Abstract
The largest family of solute transporters (major facilitator superfamily [MFS]) includes proton-motive-force-driven secondary transporters. Several characterized MFS transporters utilize essential acidic residues that play a critical role in the energy-coupling mechanism during transport. Surprisingly, we show here that no single acidic residue plays an irreplaceable role in the Escherichia coli secondary multidrug transporter MdfA.
One mechanism of bacterial drug resistance involves the extrusion of drugs from the cell by membrane transporters (13, 18, 19, 28, 31). Transporters, which handle a wide spectrum of structurally dissimilar drugs, are called multidrug (Mdr) transporters (18, 25). The substrate recognition profile of Mdr transporters usually encompasses structurally unrelated cationic lipophilic compounds, although negatively charged, neutral, or zwitterionic compounds are also substrates of Mdr transporters (7, 9, 17, 31). The bacterial Mdr transporters fall into one of five families, the largest of which is themajor facilitator superfamily (MFS) of secondary transporters (29). The MFS Mdr transporters are abundant in the genomes of many bacterial strains and pose intriguing mechanistic (5) and evolutionarily related questions (12, 21). Several of these questions are being investigated by utilizing MdfA from Escherichia coli (5) as a model for secondary MFS-related Mdr transporters.
MdfA (7), encoded by the cmr gene (22), is a 410-amino-acid-residue-long membrane protein (Fig. 1A), with close homologues in several pathogenic bacteria: Shigella flexneri (10), Salmonella enterica serovar Typhi (23), and Yersinia pestis (24). Cells expressing MdfA from a multicopy plasmid exhibit multidrug resistance against a diverse group of structurally and electrically dissimilar toxic compounds (4, 7). Transport experiments have shown that MdfA is driven by proton electrochemical potential and functions as a (multidrug) Na+,K+/proton antiporter (7, 14, 16, 20). Clearly, therefore, proton translocation by MdfA is crucial for all of its known transport activities. Of all the open mechanistic questions regarding the function of MFS Mdr transporters, the question of multidrug recognition has been characterized in detail (15), whereas the mechanism underlying active transport remained the least understood. Specifically, although crucial for their drug/proton antiport activity (14), very little is known about proton recognition and translocation by MFS Mdr transporters. In this regard, studies of several substrate-specific MFS transporters have been instrumental in providing clues for determining how protons may be recognized. The best example is the lactose/proton symporter LacY, where two carboxyl side chains play irreplaceable roles in proton-coupled sugar translocation (11). Similarly, negatively charged residues are mechanistically involved in other secondary transporters (8, 26, 27).
FIG. 1.
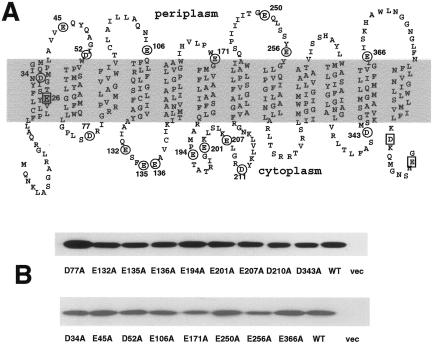
Neutralization of individual acidic residues in MdfA. (A) Acidic residues are marked on the secondary structure model of MdfA. Circled residues were analyzed in this study. Squares indicate residues that were analyzed previously (2, 4, 6). (B) Membranes were prepared from cells expressing the indicated His6-tagged mutants and analyzed by Western blotting. WT, wild type; vec, vector.
Stimulated by the studies of other MFS transporters, we have examined the role of acidic residues in MdfA by site-directed mutagenesis. As predicted from theoretical and experimental approaches (1, 2, 6, 7, 30), the putative 12 transmembrane (TM) helices of MdfA contain two membrane-embedded acidic residues, E26 and D34, in TM1 (Fig. 1A). Previously, we have shown that E26 constitutes an important part of the drug recognition pocket in MdfA, although it is not essential for transport activity (3, 4, 6). Additional, nonessential acidic residues had previously been identified in the C-terminal region of the protein (2) (Fig. 1A). Therefore, the remaining uncharacterized 17 acidic residues in MdfA were replaced, one by one, with alanines. All the mutations were constructed in a template harboring a His6 tag to enable clear detection and comparison of expression and were confirmed by DNA sequencing. In all the mutants, MdfA expression is regulated by the native, constitutive promoter (6), and Fig. 1B shows a relatively lower expression level only in the case of the E171A and E256A mutants.
