Abstract
Site-specific recombinases of the integrase family usually require cofactors to impart directionality in the recombination reactions that they catalyze. The FimB integrase inverts the Escherichia coli fim switch (fimS) in the on-to-off and off-to-on directions with approximately equal efficiency. Inhibiting DNA gyrase with novobiocin caused inversion to become biased in the off-to-on direction. This directionality was not due to differential DNA topological distortion of fimS in the on and off phases by the activity of its resident PfimA promoter. Instead, the leucine-responsive regulatory (Lrp) protein was found to determine switching outcomes. Knocking out the lrp gene or abolishing Lrp binding sites 1 and 2 within fimS completely reversed the response of the switch to DNA relaxation. Inactivation of either Lrp site alone resulted in mild on-to-off bias, showing that they act together to influence the response of the switch to changes in DNA supercoiling. Thus, Lrp is not merely an architectural element organizing the fim invertasome, it collaborates with DNA supercoiling to determine the directionality of the DNA inversion event.
Site-specific recombinases of the integrase family are usually associated with the integration and excision of DNA sequences such as bacteriophage genomes from bacterial chromosomes or other replicons. The best-studied integrase is Int, the prototypic member of the family that catalyzes the integration and excision of bacteriophage lambda from the chromosome of Escherichia coli (33, 40). Although the integration and excision reactions both require Int, an additional phage-encoded factor called the excisionase (Xis) confers directionality by being specific for the excision reaction. Despite its name, the excisionase has no enzymatic activity. Instead, it is an architectural element that helps to organize the local structure of lambda DNA in a way that favors the excision reaction. The Xis protein has been classified as a recombination directionality factor (RDF), and several other proteins have been identified that provide, or may provide, an analogous function in other integrase-dependent site-specific recombination reactions (23-25). The requirement for the RDFs arises due to the similarities of the DNA substrates and products of the integration and excision reactions. The RDF confers directionality by stimulating one reaction while inhibiting the other.
An integrase-mediated site-specific recombination event controls the phase-variable expression of type 1 fimbriae in E. coli. A key difference between the fimbrial and phage recombination mechanisms is that the fimbrial system involves DNA inversion and not integration/excision. The promoter for fim operon transcription (PfimA) is carried on a 314-bp invertible DNA element called the fim switch (fimS), and expression of the fim structural genes depends on its orientation (1, 15). With PfimA directed toward the fim operon, the genes are transcribed, and when it is inverted to the opposite orientation, the fim operon is silent (Fig. 1A). The FimB and FimE site-specific recombinases catalyze inversion of fimS. FimB inverts the switch in the on-to-off and off-to-on directions with approximately equal efficiency, while FimE shows a strong preference for switching in the on-to-off direction (16, 27, 37). The activity of FimE dominates that of FimB, making on-to-off switching predominant under many growth conditions (4, 27). The FimB protein binds equally well in vitro and in vivo to its target sites located at the functionally equivalent left (IRL) and right (IRR) inverted repeats that flank the switch (7, 12). Several widely studied laboratory strains of E. coli K-12 harbor knockout mutations in the fimE gene and exhibit a two-way inversion of fimS that is catalyzed by FimB alone (4). This form of the switch is reminiscent of the phage integration/excision systems in being catalyzed by a single integrase protein.
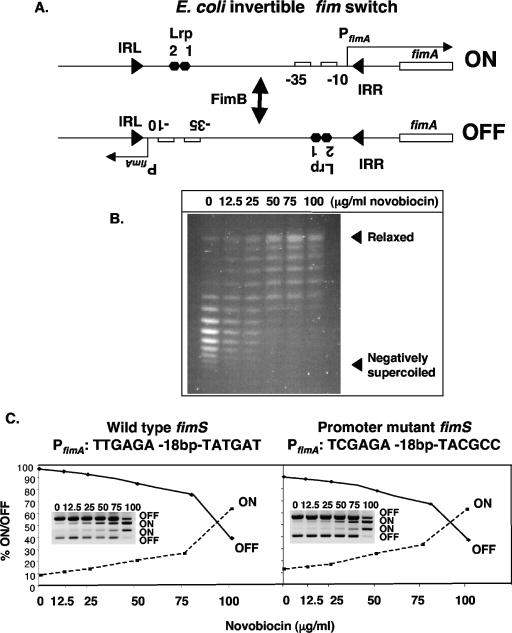
FIG. 1.
DNA relaxation and fim switch inversion preferences. (A) Phase-on and -off fim switches, showing inverted repeats IRL and IRR, the PfimA promoter (−10, −35), the fimA transcription start site, and Lrp binding sites 1 and 2. (B) Topoisomers of plasmid pUC18 isolated from E. coli K-12 strain VL386 treated with novobiocin at the concentrations shown. (C) Switch inversion preferences with and without a functional PfimA promoter (structures summarized above each panel) at increasing concentrations of novobiocin. Densitometric data from PCR switch assay gels (insets) were used to plot the graphs. Numbers above each gel lane are μg/ml of novobiocin. Bands corresponding to phase-on or -off are labeled.
