Abstract
The membrane-bound protein EIICBGlc encoded by the ptsG gene is the major glucose transporter in Escherichia coli. This protein is part of the phosphoenolpyruvate:glucose-phosphotransferase system, a very important transport and signal transduction system in bacteria. The regulation of ptsG expression is very complex. Among others, two major regulators, the repressor Mlc and the cyclic AMP-cyclic AMP receptor protein activator complex, have been identified. Here we report identification of a novel protein, YeeI, that is involved in the regulation of ptsG by interacting with Mlc. Mutants with reduced activity of the glucose-phosphotransferase system were isolated by transposon mutagenesis. One class of mutations was located in the open reading frame yeeI at 44.1 min on the E. coli K-12 chromosome. The yeeI mutants exhibited increased generation times during growth on glucose, reduced transport of methyl-α-d-glucopyranoside, a substrate of EIICBGlc, reduced induction of a ptsG-lacZ operon fusion, and reduced catabolite repression in lactose/glucose diauxic growth experiments. These observations were the result of decreased ptsG expression and a decrease in the amount of EIICBGlc. In contrast, overexpression of yeeI resulted in higher expression of ptsG, of a ptsG-lacZ operon fusion, and of the autoregulated dgsA gene. The effect of a yeeI mutation could be suppressed by introducing a dgsA deletion, implying that the two proteins belong to the same signal transduction pathway and that Mlc is epistatic to YeeI. By measuring the surface plasmon resonance, we found that YeeI (proposed gene designation, mtfA) directly interacts with Mlc with high affinity.
In Escherichia coli K-12, as in many other gram-positive and gram-negative bacteria, the phosphoenolpyruvate-dependent carbohydrate phosphotransferase systems (PTSs) are the major transport and sensor systems for carbohydrates. All of the PTSs except the mannose-specific PTS consist of five conserved functional domains, designated enzyme I (EI) (gene, ptsI), the histidine-containing phosphoryl carrier protein HPr (gene, ptsH), enzyme IIA (EIIA), enzyme IIB (EIIB), and enzyme IIC (EIIC) (for reviews see references 30 and 31). Depending on the organism or system, these functional PTS domains exist as single or multidomain proteins. The two cytoplasmic proteins, EI and HPr, are the general components of all PTS, whereas the EII complexes are carbohydrate specific. The protein kinase EI uses phosphoenolpyruvate in an autophosphorylation reaction, and the phosphoryl group is subsequently transferred to HPr, EIIA, and EIIB. Finally, the carbohydrate substrate, which is bound by the integral membrane domain of EIIC, is phosphorylated and concomitantly translocated across the membrane. The preferred carbon source of E. coli, d-glucose, is taken up by two different EIIs, the high-affinity glucose-specific molecule EIIGlc (Glc-PTS) and the low-affinity mannose-specific molecule EIIMan (Man-PTS). The Glc-PTS consists of the cytoplasmic protein EIIAGlc, encoded by the crr gene (part of the ptsHI crr operon), and the membrane protein EIICBGlc (gene, ptsG). In addition to its transport function, EIIAGlc has a central regulatory role in carbon catabolite repression involving activation of the adenylate cyclase in its phosphorylated state (in the absence of glucose) and inducer exclusion by binding to several non-PTS carbohydrate systems in its unphosphorylated state (in the presence of glucose) (for a review see reference 31). In contrast to other PTSs, the Man-PTS is composed of the cytoplasmic protein EIIABMan and two membrane proteins, EIICMan and EIIDMan. The d-mannose-specific EII is encoded by the manXYZ operon. E. coli K-12 derivatives lacking the manXYZ genes are not capable of transporting and utilizing mannose as a sole carbon source. However, selection for Man+ suppression mutants revealed that several different single-amino-acid substitutions in EIICBGlc result in changes in the substrate specificity of the glucose transporter (24, 48; for a review see reference 9). These mutations result in a so-called “relaxed” conformation of the EIICBGlc (24), which, in contrast to the wild-type protein, enables the protein to transport and phosphorylate mannose, d-glucosamine, or the pentitols d-arabinitol and ribitol.
Many recent studies have revealed that the regulation of ptsG expression is very complex and takes place at both the transcriptional and posttranscriptional levels (for reviews see references 4 and 29). Glucose uptake derepresses ptsG expression by inactivation of the glucose repressor Mlc (makes large colonies) (11). The mlc gene has been mapped at 35 min on the E. coli chromosome. There is good evidence that mlc is identical to the previously identified dgsA gene (27, 33); the latter designation is used here.
The current model is that dephosphorylated EIICBGlc generated during glucose uptake binds Mlc and sequesters the repressor away from its DNA-binding sites (16, 19, 23, 43, 48; for reviews see references 4 and 29). Membrane localization, and not binding to EIICBGlc, seems to be responsible for the inactivation of Mlc in this process (39, 44). Furthermore, it has been shown that Mlc is also involved in the glucose-dependent regulation of the ptsHI crr operon (15, 28, 42, 48), the malT gene encoding the transcriptional activator of the maltose regulon (7), and the manXYZ operon (27). Mlc is therefore considered to be a global transcription factor responsible for the induction of genes in the presence of glucose. The second major regulator of ptsG transcription is the cyclic AMP (cAMP)-cAMP receptor protein (CRP) complex, which activates transcription. The two global regulators work antagonistically, since cAMP levels should be low during growth on glucose. This indicates that precise control of ptsG expression is necessary under various growth conditions, since EIICBGlc activity has a major effect on the levels of phosphorylation of all other PTS proteins, especially EIIAGlc (34, 35, 46). In this context it was not surprising that several more transcription factors which are also involved in ptsG gene regulation were identified recently. Among these factors are ArcA, a major transcription factor for the switch between aerobic growth and anaerobic growth in E. coli (13), two alternative sigma factors, σ32 for the heat shock response (40) and σS for expression of genes in the stationary growth phase (38), and the small DNA-binding protein Fis (41). In addition to these regulation mechanisms at the transcriptional level, ptsG expression is posttranscriptionally regulated by modulation of ptsG mRNA stability in response to the glycolytic flux in the cells (14, 17, 21, 45).
The aim of this study was to develop a genetic screen based on transposon mutagenesis in order to search for new factors which influence ptsG gene expression. We found that one open reading frame having an unknown function, yeeI (b1976), encodes a new Mlc-binding protein, and we characterized the physiological properties of yeeI mutants and compared these mutants to wild-type cells.
MATERIALS AND METHODS
Media and growth conditions.
Cells were routinely grown either in the standard phosphate minimal medium described previously (48) supplemented with carbon sources at a concentration of 0.2%, in Lennox broth without glucose and calcium ions (LB0 medium), or in 2× TY medium, as described in Ausubel et al. (1). The utilization of various carbohydrates was examined by using MacConkey agar plates (Difco, Detroit, MI) containing carbon sources at a concentration of 1% (all carbon sources except mannose) or 0.5% (mannose). Antibiotics were used at the following concentrations: tetracycline, 10 mg/liter; ampicillin, 50 mg/liter; and chloramphenicol, 25 mg/liter.
Bacterial strains and plasmids.