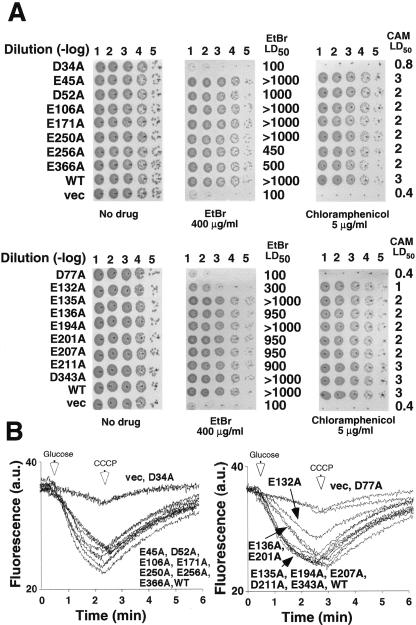
The biological function of each MdfA mutant was examined in E. coli UT mdfA::kan cells (R. Edgar and E. Bibi, unpublished data) by growth inhibition experiments using two methods in parallel. One approach utilized LB agar plates supplemented with the indicated toxic compound at concentrations significantly higher than those that prevent the growth of cells harboring an empty vector. The ability of the transformed cells to form single colonies after 20 h of incubation at 37°C served as an indicator for drug resistance. The results show that all the transformants, except for D34A and D77A, grew relatively well on ethidium bromide (EtBr) and chloramphenicol (Fig. 2A). The second approach utilized growth in LB broth and 50% lethal dose (LD50) measurements with chloramphenicol and EtBr (4). As expected, cells expressing MdfA mutants D34A and D77A grew as cells harboring empty vector. In contrast, the LD50 of cells expressing all the other mutants was substantially higher than that of cells harboring empty vector (Fig. 2A).
FIG. 2.
Functional analyses of MdfA mutants. E. coli cells transformed with empty vector (vec) or plasmids encoding wild-type MdfA (WT) or the indicated MdfA mutants were diluted as marked [Dilution (-log)] and spotted onto LB agar plates with or without the test drug, and plates were visualized after ∼20 h at 37°C. LD50 measurements with chloramphenicol (CAM) and EtBr (4) are indicated (in micrograms/milliliter) on the right. (B) Cells expressing the indicated mutants were loaded with EtBr and energized with glucose (left arrow), and the EtBr efflux was monitored continuously in the fluorimeter. As expected, the addition of carbonyl cyanide m-chlorophenylhydrazone (CCCP) (right arrow) abolished efflux. The experiments were repeated at least three times, and the results shown are representative. a.u., arbitrary units.
In order to evaluate the resistance results further, the activity of the mutants was tested directly by EtBr transport assays as described previously (30). Figure 2B shows a rapid efflux of EtBr with cells expressing the wild-type transporter and most of the mutants, except for MdfA E132A, which exhibited low but significant EtBr efflux activity, and D34A and D77A, which exhibited no transport activity whatsoever.
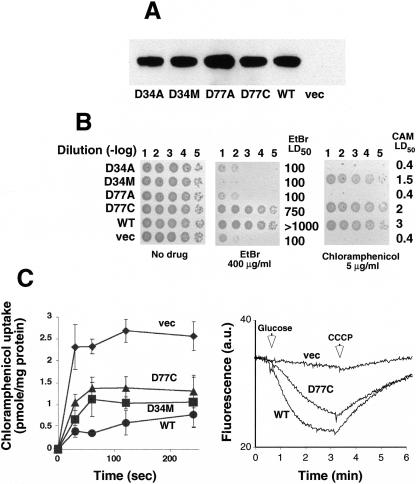
Taken together, the results showed that only mutants D77A and D34A lost multidrug resistance and transport activity, suggesting that these acidic residues may have important roles. Therefore, we constructed and analyzed additional mutations at positions 34 and 77, and the expression (Fig. 3A), drug resistance, and transport activity of the mutants were tested as described above. The results with selected active and inactive variants (Fig. 3B and C) demonstrate that both MdfA mutants D34M and D77C support growth of the transformed cells on chloramphenicol, and cells expressing mutant D77C grew also on EtBr (Fig. 3B). Next, we tested the function of the mutants by transport experiments with radiolabeled chloramphenicol and EtBr as described previously (30). Figure 3C (left panel) shows that mutants D34M and D77C behave almost as cells harboring wild-type MdfA, where very low chloramphenicol accumulation was observed, due to active efflux of the antibiotic. Unlike mutant D34M (not shown), mutant D77C was also able to catalyze EtBr efflux (Fig. 3C, right panel).