Inversion of fimS requires the Lrp accessory protein in addition to the recombinases. Lrp binds to two sites within the switch, where it acts positively on DNA inversion (5, 17, 22, 34) (Fig. 1a). It is thought that Lrp alters the trajectory of the fimS DNA to enhance the formation of a synaptic complex for recombination. Similarly, DNA supercoiling might be expected to influence recombination efficiency through its effects on the topology of the switch DNA by analogy with other site-specific recombination systems (3, 21, 31). In the absence of FimE, bacteria growing in LB medium exhibit FimB-mediated on-to-off and off-to-on switching at approximately equal rates of ∼10−3 per cell per generation (18, 27, 37). This pattern is disturbed following inhibition of DNA gyrase, the type II topoisomerase that introduces negative supercoils into DNA in an ATP-dependent manner. If phase-off bacteria are treated with the DNA gyrase-inhibiting antibiotic novobiocin, DNA becomes relaxed (Fig. 1B), switching becomes biased in the off-to-on direction, and the bias becomes more pronounced as the dosage of the drug is increased (12).
The most straightforward explanation for these observations is that the phase-off switch and the phase-on switch become distinct as substrates for FimB following gyrase inhibition: relaxed phase-on switches are poor substrates for FimB, whereas relaxed phase-off switches make suitable substrates. Thus, when the switch inverts to the ON phase, it is difficult for FimB to catalyze the reverse reaction if the DNA is too relaxed, and fimS becomes trapped in phase-on. To account for the observed difference in inversion preferences when gyrase is inhibited, one must consider local features within or close to fimS that might collaborate with DNA relaxation to influence inversion bias. Such features would have to adopt phase-specific configurations to enable on and off switch orientations to be distinguished.
The effect of transcription initiated from the promoter within fimS is an attractive candidate for the role of a phase-specific feature capable of imparting directionality to the inversion reaction. Transcription alters local DNA topology by creating differentially supercoiled domains that flank the moving RNA polymerase (10, 14, 26, 32, 43). The fim promoter is active in on and off switches, the same transcription start site is used in both phases, and the promoter is active to a similar level regardless of switch orientation or the degree of negative supercoiling of the DNA (11, 29, 30). Moreover, the domains of relaxed (or even positively supercoiled) DNA that are created by the movement of RNA polymerase may not be constant in both orientations of fimS. This is because in on and off switches the supercoiled domains are propagated at opposite ends of fimS where the facility with which they can dissipate by rotational diffusion may be distinct. Any such inequality would be exacerbated by the inhibition of DNA gyrase because this topoisomerase would be less able to supercoil relaxed DNA negatively or to relax positively supercoiled DNA. Since transcription from the fim promoter always reads across one of the inverted repeats of fimS (Fig. 1A), the associated topological disturbance could interfere with the ability of FimB to utilize these DNA sequences.
Alternatively, a DNA-binding protein could sustain a phase-specific nucleoprotein complex at the switch that disfavors FimB-mediated inversion in phase-on but not phase-off following gyrase inhibition. Obviously, this second possibility does not exclude the first. Here we demonstrate that the leucine-responsive regulatory protein (Lrp), a DNA binding and bending protein (42), plays such a role and is a directionality determinant in the fim site-specific recombination system.
MATERIALS AND METHODS
Media, growth conditions, and genetic techniques.
All strains were derivatives of E. coli K-12 (Table 1). VL386 lrp-201::Tn10 was constructed by P1vir-mediated phage transduction (28, 38) using a CSH50 lrp-201::Tn10 lysate (16). Complementation was carried out with plasmid pKMC102 (lrp+), a single-copy plasmid derived from pZC320 (36). Bacteria were cultured in L broth (Difco) or L agar (containing agar at 1.5% wt/vol). MacConkey-lactose agar plates (28) were used for Lac phenotype determination. Unless otherwise stated, liquid cultures were grown overnight at 37°C with aeration. Antibiotics were used at the following concentrations: carbenicillin, 100 μg ml−1; kanamycin, 20 μg ml−1; chloramphenicol, 25 μg ml−1; tetracycline, 12.5 μg ml−1. Plasmid DNA was introduced to bacterial cells by CaCl2 transformation (35) or electroporation (19) using a Bio-Rad Gene Pulser.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| E. coli strains | ||
| VL386 | φ(fimA-lacZ)λ pL(209)fimE::IS1 | 1 |
| VL386 recD | VL386 recD::Tn10 | 37 |
| VL386 lrp | VL386 lrp::Tn10 | This work |
| AK13 | VL386 recD::Tn10 ΔfimB::kan | This work |
| AK14 | VL386 recD::Tn10 ΔfimB::kan Lrp binding site 1 mutation | This work |
| AK15 | VL386 recD::Tn10 ΔfimB::kan Lrp binding site 2 mutation | This work |
| AK16 | VL386 recD::Tn10 ΔfimB::kan Lrp binding sites 1 and 2 mutation | This work |
| BL21(DE3) | F−dcm ompT hsdS gal | 39 |
| Plasmids | ||
| pSLD203 | fimB gene cloned in pUC18 | 13 |
| pKMC102 | lrp gene cloned in pZC320 | K. McFarland |
| pSGS501 | fimB::kan fimE::IS1 φ(fimA-lacZ) cloned in pAYC184, switch phase off | 37 |
| pSGS501Lrp-1 | Lrp binding site 1 mutated in pSGS501 | This work |
| pSGS501Lrp-2 | Lrp binding site 2 mutated in pSGS501 | This work |
| pSGS501Lrp-1/2 | Lrp binding sites 1 and 2 mutated in pSGS501 | This work |
| pKMC301 | lrp gene cloned in pET22b | K. McFarland |
| pMMC108 | fimS cloned as 550-bp fragment in the PstI site of pMMC106 | M. McCusker |
| pMMC108Lrp-1 | Lrp binding site 1 mutated in pMMC108 | This work |
| pMMC108Lrp-2 | Lrp binding site 2 mutated in pMMC108 | This work |
| pMMC108Lrp-1&2 | Lrp binding sites 1 and 2 mutated in pMMC108 | This work |
| pUC18 | Cloning vector, Apr | 45 |
Molecular biological techniques.
