN Corp., Valencia, Calif.) according to the manufacturer's instructions. Competent cells were prepared and transformed by electroporation, using a Gene Pulser apparatus from Bio-Rad (Hercules, Calif.). Chromosomal DNA for PCR amplification was prepared by using a DNeasy tissue kit (QIAGEN Corp.). DNA agarose gel electrophoresis was performed as previously described (38). DNA purifications from agarose gels were performed with a QIAquick gel extraction kit (QIAGEN Corp.). Restriction digests and ligations were performed with enzymes from New England Biolabs (Beverly, Mass.). Amplification by PCR was performed in a 2400 GeneAmp Thermal cycler (Perkin Elmer, Boston, Mass.), and oligonucleotide primers for PCR were purchased from MWG Biotech (Highpoint, NC). Deoxynucleotide triphosphates were from Promega (Madison, Wis.,), and Deep Vent DNA polymerase was from New England Biolabs (Beverly, Mass.). PCR was carried out as described previously (25, 26). The res-npt-res kanamycin resistance cassette was amplified from plasmid pBMM1 by using a MasterAmp Extra-long PCR kit from Epicenter (Madison, Wis.).
Construction of amidase deletion mutants.
Strain MHD41 was obtained from Heidrich et al. (10) and carries a deletion within amiB that begins 33 nucleotides downstream of the start codon and ends at nucleotide 1307 of the amiB gene. Because this strain did not contain a selectable marker within the amiB gene, a kanamycin resistance marker was inserted nearby so that the mutation could be moved by P1 transduction into new strains. For this purpose, a kanamycin gene cassette flanked by two res sites was inserted 13,838 bp upstream of the chromosomal position of amiB, into the noncoding region between frdA and yjeA, 25 bp downstream from the stop codon of the frdA gene. The res-npt-res cassette from plasmid pBMM1 (25, 26) was amplified by PCR using the following primers: 5′-CCCACAGCCACGTACTTCAGGGTAAGTACCTGAAAGTTACGGAATTCGAGCTCTGCAGTCC-3′ and 5′-GTCAAATTTCAGACTTATCCATCAGACTATACTGTTGTACGATAAGCTTGCATGCCTGCAG-3′.
The primer pairs amplified the res-npt-res cassette and added sequences to the 5′ and 3′ ends that were identical to the chromosomal insertion site upstream of amiB.
The amplified PCR products were transferred by electroporation into E. coli KM-32, where they were incorporated into the chromosome via homologous recombination mediated by the λ phage recombination system, as previously described (26, 28, 40). Kanamycin-resistant colonies were selected, and colonies having correct insertions were identified by diagnostic PCR of chromosomal DNA by using the following primers: 5′-GGCAGCAATTGCAGCACGTA-3′ and 5′-TAGCCCTCTTGCGCACTAAA-3′. This insertion mutation was named yje25::res-npt-res. The resulting strain was named KMRP-1K, and a phage P1 lysate was prepared on this strain and used to transduce strain MHD41. Transductants were screened by PCR for the presence of the kanamycin cassette by using the above primers and were also screened for the presence of the amiB deletion by using the following primers: 5′-GTTTATGGGGATCCGCGATT-3′ and 5′-TCGTCCTTAGTTTGGCAGCG-3′. One strain containing both the kanamycin cassette and the amiB deletion was selected and named MHD41-12K. A P1 lysate from this organism was used to transfer the res-npt-res-linked amiB deletion to other strains.
An amiC deletion mutant carrying res-npt-res within the amiC gene was also created. The res-npt-res cassette from plasmid pBMM1 was amplified by PCR, using the following primers: 5′-TCATCCCCTTCTCGCCAGCGTCGCCCCATCGGCAAAATACGCGGAATTCGAGCTCTGCAGTCC-3′ and 5′-GATAAGCTTGCATGCCGTCAGGATGACAGATTATGCGTCTTTCGCTAAAGTTTCCGGTCAAA-3′. The primer pairs amplified the res-npt-res cassette and added sequences to its 5′ and 3′ ends that were identical to chromosomal sites 41 bases upstream of the amiC stop codon and 40 bases downstream of the amiC start codon. Kanamycin-resistant colonies were selected, and colonies having correct insertions were identified by diagnostic PCR of chromosomal DNA by using the following primers: 5′-CCATAAAAAAGCGCCATTCA-3′ and 5′-GCGACTTTTTTATCAGTAATGCCGTG-3′. One strain carrying res-npt-res within amiC was selected and named RPKM179-1K.
Microscopy.
Cultures were grown in LB medium at 37°C to an optical density (OD) of approximately 0.6. Cell samples (5 μl) were placed on agarose-coated microscope slides and incubated at room temperature for 5 to 10 min to immobilize the cells. Bright-field microscopy was performed by using a ×100 oil immersion objective on a Nikon EFD-3 microscope. Photographs were obtained with an attached Sensys charged-coupled-device camera and capture software (Photometrics Ltd., Tucson, Ariz.). Cells were counted and grouped into three categories: single cells, two cells attached as double cells, and three or more cells attached in chains. Each category was expressed as the percentage of the total number of cells present, whether in chains or not, to give the equivalent of total mass or units in each group. Data were plotted in Sigma Plot 8.0 and analyzed by a paired Student's t test.
Peptidoglycan preparation.
Peptidoglycan was prepared as previously described (22). Bacteria from a fresh overnight culture were diluted 1:200 into 400 ml of LB medium and grown at 37°C to an OD at 600 nm (OD600) of 0.5 to 0.7. Cells were cooled rapidly to 4°C, harvested, and resuspended to 0.2 g/ml in distilled water. Resuspended cells were added dropwise, with vigorous stirring, to an equal volume of boiling 8% sodium dodecyl sulfate (SDS) solution. The solution was boiled for 30 min, after which the lysate was allowed to cool overnight to room temperature. Insoluble peptidoglycan was pelleted by ultracentrifugation at 100,000 × g for 60 min at 20°C, and the pellet was washed and resuspended in distilled water. This step was repeated six times until the SDS concentration fell below 1 μg/ml, as determined by the methylene blue assay (9). Pelleted peptidoglycan was resuspended in 1 ml of distilled water and disaggregated by sonication (1.5 min; 90% duty cycle; 4.5 output), using a Branson sonifier 450. Glycogen contamination was removed by adding α-amylase (final concentration, 100 μg/ml) and imidazole (final concentration, 0.32 M) and incubating at 37°C for 2 h. Afterwards, pronase was added to 200 μg/ml, and the sample was incubated at 60°C for 1.5 h to remove lipoprotein. After pronase treatment, the solution was added to an equal volume of boiling SDS with vigorous stirring and boiled for 15 min. Insoluble peptidoglycan was pelleted, washed free of SDS as described above, and resuspended in 200 to 300 μl of distilled water containing 0.02% NaNO3. Samples were stored at −20°C.