FIG. 3.
Characterization of MdfA mutants at positions D77 and D34. (A) Membranes were prepared from cells expressing the indicated His6-tagged mutants and analyzed by Western blotting. (B) E. coli was transformed with empty vector (vec) or plasmids encoding wild-type MdfA (WT) or the indicated MdfA mutants. Cells were diluted as marked [dilution (-log)] and spotted onto LB agar plates with or without the test drug, and plates were visualized after ∼20 h at 37°C. (C) Uptake of [3H]chloramphenicol by cells expressing the indicated mutants was assayed by rapid filtration (left panel). The experiment was repeated at least three times. The results shown are the average of triplicates. For the right panel, EtBr efflux was assayed as described in the legend of Fig. 2. a.u., arbitrary units.
In conclusion, the results demonstrate that there is no single acidic residue whose mutation to a neutral residue abolished MdfA function. Interestingly, in this regard, studies of the putative membrane-embedded acidic residues of another MFS Mdr transporter, LmrP from Lactococcus lactis, suggested that none of them is irreplaceable. Therefore, we hypothesize that MdfA and possibly other MFS Mdr transporters might utilize different proton recognition strategies than those used by substrate-specific ones.
Acknowledgments
This work was supported by a grant from the Y. Leon Benoziyo Institute for Molecular Medicine at the Weizmann Institute of Science and by the Israel Cancer Research Foundation (ICRF).
REFERENCES
- 1.Adler, J., and E. Bibi. 2004. Determinants of substrate recognition by the Escherichia coli multidrug transporter MdfA identified on both sides of the membrane. J. Biol. Chem. 279:8957-8965. [DOI] [PubMed] [Google Scholar]
- 2.Adler, J., and E. Bibi. 2002. Membrane topology of the multidrug transporter MdfA: complementary gene fusion studies reveal a nonessential C-terminal domain. J. Bacteriol. 184:3313-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adler, J., and E. Bibi. 2005. Promiscuity in the geometry of electrostatic interactions between the Escherichia coli multidrug resistance transporter MdfA and cationic substrates. J. Biol. Chem. 280:2721-2729. [DOI] [PubMed] [Google Scholar]
- 4.Adler, J., O. Lewinson, and E. Bibi. 2004. Role of a conserved membrane-embedded acidic residue in the multidrug transporter MdfA. Biochemistry 43:518-525. [DOI] [PubMed] [Google Scholar]
- 5.Bibi, E., J. Adler, O. Lewinson, and R. Edgar. 2001. MdfA, an interesting model protein for studying multidrug transport. J. Mol. Microbiol. Biotechnol. 3:171-177. [PubMed] [Google Scholar]
- 6.Edgar, R., and E. Bibi. 1999. A single membrane-embedded negative charge is critical for recognizing positively charged drugs by the Escherichia coli multidrug resistance protein MdfA. EMBO J. 18:822-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgar, R., and E. Bibi. 1997. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J. Bacteriol. 179:2274-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujihira, E., T. Kimura, Y. Shiina, and A. Yamaguchi. 1996. Transmembrane glutamic acid residues play essential roles in the metal-tetracycline/H+ antiporter of Staphylococcus aureus. FEBS Lett. 391:243-246. [DOI] [PubMed] [Google Scholar]
- 9.Jack, D. L., M. L. Storms, J. H. Tchieu, I. T. Paulsen, and M. H. Saier, Jr. 2000. A broad-specificity multidrug efflux pump requiring a pair of homologous SMR-type proteins. J. Bacteriol. 182:2311-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin, Q., Z. Yuan, J. Xu, Y. Wang, Y. Shen, W. Lu, J. Wang, H. Liu, J. Yang, F. Yang, X. Zhang, J. Zhang, G. Yang, H. Wu, D. Qu, J. Dong, L. Sun, Y. Xue, A. Zhao, Y. Gao, J. Zhu, B. Kan, K. Ding, S. Chen, H. Cheng, Z. Yao, B. He, R. Chen, D. Ma, B. Qiang, Y. Wen, Y. Hou, and J. Yu. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaback, H. R., M. Sahin-Toth, and A. B. Weinglass. 2001. The kamikaze approach to membrane transport. Nat Rev. Mol. Cell Biol. 2:610-620. [DOI] [PubMed] [Google Scholar]