Plasmid DNA was isolated using QIAGEN Midi columns or Wizard mini prep columns (Promega). DNA fragments were purified from agarose gels using the High Pure PCR product purification kit (Roche Applied Science). Restriction enzymes were purchased from New England Biolabs and used according to the manufacturer's directions. Automated sequencing was carried out at GATC Biotech. Oligonucleotide synthesis was by MWG Biotech. Plasmid topoisomer distributions were analyzed by agarose-chloroquine gel electrophoresis as described previously (20). At the concentration of chloroquine used (2.5 μg/ml), the more negatively supercoiled topoisomers ran fastest in the gel.
Determination of fim switch orientation.
The orientation of the fim switch on the chromosome was determined as previously described (37). This method exploited a unique BstUI restriction site in the fim switch that results in restriction fragment length dimorphism among BstUI-digested PCR products that are characteristic of phase-on and -off switches.
Fifty microliters of culture was boiled following overnight incubation at 37°C. Oligonucleotides OL4 and OL20 (Table 2) were used with Taq polymerase (New England Biolabs) to amplify the switch region as a 726-bp DNA product. The PCR cycle involved denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min, followed by a final extension for 10 min at 72°C. Samples were cooled to 60°C, and 10 units of BstUI were added and incubated at 60°C for 3 h. Digested PCR products were electrophoresed on 2% agarose gels. Phase-off switches yielded DNA fragments of 539 and 187 bp, while phase-on switches gave fragments of 433 and 293 bp. The proportions of on- and off-specific fragments in the population was determined using QUANTITY ONE image analysis software.
TABLE 2.
Oligonucleotides
| Primer name | Sequence (5′-3′)a |
|---|---|
| OL4 | GACAGAACAACGATTGCCAG |
| OL20 | CCGTAACGCAGACTCATCCTC |
| AKBSFORBIO | CTCCAAAAACCACCTCATGC |
| BSREVBIO | CCCCCAAAAGATGAAACATTT |
| SDMlrp1fw | CCAAAAACCACCTCATGCAACTCGAGCATCTATAAATAAAGATAAC |
| SDMlrp1rv | GTTATCTTTATTTATAGATGCTCGAGTTGCATGAGGTGGTTTTTGG |
| SDMlrp2fw | GATACCAATAGAATCTCGAGCCAACAAATAAAC |
| SDMlrp2rv | GTTTATTTGTTGGCTCGAGATTCTATTGTTATC |
| ARLCLOfwb | AAGACAATTGGGGCCAAACTGTCC |
| ARLCLOrvc | GGGCAGTCGTTCTGTACACTTT |
| FimBfw | GCGCGTCTGTAATTATAAGGG |
| FimBrv | CCCTGGTATCTCAACTAT |
Restriction enzyme cleavage sites are underlined.
Primer tailed with restriction site MfeI.
Primer tailed with restriction site BsrGI.
Site-directed mutagenesis.
Site-directed mutagenesis was performed using the Quikchange II (Stratagene) kit according to the manufacturer's recommendations. Oligonucleotide pairs (MWG Biotech) used to mutate the Lrp binding sites Lrp-1 (SDMlrp1fw and SDMlrp1rv) and Lrp-2 (SDMlrp2fw and SDMlrp2rv), individually and in combination, are described in Table 2. Plasmid pMMC108 was used as the substrate to generate plasmids pMMC108Lrp-1, pMMC108Lrp-2, and pMMC108Lrp-1+2. Lrp binding site 1 was replaced with 5′-AACTCGAGCATCT-3′ and site 2 was replaced with 5′-AGAATCTCGAGCC-3′ as previously described (17). The presence of either site 1 or 2 mutations introduced an XhoI restriction site. Binding site alterations were confirmed by sequencing, PCR analysis using oligonucleotides OL4 and OL20, and XhoI digestion of PCR products.
Purification of Lrp.
Expression of N-terminally His-tagged Lrp was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in exponentially growing 100-ml BL21(DE3) cultures harboring pKMC301. After 2 h, the cells were harvested and lysed by sonication. Lysates (∼15 ml) were applied to a His-Bind Quick column (Novagen) (2) preequilibrated with 10 ml binding buffer (4 M NaCl, 160 mM Tris-HCl, 40 mM imidazole, pH 7.9). The column was then washed with 10 ml wash buffer (4 M NaCl, 480 mM imidazole, 160 mM Tris-HCl, pH 7.9) and the protein eluted in 5,001-μl fractions of 5 ml of elution buffer (4 M imidazole, 2 M NaCl, 80 mM Tris-HCl, pH 7.9). Glycerol was added to a final concentration of 10% to each fraction. Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and those containing Lrp were pooled and dialyzed against 1 liter of 100 mM phosphate buffer, pH 8.0, 10% glycerol, 1 mM EDTA, pH 8.0, 300 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol. Lrp was estimated to be approximately 95% pure.
