Abstract
Alginate is a linear copolymer of β-d-mannuronic acid and its C-5-epimer, α-l-guluronic acid. During biosynthesis, the polymer is first made as mannuronan, and various fractions of the monomers are then epimerized to guluronic acid by mannuronan C-5-epimerases. The Azotobacter vinelandii genome encodes a family of seven extracellular such epimerases (AlgE1 to AlgE7) which display motifs characteristic for proteins secreted via a type I pathway. Putative ATPase-binding cassette regions from the genome draft sequence of the A. vinelandii OP strain and experimentally verified type I transporters from other species were compared. This analysis led to the identification of one putative A. vinelandii type I system (eexDEF). The corresponding genes were individually disrupted in A. vinelandii strain E, and Western blot analysis using polyclonal antibodies against all AlgE epimerases showed that these proteins were present in wild-type culture supernatants but absent from the eex mutant supernatants. Consistent with this, the wild-type strain and the eex mutants produced alginate with about 20% guluronic acid and almost pure mannuronan (≤2% guluronic acid), respectively. The A. vinelandii wild type is able to enter a particular desiccation-tolerant resting stage designated cyst. At this stage, the cells are surrounded by a rigid coat in which alginate is a major constituent. Such a coat was formed by wild-type cells in a particular growth medium but was missing in the eex mutants. These mutants were also found to be unable to survive desiccation. The reason for this is probably that continuous stretches of guluronic acid residues are needed for alginate gel formation to take place.
Alginate is an industrially important linear copolymer composed of β-d-mannuronic acid (M) and its C-5-epimer, α-l-guluronic acid (G). It is produced by brown algae (25) and by bacteria belonging to the genera Pseudomonas (17, 22, 41) and Azotobacter (10, 32). Azotobacter vinelandii is a nitrogen-fixing soil bacterium belonging to the γ-group of purple bacteria. Vegetatively growing cells of this organism produce alginates that are mostly released from the cell surface. In addition, A. vinelandii synthesizes alginates as part of a reversible differentiation process which is activated under adverse environmental conditions (49). After completion of this process, the cells are at a resting stage designated cyst, and in these cysts the cells are surrounded by a rigid coat consisting of layers (intine and exine) in which alginate is a major component. As expected, mutants unable to produce alginate also fail to form cysts (6).
Alginates isolated from natural sources are, unlike most other polysaccharides, not composed of a uniform pattern of repeating sugar units. In contrast, the fractional contents and sequence distributions of the M and G residues vary widely (54), and alginate structures are described as being composed of G blocks, M blocks, and MG blocks of various lengths. G blocks are of great biological and applied significance since they are a prerequisite for the formation of strong polymer gels in the presence of divalent cations like Ca2+ (26).
All genes involved in bacterial alginate biosynthesis, except algC (20), are clustered on the chromosome and are transcribed in one or several units (6, 8, 34, 36). Polymerization of the sugar monomers takes place in the cytoplasmic membrane, using GDP-mannuronic acid as a precursor. The resulting mannuronan is then transported into the periplasmic space where a fraction of the residues are then epimerized to G at the polymer level by the mannuronan C-5-epimerase (ME) AlgG (7, 21, 48). The alginate is then exported to the extracellular environment, and in A. vinelandii the polymer is further epimerized by a family of MEs designated AlgE1 to AlgE7 (see below). The AlgG-type MEs are believed not to form G blocks, as alginates isolated from Pseudomonas species are devoid of such structural units. However, recently it was shown that Pseudomonas aeruginosa AlgG can form G blocks in vitro (30). An enzyme homologous to the Pseudomonas AlgG is also encoded in the A. vinelandii genome (48), but its epimerization pattern has not been experimentally determined, due to low in vitro activity. Bacterial alginates are to various extents also acetylated in the O-2 and/or O-3 position (54) by periplasmic enzymes (19). After these modifications, the alginate polymer is exported out of the cell, probably via an outer membrane channel encoded in the alginate biosynthetic cluster (46, 47). The algU mucABCD cluster, which is a σE-type cooperative regulatory unit, controls expression of the alginate biosynthesis genes in P. aeruginosa (45) and A. vinelandii (20, 35, 39).
Alginates from Azotobacter species and brown seaweeds contain all three types of block structures, and in A. vinelandii, this appears to be caused by the presence of a set of seven MEs, designated AlgE1 to AlgE7 (14, 15, 55). A similar ME from Pseudomonas syringae (PsmE) has also been reported (4). The AlgE MEs are extracellular, Ca2+-dependent enzymes which generate a variety of epimerization patterns, including G blocks of various lengths. Sequence analyses of all of the A. vinelandii AlgE proteins have revealed that each of them is composed of two types of protein modules. The amino acid sequences of these modules, designated A (one or two in each enzyme) and R (one to seven copies), are highly related but in no case completely identical.
Since the AlgE MEs are found extracellularly or on the outer cell surface, there must be a pathway for their export. The R modules in the AlgE MEs display glycine-rich tandem nonameric repeats of the putative Ca2+-binding motif (GGXGXDXXX) which is characteristic for proteins secreted by type I export systems (11, 12). These repeats are, for historical reasons, referred to as RTX (repeats in toxin) repeats, and proteins displaying them are referred to as RTX proteins. Sequence analyses of AlgE1 to AlgE7 and PsmE by InterProScan (http://www.ebi.ac.uk/InterProScan/) confirm that these proteins display the putative hemolysin-type calcium-binding regions (IPR001343) characteristic for type I allocrites (the term “allocrite” is used in this paper for “transported compound”). The signal sequences for type I allocrites are found C terminally and remain uncleaved upon export. Type I secretion machineries are composed of an ABC transporter, a membrane fusion protein (MFP), and an outer membrane protein (OMP) (11). During secretion, these components are associated in a complex which forms a channel extending from the inner membrane through the periplasmic space to the outer membrane (56). In this complex, the OMPs and MFPs are thought to function as trimers, while the ABC transporters probably exist as dimers. The ABC transporters are members of the ABC protein superfamily, which encodes a characteristic ATP-binding cassette domain (ABC), and in previously described type I systems the ABC is fused to a membrane domain (MD). The transport mediated by ABC transporters is energized by ATP hydrolysis.
The ability of the A. vinelandii AlgE MEs to form G blocks in alginates suggests that they are most likely involved in the formation of the gel-like protective coat surrounding cells that have entered the cyst stage. A draft genome sequence of the A. vinelandii strain OP is now available, and we saw this as an opportunity to use computer searches to identify a candidate type I transport system putatively involved in AlgE secretion. Most of our previous studies of these enzymes have been carried out with another A. vinelandii strain (strain E), but we have found that the ME system appears to be remarkably conserved among different isolates of this species (14). In this report, we show that a type I secretion system is present in the OP strain, and based on this information, we were able to clone it from strain E and demonstrate its importance for the control of alginate structure and cyst function in A. vinelandii.
MATERIALS AND METHODS
Growth of bacteria.