All of the strains used were E. coli K-12 derivatives. The genotypes and sources of the relevant bacterial strains and plasmids are shown in Table 1. P1 transduction was performed as described previously (48). Strain LJB17 was obtained by disruption of the complete dgsA gene using the method described by Datsenko and Wanner (6). To do this, a PCR product was generated using pKD3 template DNA and primers 5′-TTA ACC CTG CAA CAG ACG AAT CAA CAA AGA ACC GTT GTG TAG GCT GGA GCT GCT TC-3′ and 5′-GAT TAT TTC GGA GCG CGA AAA TAT AGG GAG TAT GCG CAT ATG AAT ATC CTC CTT AG-3′. DNA amplification was performed as described by Saiki et al. (36), using Taq DNA polymerase from Roche Diagnostics (Mannheim, Germany) or Herculase from Stratagene (La Jolla, CA). Strain LMH111, a Tets derivative of LZ110, was obtained using the selection method described by Bochner et al. (3). For construction of the yeeI expression vector pTMByeeI-S, primers 5′-ACG CTG CAG AAG TGG CCC TGG AAA GTA CAA G-3′ and 5′-AAG CTT GTG GTG GTG GTG GTG ATG AAC ATT CGT CGC CGA AAA C-3′ and genomic DNA from E. coli wild-type cells were used. The original start codon, ATG, was changed to CTG. Artificial PstI (upstream) and HindIII (downstream) restriction sites were introduced (underlined), and codons for five carboxy-terminal histidine residues were added. The vector pTMByeeI-L was constructed in a similar way, using primers 5′-TCC CTG CAG TTT TGT TCA AGT GAC G-3′ and 5′-AAG CTT GTG GTG GTG GTG GTG ATG AAC ATT CGT CGC CGA AAA C-3′. The PCR products obtained were treated with PstI and HindIII and cloned into identically treated expression vector pTM30. Using the PstI site in the multiple-cloning site of the pTM30 expression vector always leads to addition of a new, artificial ATG start codon and therefore one additional amino-terminal amino acid.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| JM109 | thi-1 Δ(lac-proAB) recA1 hsdR1/F′ traD36 proA+B+ lacIqZΔM15 | 47 |
| LJ110 | F− (wild type) | 48 |
| LJ121 | LJ110 Δ(ptsG::cat) man-8 zea-225::Tn10 | 48 |
| LZ1 | LJ110 Δ(manXYZ::cat) ptsG1 | 48 |
| LZ110 | LJ110 Δ(argF-lac)169 zah-735::Tn10Tetr | 48 |
| LZ150 | LZ110 Δ(ptsG::cat) | 48 |
| LJ231-3 | LJ110 Δ(manXYZ::cat) ptsG3csc+ | 12 |
| LMH111 | LZ110 Tets | This study |
| LZ17-3 | LJ231-3 yeeI77::mini-Tn10Kanr | This study |
| LJB33 | LJ231-3 yeeI161::mini-Tn10Kanr | This study |
| LJB43 | LJ231-3 yeeI210::mini-Tn10Kanr | This study |
| LJB41 | LJ110 yeeI210::mini-Tn10Kanr | This study |
| LJB17 | LJ110 dgsA::cat | This study |
| LJB61-1 | LMH111 yeeI210::mini-Tn10Kanr Δ(ptsG::cat) dgsA::mini-Tn10Tetr | This study |
| LJB70 | LJB17 yeeI210::mini-Tn10Kanr | This study |
| LJB110 | LZ110 yeeI210::mini-Tn10Kanr | This study |
| Plasmids | ||
| pACYC177 | Apr Knr | 5 |
| pBluescript SK(+) | Apr | 1 |
| pKD3 | Apr Cmr | 6 |
| pNK2859 | Apr, mini-Tn10Kanr | 18 |
| pSU18 | Cmr | 20 |
| pSU19 | Cmr | 20 |
| pTM30 | Apr | 22 |
| pMAN1 | pACYC177, Apr, manXYZ+ | This study |
| pTMByeeI-S | pTM30, Apr, yeeI-L | This study |
| pTMByeeI-L | pTM30, Apr, yeeI-S | This study |
| F′8::TnΦ(ptsGop-lacZ) | 48 |
The genetic nomenclature is that of Berlyn et al. (2).
For construction of pMAN1, primers 5′-CGC GAA ACG CCC GGG TTT TTG GTT GTA GCC-3′ and 5′-CTT ACA GTC CCG GGA GGC CGC AAG CGT AA-3′ and genomic DNA from E. coli wild-type cells were used. The two primers, which are complementary to the immediate upstream and downstream regions of the manXYZ operon in E. coli K-12, introduced artificial SmaI sites into the PCR product. After treatment with SmaI the PCR product was purified and ligated into identically treated plasmid pACYC177.
Isolation of chromosomal and plasmid DNA, restriction analysis, and DNA sequencing.
All manipulations with chromosomal or recombinant DNA were carried out using standard procedures, as described previously (1). Plasmid DNA was prepared by using the JETstar DNA purification system (Genomed, Bad Oeynhausen, Germany). Restriction enzymes were purchased from New England Biolabs (Schwalbach, Germany) and were used according to the recommendations of the supplier. The oligonucleotides used for PCR were purchased from Thermo Electron (Ulm, Germany). All DNA sequencing reactions were performed by the dideoxy chain termination method using an ALFexpress AutoRead or dATP Labeling Mix sequencing kit from Amersham Biosciences (Freiburg, Germany).
Transport and β-galactosidase assays.
Transport of α-d-[methyl-14C]glucopyranoside (α-MG) or [14C]sucrose (final concentration, 25 μM) was determined using exponentially grown cells, as described previously (12, 48). Samples were taken after 10, 20, 30, and 40 s. The β-galactosidase assay was performed as described by Pardee and Prestige (25).
Western blot analyses.
Bacterial cells grown in LB0 medium were harvested when the absorption at 600 nm was 0.6 and were resuspended in 60 μl of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.1% bromophenol blue). Unless indicated otherwise, the samples were heated at 100°C for 5 min. Ten microliters of total cellular proteins was separated by electrophoresis on 0.1% SDS—12% polyacrylamide gels and transferred to an Immobilon membrane (Millipore, Eschborn, Germany). For detection of YeeI protein derivatives, a penta-His antibody (QIAGEN, Hilden, Germany) was used. For detection of EIIAGlc, EIICBGlc, and Mlc polyclonal anti-EIIAGlc, anti-EIIBGlc, anti-EI, and anti-Mlc antibodies were used as described previously (43). Antibody binding was visualized by using the enhanced chemiluminescence system of Amersham Bioscience (Braunschweig, Germany). For quantification films were scanned with a Hewlett-Packard Scanjet 7400c, and bands were quantified using the Metamorph 6.1 software (Universal Software Corporation, Nashua, NH).
Protein purification.
Mlc derivatives were constructed and purified as described previously (39). For purification of YeeI-His5, cells harboring pTMByeeI-S were grown in LB0 medium with ampicillin and 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG). When cultures reached an optical density at 650 nm of 1, cells were harvested by centrifugation. The cells were resuspended in cold buffer A1 (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole; pH 8) and disrupted by sonication using a W-250D Sonifier (Branson, Danbury, Conn.). The cell debris was removed by low-speed centrifugation (14,000 rpm, 15,000 × g). The supernatant was incubated with Ni-nitrilotriacetic acid-agarose (QIAGEN, Hilden, Germany) for 1 h at 5°C. The Ni-nitrilotriacetic acid was washed eight times with buffer B1 (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole; pH 8), and the bound protein was eluted with buffer C (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole; pH 8).
Preparation of membrane and cytoplasmic fractions.
Cells were grown on LB0 medium with ampicillin in the presence of inducers until the early stationary phase. Cells were collected by low-speed centrifugation, washed with buffer A2 (50 mM NaH2PO4, 300 mM NaCl; pH 8), resuspended in buffer A2, and disrupted by sonication (W-250D Sonifier; Branson, Danbury, Conn.). The cell debris was removed by low-speed centrifugation. Subsequently, cytoplasmic and membrane fractions were separated by high-speed centrifugation (80,000 × g, 1 h). For solubilization of membrane-associated proteins, membranes were washed with low-ionic-strength buffer B2 (1 mM Tris-HCl, 3 mM EDTA; pH 6) and again precipitated by high-speed centrifugation. The distributions of EIICBGlc, EI, and YeeI in the different fractions were determined by performing Western blot analyses with the appropriate antibodies.
Surface plasmon resonance experiments.