DNA binding and gel retardation.
Binding of purified Lrp protein to the wild-type and Lrp binding site mutant alleles of the switch was tested by electrophoretic mobility shift assay. Probes (144-bp) were amplified by PCR with Pfu polymerase (Stratagene) using the primer pair AKBSFORBIO and BSREVBIO (Table 2) and using pMMC108, pMMC108Lrp-1, pMMC108Lrp-2, and pMMC108Lrp-1+2 as templates. Amplified probes were gel purified (Roche Applied Science). The oligonucleotide AKBSFORBIO had a 5′ biotinylated end allowing detection of protein-DNA complexes. Amplified probe was incubated with increasing concentrations (0 to 50 nM) of purified Lrp protein for 15 min at room temperature as recommended by manufacturers of the electrophoretic mobility shift assay kit (Pierce). Protein-DNA complexes were resolved by electrophoresis through a 7.5% polyacrylamide gel for 1 h at room temperature. The gel was then electrophoretically blotted and developed.
Allele replacement.
The allele replacement method used to eliminate Lrp sites 1 and 2 was based on one described previously (37). A DNA fragment containing the Lrp binding site(s) mutation(s) (PCR amplified using the primer pair Arlclofw and Arlclorv and the plasmids pMMC108Lrp-1, pMMC108Lrp-2, and pMMC108Lrp-1+2 as template DNA) was cloned into pSGS501 following digestion with MfeI and BsrGI, to give pSGS501Lrp-1, pSGS501Lrp-2, and pSGS501Lrp-1+2. pSGS501, pSGS501Lrp-1, pSGS501Lrp-2, and pSGS501Lrp-1+2 were subsequently digested with EcoRV. An 8.5-kb fragment that included the fimB gene interrupted by a Kanr cassette with or without the Lrp site mutations was gel extracted. Two micrograms of this fragment was electroporated into strain VL386recD, generating strains AK13 to AK16 (Table 1). Transformants were selected on L agar containing kanamycin. Oligonucleotides FimBfw and FimBrv were used to verify fimB::kan by PCR analysis and DNA sequencing.
RESULTS
Eliminating the fimA promoter does not rescue biased fimS inversion.
The movement of RNA polymerase during transcription creates differentially supercoiled domains in double-stranded DNA (10, 14, 26, 32, 43). This suggests an attractive explanation for the directional biasing seen in FimB-mediated inversion of fimS when DNA gyrase activity is inhibited by novobiocin treatment (11). The transcription start site of the fimA promoter (PfimA) has been mapped previously, is constant in both phase-on and phase-off switches, and is unaffected by changes in DNA supercoiling (8, 11, 29, 30). Transcription originating at PfimA is directed toward distinct regions of the chromosome in phase-on and phase-off switches (Fig. 1A). These different regions of the chromosome may have distinct abilities to absorb positive supercoils emanating from the switch. Normally, gyrase eliminates these transcriptionally generated positive supercoils, but when the topoisomerase is inhibited, the supercoils may persist to different extents in phase-on and phase-off orientations of the switch, making the two forms of the switch distinguishable as suitable recombination substrates for FimB.
To test this hypothesis, we introduced base pair substitution mutations into the chromosomal copy of fimS at both the −10 and −35 motifs of PfimA that completely abolished promoter activity without altering the length of the fim switch. The effect of novobiocin on DNA supercoiling was confirmed by high-resolution agarose gel electrophoresis of reporter plasmid topoisomers. The plasmid was seen to be increasingly relaxed as the dose of novobiocin increased (Fig. 1B). We then compared fim switch inversion in the wild type and its PfimA promoter knockout derivative for sensitivity to novobiocin treatment. In both cases, the phase-off switch was seen to invert progressively in the off-to-on direction when gyrase activity was inhibited (Fig. 1C). Had transcription from PfimA been the generator of distinct substrates for FimB through an effect on local DNA supercoiling, one would have anticipated that the promoter knockout mutant would have shown either no bias or altered bias when gyrase activity was inhibited. Our findings showed that PfimA promoter activity was not responsible for the biasing of fimS inversion outcomes in relaxed DNA, so we sought an alternative explanation.
The switching bias of fimS is reversed in an lrp mutant.