HPLC analysis.
Muropeptides were produced by digesting peptidoglycan with N-acetylmuramidase SG (USB Specialty Biochemicals, Amersham Life Sciences, Cleveland, Ohio). The reaction mixture consisted of the following: 50 μl peptidoglycan (A208 = 0.3), 7.5 μl of 0.2 M phosphate buffer, pH 5.5, 2.5 μl of 0.2% NaNO3, 5.0 μl of H2O, and 10 μl of N-acetylmuramidase SG (1 mg/ml stock in 20 mM NaPO4 buffer, pH 5.5). Samples were digested by incubation at 37°C overnight, after which the sample was dried under centrifugal vacuum. The resulting muropeptides were reduced with sodium borohydride and fractionated by reverse-phase high-pressure liquid chromatography (HPLC), as previously described (8, 36). The peaks were collected, purified by HPLC using a trifluoroacetic acid-acetonitrile gradient system, identified by amino acid-amino sugar analysis (24), and quantified as previously described (8).
Scanning electron microscopy (SEM).
Cells were grown to mid-log phase in LB medium, collected, resuspended in phosphate-buffered saline, spotted onto glass coverslips coated with poly-l-lysine, fixed in Karnovsky's fixative overnight, postfixed in 1% OsO4, and prepared as described previously (2, 18). Cells were observed and photographed with a Hitachi 4700 field emission scanning electron microscope.
RESULTS
LMW PBPs contribute to cell separation in amiC mutants.
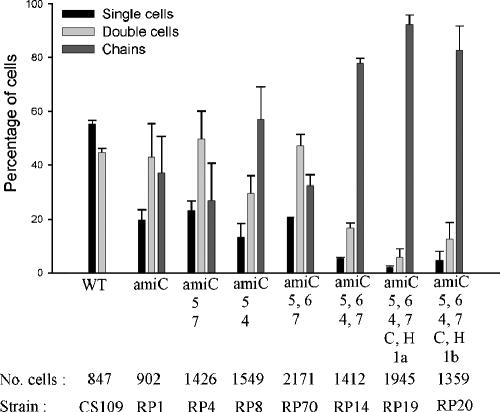
To test whether PBPs play a role in cell separation, we deleted amiC from strains lacking various PBPs. We chose this route because deleting amiC produces a higher percentage of cells in chains than the deletion of amiA or amiB alone (10). In amiC mutants, the numbers of cells in chains increased when certain sets of multiple PBPs were deleted (Fig. 2). At the extremes, 37% of the cell population grew in chains in the control strain RP1 (ΔAmiC), whereas the numbers of cells in chains reached 78% in strain RP14, with deletions of PBPs 4, 5, 6, and 7 and AmiC (ΔPBP4,5,6,7 ΔAmiC), and 92% in strain RP19 (ΔPBP1a,4,5,6,7 ΔAmpC ΔAmpH ΔAmiC) (Fig. 2). Since deleting the high-molecular-weight PBPs 1a or 1b had no differential effects (Fig. 2, strains RP19 and RP20), the results indicated that one or more LMW PBPs played a role in cell separation in amiC mutants. Note that all these results depended on mutating one or more of the ami genes, because no chaining was observed in any PBP mutant that expressed all three wild-type amidases.
FIG. 2.
Multiple PBP deletions increase chaining in amiC mutants. Strains were grown in LB medium at 37°C to an OD600 of 0.6. Cells were counted as described in Materials and Methods. Data are from three independent experiments, and the error bars represent the standard deviation. Deleted genes are indicated on the x axis, and numbers refer to the respective PBPs (e.g., 1a is PBP 1a). C, AmpC; H, AmpH; WT, wild-type parent, CS109; No. cells, the total number of cells counted for each strain. The respective strain names are given at the bottom of the figure. The relevant paired t test results were as follows: RP1 versus RP4, P > 0.05 (not significant); RP1 versus RP8, P = 0.002 (significant); RP4 versus RP8, P = 0.002 (significant).
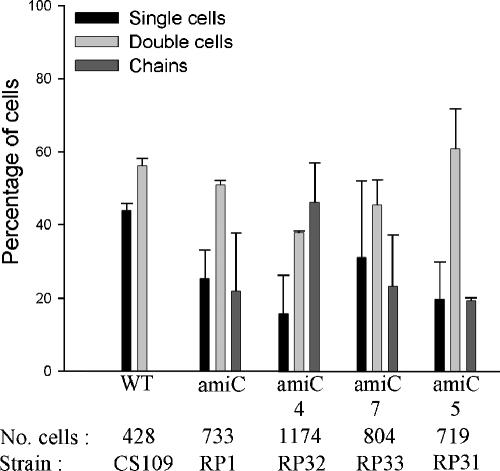
Because strains showing increased chaining were missing four or more LMW PBPs, including the endopeptidases PBP 4 and PBP 7, we analyzed amiC mutants lacking PBPs alone and in combination to determine which were most important for cell separation. In strain RP8 (ΔPBP4,5 ΔAmiC) 57% of cells grew in chains (P = 0.002 compared to either RP1 or RP4), but in RP4 (ΔPBP5,7 ΔAmiC) only 27% of the cells were in chains (P > 0.05 compared to RP1) (Fig. 2). Likewise, for strain RP14 (ΔPBP4,5,6,7 ΔAmiC) 78% of the cells grew in chains, but only 32% were in chains for strain RP70 (ΔPBP5,6,7 ΔAmiC) (Fig. 2). Two strains equivalent to RP14 and RP70 (RP184-1K and RP185-1K, respectively) were generated independently by inserting a different cassette into the amiC gene. Although the absolute values of cells in chains (66% and 42%) (Table 2) were slightly different from those of RP14 and RP70 (Fig. 2), the trend was the same as in all other cases. In each pairwise comparison, deletion of PBP 4 was associated with an increase in chaining, whereas deletion of PBP 7 had little or no effect under the conditions employed. However, when PBPs were removed individually from an amiC mutant, chaining was not increased to a significant degree (P > 0.05) in mutants lacking the endopeptidases PBP 4 (Fig. 3, strain RP32), PBP 7 (Fig. 3, strain RP33), or MepA (data not shown), nor was chaining increased in amiC mutants lacking the dd-carboxypeptidases PBP 5 (Fig. 3, strain RP31) or DacD (data not shown).