The plasmids and bacterial strains used are described in Table 1. Escherichia coli strains were routinely grown in L broth (containing, per liter, 10 g tryptone, 5 g yeast extract, and 5 g NaCl) or on L agar at 37°C. A. vinelandii was routinely grown in liquid Burk medium (pH 7.2) containing, per liter, glucose (10 g), CaCl2 (15.1 mg), MgSO4 (98 mg), FeSO4 (3.28 mg), NaCl (200 mg), Na2MO4 (0.179 mg), K2HPO4 (0.16 g), and KH2PO4 (0.64 g) or on Burk agar. Production of A. vinelandii alginate was performed in liquid RA1 medium (pH 7.0) containing the following ingredients per liter: MgSO4 · 7H2O (2.0 g), NH4NO3 (1.5 g), peptone (2.0 g), MOPS (morpholinepropanesulfonic acid) (10.5 g), CaCl2 · 2H2O (0.29 g), K2HPO4 (0.5 g), and fructose (20 g). When used, alkalase 2.4L and neutrase 0.5L (proteases) from Novo Nordisk were added at 0.1 ml/liter of each. Antibiotics were supplemented at the following concentrations: ampicillin, 100 to 200 μg/ml (E. coli), and tetracycline, 12.5 μg/ml (E. coli) and 10 to 50 μg/ml (A. vinelandii).
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17.1 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thiΔ (lac-proAB) | 53 |
| DH5α | endA1 hsdR17 supE44 thi-1 λ− recA1 gyrA96 relA1ΔlacU169 (φ80dlacZΔM15) | Bethesda Research Laboratories |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tcr)] | Stratagene |
| A. vinelandii | ||
| E | Wild type | 32 |
| AvE109 | Strain E with eexD disrupted | This work |
| AvE152 | Strain E with eexE disrupted | This work |
| AvE153 | Strain E with eexF disrupted | This work |
| Plasmids | ||
| pGEM7 | ColE1; Apr | Promega |
| pUC7Tc | pUC7 with tetA and tetR inserted; Apr Tcr | 5 |
| Litmus28 | ColE1; Apr | New England Biolabs |
| pLitmus28Tc | Litmus28(BamHI) with an insertion of a 2.3-kb fragment (BamHI) containing tetA and tetR from pUC7Tc; Apr Tcr | 1 |
| pJT19 | RK2 replicon-based expression vector; Kmr | 60 |
| pMG101a | Derivative of pGem7(BamHI/EcoRI) with insertion of a 2.2-kb PCR fragment (BamHI/EcoRI) encoding eexDE′; Apr | This work |
| pMG109 | Derivative of pMG101 in which eexD was disrupted by insertion of a 2.3-kb fragment (NcoI/BstBI) encoding tetA and tetR from Litmus28Tc; Apr | This work |
| pMG124b | Derivative of pJT19(NdeI/EcoRI) with an insertion of an 1.8-kb PCR fragment (NdeI/EcoRI) encoding eexDE′; Kmr | This work |
| pMG125c | Derivative of pMG124(ApaLI/EcoRI) with an insertion of a 3.3-kb PCR fragment (ApaLI/EcoRI) encoding eexDEF; Kmr | This work |
| pMG145 | Derivative of pGem7(XbaI/EcoRI) with an insertion of a 4.7-kb fragment (XbaI/EcoRI) encoding gene eexDEF from pMG125; Apr | This work |
| pMG146 | Derivative of pMG145(Eco47III/HindIII) blunted with Klenow fill-in and religated (6.5 kb); Apr Tcr | This work |
| pMG152d | Derivative of pMG146(XhoI) in which a 0.5-kb fragment internal to eexE was replaced by a PCR fragment (XhoI) encoding tetA and tetR; Apr Tcr | This work |
| pMG153 | Derivative of pMG145(Eco47III) in which eexF was disrupted by insertion of a 2.3-kb fragment (BamHI, blunted by Klenow fill-in) encoding tetA and tetR from Litmus28Tc; Apr Tcr | This work |
Primers ABC1FwBamHI and ABC1RevEcoRI were used for PCR amplification from chromosomal DNA.
Primers ABC1Fw(2)NdeI and ABC1RevEcoRI were used for PCR amplification from chromosomal DNA.
Primers ABC1OpstrApaLI(2) and OMP1RevEcoRI were used for PCR amplification from chromosomal DNA.
PCR amplification was performed with primers L28TetRFw and L28TetARevXhoI and Litmus28Tc as the template.
DNA techniques, transformations, and Western blotting.
Plasmid isolation, enzymatic manipulations of DNA, and agarose gel electrophoresis were performed by the methods of Sambrook and Russell (51). A QIAquick gel extraction kit and a QIAquick PCR purification kit (QIAGEN) were used for DNA purifications from agarose gels and enzymatic reactions, respectively. Transformations of E. coli were performed using a modified RbCl protocol (www.promega.com). The E. coli strains S17.1, DH5α, and XL1-Blue were used for standard cloning procedures. PCR amplifications were performed using an Expand high-fidelity PCR system (Boehringer Mannheim). Site-directed mutagenesis was performed using a QuickChange site-directed mutagenesis kit (Stratagene). Isolation of chromosomal DNA was performed with a bacterial genomic DNA purification kit (Edge Biosystems). A Big-Dye Terminator v1.1 cycle kit (Applied Biosystems) was applied for DNA sequencing. Labeling of DNA probes was performed with a PCR digoxigenin probe synthesis kit (Roche), and signals were detected with a digoxigenin luminescence detection kit (Roche). The partial sequencing of the eex genes was performed on PCR products amplified from pMG101 (primer pair, ABC1RevEcoRI and ABC1FwBamHI) and pMG145 [primer pairs, ABC1Fw(2)NdeI and OMP1RevEcoRI and OMP1FwPciI and OMP1RevEcoRI]. The PCR products were purified with ExoSAP-IT (USB Corporation) prior to sequencing. The primers used for DNA sequencing were the following: ABC1FwBamHI, ABC1RevEcoRI, IntABC1FwBamHI, IntABC1RevXhoI, MFP1Fw, ABC1OpstrApaLI(2), OMP1FwPciI, OMP1RevEcoRI, and ABC1Fw(2)NdeI. The sequences of the oligonucleotides used in this work can be obtained upon request. The transformation procedure for A. vinelandii is based on previously described procedures (42, 43). A colony of A. vinelandii E was inoculated in liquid CM medium (Burk medium without FeSO4 and Na2MoO4) and grown for 48 h to induce competence. The competent cells were washed and finally resuspended in 16 mM MgSO4 (1% of the original volume). An aliquot of cells (100 μl) was transformed with linearized plasmid DNA (10 to 20 μg) by incubation at 30°C for 1 h. The transformed cells were plated on CM agar for 48 h, collected, and plated on Burk agar with antibiotic selection of double-crossover mutants. Detection of AlgE proteins in culture supernatants was performed by Western blotting using rabbit anti-AlgE4 antiserum as described by Høidal et al. (28). Stationary-phase cultures were grown in Burk glucose medium to an optical density at 600 nm of about 0.5, collected by centrifugation, washed thoroughly in Burk medium containing 0.2% β-hydroxybutyrate (Burk-BHB), and inoculated into 100 ml new Burk-BHB in which encystment took place. Proteins in the culture supernatants were precipitated by acetone and subjected to Western blotting.
Construction of the eex mutants.