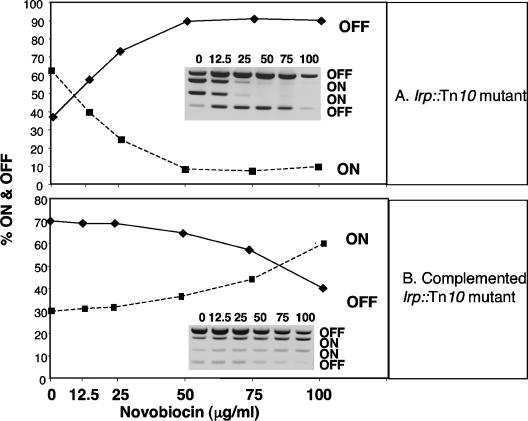
Genetic analysis revealed a role for the Lrp protein in determining fimS inversion outcomes. Lrp is essential for fimS inversion when the fimB gene is present in single copy but is dispensable in the presence of multicopy fimB (13). An lrp knockout mutation was introduced into strain VL386 by bacteriophage P1 transduction. Inversion of fimS was restored in this mutant following introduction of a multicopy plasmid encoding the FimB recombinase. When DNA gyrase activity in this strain was inhibited progressively with increasing concentrations of novobiocin, the orientation of fimS became biased in the off orientation (Fig. 2A). The culture was 63% on and 37% off in the absence of novobiocin, and this shifted to 10% on and 90% off at the highest concentration of the antibiotic used. This was the reverse of the situation seen in the wild-type strain that expressed Lrp, where novobiocin treatment resulted in biasing in favor of the on orientation (Fig. 1C). Complementation of the lrp::Tn10 mutation with a plasmid-borne lrp gene restored the wild-type inversion pattern of fimS (biased off-to-on) in response to gyrase inhibition (Fig. 2B). These data showed that the Lrp protein played a pivotal role in determining the outcome of FimB-mediated fimS site-specific recombination with relaxed DNA templates.
FIG. 2.
Inactivation of the lrp gene reverses the response of the fim switch to DNA relaxation. (A) Treatment with increasing concentrations of novobiocin results in a strong on-to-off bias in an lrp::Tn10 knockout mutant, the opposite of the pattern seen in the wild type. (B) Complementation of the lrp knockout mutation with a functional copy of the lrp gene restores the wild-type pattern of response to novobiocin treatment. Densitometric data from PCR switch assay gels (insets) were used to plot the graphs. Numbers above each gel lane are μg/ml of novobiocin. Bands corresponding to phase-on or -off are labeled.
The Lrp binding sites determine the response of fimS to DNA relaxation.
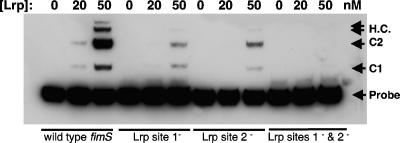
The Lrp binding sites 1 and 2 were removed from the switch by base substitution without altering the switch length to yield derivatives lacking just site 1 or just site 2 or lacking both sites. The altered sequences were tested in vitro for binding of purified Lrp protein. Binding of Lrp to sites 1 and 2 is known to be cooperative (34). An electrophoretic mobility shift assay showed that removal of either site strongly reduced Lrp-mediated shifting of a labeled switch probe while removal of both sites completely abrogated Lrp binding (Fig. 3).
FIG. 3.
Inactivation of the Lrp binding sites 1 and 2 within the fim switch. Data are shown from an electrophoretic mobility shift assay monitoring interaction of purified Lrp protein with the wild-type fim switch and mutants deficient in site 1, site 2, or sites 1 and 2. The protein concentrations used are given at the top of each lane. Lrp-fimS complexes are labeled at the right: complexes C1 and C2 and higher-order complexes (H.C.).
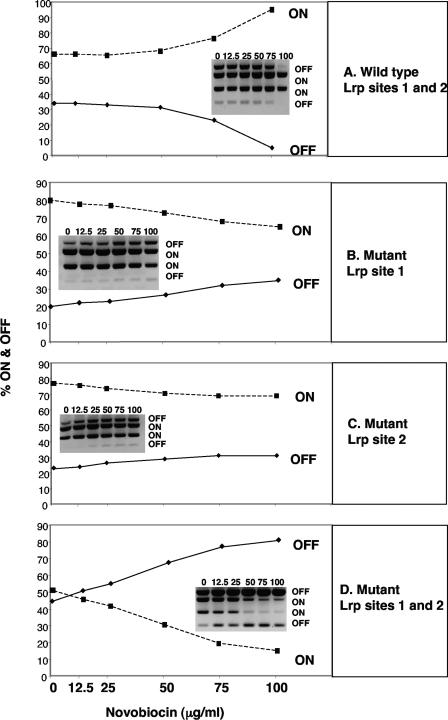
The individual Lrp site mutations and the double mutation were placed within the fimS element on the chromosome by homologous recombination. When a multicopy plasmid carrying the fimB gene was introduced, fimS inversion was restored in each strain, in agreement with previous data (13). The strains were then treated with increasing concentrations of novobiocin. In the mutants lacking just site 1 or site 2, the off-to-on biasing that is characteristic of the wild-type switch was no longer seen. In fact, a very slight shift in favor of the off orientation was observed as the concentration of novobiocin increased (Fig. 4). For example, cultures that began as ∼20% phase-off in the absence of novobiocin had become ∼30% phase-off at the highest dose of antibiotic used (Fig. 4B and C). These results indicated that the Lrp binding sites were required for the switch to respond to DNA relaxation. In the switch lacking both sites 1 and 2, the effect of the mutations on DNA inversion preferences was much more dramatic. Here, a very strong bias in favor of the off phase was clearly seen (Fig. 4D), a pattern that mimicked precisely that observed in the lrp knockout mutant (Fig. 2A). A culture with approximately equal numbers of on and off switches was shifted to ∼20% on and ∼80% off at the highest concentration of antibiotic. These results showed that Lrp, acting through sites 1 and 2, is responsible for determining the directionality of the response of the fimS invertible element to DNA relaxation.
FIG. 4.