TABLE 2.
PBPs complement chaining phenotype of amiC mutants
| Straina | PBPs deletedb | PBP replacedc | Percentage of cells arranged as:
|
No. of cells counted | ||
|---|---|---|---|---|---|---|
| Single cells | Double cells | Chains | ||||
| CS109 | 41.3 ± 0.2 | 58.6 ± 0.2 | 0.00 ± 0 | 416 | ||
| RP1 | 24.1 ± 1.8 | 62.3 ± 4.3 | 13.4 ± 6.2 | 503 | ||
| RP185-1K | 5, 6, 7 | 19.0 ± 0.8 | 39.0 ± 3.0 | 41.8 ± 2.2 | 758 | |
| RP184-1K | 4, 5, 6, 7 | 10.5 ± 2.9 | 23.7 ± 2.8 | 65.7 ± 5.7 | 638 | |
| RP188 | 5, 6, 7 | 4 | 15.9 ± 8.6 | 36.2 ± 7.1 | 47.7 ± 15.9 | 437 |
| RP189 | 4, 6, 7 | 5 | 13.3 ± 5.9 | 30.8 ± 9.5 | 55.7 ± 15.7 | 786 |
| RP191 | 4, 5, 6 | 7 | 19.4 ± 1.0 | 35.3 ± 2.8 | 45.3 ± 4.2 | 535 |
| RP12 | 4, 5, 6 | 19.6 ± 0.8 | 38.5 ± 3.4 | 41.8 ± 4.3 | 1066 | |
| RP167 | 4, 6, 7 | 22.1 ± 3.7 | 44.0 ± 0.3 | 33.5 ± 3.3 | 828 | |
All strains contain an amiC deletion except for E. coli CS109. Strains were grown in LB medium at 37°C until the OD600 = 0.6. Data was from two independent experiments, and the mean ± standard deviation is reported.
Numbers refer to the respective PBPs deleted: 4, PBP 4; 5, PBP 5; 6, PBP 6; 7, PBP 7.
Denotes that the wild type versions of the indicated PBPs replaced genes originally deleted from the chromosome of RP184-1K.
FIG. 3.
Deleting PBP 5 does not increase chaining in amiC mutants. Growth conditions, graphical presentation, and abbreviations are described in the legend of Fig. 2. Data are from two independent sets of experiments. Paired t test results between RP1 and any of the strains lacking PBPs are not significant (P > 0.05).
The absolute number of cells in chains sometimes varied from day to day for any set of strains being compared, but their relative behavior remained constant. For example, the control strain RP1 (ΔAmiC) exhibited variation in chaining percentage on different days (e.g., compare RP1 in Fig. 2 with Fig. 3), but PBP mutations caused similar changes in chaining each time. The causes of this variation are unknown, but the most likely considerations are subtle differences in medium composition, growth conditions, age at harvest, rotatory flask speed, or variation in collection procedures. Though we standardized these parameters, the phenomenon of cell separation may be sensitive to unknown factors. In any case, the relative behavior of the mutants (increases or decreases in chaining) was consistent in any one experiment and was taken into account by using paired statistical tests.
To confirm the results obtained by gene deletion, we complemented the chaining phenotype by reinserting wild-type PBP genes into the chromosome of selected amiC mutants. For this purpose, multiply mutated strains were constructed by using an amiC::res-npt-res insertion mutant from which the kanamycin gene could be removed by site-specific recombination (4, 28, 29). The kanamycin cassette was removed from the amiC locus in these strains, and mutated PBP genes were replaced with their wild-type alleles by selecting for a kanamycin resistance marker inserted nearby on the chromosome.
When wild-type genes for PBPs 4, 5, or 7 were reinserted into E. coli RP184-1K (ΔPBP4,5,6,7 ΔAmiC), each resulting strain exhibited a decrease in chaining (Table 2). When wild-type endopeptidases PBP 4 or PBP 7 were reinserted into RP184-1K, the number of cells in chains decreased by 27% and 32%, respectively, bringing the chain numbers to levels equivalent to those observed for mutants lacking only three PBPs (RP185-1K and RP12, respectively). Reinserting wild-type PBP 5 produced only a 15% decrease in chaining, and this number was 64% higher than the number of cells in chains for strain RP167, from which PBPs 4, 6, and 7 were deleted originally (Table 2).
Because the chaining phenotype was complemented completely or partially by any one of the three LMW PBPs, the results suggest that each protein affects cell separation in this genetic background. The effect of PBPs 4 and 7 indicates that the activities of these two endopeptidases are additive when multiple PBPs are missing from a single strain. Thus, even though deleting PBP 7 had no effect on cell separation when PBP 4 was present (Fig. 3), PBP 7 contributed to separation in the absence of PBP 4. The explanation for the incomplete complementation after reintroducing PBP 5 is not clear, but in light of the possibility that amidases exhibit different substrate preferences (see below), the mutants may have adapted their physiology to increase the rate of removal of pentapeptide side chains, which would be reduced when PBP 5 was reintroduced. In summary, of all the LMW PBPs, PBP 4 contributed most toward cell separation in amiC mutants, and PBP 7 played a role only if PBP 4 was inactive.
Peptidoglycan amidases may prefer different peptide substrates.