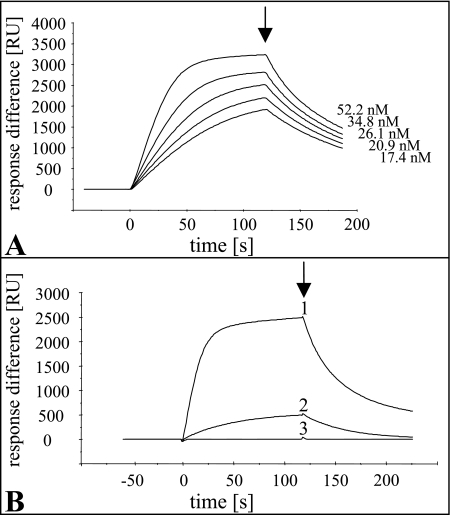
The interaction of YeeI and Mlc was analyzed by surface plasmon resonance (SPR) with a Biacore X biosensor (Amersham Pharmacia Biacore AB, Freiburg, Germany). YeeI or Mlc was routinely immobilized on the surface of a CM5 sensor chip (Pharmacia Biosensor AB) at a surface concentration of about 5 ng mm−2 (equivalent to 5,000 resonance units) by amide coupling by following the instructions of the manufacturer. The differences in binding of Mlc derivatives to immobilized YeeI were determined by subsequently pouring Mlc proteins over the sensor chip using HBS-EP buffer (Biacore AB) at a flow rate of 5 μl/min, using equimolar amounts (50 nM). To determine the dissociation constant for the interaction between Mlc and YeeI, carboxy-terminal His-tagged Mlc was injected at different concentrations at a flow rate of 10 μl/min. Calculation was performed with the Langmuir bimolecular interaction model of the BIAevaluation 3.1 software (Biacore AB).
RESULTS
Isolation of mutants with reduced activity of the Glc-PTS.
In order to identify new proteins involved in the regulation of ptsG, a genetic screen was performed to detect mutants with reduced EIICBGlc activity. Strain LZ1 (ΔmanXYZ ptsG1) was subjected to mutagenesis with a mini-Tn10Kanr insertion, using the method described by Kleckner et al. (18). LZ1 incorporates mannose at a low rate via a mutated “relaxed” PtsG variant (S169F). As mannose is a poor substrate (24, 48), the phenotype on MacConkey mannose indicator plates was suitable for searching for mutations, which decrease but do not completely suppress the transport activity of the Glc-PTS, by looking for Glc+ Man− cells. A total of 10,000 kanamycin-resistant cells from the mutant library were screened for a white Man− phenotype. In the two predominant classes of mutants either the transposon had integrated into the ptsG gene, which was indicated by simultaneous loss of glucose fermentation capability and an otherwise positive phenotype on other PTS substrates, or the cells had a pleiotropic negative phenotype for all PTS carbohydrates, resembling ptsI or ptsH mutants (data not shown).
However, three mannose-negative mutants which still fermented glucose were obtained. To avoid multiple transposon insertions, we transduced the mutations by using the kanamycin resistance marker into strain LJ231-3 (csc+ ΔmanXYZ ptsG3) (12), which expresses a similar “relaxed” PtsG (S169F). The sucrose-positive Csc+ (chromosomally encoded sucrose catabolism; a marker used to distinguish strains from LZ1) strains obtained were mannose negative like LZ1 and were designated LJB33, LJB43, and LZ17-3.
To ensure that reduced uptake of mannose by the Glc-PTS was responsible for the Man− phenotype, all three strains were transformed with the pMAN1 plasmid. This plasmid constitutively expresses the manXYZ genes for IIMan (EIIABMan-EIICMan-EIIDMan). After introduction of pMAN1 into LJB33, LJB43, or LZ17-3, the cells were Man+, confirming that the transposon insertions had no general effect on mannose utilization.
To further characterize the sites of the transposon insertions, chromosomal DNA of LZ17-3 was isolated and treated with EcoRV, and the DNA fragments were shotgun cloned into the pSU18 vector. Plasmid DNA from kanamycin-resistant colonies was isolated and sequenced. The mini-Tn10Kanr insertion of LZ17-3 was found in an open reading frame having an unknown function, yeeI (b1976), which is located between the tRNA-encoding genes serU and asnT at 44.1 min on the E. coli K-12 chromosome. The exact position was behind bp 77 (the A of the first predicted start codon, ATG, was defined as position 1). To locate the transposon insertions in LJB33 and LJB43, chromosomal DNAs of these two strains were isolated. Using the chromosomal DNAs as templates and two primers which corresponded to the immediate upstream and downstream regions of the yeeI open reading frame, the relevant DNA fragments were amplified, subcloned, and sequenced. DNA sequence analyses revealed that in LJB33 the mini-Tn10Kanr insertion was behind bp 161 of yeeI and in LJB43 it was behind bp 210 of yeeI.
yeeI cloning and complementation assays.
Two putative start sites are annotated for yeeI in the NCBI GenBank accession number U00096 sequence of the complete E. coli K-12 strain MG1655 genome.
The larger of the two putative open reading frames encodes a predicted protein having 278 amino acid residues, and the smaller one codes for a protein having 265 amino acid residues. Both open reading frames were amplified by PCR and cloned into the expression vector pTM30 (22). In both constructs codons for one additional amino-terminal histidine residue (provided by the expression vector) and five additional carboxy-terminal histidine residues were added. The recombinant plasmids were designated pTMByeeI-L (larger open reading frame) and pTMByeeI-S (smaller open reading frame). The correct sizes of the two proteins were controlled during SDS-PAGE and subsequent Western blot analysis using a penta-His antibody (data not shown). Transformants of LJB33, LJB43, or LZ17-3 with either pTMByeeI-L or pTMByeeI-S were tested on MacConkey agar indicator plates for the ability to ferment mannose. Both plasmids were capable of complementing the strains tested to a Man+ phenotype, indicating that the shorter of the two products is sufficient for YeeI activity.
Physiological characterization of YeeI− mutants.
The yeeI::mini-Tn10Kanr mutation of LJB43 was transferred by P1 transduction to wild-type strain LJ110 (48), yielding strain LJB41. The growth rates of LJ110 and LJB41 on glycerol and glucose as single carbon sources were determined (Table 2). Whereas the generation times of the two strains on glycerol were identical, the generation time of LJB41 was 14 min longer than the generation time of LJ110 on glucose. Furthermore, identical generation times for the wild-type and the yeeI mutant were obtained during growth on LB0 rich medium or minimal medium with the PTS substrate d-mannitol, N-acetyl-d-glucoseamine, or d-trehalose or with the non-PTS substrate d-lactose, d-galactose, maltose, or succinate as a sole carbon source (each carbon source was used at a concentration of 0.2%) (data not shown). These results indicate that the yeeI mutation does not cause a general growth defect.
TABLE 2.
Influence of a yeeI mutation on growth on glucose, uptake of α-MG, and induction of a ptsG-lacZ fusiona
| Parameter | Wild type
|
yeeI mutant
|
||
|---|---|---|---|---|
| Gly | Glc | Gly | Glc | |
| Generation time (min) | 95 ± 4 | 73 ± 3 | 93 ± 3 | 87 ± 3 |
| α-MG uptake (nmol mg−1 min−1) | 0.82 ± 0.1 | 2.99 ± 0.3 | 0.72 ± 0.1 | 2.05 ± 0.1 |
| Induction of ptsG-lacZ (nmol mg−1 min−1) | 109 ± 9 | 218 ± 14 | 88 ± 10 | 166 ± 11 |
For determination of generation times, LJ110 and LJB41 were grown in liquid minimal medium with 0.2% glycerol (Gly) or 0.2% glucose (Glc). Growth was determined by measuring the optical density at 420 nm. At least three samples were taken during one doubling time. To measure transport of α-MG, LJ110 and LJB41 were grown in minimal medium with 0.2% glycerol. For induction, glucose was added to a final concentration of 0.2%. The cells were grown for an additional 2 h and harvested during the exponential growth phase. The uptake of α-MG was determined by using 25 μM (final concentration). To determine the effect of the yeeI mutation on the induction of ptsG, the Δ(lac) derivative LZ110/F′8::TnΦ(ptsGop-lacZ) and the yeeI mutant LJB110/F′8::TnΦ(ptsGop-lacZ) were used. The cells used in the β-galactosidase assays were treated like the cells in the transport assay. The values are the means ± standard deviations for at least three measurements.