Removal of the Lrp binding sites alters the response of the fim switch to DNA relaxation. With sites 1 and 2 intact, fimS becomes biased in favor of phase-on in response to increasing concentrations of novobiocin (A). Elimination of site 1 (B) or 2 (C) prevents on-biasing in response to novobiocin. When Lrp sites 1 and 2 are both disrupted, the response of the switch to novobiocin treatment is reversed (D) and resembles that seen in the lrp::Tn10 mutant (Fig. 2). The y axes of the graphs report the percentage of on and off switches in the population. The dosage of novobiocin is reported on the x axes. Densitometric data from PCR switch assay gels (insets) were used to plot the graphs. Numbers above each gel lane are μg/ml of novobiocin. Bands corresponding to phase-on or -off are labeled.
DISCUSSION
When DNA gyrase activity is unperturbed by novobiocin treatment, FimB inverts the fim switch in either direction with approximately equal efficiency. As increasing concentrations of novobiocin progressively inhibit gyrase activity with a concomitant relaxation of DNA supercoiling, fimS biasing in the off-to-on direction becomes apparent (Fig. 1). Thus, DNA relaxation creates a situation in which the on form of the switch becomes a trap that is difficult to escape. An attractive hypothesis proposed previously to account for this observation (12) envisaged a role for the PfimA promoter within the fim switch as a generator of topologically distinct substrates through the well-established ability of RNA polymerase movement on a DNA substrate to create differentially supercoiled domains (10, 14, 26, 32, 43). In this model, differential diffusion of supercoils created by PfimA activity in phase-on and phase-off would result in distinct substrates for FimB. However, we show here that inactivation of the PfimA promoter has no influence on the off-to-on-biased response of the switch to DNA relaxation.
On the other hand, we have now established cooperative roles for the Lrp DNA-binding protein and DNA supercoiling in determining the directionality of FimB-mediated fim switch inversion. In particular, we have shown that Lrp protein is required to maintain the phase-on trap. When this protein is removed from the cell by inactivation of the lrp gene, or when the sites within fimS to which it binds are disrupted, the phase-on trap is relieved. Thus, Lrp performs a role that is analogous to the RDFs that have been described in integrase-mediated integration and excision events (23-25). It allows two otherwise similar recombination substrates to become distinguishable, and like an RDF of the excisionase type, it probably does so by acting as a DNA-bending architectural element.
What is the nature of the phase-on trap? It seems clear that the recombination machinery can distinguish between phase-on and -off switches and that the Lrp protein bound to sites 1 and 2 plays a role in this discriminatory mechanism. The most straightforward model envisages a fixed reference point (Ref), currently of unknown molecular composition, in a constant (i.e., noninverting) region of the chromosome (Fig. 5). Bioinformatic analysis reveals no evidence of another Lrp binding site to the left of IRL where it might contribute the Ref function. However, a previously characterized integration host factor (IHF) binding site in this area (6), with the anticipated DNA bend that is a characteristic of occupied IHF sites, might fulfill the role of Ref. Communication between Ref and the mobile (i.e., inverting) and fully occupied Lrp binding sites 1 and 2 constitutes the on/off discriminatory mechanism. This communication influences the quality of the recombination substrate (the inverted repeats) and determines the efficiency with which it can be processed by the FimB recombinase. Further modulation of recombination is imposed by supercoiling or relaxation of the DNA.
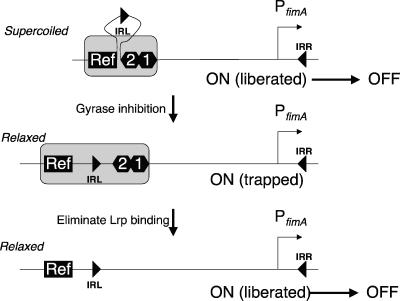
FIG. 5.
A model to account for trapping of the switch in the on orientation following DNA relaxation. The relevant features of the fim switch are shown: inverted repeats IRL and IRR, Lrp sites 1 and 2 (numbered hexagons), the PfimA promoter, and a putative reference point (Ref) located in the noninvertible region of the chromosome adjacent to IRL. Communication between Ref and the occupied Lrp binding sites facilitates the formation of an inhibitory complex (gray box). Negative supercoiling of the DNA assists extrusion of IRL from the inhibitory complex (top). Removal of negative superhelicity following gyrase inhibition abrogates extrusion of IRL, and the switch is trapped in the on orientation (middle). Elimination of Lrp binding disrupts the inhibitory complex, and rapid on-to-off switching occurs (bottom).
The proposed communication between Ref and the occupied Lrp binding sites 1 and 2 has the potential to inhibit the ability of IRL to participate with IRR in recombination (Fig. 5). Negative supercoiling facilitates the presentation of the IRL sequence, allowing it to interact with IRR in the FimB-mediated DNA inversion reaction. Relaxation of the DNA due to attenuation of gyrase activity inhibits IRL presentation, and the on orientation of the switch becomes increasingly disfavored as a recombination substrate. This results in the experimentally observed build-up in the population of bacteria with phase-on switches following novobiocin treatment.
The trapping of IRL in an inert complex within the relaxed phase-on switch requires sustained communication between Ref and the Lrp proteins at sites 1 and 2. When the Lrp binding sites are disrupted (or expression of Lrp protein is eliminated by inactivation of the lrp gene), the trap is relieved. Partial relief is seen when only one site (either 1 or 2) is disrupted. In the complete absence of Lrp protein binding, the relaxed switch is an excellent substrate for FimB-mediated recombination and shows a strong on-to-off inversion preference.