Deleting PBP 5 by itself did not significantly increase chaining in E. coli RP31 (ΔPBP5 ΔAmiC) compared to RP1 (ΔAmiC) (Fig. 3). However, the AmiA and AmiB amidases remained active in these strains. The only known activity of PBP 5 is to remove the terminal d-alanine from pentapeptides, thereby creating tetrapeptide side chains; so in the absence of PBP 5, a surplus of pentapeptides accumulates (20, 22). It was possible that the loss of PBP 5 had no effect on cell separation because AmiA or AmiB were able to substitute for AmiC by removing pentapeptides from the glycan chains in strain RP31 (Fig. 3) as well as tetrapeptides from strain RP1 (Fig. 3). This led us to speculate that AmiC acted on a different spectrum of peptide side chains than did AmiA and AmiB.
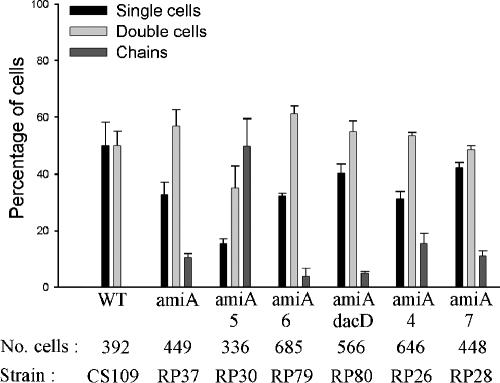
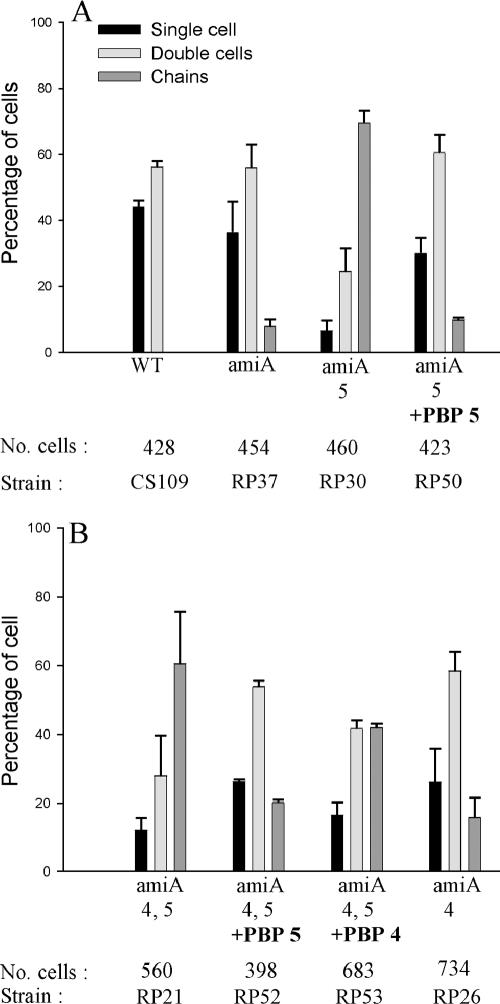
To test the idea that the amidases process different substrates, we deleted combinations of PBPs from amiA or amiB mutants. If AmiA and AmiB cleaved pentapeptide side chains almost as efficiently as tetrapeptides and if AmiC cleaved tetrapeptides more efficiently, then a mutant lacking PBP 5 and either AmiA or AmiB should exhibit increased chaining because pentapeptides would accumulate but would not be removed by AmiC. In fact, only ∼10% of the population of E. coli RP37 (ΔAmiA) was composed of chains, indicating that AmiC was actively removing tetrapeptides, but deletion of PBP 5 from strain RP30 (ΔPBP5 ΔAmiA) resulted in ∼50% (Fig. 4) and up to 70% (Fig. 5) of the population growing in chains, suggesting that AmiC was much less effective in removing pentapeptides than was AmiA. Deletion of no other single PBP increased chaining significantly in amiA mutants (Fig. 4).
FIG. 4.
Deleting PBP 5 increases chaining in amiA mutants. Growth conditions, graphical presentation, and abbreviations are described in the legend of Fig. 2. Data are from two independent sets of experiments.
FIG. 5.
PBPs complement chaining phenotype in amiA mutants. Growth conditions, graphical presentation, and abbreviations are described in the legend of Fig. 2. Data are from two independent sets of experiments. PBPs replaced by the wild-type allele are noted in bold. (A) Effect of replacing wild-type PBP 5 in amiA mutants lacking PBP 5. (B) Effect of replacing wild-type PBP 5 or PBP 4 in an amiA mutant lacking PBPs 4 and 5.
To confirm that the increase in chaining was due to the activity of PBP 5, we reinserted the wild-type allele of the PBP 5 gene into the chromosome of E. coli RP30 (ΔPBP5 ΔAmiA). This new strain, RP50 (ΔAmiA) exhibited ∼10% chaining as opposed to ∼70% chaining for the parent strain RP30 (ΔPBP5 ΔAmiA) (Fig. 5A). We also reinserted the wild-type PBP 4 or PBP 5 genes into the chromosome of E. coli RP21 (ΔPBP4,5 ΔAmiA). Adding back PBP 5 reduced chaining to background levels in strain RP52 (ΔPBP4 ΔAmiA), whereas adding back PBP 4 reduced chaining only slightly in strain RP53 (ΔPBP5 ΔAmiA) (Fig. 5B), perhaps because PBP 4 exhibits a very low dd-carboxypeptidase activity. The results indicate that the chaining phenotype was controlled predominantly by the presence or absence of PBP 5.
As for AmiB, 6.5% of the cells were in chains in E. coli RP49 (ΔAmiB), but deletion of PBP 5 increased this proportion to 23% in E. coli RP29-1K (ΔPBP5 ΔAmiB) (data not shown). This increase in chaining was less than that observed in an amiA background but could be explained if AmiA cleaved pentapeptides more readily than did AmiB. In any case, the results support the hypothesis that AmiC was less effective in removing pentapeptides than were AmiA or AmiB.
Pentapeptide levels increase in amiA and amiB deletion mutants.
If the substrate preferences of the amidases differ, then overall peptidoglycan structure would be expected to vary in the presence or absence of different amidases. To test this, we compared the muropeptide composition of mutants by growing the cells to mid-log phase, isolating the sacculi from each strain, digesting with muramidase, and analyzing the products by reverse-phase HPLC. In the wild-type strain CS109 where dd-carboxypeptidases were active, few pentapeptides were present in the cell wall (∼0.4%) (Table 3). However, E. coli CS703-1, which lacks the major dd-carboxypeptidases, had a much higher pentapeptide content (∼34%) (Table 3). The increase in pentapeptides was accompanied by a roughly equivalent decrease in the amounts of tetrapeptides and tripeptides (Table 3), because PBP 5 does not process the pentapeptide side chains.