Genes eexD to eexF were individually inactivated by a homologous recombination procedure. As part of this strategy, we first used PCR to clone the genes from the A. vinelandii E strain, using primers designed on the basis of the OP strain draft genome sequence. Two independent constructs were made by cloning eexD and all three genes into the vector pGEM7, generating plasmids pMG101 and pMG145, respectively (Table 1). Restriction endonuclease mapping and partial sequencing confirmed the identities of the inserts. The tetracycline resistance cassette from pLitmus28Tc was then inserted into gene eexD in pMG101 (generating pMG109) and into genes eexE and eexF in pMG145 (yielding pMG152 and pMG153, respectively). During the construction of pMG109 and pMG152, care was taken to avoid apparent ρ-independent transcription terminators downstream of the resistance cassette insertions in eexD and eexE, respectively. The A. vinelandii E strain was then separately transformed with SphI-digested pMG109, NsiI-digested pMG152, and NcoI/EcoRI-digested pMG153, followed by selection of double-crossover mutants on Burk agar with tetracycline. The mutant strains (with the disrupted gene in parentheses) were designated AvE109 (eexD mutant), AvE152 (eexE mutant), and AvE153 (eexF mutant). These three mutant strains were grown in liquid Burk glucose supplemented with increasing concentrations of tetracycline (10 to 50 μg/ml) in a series of overnight cultures to obtain genetically pure mutants. The presence of tetracycline resistance in AvE109, AvE152, and AvE153 was verified by PCR amplification of an internal region of tetA by using the primers TetAFw and TetARev, resulting in a 1.1-kb fragment, and the chromosomal disruptions and the genetic purity of each mutant were confirmed by Southern hybridization or PCR. Chromosomal disruptions of gene eexD (strain AvE109) and eexF (strain AvE153) were verified by hybridization against the respective genes. The probe against eexD was made using the primers ABC1intSH1 and ABCintSH2 (pMG101 as template), and the probe against gene eexF was made using the primers OMP1FwPciI and OMP1RevEcoRI (pMG145 as template). Chromosomal DNA from AvE109 (SacI/XcmI digested), AvE153 (SfiI digested), and the wild-type strain was analyzed. As predicted, the hybridizing fragments from AvE109 and the wild type were 3.2 kb and 1.2 kb, respectively. Correspondingly, the hybridizing fragments containing eexF in AvE153 and the wild type were 4.4 kb and 2.2 kb, respectively. Chromosomal disruption of eexE (strain AvE152) was verified by PCR amplification of this gene from wild-type and AvE152 chromosomal DNA, using the primers MFP1Fw and MFP1IntRev1. As predicted, this resulted in amplification of 1.6- and 3.4-kb fragments, respectively.
Measurements of downstream transcriptional effects resulting from insertion of the tetracycline resistance cassette into the eex genes.
Stationary-phase A. vinelandii cultures were inoculated (1%) into Burk medium containing 0.01 U alginate lyase (AlgL) (13) and grown for 48 h. Three milliliters of culture was diluted by 2 volumes cold NaCl (0.2 M), and the cells were pelleted by centrifugation and resuspended in 0.5 ml NaCl (0.2 M). RNA was stabilized by RNAprotect (QIAGEN) and isolated using an RNAqueous kit (Ambion, Inc). Residual DNA was removed using a TURBO DNA-free kit (Ambion, Inc). cDNA was synthesized using a first-strand cDNA synthesis kit (GE Healthcare). Real-time PCR was performed using an Applied Biosystems 7500 real-time PCR system instrument and Power SYBR green PCR master mix. The primer pairs were designed to amplify a 60- to 70-nucleotide fragment by use of the instrument's default parameters. For eexD, two primer pairs amplifying a fragment upstream and downstream of the insertion point of the tetracycline resistance genes were used. For eexE and eexF, primer pairs amplifying fragments downstream of the insertion points were made. All four primer pairs were used for all eight RNA isolates (two independently grown cultures of each strain).
Isolation of alginate and preparation of samples for 1H NMR and SEC-MALLS.
A. vinelandii was inoculated (1%) to RA1 medium from stationary-phase Burk glucose cultures and grown for 68 to 72 h. The cells were removed by centrifugation (4,000 × g, ≥20 min). If necessary, the culture was diluted 10- to 20-fold (0.2 M NaCl) prior to centrifugation in order to reduce viscosity. Samples for 1H nuclear magnetic resonance (NMR) analysis were deacetylated directly in the supernatant (NaOH to pH of ≥12, gentle stirring for 1 h, neutralization with HCl) prior to precipitation. Samples for size exclusion chromatography with online multiangle laser light scattering (SEC-MALLS) analysis were filtered through 0.22-μm filters prior to precipitation. Alginate was precipitated with equal volumes of isopropanol and washed with 70% and 96% ethanol. The precipitates were dissolved in water, and if necessary, a few drops of Na2EDTA solution (0.5 M, pH 8.0) were added to increase solubility. Samples for 1H NMR analysis were further dialyzed (Spectra/Por membrane; molecular weight cutoff, 12,000 to 14,000) three times against 0.05 M NaCl and six times against ion-free water (>3 h for each shift). The viscosities of the samples were then reduced by acid hydrolysis, and the samples were finally neutralized (NaOH) and lyophilized (16).
1H NMR spectroscopy.
The 1H NMR spectra were obtained using Bruker 300- and 400-MHz spectrometers. Assignment of peaks and integration of the spectra were performed as described by Grasdalen (23), and the fractions of monomer monads and diads (internal residues) were independently calculated from the 1H NMR spectra. The sum of monad or diad fractions adds up to 1 only when the nonidentifiable end signals are included (e.g., FG + FM ≤ 1 and FG + FM + Freducing ends = 1, where FG is the fraction of G residues, FM is the fraction of M residues, and Freducing ends is the fraction of reducing ends).
Determination of alginate molecular mass by SEC-MALLS.
Alginate molecular mass determinations were performed using SEC-MALLS, as described by Christensen et al. (9). The eluent contained 0.05 M Na2SO4 and 0.01 M Na2EDTA (pH adjusted to 6). Consistent with the random coil polymer structure of alginate, linear relationships (slope, 0.6) between the logarithm of root mean square radius and the logarithm of the molecular mass were observed with all molecular mass determinations.
Collection and alignment of A regions of putative ABC proteins in the A. vinelandii genome.
The A. vinelandii genome sequence data were obtained from the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/). At the start of this project, the BLAST engine searches for the A. vinelandii genome sequence were not accessible. Therefore, the initial scaffold sequences were searched in silico for exact matches to the ABC protein signature sequence (LSGGQ) by use of the corresponding degenerate probes. When the BLAST searches became accessible, additional putative ABC proteins were collected by TBLASTN searches (assembly of 28 February 2003) using residues 326 to 498 in PrtD from Erwinia chrysanthemi (GenBank accession no. CAA37342). The collected protein sequences were designated in accordance with the genome draft and tested for the ABC identifier IPR003439 by InterProScan (http://www.ebi.ac.uk/InterProScan/). Only sequences passing this test were subjected to further analysis. The A regions from these sequences (a region comprising approximately 30 residues upstream of Walker A [Ge-valvGpsGsGKstll] to the end of Walker B [illlDEptsalD]) were extracted from each ABC protein as described by Saurin et al. (52). The glycine in Walker A, indicated in bold above, was generally positioned as residue 43 in the extracted sequences, and the residues shown in capital letters are those that are most conserved. When the deduced protein sequences contained two ABCs in tandem, these were indicated with α and β of the same gene number. All extracted sequences were aligned in Clone Manager 5 (Sci Ed software) by exhaustive pairwise alignments and progressive assembly using neighbor-joining phylogeny and the BLOSUM62 scoring matrix. The set of reference A regions (from experimentally verified type I ABC transporters) that was included in the alignment was extracted from sequences with the following GenBank accession numbers: S12525, S26696, BAA88492, BAA08631, CAA73292, AAC38666, AAC97164, and P08716. Transport Classification Database (TCDB) searches (http://www.tcdb.org/) with A. vinelandii ABC protein were performed with default settings. The data sets used in this paper (A regions and ABC proteins) can be obtained upon request.