DNA supercoiling and nucleoid-associated proteins collaborate in many transposition and site-specific recombination systems (3, 21, 31). In transposon Tn10, supercoiling and the IHF protein cooperate to guide the transposition event down one of two possible pathways (9). The role of IHF has been described as that of a molecular spring that helps to modulate transposition by influencing the proficiency of the transposon ends for interaction with the transposase (9). This mechanism has some similarities to the role proposed here for the Lrp protein in the presentation of recombination substrates in fim DNA inversion (Fig. 5).
Why has the fim switch evolved such an elaborate mechanism for determining the directionality of DNA inversion? The fim operon can acquire fimE knockout mutations quite readily due to insertion of insertion elements and via other mechanisms (4). The resulting mutants express type 1 fimbriae in an apparently random on-off manner due to the action of the FimB recombinase on the switch. Perhaps a completely stochastic switching mechanism might not meet the requirements of the bacterial population under all circumstances, making it advantageous to bias switching in one direction or the other under certain conditions. DNA relaxation is a prerequisite for on-biased fimS inversion, and relaxation is associated with low rates of metabolic flux in the cell (41). Moreover, novobiocin treatment mimics specifically the effect of an unfavorable [ATP]/[ADP] ratio on DNA gyrase (44). This may represent a signal that the bacterium lacks energy, and a transition from a planktonic to an attached lifestyle might be advantageous. An enhanced tendency to express type 1 fimbriae would offer an attractive solution and is consistent with the known contributions of these cell surface appendages to niche colonization and the formation of biofilms (7).
Acknowledgments
We are grateful to Craig J. Benham and Georgi Muskhelishvili for stimulating discussions and Matthew McCusker and Kirsty McFarland for plasmids.
This work was supported by grant 061796 from the Wellcome Trust and grant 02/IN.1/B65 from Science Foundation Ireland.
REFERENCES
- 1.Abraham, J. M., C. S. Freitag, J. R. Clements, and B. I. Eisenstein. 1985. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. USA 82:5724-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beloin, C., R. Exley, A. L. Mahe, M. Zouine, S. Cubasch, and F. Le Hegarat. 2000. Characterization of LrpC DNA-binding properties and regulation of Bacillus subtilis lrpC gene expression. J. Bacteriol. 182:4414-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin, K. R., A. P. Abola, R. Kanaar, and N. R. Cozzarelli. 1996. Contributions of supercoiling to Tn3 resolvase and phage Mu Gin site-specific recombination. J. Mol. Biol. 256:50-65. [DOI] [PubMed] [Google Scholar]
- 4.Blomfield, I. C., M. S. McClain, J. A. Princ, P. J. Calie, and B. I. Eisenstein. 1991. Type 1 fimbriation and fimE mutants of Escherichia coli K-12. J. Bacteriol. 173:5298-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomfield, I. C., P. J. Calie, K. J. Eberhardt, M. S. McClain, and B. I. Eisenstein. 1993. Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J. Bacteriol. 175:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomfield, I. C., D. H. Kulasekara, and B. I. Eisenstein. 1997. Integration host factor stimulates both FimB- and FimE-mediated site-specific DNA inversion that controls phase variation of type 1 fimbriae expression in Escherichia coli. Mol. Microbiol. 23:705-717. [DOI] [PubMed] [Google Scholar]
- 7.Bollinger, R. R., M. L. Everett, S. D. Wahl, Y. H. Lee, P. E. Orndorff, and W. Parker. 2006. Secretory IgA and mucin-mediated biofilm formation by environmental strains of Escherichia coli: role of type 1 pili. Mol. Immunol. 43:378-387. [DOI] [PubMed] [Google Scholar]
- 8.Burns, L. S., S. G. J. Smith, and C. J. Dorman. 2000. Interaction of the FimB integrase with the fimS invertible DNA element in Escherichia coli in vivo and in vitro. J. Bacteriol. 182:2953-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalmers, R., A. Guhathakurta, H. Benjamin, and N. Kleckner. 1998. IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell 93:897-908. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C. C., and H. Y. Wu. 2003. Transcription-driven DNA supercoiling and gene expression control. Front. Biosci. 8:d430-439. [DOI] [PubMed] [Google Scholar]
- 11.Dove, S. L. 1994. A transcriptional switch controlled by site-specific recombination: regulation of fimA promoter strength and inversion in Escherichia coli. Ph.D. thesis. University of Dundee, Dundee, Scotland.