To test the activity of the Glc-PTS, the rates of α-MG uptake by uninduced and induced wild-type and YeeI− cells were determined (Table 2). The basal transport activity of the Glc-PTS in uninduced yeeI mutants was 87% of the wild-type activity, whereas the induced α-MG uptake activity was only 68% of the wild-type activity. These results were confirmed by determining the patterns of expression of a single-copy ptsG-lacZ operon fusion in the wild-type strain and in the YeeI-negative strain. Both the basal and induced β-galactosidase activities were reduced in the yeeI mutant. The LacZ activities of the yeeI mutants were 81% (not induced) and 76% (induced with glucose) of the wild-type activities (Table 2).
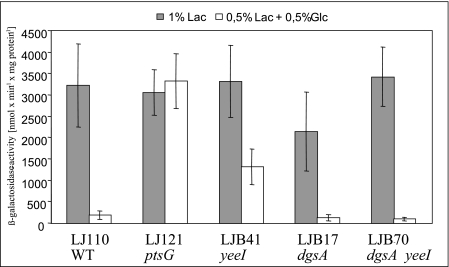
Reduced activity of the Glc-PTS in a yeeI genetic background should have an effect on the glucose-dependent carbon catabolite repression mechanisms, since the level of EIIAGlc phosphorylation, the key regulator for inducer exclusion and activation of the adenylate cyclase, depends directly on the amount of EIICBGlc (34, 35, 46). To test this hypothesis, several strains with defined mutations in relevant genes were examined for induction of lacZ by lactose in the presence of glucose (Fig. 1). In wild-type strain LJ110 induction of the lac operon by 0.5% lactose was reduced almost 20-fold in the presence of 0.5% glucose compared to cells which were grown in the presence of lactose alone. On the other hand, disruption of ptsG (in LJ121) resulted in a level of expression of lacZ that was independent of the presence of glucose in the medium, confirming the strong influence of EIICBGlc on inducer exclusion and cAMP-CRP-dependent carbon catabolite repression. As expected, constitutive expression of ptsG caused by inactivation of the Mlc-encoding dgsA gene (LJB17) had no effect on the “glucose effect” on lacZ expression. However, in yeeI mutant LJB41 cultures addition of glucose resulted in only a twofold reduction in β-galactosidase activity, confirming that the absence of YeeI led to a reduction in the Glc-PTS activity. Interestingly, disruption of dgsA (in LJB70) could suppress the effect of the yeeI mutation, implying that the two proteins belong to the same signal transduction pathway and that Mlc is epistatic to Yee. Moreover, LJB17 (dgsA) and LJB70 (dgsA yeeI) had identical generation times (75 min) in minimal medium with 0.2% glucose.
FIG. 1.
β-Galactosidase activities of various strains after induction with lactose in the presence or absence of glucose. Cells were grown in LB0 medium supplemented either with 1% lactose (Lac) or with a mixture of 0.5% lactose and 0.5% glucose (Glc). Cells were harvested during exponential growth. The values are the means of at least four measurements of β-galactosidase activity. WT, wild type.
Quantity of EIICBGlc is reduced in a yeeI background.
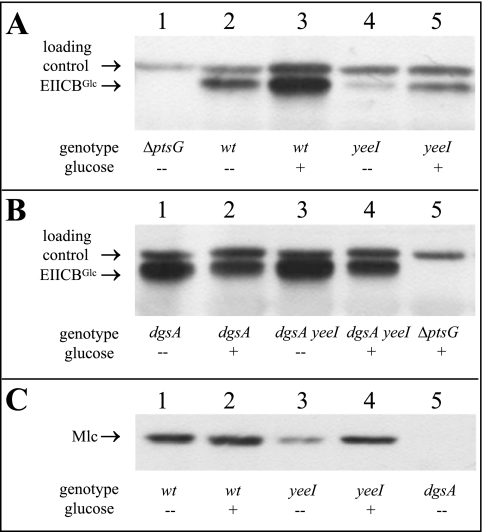
All physiological effects of the yeeI mutation measured so far suggest that there is reduced synthesis of the glucose transporter. Therefore, the quantities of EIICBGlc in different strains in response to glucose induction were determined directly by Western blot analysis (Fig. 2). The gels were digitalized, and the bands were quantified. No EIICBGlc was detected in ΔptsG strain LJ121 (Fig. 2A, lane 1). The signal in lane 1 was generated by a nonspecifically stained protein that was simultaneously recognized by the polyclonal antiserum. We used this signal to standardize the amounts of total proteins in all samples. Based on the relatively high basal level of expression of ptsG, EIICBGlc was detectable even in the absence of glucose in wild-type strain LJ110 (Fig. 2A, lane 2). However, the synthesis of EIICBGlc was increased 6.2-fold after addition of 0.2% glucose (Fig. 2A, lane 3). Addition of glucose had the same induction effect in yeeI strain LJB41 (synthesis was increased 5.8-fold) (Fig. 2A, lanes 4 and 5), but compared to the wild type, the synthesis of EIICBGlc was reduced by 5-fold both in the presence and in the absence of glucose. Moreover, as observed previously, introduction of a dgsA mutation could suppress the yeeI effect (Fig. 2B). A lack of Mlc caused constitutive ptsG expression in LJB17 (Fig. 2B, lane 1), which was reduced in the presence of glucose due to a decrease in the intracellular cAMP level (Fig. 2B, lane 2). The dgsA yeeI double mutant LJB70 exhibited exactly the same profile as LJB17 (Fig. 2B, lanes 3 and 4), indicating that YeeI does not have a direct effect on ptsG expression.
FIG. 2.
Western blot analyses of EIICBGlc and Mlc expression. Cells were grown in LB0 medium with (+) or without (−) 1% glucose. Samples were prepared from LJ121 (ΔptsG), LJ110 (wild type), LJB41 (yeeI::mini-Tn10Kanr), LJB17 (ΔdgsA::cat), and LJB70 (ΔdgsA::cat yeeI::mini-Tn10Kanr) as described in Materials and Methods and were analyzed by using anti-IIBGlc and anti-Mlc antibodies. wt, wild type.
These results raised the question whether a yeeI mutation also has an effect on the dgsA expression pattern. It was shown previously (7, 23) that dgsA expression is negatively autoregulated, and the dgsAp promoter is relatively weak. This mechanism ensures that sequestration of Mlc by unphosphorylated EIICBGlc cannot be compensated for by an increase in Mlc synthesis. In agreement with this model, the addition of glucose to wild-type cells of LJ110 resulted in no change in the quantity of the repressor protein detectable (Fig. 2C, lanes 1 and 2). Interestingly, on the other hand, introduction of a yeeI mutation reduced the amount of Mlc (Fig. 2C, lane 3). In contrast, addition of glucose, which leads to EIICBGlc dephosphorylation and thus titration of Mlc, resulted in an increase in the amount of Mlc in a yeeI background (Fig. 2C, lane 4). These results, combined with the fact that the basal level of ptsG expression was reduced even in yeeI strains, indicated that the steady-state level of Mlc is lower in the absence of YeeI but is still sufficient for ptsG repression.
YeeI overproduction leads to titration of Mlc, even in the absence of EIICBGlc.