Light and electron transmission microscopy.
Light microscopy was performed with a Leica DMLB microscope at a ×100 magnification. Staining of A. vinelandii cells was performed as described by Vela and Wyss (59). Electron microscopy was performed essentially as described by Mejía-Ruíz et al. (37), except that the cells were grown in RA1 medium and the fixation with glutaraldehyde was performed for 16 h.
Determination of cyst resistance to desiccation.
Resistance to desiccation was measured with bacterial cultures grown for 18 to 24 h on liquid Burk medium supplemented with 2% sucrose as the carbon source. These cultures were centrifuged, and the pellet was washed and resuspended in Burk medium. An aliquot of washed cells was plated on solid Burk medium with 0.2% N-butanol as the carbon source (encysting medium) and incubated for 5 days at 30°C. Cells harvested from these plates were resuspended in Burk medium. Samples of approximately 106 to 107 CFU of each strain were applied to Millipore 0.2-μm-pore-size membranes and placed in sterile Eppendorf tubes. These tubes were colocated in sterile petri dishes and incubated for 5 more days. Dried cells (cysts) in the filters were resuspended in 1 ml Burk medium, and viable counts were made to determine their numbers.
RESULTS AND DISCUSSION
In silico identification of the secretion machinery for the AlgE MEs.
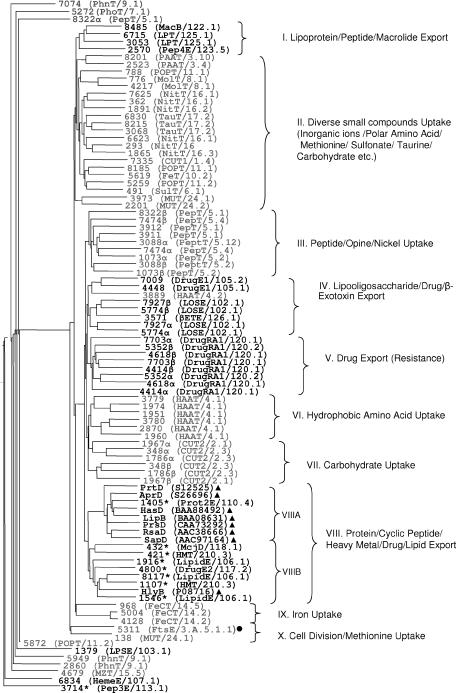
It has been shown that alignments of ABC proteins or conserved subregions of these sequences tend to cluster transporters of similar allocrite specificities (33, 40, 52). To obtain a complete overview of the putative ABC protein-encoding genes in the A. vinelandii genome, we first searched (at the protein level) the A. vinelandii OP genome for such sequences, and a total of 85 putative ABC proteins were detected. This set could be divided into proteins containing ABCs only (50 cases), ABCs fused to an MD (9 cases), and pairs of two ABCs fused in tandem (13 cases). Generally, ABCs of type I secretion systems are expected to be fused to an MD, but a possible exception to this rule has been identified (18). Therefore, to ensure an open-minded genome survey, no ABC proteins were initially excluded from the analysis. To analyze the 85 sequences in more detail, we then aligned the most conserved subregions in their ABCs (in this text referred to as the A regions), essentially as described by Saurin et al. (52). A set of A regions of experimentally verified type I ABC transporters from other bacteria was also included in the alignment to facilitate the identification of phylogenetic branches with potential type I sequences. This procedure would most likely make it easier to identify a subset of candidates with a high probability of being involved in secretion of the AlgE proteins. The resulting tree displayed 10 major branches (branches I to X) of highly related sequences and 10 divergent sequences (Fig. 1) . All type I reference sequences appeared in cluster VIII, which further branched into subclusters A and B. The A regions of protein 1405 from A. vinelandii and all type I references except HlyB constituted cluster VIIIA, while cluster VIIIB was comprised of HlyB and a set of ABC transporters which, according to BLAST searches against the TCDB, were assumed to transport allocrites of a less hydrophilic nature (e.g., the DrugE2 and LipidE classes). In addition, protein 1405 was the only A. vinelandii ABC protein that seemed to belong to a protein transporter class in the TCDB (the Prot2E family). Consistent with this analysis, protein 1405 displayed the InterPro identifier of type I ABC transporters (IPR010128). These analyses strongly suggested that protein 1405 is a type I ABC transporter and possibly the only one in the A. vinelandii OP genome.
FIG. 1.
Phylogenetic analysis of A regions of putative ABC proteins in the A. vinelandii OP genome. Individual A regions are indicated with gene/protein numbers as annotated in the draft genome sequence. Both A regions of experimentally verified type I protein exporters from other bacteria (▴) and A regions of putative A. vinelandii exporters are typed in black. One putative A region appears not to be involved in membrane transport (typed in gray and marked with •), and putative ABC proteins predicted to be involved in import are typed in gray. Individual A regions from proteins with two ABCs fused in tandem are indicated with α and β. A regions from ABCs fused to an MD are indicated with an asterisk. Classifications of the closest BLAST hit for each protein in TCDB (50), when blasting with the full protein sequence, are indicated in parentheses. Clusters of similar A regions are numbered (I to X), and expected substrate specificities within each cluster, according to closest hits in the TCDB, are indicated after each cluster number.
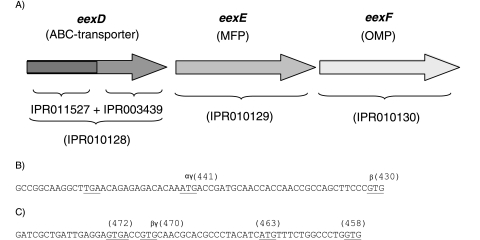
Interestingly, gene 1405, here designated eexD (epimerase exporter), is probably the first gene in a three-membered operon including open reading frames (ORFs) 1406 and 1407 (Fig. 2A). The two downstream ORFs display the InterPro identifiers for type I MFPs (gene 1406, designated gene eexE) and type I OMPs (gene 1407, designated gene eexF). These three ORFs are located close to each other and are oriented in the same direction. Furthermore, inspection of the short intergenic sequences supports the assumption that they are organized as an operon (Fig. 2B and C).
FIG. 2.