TABLE 3.
Muropeptide profiles of amidase mutants
| Straina | Amidase(s) deleted | Percentage of muropeptideb
|
||
|---|---|---|---|---|
| Tripeptide | Tetrapeptide | Pentapeptide | ||
| CS109 | 13.5 ± 0.3 | 86.1 ± 0.4 | 0.4 ± 0.1 | |
| CS703-1 | 5.7 ± 0.6 | 60.0 ± 0.5 | 34.3 ± 0.9 | |
| RP169 | AmiA | 4.4 ± 0.3 | 55.9 ± 0.3 | 39.7 ± 0 |
| RP64-1K | AmiB | 4.6 ± 1.1 | 54.0 ± 2.2 | 41.4 ± 3.2 |
| RP19 | AmiC | 4.7 ± 0.9 | 60.1 ± 2.2 | 35.2 ± 3.1 |
| RP102 | AmiA AmiC | 4.1 ± 0.3 | 58.1 ± 0.3 | 37.9 ± 0 |
All strains are derived from CS703-1 (except for CS109) and carry deletions eliminating PBPs 1a, 4, 5, 6, 7, AmpC, and AmpH (4, 26). CS109 is the original parent strain.
Muropeptide percentages were determined by HPLC and are reported as the mean ± standard deviation of two independent measurements. Bolded results are significantly different from the respective measurements for CS703-1 (P < 0.05).
To determine the effects of amidases on muropeptide composition, we deleted the amiA, amiB, or amiC genes from CS703-1. Compared to this strain, the proportions of pentapeptides in E. coli RP169 (ΔAmiA) and RP64-1K (ΔAmiB) rose from 34.3 to 39.7 (a 16.7% increase) and from 34.3 to 41.4 (a 20.6% increase), respectively, while the proportions of tetrapeptides declined by similar amounts (Table 3). Also, the proportion of pentapeptides rose from 34.3 to 37.9 (a 10.5% increase) in strain RP102 (ΔAmiA ΔAmiC) (Table 3). In each case, in the absence of AmiA or AmiB, the level of pentapeptides increased. These changes are consistent with the prediction that deleting either AmiA or AmiB reduces a strain's ability to remove pentapeptides from the glycan chains of peptidoglycan. Conversely, deleting AmiC produced a different effect: the proportion of pentapeptides was not altered in strain RP19 (ΔAmiC) but instead remained equal to the levels observed in CS703-1 (Table 3). These results are consistent with the prediction that AmiC may remove pentapeptides poorly or not at all.
Deleting PBP 5 produces twisted chains in amidase mutants.
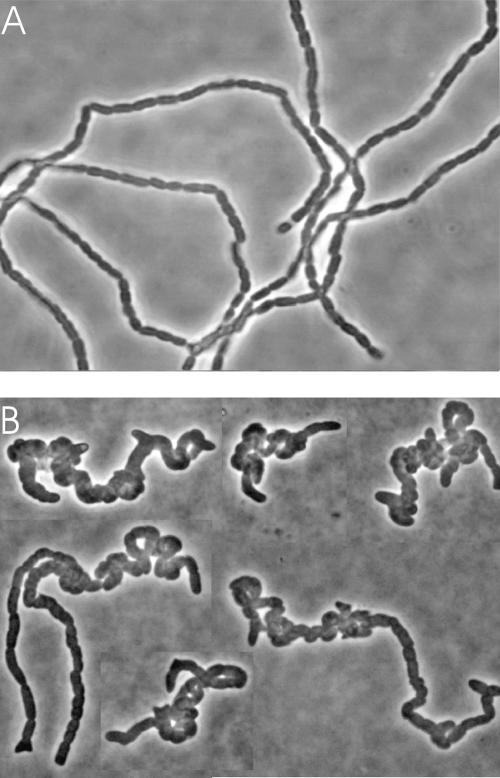
While constructing double amidase deletion mutants, we observed that strain RP139 (ΔPBP4,5,6,7 ΔAmiA ΔAmiC) produced coiled chains but that strain RP75 (ΔAmiABC) grew in long straight chains (Fig. 6). This suggested that the LMW PBPs are not only involved in cell separation but also contribute to the gross morphology of the chains. To determine which PBP contributed to the formation of this twisted phenotype, we constructed double and triple amidase mutants lacking combinations of PBPs. Twisted chains appeared only when the PBP 5 gene, dacA, was deleted in concert with mutations of amiA and amiC. The phenotype was never observed in strains carrying wild-type PBP 5 (Fig. 6, and data not shown). Also, twisted chains were not observed in mutants lacking a single amidase even when PBP 5 was deleted.
FIG. 6.
Deletion of PBP 5 produces spirally coiled chains in amidase mutants. Strains were grown in LB medium at 37°C to mid-log phase. Cells were affixed to agarose-coated slides and observed by phase-contrast microscopy with a ×100 oil immersion objective. (A) RP164 (genes deleted: PBPs 4, 6 and 7; AmpC, AmiA, and AmiC). (B) RP139 (genes deleted: PBPs 4, 5, 6 and 7; AmiA and AmiB). Image B is a composite from different fields.