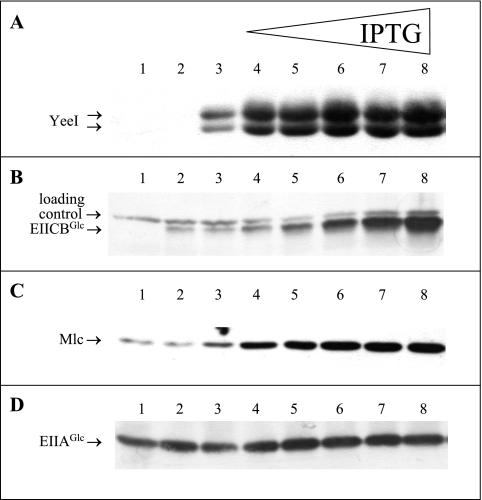
The results described above indicated that yeeI mutations cause a reduction in ptsG expression. One possible explanation for these results is that YeeI directly interacts with Mlc. To obtain more evidence for this hypothesis, we examined whether overproduction of YeeI was able to derepress ptsG transcription. Strain LJB41(yeeI::mini-Tn10Kanr) was transformed with pTMByeeI-S and grown in LB0 medium with ampicillin in the absence or presence of different amounts of IPTG. Samples were taken and analyzed in parallel by Western blot analysis to determine the amounts of YeeI-His5, EIICBGlc, Mlc, and EIIAGlc (Fig. 3A to D). LJ121 (ΔptsG; no EIICBGlc; no His-tagged YeeI) and LJB41/pTM30 (ptsG+; no His-tagged YeeI) were used as controls. The gels were digitalized, and the bands were quantified. As shown in Fig. 3A, even in the absence of IPTG there was a measurable basal level of expression of yeeI in this lacI/tacp-based pTM30 expression system (Fig. 3, lane 3). However, increasing the amount of IPTG (from 10 to 100 μM IPTG) (lanes 4 to 8) resulted in increases in the YeeI protein concentration. Interestingly, in Western blot analyses of YeeI we frequently observed the presence of two bands (see Discussion). The overexpression of YeeI, in turn, led to a significant increase in EIICBGlc levels (Fig. 3B). Quantification of the Western blots indicated that there was at least 10-fold overproduction of EIICBGlc in this process. In accordance with the autoregulation of dgsA (7, 23), YeeI overproduction resulted in a clear 3.8-fold increase in the quantity of Mlc (Fig. 3C). However, due to the relatively weak dgsAp promoter, fast saturation of Mlc production (at 30 μM IPTG and higher concentrations) was observed. It was shown previously (15, 28, 42, 48) that Mlc also has a slightly negative effect on the expression of the ptsHI crr operon. There are two promoters (P0 and P1) upstream of ptsH, and there is another internal promoter (P2) at the 3′ end of ptsI, which is the major promoter for crr (8, 10). Only P0 is subject to Mlc repression, and the overall expression of ptsH and ptsI is induced about fourfold during growth on glucose (28), whereas transcription from the P2 promoter in front of crr is constitutive. Therefore, overexpression of yeeI and subsequent titration of Mlc had only a twofold inducing effect on EIIAGlc (Fig. 3D, compare lane 3 to lanes 4 to 8).
FIG. 3.
Effects of different amounts of YeeI on the quantities of EIICBGlc, Mlc, and EIIAGlc. Cells were grown in LB0 medium with ampicillin and different concentrations of IPTG (lanes 1, 2, and 3, no IPTG; lane 4, 10 μM IPTG; lane 5, 20 μM IPTG; lane 6, 30 μM IPTG; lane 7, 50 μM IPTG; lane 8, 100 μM IPTG). Samples were prepared as described in Materials and Methods from LJ121/pTM30 (ΔptsG) (lane 1), LJB41/pTM30 (yeeI::mini-Tn10Kanr) (lane 2), and LJB41/pTMByeeI (lanes 3 to 8). Ten microliters of each sample was analyzed simultaneously using anti-penta-His (A), anti-EIIBGlc (B), anti-Mlc (C), and anti-EIIAGlc (D) antibodies.
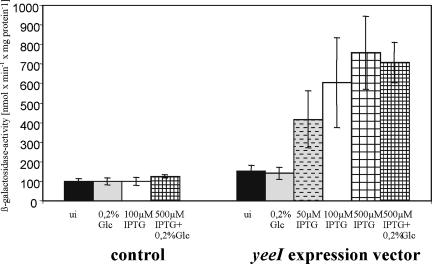
In the next set of experiments we examined whether YeeI is capable of titrating Mlc in the absence of EIICBGlc. To do this, strain LZ150 (ΔptsG Δlac dgsA+)/F′8::TnΦ(ptsGop-lacZ) (48) was transformed with either pTM30 (control) or pTMByeeI-S. Cells were grown in LB0 medium with ampicillin in the presence or absence of glucose and/or IPTG. Deletion of ptsG completely prevented induction of the ptsG-lacZ fusion by addition of glucose in strains harboring pTM30 or pTMByeeI-S, since Mlc cannot be sequestered by EIICBGlc in this background (Fig. 4). However, increasing the concentration of IPTG resulted in increased β-galactosidase activity, but only in yeeI-expressing cells. With 500 μM IPTG the maximum β-galactosidase activity was about 700 nmol min−1 mg protein−1 for LZ150/F′8::TnΦ(ptsGop-lacZ)/pTMByeeI-S. Because of the missing EIICBGlc, simultaneous addition of glucose and IPTG resulted in no reduction in ptsG-lacZ transcription by cAMP-CRP-dependent catabolite repression. In turn, after introduction of an additional dgsA mutation, which resulted in strain LJB61-1 [ΔptsG Δlac yeeI dgsA/F′8::TnΦ(ptsGop-lacZ)], the YeeI-independent constitutive β-galactosidase activity was 685 ± 110 nmol min−1 mg protein−1 under these growth conditions. This means that in accordance with the current model of ptsG regulation, Mlc is epistatic to both EIICBGlc and YeeI. These results support the idea that YeeI, like EIICBGlc, may act as an Mlc-binding protein that can sequester the repressor even in the absence of the glucose transporter and, when overexpressed, can lead to increased expression of a ptsG-lacZ fusion.
FIG. 4.
β-Galactosidase activities of LZ150/F′8::TnΦ(ptsGop-lacZ) with pTM30 (control) or pTMByeeI-S (yeeI expression vector) after addition of glucose and/or IPTG. Cells were grown in LB0 medium containing ampicillin with no supplement (uninduced [ui]) or supplemented with 0.2% glucose (Glc) and/or various amounts of IPTG. Cells were harvested during exponential growth. The values are mean β-galactosidase activities for at least four different experiments.
YeeI binds to Mlc in surface plasmon resonance experiments.
To obtain proof that there is a direct interaction between YeeI and Mlc, SPR experiments with purified proteins were performed. A similar approach was used previously to detect the interaction between Mlc and the separately expressed, soluble B-domain of EIICBGlc (23). Purified YeeI was immobilized on a Biacore sensor chip by amide coupling. When purified Mlc was exposed to immobilized YeeI, a high-affinity interaction was detected (Fig. 5). To determine the strength of the YeeI-Mlc interaction, various concentrations of Mlc (between 17.4 nM and 52.2 nM) were used for injection (Fig. 5A). From the sensorgram data an apparent dissociation constant (Kd) of 14 ± 0.3 nM for the binding of Mlc to YeeI was calculated using the BIAevaluation software. Correspondingly, in the reverse experiment purified YeeI exhibited similar binding to immobilized Mlc (data not shown). For further characterization of the interaction between the two proteins, we examined the binding of purified Mlc derivatives with 9 or 18 carboxy-terminal amino acids deleted (Δ9C-Mlc and Δ18C-Mlc, respectively) (39) to immobilized YeeI (Fig. 5B). Interestingly, whereas full-length Mlc resulted in the expected strong response signal, the Δ9C-Mlc protein gave a significantly weaker signal (20% of the wild-type protein signal) and theΔ18C-Mlc protein gave no signal in the SPR experiment. These results provide further strong evidence that there is a highly specific interaction between YeeI and Mlc. Moreover, the carboxy-terminal part of Mlc seems to be important for this interaction.