Organization of eexD, eexE, and eexF genes. (A) Orientations and sequential arrangement of eexD, eexE, and eexF. InterPro identifiers detected by InterProScan are indicated (IPR011527, ABC transporter transmembrane region; IPR003439, ABC transporter-related domain; IPR010128, type I secretion system ATPase PrtD; IPR010129, type I secretion membrane fusion protein HlyD; IPR010130, type I secretion outer membrane protein TolC). (B and C) Nucleotide sequences between genes eexD and eexE and between genes eexE and eexF, respectively. Sequences corresponding to possible start (ATG and GTG) and stop codons (TGA) are underlined, and the numbers of amino acid residues of the corresponding deduced proteins are indicated in parentheses above each start. Start sites were predicted by GeneMarkS analysis (3), signal sequence analysis (SignalP 3.0) (2), and when applicable, alignment with the closest homologues when protein-protein blasted against the nonredundant database. The corresponding start sites are indicated by γ, β, and α, respectively. All of these approaches indicated that an eexD ORF corresponds to a protein of 580 amino acids.
Disruptions of the eex genes abolish secretion of AlgE MEs.
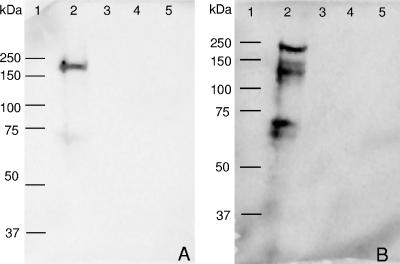
To provide experimental evidence for the involvement of the eex genes in export of the AlgE MEs, we inactivated each of the eex genes by insertion of a tetracycline resistance cassette in each gene, resulting in the mutant strains AvE109 (eexD mutant), AvE152 (eexE mutant), and AvE153 (eexF mutant) (Table 1). Western blotting was applied to evaluate the presence of AlgE MEs in cyst-induced culture supernatants. Supernatant samples were collected 48 and 176 h after cyst induction and subjected to Western blot analysis using anti-AlgE4 antibodies for detection of AlgE MEs (Fig. 3). In the 48-h sample one major band could be detected in the wild-type samples, while in the 176-h sample several MEs were detected. These results are consistent with those obtained in previously reported similar experiments (28). In contrast, no MEs were detected in the eex mutant samples. These results therefore clearly confirmed that one or more of the eex genes are required for AlgE secretion.
FIG. 3.
Western blot detection of AlgE MEs in culture supernatants at different stages of growth after replacement of Burk glucose medium with Burk BHB (0.2%). (A and B) Signals after 24 and 176 h of growth, respectively. Lane 1, molecular mass standard (Bio-Rad Precision Plus Dual Color); lane 2, A. vinelandii E wild-type strain; lane 3, AvE109; lane 4, AvE152; lane 5, AvE153. The identities of individual bands cannot be determined directly due to the abnormal mobility of the AlgE MEs (27).
Since the eex genes appear to be organized as an operon, the insertions of the tetA-tetR genes could lead to polar effects on transcription of the downstream genes (eexE and eexF). To analyze this, RNA from the wild type and the three mutant strains grown for 48 h was isolated and analyzed for the presence of mRNA encoding a C-terminal part of the three proteins. The upper part of eexD was used for normalization of the data. Transcripts encoding all three proteins were found, and the quantities were not strongly reduced (30 to 60%) relative to those in the wild type. The apparent absence of strong polar effects may be due to the lack of transcriptional terminators downstream of tetA (AvE109 and AvE153) and tetR (MG152). Thus, based on the InterPro protein sequence analyses, the physical organization of the genes, and the results of the real-time PCR analysis, it is likely that all three eex genes are required for epimerase secretion. However, more experiments are needed to completely verify this conclusion.
Disruptions of the eex genes lead to production of alginates with strongly reduced G content.
Based on current knowledge, the G residues in bacterial alginates originate either from the activity of the periplasmic AlgG or from the secreted AlgE MEs. However, their relative contributions to epimerization in A. vinelandii are unknown. If the eex genes are required for AlgE ME secretion, it seems likely that their disruption would lead to production of alginates with reduced G content. To study the structures of the alginates produced by AvE109, AvE152, and AvE153, the polymer was produced and harvested from these strains and the wild type. 1H NMR analysis of the resultant alginate (Fig. 4) showed that the fraction of G residues (FG) in the alginates obtained from the wild-type strain was 0.20, a large proportion of which had a G as a nearest neighbor (the fraction of GG, or FGG, was 0.09). In contrast, the G content in the alginates produced by all three eex mutants was very low, but apparently not zero (FG ≤ 0.02). The strongly reduced G content is consistent with the absence of AlgE MEs in the supernatants of the eex mutants and provides further evidence for the involvement of the eex cluster in export of the AlgE MEs.
FIG. 4.
1H NMR spectra of alginates harvested from the A. vinelandii E wild-type strain and the eex mutants (AvE109, AvE152, and AvE153) grown in RA1 medium. The GG-5M, GG-5G, and MG-5G signals, which identify G block alginates, are visible in wild-type alginate and absent from all of the mutant alginates. All spectra were expanded horizontally, and the tops of the M-1M peaks are not shown in order to reduce the picture size. The monad and diad frequencies (internal residues) of each alginate are summarized at the top of the figure. FGM, fraction of G residues with M as the nearest neighbor; FMM, fraction of M residues with M as the nearest neighbor.
The residual fraction of G residues in the mutant alginates is likely to originate from AlgG. These data show that, at least under the growth conditions used here, A. vinelandii AlgG contributes very little to epimerization. In comparison, Pseudomonas AlgG normally introduces more than 20% G in its alginates (21, 54). It seems possible that the A. vinelandii AlgG activity has become reduced in evolution due to the development of key roles for the AlgE MEs. Since the control of G block lengths by these enzymes is probably critical in A. vinelandii biology, a high level of single-G epimerization at an earlier stage (in the periplasm) may disturb the delicate control of alginate structure needed for this species. An AlgG protein structure may still be needed since this protein has been found to display an additional role (probably structural) in protecting the newly synthesized polymer from AlgL-mediated degradation (1, 21, 29, 38).
The eex mutants do not form stable cell coats in RA1 medium.
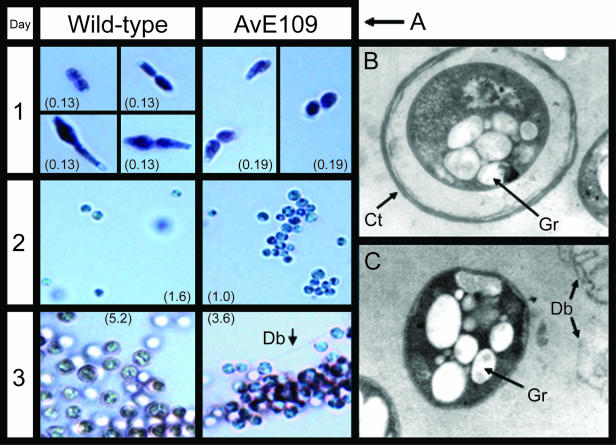
The very low level of G residues in the alginates produced by the eex mutants could be predicted to strongly affect polymer gel formation and therefore the ability to form the characteristic coats surrounding A. vinelandii cysts. When wild-type cells pregrown in Burk medium were inoculated for further growth in RA1 medium, we noticed that they first were converted to rods of different shapes, including almost-spherical and pear-shaped structures (Fig. 5A, day 1). These “fungoid” structures were assumed to originate from the peptone in RA1, as reported by Vela and Rosenthal (58). Later in the growth phase, spherical precyst-like structures, displaying a mosaic of dark and light zones, appeared (Fig. 5A, day 2). Finally, these structures became surrounded by pink staining material (cell coat), as previously described for stained cyst capsules (Fig. 5A, day 3) (59). The eex mutants developed into precyst-like structures as the wild-type strain (Fig. 5A, day 1 and 2) but did not accumulate a stable cell coat. Instead, the aggregates of pink staining material (released coat material) were observed scattered around the cells (Fig. 5A, day 3). After 10 days, both wild-type and eex mutant morphologies remained unchanged (data not shown). These observations therefore were in agreement with the hypothesis that the proteins secreted by eex proteins are involved in stabilization of A. vinelandii cell coats.