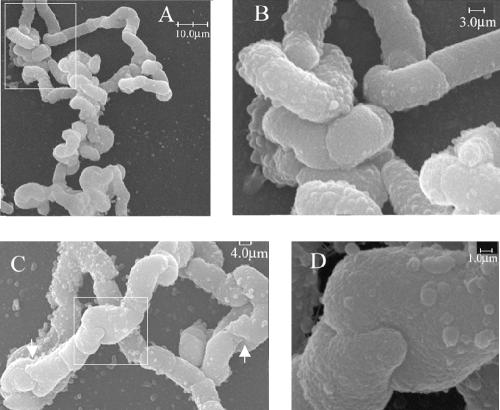
In E. coli RP108 (ΔPBP5 ΔAmiABC) only 1% of the cell population grew in twisted chains, whereas in RP102 (ΔPBP1a,4,5,6,7 ΔAmpC ΔAmpH ΔAmiA ΔAmiC) and RP152 (ΔPBP1b,4,5,7 ΔAmpH ΔAmpC ΔAmiA ΔAmiC), which each lack multiple LMW PBPs, 20 to 25% of the chains were twisted dramatically and extensively (Fig. 7), and ∼70% of the chains were twisted to a lesser degree (data not shown). Furthermore, when AmiB was deleted so that all three amidases were absent, virtually 100% of the cells were present in extraordinarily long, convoluted and twisted chains (data not shown). This suggested that although the absence of PBP 5 was essential for forming the twisted coils, the absence of other LMW PBPs also contributed to the phenotype. SEM of strain RP164 (ΔPBP4,6,7 ΔAmpC ΔAmiA ΔAmiC) revealed rod-shaped cells arrayed in chains, while strains RP102 and RP152 produced coiled chains (Fig. 7A and B and data not shown). These twisted chains did not have a specific pattern and were coiled in random directions. Curiously, neighboring cells in some of the chains were connected to one another in a novel manner. Instead of being separated by a perpendicular septum, the cells were connected by a “saddle joint,” an interlocking pair of saddle-shaped surfaces at the poles of each cell in which finger-like projections were interdigitated (Fig. 7C and D). The implication is that a combination of amidase and PBP mutations impairs the proper shape of the septum, leading to asymmetric division planes, which upon further growth and division produce twisted chains of unseparated cells.
FIG. 7.
SEM analysis of twisted chains. (A) RP102 (genes deleted: PBPs 1a, 4, 5, 6 and 7; AmpC, AmpH, AmiA, and AmiC). (B) Higher magnification of boxed area shown in panel A. (C) Examples of “saddle joint” cell junctures, one boxed and two others indicated by white arrows. (D) Higher magnification of saddle joint in boxed area of panel C.
Deleting tatC produces the twisted chain phenotype in PBP mutants.
AmiA and AmiC are transported to the periplasm via the twin arginine (Tat) pathway (1, 16). Therefore, because amidase mutants grow as chains of unseparated cells, cells defective in the Tat pathway exhibit the same phenotype (1). To confirm that the twisted chain phenotype depended on the absence of AmiA, AmiC, and PBPs, we deleted tatC from E. coli strains lacking multiple LMW PBPs. E. coli RP175 (ΔPBP1a,4,5,6,7 ΔAmpC ΔAmpH ΔTatC) grew as twisted chains, which were not observed in a strain lacking tatC by itself (data not shown). Again, twisted chains only appeared when PBP 5 was deleted and most often when additional PBPs were absent (data not shown). These results are consistent with the interpretation that the phenotype depends on the absence of AmiA and AmiC activity in cells lacking PBP 5 and other LMW PBPs.
DISCUSSION
Following bacterial division, daughter cell separation requires that the common peptidoglycan wall be cut carefully and precisely; to do otherwise risks breaking structurally essential bonds and triggering cell lysis. To accomplish this feat, the relevant hydrolytic enzymes must somehow choose and cleave a select group of chemical bonds among what seem to be equivalent substrates. Somewhat surprisingly, this process includes the activities of several different classes of hydrolytic enzymes (10, 11), each with its own slightly different substrate preferences. The ballet-like balancing of these activities is critical, highlighted by the extreme sensitivity of dividing cells to lysis by antibiotics specific for these enzymes (5, 10, 19). The results presented here represent an extension of what we need to know about how these hydrolases interact with one another and choose their substrates.
Endopeptidase functions.
Most gram-negative bacteria produce several LMW PBPs with well established biochemical capabilities but whose biological functions are ill-defined or completely unknown (14). Of the seven LMW PBPs in E. coli, only four have been associated with a physiological function. The dd-carboxypeptidase PBP 5 plays an important role in creating the uniform rod shape of each cell (30-32), and AmpC augments the resistance of E. coli to β-lactam antibiotics if its expression is increased by mutation (17, 23). PBPs 4 and 7 play unspecified auxiliary roles in maintaining regular cellular morphology (25, 31), enhance the ability of E. coli to form normal biofilms (7), and help separate daughter cells after cell division when the amidases or lytic transglycosylases cannot do so (10, 11). Our data support and extend this last conclusion by showing that PBP 4 facilitates cell separation in the absence of AmiC, the principle peptidoglycan amidase, with PBP 7 supplying secondary assistance.
How might the LMW PBPs contribute to cell separation? Amidases remove peptide side chains from N-acetylmuramic acid by cleaving the bond between the peptide and the glycan chain (Fig. 1). The reaction removes cross-links between glycan chains, which is evidently required to separate daughter cells after septation (10, 11). Because the endopeptidases PBP 4 and PBP 7 cleave a bond internal to the peptide linkage (Fig. 1), they, too, can sever previously connected glycan chains, which appears to have the same overall effect as far as cell separation is concerned. PBP 4 was more efficient in this regard because complementation by PBP 7 was observed only in PBP 4 mutants. However, the fact that PBPs 4 and 7 can effect cell separation only when AmiC is missing reinforces the idea that amidases are the principal septum-cleaving enzymes (10, 11). Thus, PBPs 4 and 7 may have a still unrecognized primary function other than their contribution to cell separation.
A potentially important inference from the current results is that PBPs 4 and 7 may act primarily at the septum. Since the activity of these two PBPs contributes to cell shape (25, 31) and since shape seems to depend on the formation and manipulation of inert peptidoglycan at cell poles (6, 33), PBPs 4 and 7 might regulate the composition or structure of septal peptidoglycan, which would be consistent with their ability to split the septum during cell separation. Also, PBP 7 cleaves intact sacculi but not isolated muropeptides (37), which would harmonize with the idea that PBP 7 requires a particular three-dimensional septal structure as its substrate. Either improper cell shape or a defect in septation may explain why the loss of PBPs 4 and 7 affects biofilm formation (7). Aside from these suppositions, what seems clear is that the data are at odds with the idea that either PBP 4 or PBP 7 is essential for insertion of new glycan chains into preexisting cell wall (12, 14). In particular, because all the endopeptidases can be deleted without affecting cell growth or extension of the lateral wall (4, 11), the data are also inconsistent with the idea that these proteins are essential components of a multienzyme peptidoglycan synthesis complex.
Amidase substrates and functions.