FIG. 5.
Surface plasmon resonance analyses of the interaction between YeeI and Mlc. (A) Real-time interaction between immobilized YeeI-S-His5 and Mlc-His6 at various concentrations (between 17.4 nM and 52.2 nM). Injection of Mlc started at time zero, and the arrow indicates when injection ended. The sensorgrams, which showed the binding response in resonance units (RU) as a function of time, from the different experiments were combined to obtain one diagram. Purified CcpA and HPr from Bacillus subtilis, which exhibited no interaction with either YeeI or Mlc, were used as controls. (B) Interaction of immobilized YeeI-S-His5 with full-length Mlc (line 1), Δ9C-Mlc (line 2), and Δ18C-Mlc (line 3). Equal amounts of the Mlc derivatives were injected. The sensorgrams from the three different experiments were combined to obtain one diagram.
YeeI is located in the cytoplasm.
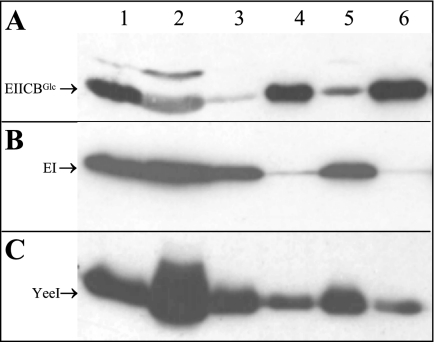
It was shown previously that the hydrophilic B-domain of EIICBGlc is capable of sequestering only Mlc, if it is connected to a membrane anchor (39, 44). Therefore, the question arose, whether YeeI is a soluble, membrane-associated, or integral membrane protein. To examine this, wild-type strain LJ110 was transformed with pYeeI-S and grown on LB0 medium in the presence of glucose and a low concentration of IPTG (50 μM) to induce EIICBGlc and the plasmid-encoded YeeI-His5, respectively. Cells were disrupted by sonication, and the cell components were separated by differential centrifugation. Different fractions with equal amounts of total proteins were tested simultaneously for the presence of the integral membrane protein EIICBGlc (membrane reference protein), the general PTS protein EI (cytoplasmic reference protein), and YeeI in Western blot analyses, using the appropriate antibodies (Fig. 6A to C). Whereas all proteins were detectable in the low-speed pellet after sonication (Fig. 6A to C, lane 1) and in the supernatant after low-speed centrifugation (Fig. 6A to C, lane 2), large amounts of membrane-bound EIICBGlc were found almost solely in the high-speed, membrane-containing pellet, and small amounts were found in the high-speed supernatant after ultracentrifugation (Fig. 6A, compare lane 4 to lane 3). Use of a low-ionic-strength buffer that usually helps to solubilize membrane-associated proteins had no effect (Fig. 6A, lanes 5 and 6). In contrast, large amounts of cytoplasmic EI were found almost exclusively in the supernatant fractions after high-speed centrifugation (Fig. 6B, compare lanes 3 and 5 to lanes 4 and 6). These results were independent of the wash buffers used. Traces of contaminating proteins in the “wrong” fractions could be detected for both of the two reference proteins, since this method of separation did not result in absolute separation. However, this affected only a minor part of the total proteins.
FIG. 6.
Western blot analyses of LJ110/pTMByeeI-S after differential centrifugation. Cells were grown after induction with glucose and IPTG in LB0 medium containing ampicillin until the early stationary phase, harvested, and disrupted by sonication. Samples were taken from the low-speed pellet after sonication (lane 1), from the supernatant after low-speed centrifugation (lane 2), from the supernatant after high-speed centrifugation in buffer A (lane 3), from the corresponding high-speed membrane pellet (lane 4), from the supernatant after high-speed centrifugation in buffer B (lane 5), and from the corresponding high-speed membrane pellet (lane 6). All samples were separated by SDS-PAGE and were simultaneously analyzed by Western blotting using antibodies against EIICBGlc (A), EI (B), and YeeI-S-His5 (C).
YeeI clearly behaved like the cytoplasmic marker protein EI, in that there was always significantly more protein in the supernatants than in the pellets (Fig. 6C, compare lanes 3 and 5 to lanes 4 and 6) after high-speed centrifugation. This provides strong evidence that YeeI is not an integral membrane protein. However, a slightly stronger signal for YeeI than for EI in the membrane fractions was observed. Although use of the different wash buffers had no effect on the distribution pattern, the possibility of membrane association of YeeI could not be completely excluded. Additionally, Mlc and several different putative docking partner proteins were tested to determine their effects on the YeeI distribution pattern. The differential centrifugation experiment was repeated using strains LJB17 (ΔdgsA), LJ121 (ΔptsG manXYZ), LJ130 (ΔmanXYZ), and LJ141 (ΔptsHI crr), all transformed with pTMByeeI-S. These strains lack the potential interaction partners Mlc, EIICBGlc, EIIXYZMan, and EI-HPr-EIIAGlc, respectively. However, none of these strain backgrounds had any influence on the distribution pattern of YeeI compared to that of wild-type strain LJ110 (data not shown).
DISCUSSION
In E. coli K-12 the functions of more than 40% of all putative open reading frames are still not known. Moreover, for more than one-half of these putative proteins, there are no obvious sequence similarities to characterized proteins. Therefore, one of the challenging tasks in E. coli genetics is elucidation of the functions of these proteins. One of the open reading frames is yeeI (also designated b1976), a single gene located at 44.1 min between serU and asnT, two genes that code for tRNAs. Both of these tRNA genes are arranged in the opposite with respect to yeeI, meaning that yeeI is a single transcriptional unit. Intensive BLAST sequence similarity searches (amino acid sequences at www.ncbi.nlm.nih.gov) revealed that there are orthologs of YeeI in many proteobacteria belonging to the beta and gamma subdivisions. Interestingly, even in the cyanobacterial group (e.g., Nostoc punctiforme), whose members might possess both glucose- and fructose-specific phosphotransferase systems, proteins homologous to YeeI that have more than 30% identical amino acids in the complete sequence can be found. In all of these cases the YeeI-like protein has been annotated basically as a “highly conserved bacterial protein of unknown function.” Additionally, a BLAST conserved domain search revealed no obvious similarities to other functionally characterized proteins.
In this work we were able to assign for the first time a function to YeeI in E. coli, which might stimulate experiments to determine the functions of similar proteins in other systems. The results described here demonstrate that YeeI is an Mlc-binding and -inactivating protein and is therefore involved in the regulation of expression of the ptsG gene encoding the major glucose transporter EIICBGlc. First, yeeI mutants exhibited an increase in generation time during growth on glucose, a decrease in the transport of methyl-α-d-glucopyranoside, a substrate of EIICBGlc, and reduced induction of a ptsG-lacZ operon fusion. Second, yeeI mutants showed reduced catabolite repression in lactose/glucose diauxic growth experiments. Third, overexpression of yeeI caused in vivo titration of Mlc even in the absence of EIICBGlc, the only other protein interaction partner of this repressor known thus far. And fourth, in SPR experiments, we demonstrated that there is a direct protein-protein interaction between YeeI and Mlc with an apparent of Kd of about 14 nM. It was shown previously that the Kd for the interaction between Mlc and EIICBGlc is close to 100 nM, whereas the Kd values for the interactions between Mlc and the ptsG or ptsHI crr operator are close to 10 nM (23). These data indicate that the strength of the interaction between YeeI and Mlc, as measured in this work, is in a physiologically relevant range.