FIG. 5.
Cell morphologies of the A. vinelandii E wild-type strain and eex mutant derivatives (here exemplified by AvE109) grown in RA1 medium. (A) Cell morphologies as a function of time (light microscopy). Samples were withdrawn from each culture and stained as described by Vela and Wyss (59). The number of days of growth is indicated in the left column. The optical densities of each culture at 600 nm are indicated in parentheses in each picture. (B and C) Electron transmission micrographs of A. vinelandii E wild type and mutant AvE109, respectively, grown in RA1 medium. Ct, cell coat; Db, coat debris; Gr, intracellular granules.
To more clearly establish the morphological differences between the wild type and the eex mutants, their cell morphologies were also observed by transmission electron microscopy. Wild-type cells obtained from 7-day-old cultures appeared as spherical central-body-like structures containing electron-transparent granules (presumably poly-β-hydroxybutyrate), which were surrounded by a coat of distinct layers (Fig. 5B), as described for mature cysts of A. vinelandii (44). In contrast, the eex mutant cells of AvE109 (Fig. 5C) appeared as naked central bodies without any coat. These observations clearly show that RA1 medium induces morphology changes similar to those observed for mature cysts and that the eex mutants are not capable of maintaining such structures. Thus, the eex genes appear to be involved in formation of the cell coat, presumably by mediating secretion of AlgE MEs which introduce G blocks into the alginates produced. In the absence of such G blocks, the cell coat can presumably not form. Even though this hypothesis seems sufficient to explain the observed cell morphologies, it cannot be excluded that the Eex proteins are also involved in secretion of proteins other than the MEs and that such proteins play an additional role in the formation of the cell coat.
Effect of the eexDEF mutations on encystment.
The lack of the outer coating in the cyst-like structures produced in RA1 medium suggested to us that the eex mutants would probably also be unable to enter a functional cyst stage. To study this, the eex mutants and the wild-type cells were induced for encystment according to a standard protocol and then subjected to a desiccation assay (see Materials and Methods). About 1% of the wild-type cells but none of the eex mutants (less than 0.000001%) survived desiccation. These results therefore showed that the eex genes are involved in formation of functional cysts, presumably also meaning that G block alginates are essential for this process.
The eex mutants produce very-high-molecular-mass alginates in the presence of externally added proteases.
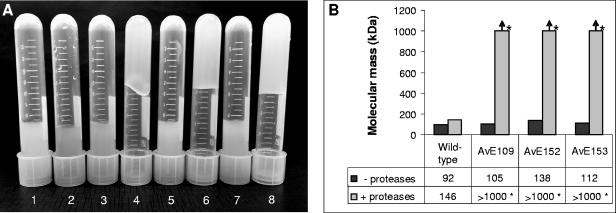
Proteases may be supplemented to culture media in order to reduce the activities of extracellular enzymes (24). An unexpected phenotype of the eex mutants appeared when they were grown in RA1 medium supplemented with proteases. The mutant cultures appeared very viscous, while the wild-type cultures did not (Fig. 6A). Since alginate molecular mass is the main factor controlling the intrinsic viscosity of alginates, we determined this parameter of the polymers produced in the presence and absence of proteases. Alginates in culture supernatants of the wild type and eex mutants grown in RA1 medium with and without proteases were precipitated, and the weight-average molecular masses (Mw) of the alginates were then determined by SEC-MALLS. The Mw values of wild-type and eex mutant alginates produced in the absence of proteases were found to be 92 and 105 to 138 kDa, respectively (Fig. 6B). In the presence of proteases, the wild-type alginates increased somewhat in Mw to 146 kDa. More interestingly, however, the Mw of the eex mutant alginates were so high that accurate SEC-MALLS measurements were not achieved. The Mw values were at some unknown level above 1 MDa, explaining the viscosity observed in the corresponding cell cultures. These data therefore clearly show that the molecular masses of eex mutant alginates, but not that of the corresponding wild-type polymer, increase considerably when proteases are present. The explanation for this phenomenon is unknown, but it appears likely that AlgL or other alginate lyases become inactivated by the added proteases. This hypothesis is consistent with the observation that a fraction of the alginate lyase activity is extracellular (31) and that disruption of the algL gene leads to production of high-molecular-mass alginate (57). Furthermore, it has been reported previously that proteases inhibit extracellular lyase activity in another alginate-producing bacterium, Pseudomonas mendocina (24). It is intriguing that the protease-dependent increase in molecular mass is seen only with the eex mutants, but this may be an indirect consequence of the observed morphological differences between the wild type and the eex mutants. The collapse of the surface coat in the eex mutants may render lyases trapped in the wild-type coat susceptible to proteolysis, thereby preventing alginate polymer degradation in the mutants.
FIG. 6.
Effect of addition of proteases to A. vinelandii E wild-type strain and eex mutant culture media. (A) Visualization of viscosity of wild-type and eex mutant RA1 culture samples with (+) and without (−) proteases. The cultures were grown in shake flasks, and samples were transferred to the tubes. Tube 1, A. vinelandii E wild type (−); tube 2, A. vinelandii E wild type (+); tube 3, AvE109 (−); tube 4, AvE109 (+); tube 5, AvE152 (−); tube 6, AvE152 (+); tube 7, AvE153 (−); tube 8, AvE153 (+). (B) Molecular masses of wild-type and eex mutant alginates produced in RA1 medium with and without proteases. Molecular masses above 1,000 kDa could not be accurately determined (marked with an asterisk).
Close EexDEF homologues are encoded in the chromosomes of alginate-producing Pseudomonas species.
The results reported in this paper show that the eex cluster is involved in secretion of the AlgE ME proteins. BLASTP searches with the EexD to EexF deduced protein sequences (580-, 441-, and 470-amino-acid versions, respectively) against the nonredundant peptide sequence database (http://www.ncbi.nlm.nih.gov/) reported that the most similar hits were identified in the genomes of the alginate-producing bacteria P. syringae pv. syringae B728a (ABC transporter, GenBank accession no. ZP_00125447; MFP, ZP_00125448; OMP, ZP_00125449; 65%, 63%, and 52% identical to the respective Eex proteins) and Pseudomonas putida KT2440 (MFP, NP_743952; OMP, NP_743953; 65% and 55% identical to the respective Eex proteins). A P. putida ABC transporter was not identified by the BLASTP search, but further investigation of the nucleotide sequence upstream of the MFP revealed an ORF (PP1796 [http://www.tigr.org/]) with an authentic frameshift mutation. Removal of a single cytosine at position 343 extends the ORF from 0.37 kb to 1.8 kb. A Bl2seq alignment (http://www.ncbi.nlm.nih.gov/BLAST/bl2seq/wblast2.cgi) reported that the deduced amino acid sequence of the extended version of PP1796 was 63% identical to EexD (580-amino-acid version). The P. syringae and P. putida genes are organized with relative positions and orientations identical to those of the eex genes (Fig. 2A). The observation that P. syringae encodes a putative type I secretion system very similar to EexDEF is not surprising, since this species has been found to encode an AlgE-type ME (4). The absence of a complete set of such genes in P. putida may indicate that these genes have lost their function in this organism, possibly because previously encoded MEs have been lost.