Our current results suggest that the amidases may prefer different substrates in vivo, with AmiC removing tetrapeptide chains more efficiently while AmiA and AmiB may remove both tetrapeptides and pentapeptides, with a possible preference for the latter. When Heidrich et al. assayed the activities of AmiA and AmiC, using peptidoglycan from a ΔamiABC ΔsltY mutant as substrate, AmiA released mostly tetrapeptides, and AmiC released roughly equal amounts of tri- and tetrapeptides (10). This is not at odds with the idea presented in this work that AmiA cleaves pentapeptides because the substrate in the experiments of Heidrich et al. was prepared from cells with functional PBP 5, meaning there was such a small amount of pentapeptide to begin with that any release would go unnoticed. Even so, their results show that the two enzymes do have different in vitro preferences: AmiA showed a preference for tetrapeptide substrates over tripeptides while AmiC acted equally on both.
Of the three amidases, AmiC is clearly the most important for separating cells after septation (10, 11), which makes sense considering the possible substrate preferences we report. Because tetrapeptides comprise the principle type of side chains in wild-type E. coli, AmiC would be expected to be more active during cell separation than AmiA or AmiB. These latter two enzymes would be more active on the less abundant pentapeptide side chains, with AmiA more active than AmiB in this regard. However, in the absence of AmiC, AmiA and AmiB clearly retain sufficient activity toward tetrapeptides to allow cell separation to proceed more slowly.
The results of Heidrich et al. (10) are intriguing in another way. In their in vitro assay, AmiA appeared to release ∼10 times more material than did AmiC when acting on same substrate, suggesting either that AmiA is a more active amidase or that AmiC is more specific in its substrate requirements. In fact, AmiA is distributed uniformly in the periplasm, but AmiC localizes to the septum in a division-specific manner (1), and, when added to isolated murein sacculi, AmiC appears to degrade the thick septal rings more effectively than sidewall peptidoglycan (10). Thus, it is quite possible that AmiC recognizes and degrades muropeptides located mostly at the septum. This would account for the smaller total amount of muropeptides released by AmiC in vitro because the septum is a minor fraction of the sacculi. It would also account for our in vivo results, where the absence of AmiA produces an increase in wall-associated pentapeptides as opposed to little change in the absence of AmiC. In this scenario, AmiC would prefer to cut tri- or tetrapeptides at the septum, whereas AmiA would cleave pentapeptides located anywhere around the sacculus.
Relationships among PBPs and peptidoglycan hydrolases.
Other than the preeminence of AmiC, we cannot assign specific roles or percentages of effort to other amidases or PBPs during septum cleavage. The possibility that the amidases have different substrate preferences suggests that LMW PBPs other than the endopeptidases might also affect cell separation by modifying the muropeptide substrates available to these hydrolytic enzymes. For example, deleting PBP 5 alters the ratio of pentapeptides to tetrapeptides in the sacculus, and mutating multiple combinations of LMW PBPs results in the accumulation of different numbers of peptide side chains (22). If the amidases have different substrate preferences, the changes probably alter the relative roles of these enzymes during cell growth and separation.
One overt sign of this relationship is that deleting PBP 5 in concert with amiA and amiC altered the morphology of chains of unseparated cells. Deleting the amidases by themselves caused E. coli to grow as straight chains, but amidase mutants lacking PBP 5 grew as grossly twisted chains. The most likely explanation for the phenotype is that the increase in pentapeptide side chains affects septum formation or processing, resulting in abnormally shaped poles (6, 31, 32). The simplest explanation for the twisted chains is that daughter cells remain attached to one another at these asymmetric poles, which may themselves be the result of an imbalance of amidase actions during septum formation or cleavage. In effect, deleting the amidases to cause chaining creates a more sensitive visual assay for the cell shape changes caused by loss of the LMW PBPs.
As for the hydrolases themselves, there is a difference in the compounds left behind by the actions of PBPs 4 and 7 and those left by amidases. Cleavage by an endopeptidase leaves two peptide side chains attached to the glycan chains which are therefore available for future cross-linking reactions during the insertion of new peptidoglycan. Conversely, amidases remove the peptides entirely, leaving no side chains at all, meaning that these particular glycan subunits cannot participate as recipients in new cross-linking reactions. Thus, the location and extent of amidase activity may dictate the future course of peptidoglycan insertion in specific areas of the sacculus. This might have a bearing on the chemical nature of “inert peptidoglycan,” which is a functional description without a structural explanation.
Peptidoglycan structure.
Because of the large differences in chaining phenotype caused by deleting various hydrolases, it seems the cells should exhibit significant cell wall differences. Unexpectedly, the muropeptide compositions of hydrolase mutants are surprisingly constant (11; data not shown). In particular, the profiles do not change significantly among mutants of differing chain length, and where the profiles do change, they do so to approximately the same degree in mutants that chain (no amidases) as in those that do not chain (no endopeptidases) (11). Thus, muropeptide profiles do not correlate with the chaining phenotype, implying that cell separation defects are related to differences in peptidoglycan structure rather than to overall composition. The nature of these structural variations is unknown, and no current techniques address this important issue.
Mysteries.
Although we are accumulating more information about what enzymes are active during septum formation and cell separation, many fundamental questions remain unanswered. How does the cell split septal peptidoglycan so that the daughter cells separate while not cutting the wall so that the cell lyses? How do the hydrolases distinguish which bonds to cut and which to leave? Why do some enzymes act only at the septum and not elsewhere, thus sparing the side wall? What is the chemical nature of the peptidoglycan that remains after cell separation? Do these areas have no peptide side chains on their outer surfaces, or is there a surplus of dangling cross-linked peptides? Why are amidases the principle splitters rather than endopeptidases? Why are multiple hydrolases required for efficient separation, and why does a lytic transglycosylase contribute to separation at all? Why does a surfeit of pentapeptides create abnormal septation and poles? It seems clear that answering these questions will describe fundamental relationships between the PBPs and hydrolases, as well as clarifying their roles in regulating bacterial growth and cell morphology.
Acknowledgments
We thank Miguel de Pedro for reading the manuscript and for his helpful suggestions, and we thank Edward Carlson and Donna Laturnus for performing the SEM.
This work was supported by grants GM061019 (to K.Y.) and GM056695 (to D.P.) from the National Institutes of Health.