- 12.Krulwich, T. A., O. Lewinson, E. Padan, and E. Bibi. 2005. Do physiological roles foster persistence of drug/multidrug-efflux transporters? A case study. Nat. Rev. Microbiol. 3:566-572. [DOI] [PubMed] [Google Scholar]
- 13.Levy, S. B. 2002. Active efflux, a common mechanism for biocide and antibiotic resistance. J. Appl. Microbiol. 92:65S-71S. [PubMed] [Google Scholar]
- 14.Lewinson, O., J. Adler, G. J. Poelarends, P. Mazurkiewicz, A. J. Driessen, and E. Bibi. 2003. The Escherichia coli multidrug transporter MdfA catalyzes both electrogenic and electroneutral transport reactions. Proc. Natl. Acad. Sci. USA 100:1667-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewinson, O., J. Adler, N. Sigal, and E. Bibi. Promiscuity in multidrug recognition and transport: the bacterial MFS Mdr transporters. Mol. Microbiol., in press. [DOI] [PubMed]
- 16.Lewinson, O., E. Padan, and E. Bibi. 2004. Alkalitolerance: a biological function for a multidrug transporter in pH homeostasis. Proc. Natl. Acad. Sci. USA 101:14073-14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis, K., V. Naroditskaya, A. Ferrante, and I. Fokina. 1994. Bacterial resistance to uncouplers. J. Bioenerg. Biomembr. 26:639-646. [DOI] [PubMed] [Google Scholar]
- 18.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159-204. [DOI] [PubMed] [Google Scholar]
- 19.Lomovskaya, O., and W. Watkins. 2001. Inhibition of efflux pumps as a novel approach to combat drug resistance in bacteria. J. Mol. Microbiol. Biotechnol. 3:225-236. [PubMed] [Google Scholar]
- 20.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1998. Evidence for chloramphenicol/H+ antiport in Cmr (MdfA) system of Escherichia coli and properties of the antiporter. J. Biochem. (Tokyo) 124:187-193. [DOI] [PubMed] [Google Scholar]
- 21.Neyfakh, A. A. 1997. Natural functions of bacterial multidrug transporters. Trends Microbiol. 5:309-313. [DOI] [PubMed] [Google Scholar]
- 22.Nilsen, I. W., I. Bakke, A. Vader, O. Olsvik, and M. R. El-Gewely. 1996. Isolation of cmr, a novel Escherichia coli chloramphenicol resistance gene encoding a putative efflux pump. J. Bacteriol. 178:3188-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 24.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 25.Poole, K. 2005. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 56:20-51. [DOI] [PubMed] [Google Scholar]
- 26.Poolman, B., J. Knol, and J. S. Lolkema. 1995. Kinetic analysis of lactose and proton coupling in Glu379 mutants of the lactose transport protein of Streptococcus thermophilus. J. Biol. Chem. 270:12995-13003. [DOI] [PubMed] [Google Scholar]
- 27.Pourcher, T., M. L. Zani, and G. Leblanc. 1993. Mutagenesis of acidic residues in putative membrane-spanning segments of the melibiose permease of Escherichia coli. I. Effect on Na(+)-dependent transport and binding properties. J. Biol. Chem. 268:3209-3215. [PubMed] [Google Scholar]
- 28.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saier, M. H., Jr., and I. T. Paulsen. 2001. Phylogeny of multidrug transporters. Semin. Cell Dev. Biol. 12:205-213. [DOI] [PubMed] [Google Scholar]
- 30.Sigal, N., E. Vardy, S. Molshanski-Mor, A. Eitan, Y. Pilpel, S. Schuldiner, and E. Bibi. 2005. 3D model of the Escherichia coli multidrug transporter MdfA reveals an essential membrane-embedded positive charge. Biochemistry 44:14870-14880. [DOI] [PubMed] [Google Scholar]
- 31.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]