The experiments reported here clearly show that at least one of the eex gene products, and most likely all of them, is required for transport of the AlgE epimerases. Another independent way of demonstrating the biological role of these enzymes would be to knock out the AlgE epimerase genes themselves. Such experiments are ongoing in our laboratory.
Acknowledgments
This work was funded by a grant from the Norwegian Research Council.
We are very grateful to Ann-Sissel Ulset (NTNU) for performing the molecular mass determinations, Wenche Iren Strand (NTNU) for recording the 1H NMR spectra, Kjell Evjen (NTNU) and Marianne Bendheim (NTNU) for supervising electron transmission microscopy, and Håvard Sletta and Trond E. Ellingsen (SINTEF Materials and Chemistry) for providing the RA1 medium recipe.
REFERENCES
- 1.Bakkevig, K., H. Sletta, M. Gimmestad, R. Aune, H. Ertesvåg, K. Degnes, B. E. Christensen, T. E. Ellingsen, and S. Valla. 2005. Role of the Pseudomonas fluorescens alginate lyase (AlgL) in clearing the periplasm of alginates not exported to the extracellular environment. J. Bacteriol. 187:8375-8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Besemer, J., A. Lomsadze, and M. Borodovsky. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjerkan, T. M., C. L. Bender, H. Ertesvåg, F. Drabløs, M. K. Fakhr, L. A. Preston, G. Skjåk-Bræk, and S. Valla. 2004. The Pseudomonas syringae genome encodes a combined mannuronan C-5-epimerase and O-acetyl hydrolase, which strongly enhances the predicted gel-forming properties of alginates. J. Biol. Chem. 279:28920-28929. [DOI] [PubMed] [Google Scholar]
- 5.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, K. Haugan, and S. Valla. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63:370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campos, M., J. M. Martínez-Salazar, L. Lloret, S. Moreno, C. Núñez, G. Espín, and G. Soberón-Chávez. 1996. Characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J. Bacteriol. 178:1793-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chitnis, C. E., and D. E. Ohman. 1990. Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J. Bacteriol. 172:2894-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chitnis, C. E., and D. E. Ohman. 1993. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol. Microbiol. 8:583-593. [DOI] [PubMed] [Google Scholar]
- 9.Christensen, B. E., A. S. Ulset, M. U. Beer, B. E. Knuckles, D. L. Williams, M. L. Fishman, H. K. Chau, and P. J. Wood. 2001. Macromolecular characterisation of three barley β-glucan standards by size-exclusion chromatography combined with light scattering and viscometry: an inter-laboratory study. Carbohydr. Polym. 45:11-22. [Google Scholar]
- 10.Cote, G. L., and L. H. Krull. 1988. Characterization of the exocellular polysaccharides from Azotobacter chroococcum. Carbohydr. Res. 181:143-152. [Google Scholar]
- 11.Delepelaire, P. 2004. Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta 1694:149-161. [DOI] [PubMed] [Google Scholar]
- 12.Ertesvåg, H., B. Doseth, B. Larsen, G. Skjåk-Bræk, and S. Valla. 1994. Cloning and expression of an Azotobacter vinelandii mannuronan C-5-epimerase gene. J. Bacteriol. 176:2846-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ertesvåg, H., F. Erlien, G. Skjåk-Bræk, B. H. Rehm, and S. Valla. 1998. Biochemical properties and substrate specificities of a recombinantly produced Azotobacter vinelandii alginate lyase. J. Bacteriol. 180:3779-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ertesvåg, H., H. K. Høidal, I. K. Hals, A. Rian, B. Doseth, and S. Valla. 1995. A family of modular type mannuronan C-5-epimerase genes controls alginate structure in Azotobacter vinelandii. Mol. Microbiol. 16:719-731. [DOI] [PubMed] [Google Scholar]
- 15.Ertesvåg, H., H. K. Høidal, H. Schjerven, B. I. Svanem, and S. Valla. 1999. Mannuronan C-5-epimerases and their application for in vitro and in vivo design of new alginates useful in biotechnology. Metab. Eng. 1:262-269. [DOI] [PubMed] [Google Scholar]
- 16.Ertesvåg, H., and G. Skjåk-Bræk. 1999. Modification of alginate using mannuronan C-5-epimerases, p. 71-78. In C. Bucke (ed.), Methods in biotechnology 10: carbohydrate bio/technology protocols. Humana Press, Inc., Totowa, N.J.
- 17.Fett, W. F., S. F. Osman, and M. F. Dunn. 1989. Characterization of exopolysaccharides produced by plant-associated fluorescent pseudomonads. Appl. Environ. Microbiol. 55:579-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiedler, G., M. Arnold, and I. Maldener. 1998. Sequence and mutational analysis of the devBCA gene cluster encoding a putative ABC transporter in the cyanobacterium Anabaena variabilis ATCC 29413. Biochim. Biophys. Acta 1375:140-143. [DOI] [PubMed] [Google Scholar]
- 19.Franklin, M. J., and D. E. Ohman. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184:3000-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaona, G., C. Núñez, J. B. Goldberg, A. S. Linford, R. Nájera, M. Castañeda, J. Guzman, G. Espín, and G. Soberón-Chavez. 2004. Characterization of the Azotobacter vinelandii algC gene involved in alginate and lipopolysaccharide production. FEMS Microbiol. Lett. 238:199-206. [DOI] [PubMed] [Google Scholar]
- 21.Gimmestad, M., H. Sletta, H. Ertesvåg, K. Bakkevig, S. Jain, S. J. Suh, G. Skjåk-Bræk, T. E. Ellingsen, D. E. Ohman, and S. Valla. 2003. The Pseudomonas fluorescens AlgG protein, but not its mannuronan C-5-epimerase activity, is needed for alginate polymer formation. J. Bacteriol. 185:3515-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govan, J. R. W., J. A. M. Fyfe, and T. R. Jarman. 1981. Isolation of alginate-producing mutants of Pseudomonas fluorescens, Pseudomonas putida and Pseudomonas mendocina. J. Gen. Microbiol. 125:217-220. [DOI] [PubMed] [Google Scholar]
- 23.Grasdalen, H. 1983. High-field, 1H-n.m.r. spectroscopy of alginate: sequential structure and linkage conformations. Carbohydr. Res. 118:255-260. [Google Scholar]
- 24.Hacking, A. J., I. W. F. Taylor, T. R. Jarman, and J. R. W. Govan. 1983. Alginate biosynthesis by Pseudomonas mendocina. J. Gen. Microbiol. 129:3473-3480. [Google Scholar]
- 25.Haug, A., B. Larsen, and O. Smidsrød. 1974. Uronic acid sequence in alginate from different sources. Carbohydr. Res. 32:217-225. [Google Scholar]
- 26.Haug, A., and O. Smidsrød. 1965. The effect of divalent metals on the properties of alginate solutions. Acta Chem. Scand. 19:341-351. [Google Scholar]
- 27.Høidal, H. K., H. Ertesvåg, G. Skjåk-Bræk, B. T. Stokke, and S. Valla. 1999. The recombinant Azotobacter vinelandii mannuronan C-5-epimerase AlgE4 epimerizes alginate by a nonrandom attack mechanism. J. Biol. Chem. 274:12316-12322. [DOI] [PubMed] [Google Scholar]