- 12.Dove, S. L., and C. J. Dorman. 1994. The site-specific recombination system regulating expression of the type 1 fimbrial subunit gene of Escherichia coli is sensitive to changes in DNA supercoiling. Mol. Microbiol. 14:975-988. [DOI] [PubMed] [Google Scholar]
- 13.Dove, S. L., and C. J. Dorman. 1996. Multicopy fimB gene expression in Escherichia coli: binding to inverted repeats in vivo, effect on fimA gene transcription and DNA inversion. Mol. Microbiol. 21:1161-1173. [DOI] [PubMed] [Google Scholar]
- 14.Dröge, P. 1993. Transcription-driven site-specific DNA recombination in vitro. Proc. Natl. Acad. Sci. USA 90:2759-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenstein, B. I. 1981. Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science 214:347-349. [DOI] [PubMed] [Google Scholar]
- 16.Free, A., and C. J. Dorman. 1997. The Escherichia coli stpA gene is transiently expressed during growth in rich medium and is induced in minimal medium and by stress conditions. J. Bacteriol. 179:909-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gally, D. L., T. J. Rucker, and I. C. Blomfield. 1994. The leucine-responsive regulatory protein binds to the fim switch to control phase variation of type 1 fimbrial expression in Escherichia coli K-12. J. Bacteriol. 176:5665-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gally, D. L., J. Leathart, and I. C. Blomfield. 1996. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol. Microbiol. 21:725-738. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 204:63-113. [DOI] [PubMed] [Google Scholar]
- 20.Higgins, C. F., C. J. Dorman, D. A. Stirling, L. Waddell, I. R. Booth, G. May, and E. Bremer. 1988. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 52:569-584. [DOI] [PubMed] [Google Scholar]
- 21.Huang, J., T. Schlick, and A. Vologodskii. 2001. Dynamics of site juxtaposition in supercoiled DNA. Proc. Natl. Acad. Sci. USA 98:968-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahooti, M., P. L. Roesch, and I. C. Blomfield. 2005. Modulation of the sensitivity of FimB recombination to branched-chain amino acids and alanine in Escherichia coli K-12. J. Bacteriol. 187:6273-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lesic, B., S. Bach, J.-M. Ghigo, U. Dobrindt, J. Hacker, and E. Carniel. 2004. Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol. Microbiol. 52:1337-1348. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, J. A., and G. F. Hatfull. 2001. Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 29:2205-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis, J. A., and G. F. Hatfull. 2003. Control of directionality in L5 integrase-mediated site-specific recombination. J. Mol. Biol. 326:805-821. [DOI] [PubMed] [Google Scholar]
- 26.Liu, L. F., and J. C. Wang. 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 84:7024-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClain, M. S., I. C. Blomfield, and B. I. Eisenstein. 1991. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J. Bacteriol. 173:5308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 29.O'Gara, J. P., and C. J. Dorman. 2000. Effects of local transcription and H-NS on inversion of the fim switch of Escherichia coli. Mol. Microbiol. 36:457-466. [DOI] [PubMed] [Google Scholar]
- 30.Olsen, P. B., and P. Klemm. 1994. Localization of promoters in the fim gene cluster and the effect of H-NS on the transcription of fimB and fimE. FEMS Microbiol. Lett. 116:95-100. [DOI] [PubMed] [Google Scholar]
- 31.Peña, C. E., J. M. Kahlenberg, and G. F. Hatfull. 1998. The role of supercoiling in mycobacteriophage L5 integrative recombination. Nucleic Acids Res. 26:4012-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruss, G. J., and K. Drlica. 1989. DNA supercoiling and prokaryotic transcription. Cell 56:521-523. [DOI] [PubMed] [Google Scholar]
- 33.Radman-Livaja, M., T. T. Biswas, A. Ellenberger, A. Landy, and H. Aihara. 2006. DNA arms do the legwork to ensure the directionality of l site-specific recombination. Curr. Opin. Struct. Biol. 16:42-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roesch, P. L., and I. C. Blomfield. 1998. Leucine alters the interaction of the leucine-responsive regulatory protein (Lrp) with the fim switch to stimulate site-specific recombination in Escherichia coli. Mol. Microbiol. 27:751-761. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Shi, J., and D. B. Biek. 1995. A versatile low-copy-number cloning vector derived from plasmid F. Gene 164:55-58. [DOI] [PubMed] [Google Scholar]
- 37.Smith, S. G. J., and C. J. Dorman. 1999. Functional analysis of the FimE integrase of Escherichia coli K-12: isolation of mutant derivatives with altered DNA inversion preferences. Mol. Microbiol. 34:965-979. [DOI] [PubMed] [Google Scholar]
- 38.Sternberg, N. L., and R. Maurer. 1991. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 204:18-43. [DOI] [PubMed] [Google Scholar]
- 39.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 40.van Duyne, G. D. 2005. Lambda integrase: armed for recombination. Curr. Biol. 15:R658-R660. [DOI] [PubMed] [Google Scholar]
- 41.van Workum, M., S. J. van Dooren, N. Oldenburg, D. Molenaar, P. R. Jensen, J. L. Snoep, and H. van Westerhoff. 1996. DNA supercoiling depends on the phosphorylation potential in Escherichia coli. Mol. Microbiol. 20:351-360. [DOI] [PubMed] [Google Scholar]
- 42.Wang, Q., and J. M. Calvo. 1993. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J. 12:2495-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Z., and P. Dröge. 1997. Long-range effects in a supercoiled DNA domain generated by transcription in vitro. J. Mol. Biol. 271:499-510. [DOI] [PubMed] [Google Scholar]
- 44.Williams, N. L., and A. Maxwell. 1999. Locking the DNA gate of DNA gyrase: investigating the effects on DNA cleavage and ATP hydrolysis. Biochemistry 38:14157-14164. [DOI] [PubMed] [Google Scholar]
- 45.Yanisch-Perron, C., J. Viera, and J. C. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]