As shown previously, the EIIBGlc domain of the glucose transporter is responsible for the interaction with Mlc (23, 39, 44). Soluble unphosphorylated EIIBGlc can bind to Mlc, as demonstrated in SPR experiments (23), but is not capable of preventing Mlc from binding to its cognate operator sites (19). Consequently, it was demonstrated that EIIBGlc could repress Mlc activity only if it was attached either to the membrane by the EIICGlc domain or to a heterologous membrane anchor (e.g., the Gp8 protein, the bacteriophage M13 major coat protein [39], or the lactose permease LacY [44]). These results suggest that under physiological conditions membrane sequestration of Mlc by unphosphorylated EIICBGlc, which builds up during the transport of glucose into the cell, is the key process in the inactivation of Mlc and leads to physical separation of Mlc from its DNA target sites. Furthermore, it was suggested by Seitz et al. (39) and Tanaka et al. (44) that in addition to the physical separation mechanism, a membrane environment-induced conformational change of Mlc might be necessary to prevent the repressor from binding to DNA. In contrast to these results, we obtained strong evidence that YeeI is not an integral membrane protein but is nevertheless capable of inactivating Mlc directly in the cytosol, even in the absence of EIICBGlc. Therefore, there must be a fundamental difference in the mode of action between YeeI and EIICBGlc during inactivation of Mlc. Native Mlc exists as a tetramer. Deletion of nine carboxy-terminal amino acids (Δ9C-Mlc) did not have a severe effect on the activity, whereas deletion of 18 carboxy-terminal amino acids (Δ18C-Mlc) removed an amphipathic helix that is necessary for tetramerization (39). In addition, as revealed by the crystal structure of Mlc (37), this carboxy-terminal amphipathic helix stabilizes the amino-terminal helix-turn-helix domain, which is responsible for the DNA binding of Mlc. Thus, Δ18C-Mlc can only form dimers that cannot repress ptsG expression. Interestingly, the carboxy-terminal part of Mlc seems to be the target site for YeeI binding, since fully active Δ9C-Mlc had only a 20% residual binding response and Δ18C-Mlc did not interact at all with YeeI in our SPR experiments. It is tempting to speculate that tetramerization of the repressor is inhibited by binding of YeeI to the carboxy-terminal region of Mlc and repression of ptsG expression cannot take place. In this case membrane sequestration might not be necessary for inactivation of Mlc by YeeI.
Another unresolved question concerns the physiological function of YeeI. During the last few years a remarkable number of regulators of ptsG expression have been found, and these regulators can be divided into different classes. The first class consists of Mlc and cAMP-CRP. Whereas Mlc is responsible for induction of ptsG (and several other genes) in the presence of glucose, the catabolite activator complex cAMP-CRP is also absolutely necessary for ptsG expression (15, 26, 32, 48). Since cAMP levels are low during growth on glucose, the two regulatory systems work antagonistically. Indeed, addition of cAMP to cells growing on glucose resulted in a significant increase in ptsG expression (48). These two systems seem to be responsible for sophisticated fine-tuning of ptsG expression under various growth conditions. The second class consists of several other factors that have minor effects on ptsG expression levels. The members of this group include ArcA, a major regulator of the switch between aerobic growth and anaerobic growth in E. coli (13), two sigma factors, σ32 for the heat shock response (40) and σS for expression of genes in the stationary growth phase (38), and the small DNA-binding protein Fis (41). There is a third class of regulatory effects, which occur at the level of ptsG mRNA stability. Several workers have provided evidence that intracellular accumulation of glucose-6-phosphate or fructose-6-phosphate leads to specific degradation of ptsG mRNA in an RNase E-dependent manner (14, 17, 21). Vanderpool and Gottesman (45) identified a transcriptional activator called SgrR, which is activated under these conditions and causes enhanced transcription of a small RNA designated SgrS. SgrS is complementary to the 5′ end of ptsG and is capable of forming Hfq-dependent RNA-RNA hybrids. This is the first step in ptsG mRNA degradation.
Several of these regulatory systems, including YeeI, cause rather small changes in the amounts of EIICBGlc under various growth conditions. However, experiments to determine the enzyme flux control coefficients of all phosphotransferase reactions of the glucose-PTS revealed that only the EIICBGlc activity controls the flux through the glucose phosphotransferase system with wild-type levels of expression of the proteins involved, EI, HPr, EIIAGlc, and EIICBGlc (34, 35, 46). Furthermore, simulation experiments showed that if the EIICBGlc activity is less than 40% of the maximum induction level, the amount of unphosphorylated EIIAGlc starts to increase even under glucose saturating conditions (unpublished results). This explains the reduced catabolite repression response of the yeeI mutant in the glucose/lactose diauxic growth experiment described here.
Another open question is, how is YeeI activity regulated? As shown in Fig. 3A, in Western blot analyses of YeeI we frequently observed the presence of two bands. This may indicate that there is protein modification. However, we were not able to correlate the occurrence of the double bands to any growth conditions that were used. Introduction of defined mutations (ΔptsHI crr, ΔptsG, and ΔdgsA) into our test strain also did not affect the appearance of YeeI double bands (data not shown). Therefore, further analyses have to be performed to identify a putative protein modification and to find out if this has some regulatory effects on YeeI activity.
In conclusion, we identified a new protein involved in ptsG expression by binding and inactivating Mlc. Consequently, we propose that the open reading frame with an unknown function identified should be renamed mtfA (Mlc titration factor A).
Acknowledgments
We gratefully acknowledge Kurt Schmid, Joseph W. Lengeler, Katja Bettenbrock, and Jürgen Heinisch for helpful discussions, Sandra Bartels for excellent technical support, Simona Hempelmann for construction of strain LMH111, and Lucille Schmieding for help with the manuscript.
This work was financially supported by the Deutsche Forschungsgemeinschaft through “Sonderforschungsbereich” 431 (Teilprojekt P14 to K.J.) and by the Deutscher Akademischer Austausch Dienst (A.-K.B.).
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmann, J. A. Smith, and K. Struhl (ed.). 1990. Current protocols in molecular biology. Greene Publishing and Wiley-Interscience, New York, NY.
- 2.Berlyn, M. K. B. 1998. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol. Mol. Biol. Rev. 62:814-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bochner, B. R., H.-C. Huang, G. L. Schieven, and B. N. Ames. 1980. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143:926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böhm, A., and W. Boos. 2004. Gene regulation in prokaryotes by subcellular relocalization of transcription factors. Curr. Opin. Microbiol. 7:151-156. [DOI] [PubMed] [Google Scholar]
- 5.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decker, K., J. Plumbridge, and W. Boos. 1998. Negative transcriptional regulation of a positive regulator: the expression of malT, encoding the transcriptional activator of the maltose regulon of Escherichia coli, is negatively controlled by Mlc. Mol. Microbiol. 27:381-390. [DOI] [PubMed] [Google Scholar]
- 8.De Reuse, H., A. Kolb, and A. Danchin. 1992. Positive regulation of the expression of the Escherichia coli pts operon. Identification of the regulatory regions. J. Mol. Biol. 226:623-625. [DOI] [PubMed] [Google Scholar]
- 9.Erni, B. 2001. Glucose transport by the bacterial phosphotransferase system (PTS): an interface between energy and signal transduction, p. 115-138. In G. Winkelmann (ed.), Microbial transport systems. Wiley-VCH, Weinheim, Germany.