- 28.Høidal, H. K., B. I. G. Svanem, M. Gimmestad, and S. Valla. 2000. Mannuronan C-5 epimerases and cellular differentiation of Azotobacter vinelandii. Environ. Microbiol. 2:27-38. [DOI] [PubMed] [Google Scholar]
- 29.Jain, S., M. J. Franklin, H. Ertesvåg, S. Valla, and D. E. Ohman. 2003. The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol. Microbiol. 47:1123-1133. [DOI] [PubMed] [Google Scholar]
- 30.Jerga, A., A. Raychaudhuri, and P. A. Tipton. 2006. Pseudomonas aeruginosa C5-mannuronan epimerase: steady-state kinetics and characterization of the product. Biochemistry 45:552-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy, L., K. McDowell, and I. W. Sutherland. 1992. Alginases from Azotobacter species. J. Gen. Microbiol. 138:2465-2471. [Google Scholar]
- 32.Larsen, B., and A. Haug. 1971. Biosynthesis of alginate. Part I. Composition and structure of alginate produced by Azotobacter vinelandii (Lipman). Carbohydr. Res. 17:287-296. [DOI] [PubMed] [Google Scholar]
- 33.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28:5-13. [DOI] [PubMed] [Google Scholar]
- 34.Lloret, L., R. Barreto, R. León, S. Moreno, J. Martínez-Salazar, G. Espín, and G. Soberón-Chávez. 1996. Genetic analysis of the transcriptional arrangement of Azotobacter vinelandii alginate biosynthetic genes: identification of two independent promoters. Mol. Microbiol. 21:449-457. [DOI] [PubMed] [Google Scholar]
- 35.Martínez-Salazar, J. M., S. Moreno, R. Nájera, J. C. Boucher, G. Espín, G. Soberón-Chávez, and V. Deretic. 1996. Characterization of the genes coding for the putative sigma factor AlgU and its regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their roles in alginate biosynthesis. J. Bacteriol. 178:1800-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mejía-Ruíz, H., J. Guzmán, S. Moreno, G. Soberón-Chávez, and G. Espín. 1997. The Azotobacter vinelandii alg8 and alg44 genes are essential for alginate synthesis and can be transcribed from an algD-independent promoter. Gene 199:271-277. [DOI] [PubMed] [Google Scholar]
- 37.Mejía-Ruíz, H., S. Moreno, J. Guzmán, R. Nájera, R. León, G. Sóberon-Chávez, and G. Espín. 1997. Isolation and characterization of an Azotobacter vinelandii algK mutant. FEMS Microbiol. Lett. 156:101-106. [DOI] [PubMed] [Google Scholar]
- 38.Morea, A., K. Mathee, M. J. Franklin, A. Giacomini, M. O'Regan, and D. E. Ohman. 2001. Characterization of algG encoding C5-epimerase in the alginate biosynthetic gene cluster of Pseudomonas fluorescens. Gene 278:107-114. [DOI] [PubMed] [Google Scholar]
- 39.Núñez, C., R. León, J. Guzmán, G. Espín, and G. Soberón-Chávez. 2000. Role of Azotobacter vinelandii mucA and mucC gene products in alginate production. J. Bacteriol. 182:6550-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Omori, K., and A. Idei. 2003. Gram-negative bacterial ATP-binding cassette protein exporter family and diverse secretory proteins. J. Biosci. Bioeng. 95:1-12. [DOI] [PubMed] [Google Scholar]
- 41.Osman, S. F., W. F. Fett, and M. L. Fishman. 1986. Exopolysaccharides of the phytopathogen Pseudomonas syringae pv. glycinea. J. Bacteriol. 166:66-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Page, W. J., and H. L. Sadoff. 1976. Control of transformation competence in Azotobacter vinelandii by nitrogen catabolite derepression. J. Bacteriol. 125:1088-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Page, W. J., and M. von Tigerstrom. 1978. Induction of transformation competence in Azotobacter vinelandii iron-limited cultures. Can. J. Microbiol. 24:1590-1594. [DOI] [PubMed] [Google Scholar]
- 44.Pope, L., and O. Wyss. 1970. Outer layers of the Azotobacter vinelandii cyst. J. Bacteriol. 102:234-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramsey, D. M., and D. J. Wozniak. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56:309-322. [DOI] [PubMed] [Google Scholar]
- 46.Rehm, B. H. 1996. The Azotobacter vinelandii gene algJ encodes an outer-membrane protein presumably involved in export of alginate. Microbiology 142:873-880. [DOI] [PubMed] [Google Scholar]
- 47.Rehm, B. H., G. Boheim, J. Tommassen, and U. K. Winkler. 1994. Overexpression of algE in Escherichia coli: subcellular localization, purification, and ion channel properties. J. Bacteriol. 176:5639-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rehm, B. H., H. Ertesvåg, and S. Valla. 1996. A new Azotobacter vinelandii mannuronan C-5-epimerase gene (algG) is part of an alg gene cluster physically organized in a manner similar to that in Pseudomonas aeruginosa. J. Bacteriol. 178:5884-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sadoff, H. L. 1975. Encystment and germination in Azotobacter vinelandii. Bacteriol. Rev. 39:516-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saier, M. H. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Saurin, W., M. Hofnung, and E. Dassa. 1999. Getting in or getting out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J. Mol. Evol. 48:22-41. [DOI] [PubMed] [Google Scholar]
- 53.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 54.Skjåk-Bræk, G., H. Grasdalen, and B. Larsen. 1986. Monomer sequence and acetylation pattern in some bacterial alginates. Carbohydr. Res. 154:239-250. [DOI] [PubMed] [Google Scholar]
- 55.Svanem, B. I., G. Skjåk-Bræk, H. Ertesvåg, and S. Valla. 1999. Cloning and expression of three new Azotobacter vinelandii genes closely related to a previously described gene family encoding mannuronan C-5-epimerases. J. Bacteriol. 181:68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thanabalu, T., E. Koronakis, C. Hughes, and V. Koronakis. 1998. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 17:6487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trujillo-Roldan, M. A., S. Moreno, D. Segura, E. Galindo, and G. Espin. 2003. Alginate production by an Azotobacter vinelandii mutant unable to produce alginate lyase. Appl. Microbiol. Biotechnol. 60:733-737. [DOI] [PubMed] [Google Scholar]
- 58.Vela, G. R., and R. S. Rosenthal. 1972. Effect of peptone on Azotobacter morphology. J. Bacteriol. 111:260-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vela, G. R., and O. Wyss. 1964. Improved stain for visualization of Azotobacter encystment. J. Bacteriol. 87:476-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winther-Larsen, H. C., K. D. Josefsen, T. Brautaset, and S. Valla. 2000. Parameters affecting gene expression from the Pm promoter in gram-negative bacteria. Metab. Eng. 2:79-91. [DOI] [PubMed] [Google Scholar]