REFERENCES
- 1.Bernhardt, T. G., and P. A. de Boer. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson, E., J. Audette, L. Klevey, H. Nguyen, and P. Epstein. 1997. Ultrastructural and functional analyses of nephropathy in calmodulin-induced diabetic transgenic mice. Anat. Rec. 247:9-19. [DOI] [PubMed] [Google Scholar]
- 3.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 4.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Pedro, M. A., J.-V. Höltje, and H. Schwarz. 2002. Fast lysis of Escherichia coli filament cells requires differentiation of potential division sites. Microbiology 148:79-86. [DOI] [PubMed] [Google Scholar]
- 6.de Pedro, M. A., K. D. Young, J. V. Holtje, and H. Schwarz. 2003. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J. Bacteriol. 185:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallant, C. V., C. Daniels, J. M. Leung, A. S. Ghosh, K. D. Young, L. P. Kotra, and L. L. Burrows. 2005. Common β-lactamases inhibit bacterial biofilm formation. Mol. Microbiol. 58:1021-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glauner, B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172:451-464. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi, K. 1975. A rapid determination of sodium dodecyl sulfate with methylene blue. Anal. Biochem. 67:503-506. [DOI] [PubMed] [Google Scholar]
- 10.Heidrich, C., M. F. Templin, A. Ursinus, M. Merdanovic, J. Berger, H. Schwarz, M. A. de Pedro, and J. V. Höltje. 2001. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41:167-178. [DOI] [PubMed] [Google Scholar]
- 11.Heidrich, C., A. Ursinus, J. Berger, H. Schwarz, and J. V. Höltje. 2002. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J. Bacteriol. 184:6093-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Höltje, J. V. 1996. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology 142:1911-1918. [DOI] [PubMed] [Google Scholar]
- 13.Höltje, J.-V. 1995. From growth to autolysis: the murein hydrolases in Escherichia coli. Arch. Microbiol. 164:243-254. [DOI] [PubMed] [Google Scholar]
- 14.Höltje, J.-V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Höltje, J.-V. 1993. “Three for one”—a simple growth mechanism that guarantees a precise copy of the thin, rod-shaped murein sacculus of Escherichia coli, p. 419-426. In M. A. de Pedro, J.-V. Höltje, and W. Löffelhardt (ed.), Bacterial growth and lysis. Plenum Press, New York, N.Y.
- 16.Ize, B., N. R. Stanley, G. Buchanan, and T. Palmer. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol. Microbiol. 48:1183-1193. [DOI] [PubMed] [Google Scholar]
- 17.Jacoby, G. A. 1994. Extrachromosomal resistance in Gram-negative organisms: the evolution of β-lactamase. Trends Microbiol. 2:357-360. [DOI] [PubMed] [Google Scholar]
- 18.Karnovsky, M. J.1965. A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 27:137-138A. [Google Scholar]
- 19.Korsak, D., S. Liebscher, and W. Vollmer. 2005. Susceptibility to antibiotics and β-lactamase induction in murein hydrolase mutants of Escherichia coli. Antimicrob. Agents Chemother. 49:1404-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraus, W., and J.-V. Höltje. 1987. Two distinct transpeptidation reactions during murein synthesis in Escherichia coli. J. Bacteriol. 169:3099-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kristensen, C. S., L. Eberl, J. M. Sanchez-Romero, M. Givskov, S. Molin, and V. de Lorenzo. 1995. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J. Bacteriol. 177:52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, S. Y., J.-V. Höltje, and K. D. Young. 2004. Comparison of high-performance liquid chromatography and fluorophore-assisted carbohydrate electrophoresis methods for analyzing peptidoglycan composition of Escherichia coli. Anal. Biochem. 326:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindberg, F., and S. Normark. 1986. Contribution of chromosomal β-lactamases to β-lactam resistance in enterobacteria. Rev. Infect. Dis. 8 (Suppl. 3):S292-S304. [DOI] [PubMed] [Google Scholar]
- 24.Meador-Parton, J., and D. L. Popham. 2000. Structural analysis of Bacillus subtilis spore peptidoglycan during sporulation. J. Bacteriol. 182:4491-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meberg, B. M., A. L. Paulson, R. Priyadarshini, and K. D. Young. 2004. Endopeptidase penicillin-binding proteins 4 and 7 play auxiliary roles in determining uniform morphology of Escherichia coli. J. Bacteriol. 186:8326-8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meberg, B. M., F. C. Sailer, D. E. Nelson, and K. D. Young. 2001. Reconstruction of Escherichia coli mrcA (PBP 1a) mutants lacking multiple combinations of penicillin binding proteins. J. Bacteriol. 183:6148-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Murphy, K. C. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene. 246:321-330. [DOI] [PubMed] [Google Scholar]
- 30.Nelson, D. E., A. S. Ghosh, A. L. Paulson, and K. D. Young. 2002. Contribution of membrane-binding and enzymatic domains of penicillin binding protein 5 to maintenance of uniform cellular morphology of Escherichia coli. J. Bacteriol. 184:3630-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson, D. E., and K. D. Young. 2001. Contributions of PBP 5 and dd-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 183:3055-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson, D. E., and K. D. Young. 2000. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J. Bacteriol. 182:1714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsen, T., A. S. Ghosh, M. B. Goldberg, and K. D. Young. 2004. Branching sites and morphological abnormalities behave as ectopic poles in shape-defective Escherichia coli. Mol. Microbiol. 52:1045-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, J. T. 1996. The murein sacculus, p. 48-57. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 35.Park, J. T. 1995. Why does Escherichia coli recycle its cell wall peptides? Mol. Microbiol. 17:421-426. [DOI] [PubMed] [Google Scholar]
- 36.Popham, D. L., and P. Setlow. 1996. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin-binding proteins. J. Bacteriol. 178:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romeis, T., and J.-V. Höltje. 1994. Penicillin-binding protein 7/8 of Escherichia coli is a DD-endopeptidase. Eur. J. Biochem. 224:597-604. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Shockman, G. D., and J.-V. Höltje. 1994. Microbial peptidoglycan (murein) hydrolases, p. 131-166. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier Science B.V., Amsterdam, The Netherlands.
- 40.Yu, D., H. M. Ellis, E.-C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]