- 10.Fox, D. K., A. Presper, S. Adhya, S. Roseman, and S. Garges. 1992. Evidence for two promoters upstream of the pts operon: regulation by the cAMP receptor protein regulatory complex. Proc. Natl. Acad. Sci. USA 89:7056-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosono, K., H. Kakuda, and S. Ichihara. 1995. Decreasing accumulation of acetate in rich medium by Escherichia coli on introducing of genes on a multicopy plasmid. Biosci. Biotechnol. Biochem. 59:256-261. [DOI] [PubMed] [Google Scholar]
- 12.Jahreis, K., L. Bentler, J. Bockmann, S. Hans, A. Meyer, J. Siepelmeyer, and J. W. Lengeler. 2002. Adaptation of sucrose metabolism in the Escherichia coli wild-type strain EC3132. J. Bacteriol. 184:5307-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong, J.-Y., Y.-J. Kim, N. Cho, D. Shin, T.-W. Nam, S. Ryu, and Y.-J. Seok. 2004. Expression of ptsG encoding the major glucose transporter is regulated by ArcA in Escherichia coli. J. Biol. Chem. 279:38513-38518. [DOI] [PubMed] [Google Scholar]
- 14.Kawamoto, H., T. Morita, A. Shimizu, T. Inada, and H. Aiba. 2005. Implication of membrane localization of target mRNA in the action of a small RNA: mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 19:328-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, S.-Y., T.-W. Nam, D. Shin, B.-M. Koo, Y.-J. Seok, and S. Ryu. 1999. Purification of Mlc and analysis of its effects on the pts expression in Escherichia coli. J. Biol. Chem. 274:25398-25402. [DOI] [PubMed] [Google Scholar]
- 16.Kimata, K., T. Inada, H. Tagami, and H. Aiba. 1998. A global repressor (Mlc) is involved in glucose induction of the ptsG encoding major glucose transporter in Escherichia coli. Mol. Microbiol. 29:1509-1519. [DOI] [PubMed] [Google Scholar]
- 17.Kimata, K., Y. Tanaka, T. Inada, and H. Aiba. 2001. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli. EMBO J. 20:3587-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 19.Lee, S.-J., W. Boos, J.-P. Bouche, and J. Plumbridge. 2000. Signal transduction between a membrane-bound transporter, PtsG, and a soluble transcription factor, Mlc, of Escherichia coli. EMBO J. 19:5353-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez, E., B. Bartolome, and F. de la Cruz. 1988. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ, a reporter gene of pUC8/9 and pUC18/19 plasmids. Gene 68:159-162. [DOI] [PubMed] [Google Scholar]
- 21.Morita, T., W. El-Kazzaz, Y. Tanaka, T. Inada, and H. Aiba. 2003. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J. Biol. Chem. 278:15608-15614. [DOI] [PubMed] [Google Scholar]
- 22.Morrison, T. B., and J. S. Parkinson. 1997. A fragment liberated from the Escherichia coli CheA kinase that blocks stimulatory, but not inhibitory, chemoreceptor signaling. J. Bacteriol. 179:5543-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nam, T.-W., S.-H. Cho, D. Shin, J.-H. Kim, J.-Y. Jeong, J.-H. Lee, J.-H. Roe, A. Peterkofsky, S.-O. Kang, S. Ryu, and Y.-J. Seok. 2001. The Escherichia coli glucose transporter enzyme IICBGlc recruits the global repressor Mlc. EMBO J. 20:491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Notley-McRobb, L., and T. Ferenci. 2000. Substrate specificity and signal transduction pathways in the glucose-specific enzyme II (EIIGlc) component of Escherichia coli phosphotransferase system. J. Bacteriol. 182:4437-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pardee, A. B., and L. S. Prestige. 1961. The initial kinetics of enzyme induction. Biochim. Biophys. Acta 49:77-88. [DOI] [PubMed] [Google Scholar]
- 26.Plumbridge, J. 1998. Expression of ptsG, the gene for the major glucose PTS transporter in Escherichia coli, is repressed by Mlc and induced by growth on glucose. Mol. Microbiol. 29:1053-1063. [DOI] [PubMed] [Google Scholar]
- 27.Plumbridge, J. 1998. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol. Microbiol. 27:369-380. [DOI] [PubMed] [Google Scholar]
- 28.Plumbridge, J. 1999. Expression of the phosphotransferase system both mediates and is mediated by Mlc regulation in Escherichia coli. Mol. Microbiol. 33:260-273. [DOI] [PubMed] [Google Scholar]
- 29.Plumbridge, J. 2002. Regulation of gene expression in the PTS in Escherichia coli: the role and interactions of Mlc. Curr. Opin. Microbiol. 5:187-193. [DOI] [PubMed] [Google Scholar]
- 30.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1996. Phosphoenolpyruvate:carbohydrate phosphotransferase systems, p. 1149-1174. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 32.Rephaeli, A. W., and M. H. Saier, Jr. 1980. Regulation of genes coding for enzyme constituents of the bacterial phosphotransferase system. J. Bacteriol. 141:658-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roehl, R. A., and R. T. Vinopal. 1980. Genetic locus, distant from ptsM, affecting enzyme IIA/IIB function in Escherichia coli K-12. J. Bacteriol. 142:120-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohwer, J. M., N. D. Meadow, S. Roseman, H. V. Westerhoff, and P. W. Postma. 2000. Understanding glucose transport by the bacterial phosphoenolpyruvate:glycose phosphotransferase system on the basis of kinetic measurements in vitro. J. Biol. Chem. 275:34909-34921. [DOI] [PubMed] [Google Scholar]
- 35.Ruyter, G. J. G., P. W. Postma, and K. van Dam. 1991. Control of glucose metabolism by enzyme IIGlc of the phosphoenolpyruvate-dependent phosphotransferase system in Escherichia coli. J. Bacteriol. 173:6184-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saiki, R. K., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-491. [DOI] [PubMed] [Google Scholar]
- 37.Schiefner, A., K. Gerber, S. Seitz, W. Welte, K. Diederichs, and W. Boos. 2005. The crystal structure of Mlc, a global regulator of sugar metabolism in Escherichia coli. J. Biol. Chem. 280:29073-29079. [DOI] [PubMed] [Google Scholar]
- 38.Seeto, S., L. Notley-McRobb, and T. Ferenci. 2004. The multifactorial influences of RpoS, Mlc, and cAMP on ptsG expression under glucose-limited and anaerobic growth conditions. Res. Microbiol. 155:211-215. [DOI] [PubMed] [Google Scholar]
- 39.Seitz, S., S.-L. Lee, C. Pennetier, W. Boos, and J. Plumbridge. 2003. Analysis of the interaction between the global regulator Mlc and EIIBGlc of the glucose-specific phosphotransferase system in Escherichia coli. J. Biol. Chem. 278:10744-10751. [DOI] [PubMed] [Google Scholar]
- 40.Shin, D., S. Lim, Y.-J. Seok, and S. Ryu. 2001. Heat shock RNA polymerase (Eσ32) is involved in the transcription of mlc and crucial for induction of the mlc regulon by glucose in Escherichia coli. J. Biol. Chem. 276:25871-25875. [DOI] [PubMed] [Google Scholar]
- 41.Shin, D., N. Cho, S. Heu, and S. Ryu. 2003. Selective regulation of ptsG expression by Fis. J. Biol. Chem. 278:14776-14781. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka, Y., K. Kimata, T. Inada, H. Tagami, and H. Aiba. 1999. Negative regulation of the pts operon by Mlc: mechanisms underlying glucose induction in Escherichia coli. Genes Cells 4:391-399. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka, Y., K. Kimata, and H. Aiba. 2000. A novel regulatory role of glucose transporter of Escherichia coli: membrane sequestration of a global repressor Mlc. EMBO J. 19:5344-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka, Y., F. Itoh, K. Kimata, and H. Aiba. 2004. Membrane localization itself but not binding to IICBGlc is directly responsible for the inactivation of the global repressor Mlc in Escherichia coli. Mol. Microbiol. 53:941-951. [DOI] [PubMed] [Google Scholar]
- 45.Vanderpool, C. K., and S. Gottesman. 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 54:1076-1089. [DOI] [PubMed] [Google Scholar]
- 46.Van der Vlag, J., R. Van′t Hof, K. Van Dam, and P. W. Postma. 1995. Control of glucose metabolism by the enzymes of the glucose phosphotransferase system in Salmonella typhimurium. Eur. J. Biochem. 230:170-182. [DOI] [PubMed] [Google Scholar]
- 47.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 48.Zeppenfeld, T., C. Larisch, J. W. Lengeler, and K. Jahreis. 2000. Glucose transporter mutants of Escherichia coli K-12 with changes in substrate recognition of IICBGlc and induction behavior of the ptsG gene. J. Bacteriol. 182:4443-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]