Abstract
In metazoa and fungi, the catabolic dissimilation of cysteine begins with its sulfoxidation to cysteine sulfinic acid by the enzyme cysteine dioxygenase (CDO). In these organisms, CDO plays an important role in the homeostatic regulation of steady-state cysteine levels and provides important oxidized metabolites of cysteine such as sulfate and taurine. To date, there has been no experimental evidence for the presence of CDO in prokaryotes. Using PSI-BLAST searches and crystallographic information about the active-site geometry of mammalian CDOs, we identified a total of four proteins from Bacillus subtilis, Bacillus cereus, and Streptomyces coelicolor A3(2) that shared low overall identity to CDO (13 to 21%) but nevertheless conserved important active-site residues. These four proteins were heterologously expressed and purified to homogeneity by a single-step immobilized metal affinity chromatography procedure. The ability of these proteins to oxidize cysteine to cysteine sulfinic acid was then compared against recombinant rat CDO. The kinetic data strongly indicate that these proteins are indeed bona fide CDOs. Phylogenetic analyses of putative bacterial CDO homologs also indicate that CDO is distributed among species within the phyla of Actinobacteria, Firmicutes, and Proteobacteria. Collectively, these data suggest that a large subset of eubacteria is capable of cysteine sulfoxidation. Suggestions are made for how this novel pathway of cysteine metabolism may play a role in the life cycle of the eubacteria that have it.
Cysteine is an indispensable amino acid for all forms of life. It serves as a precursor for protein synthesis, and the unique chemistry of its free thiol group forms the active moiety of several essential metabolites such as coenzyme A, glutathione and its derivatives, and mycothiol. Because many basic cellular processes are dependent upon a steady supply of cysteine, there has been extensive work to identify in both prokaryotes and eukaryotes the pathways that ultimately determine the steady-state free intracellular cysteine pool (26). The generic pathways that increase the levels of free intracellular cysteine are those of de novo biosynthesis and transport of preformed cysteine from the extracellular milieu. On the other hand, the pathways responsible for depleting free cysteine include those that incorporate cysteine into other molecules and those that physically degrade the cysteine molecule.
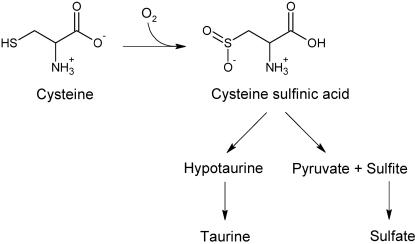
Among the specific pathways that contribute to the metabolic economy of intracellular cysteine, one of the major differences between eukaryotes and prokaryotes has been believed to be the capacity to degrade cysteine to cysteine sulfinic acid (21). This reaction, catalyzed by the Fe2+-dependent enzyme cysteine dioxygenase (CDO; EC 1.13.11.20), irreversibly oxidizes the sulfhydryl group of cysteine (Fig. 1). CDO shows a high degree of specificity for cysteine since structurally related thiols are neither substrates for oxidation nor competitive inhibitors of activity (31). In mammals—eukaryotes for which the biology of CDO has been best described—a significant amount of cysteine is converted to cysteine sulfinic acid and used for the production of inorganic sulfate, pyruvate, hypotaurine, and taurine (27). The rate of flux through this sulfoxidation pathway is regulated by the concentration of free cysteine and the level of CDO. Indeed, in vivo data from the rat have shown that the level of hepatic CDO protein is directly controlled by the dietary availability of cysteine and may be part of a homeostatic response by the liver to keep intracellular cysteine levels within a narrow range (4, 9). To date, there have been no reports in the literature on the ability of prokaryotes to enzymatically oxidize cysteine to cysteine sulfinic acid, and this pathway of cysteine dissimilation has consequently not been considered in models of prokaryotic cysteine metabolism.
FIG. 1.
CDO uses molecular oxygen to catalyze the first step in the eukaryotic cysteine sulfoxidation pathway to produce cysteine sulfinic acid. This pathway irreversibly shunts cysteine to the production of pyruvate and sulfite/sulfate or hypotaurine and taurine.
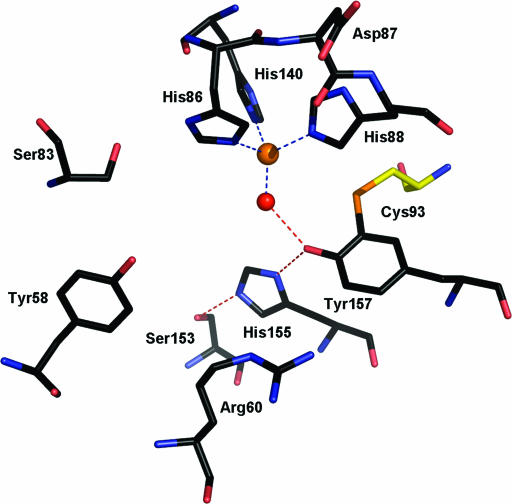
Recently, the crystal structures for mouse and rat CDO have been determined (PDBs: 2ATF [17] and 2B5H [22]). These structures have definitively confirmed that CDO belongs to the cupin superfamily, which encompasses a group of functionally diverse proteins including auxin-binding protein, mannose-6-phosphate isomerase, and hydroxyanthranilate dioxygenase. The members of this superfamily have a β-barrel central domain with a jelly roll topology and two primary consensus sequence motifs (motif 1: Gx5HxHx3,4Ex6G and motif 2: Gx5PxGx2Hx3N) separated by an intermotif distance of 15 to 50 amino acids (10). Eukaryotic CDO is unusual among the cupins because it lacks the conserved glutamate of motif 1, a residue generally used for metal cofactor coordination in other cupins. Instead, CDO uses only the three histidine residues of its cupin motifs to bind a mononuclear iron center at the base of a well-conserved active site (Fig. 2) (23). A remarkable feature of the active site of rat/mouse CDO is an intramolecular thioether bond that forms between residues Cys93 and Tyr157. These two residues are strictly conserved among eukaryotic CDOs and are presumed to be important participants in either the catalytic mechanism and/or the structural stability of the protein's active site.
FIG. 2.
Stick representation of the R. norvegicus CDO active site (PDB 2B5H) (23). The four coordinate iron metallocenter (orange) is chelated by three histidine residues and a water molecule (red). The active-site pocket is lined by residues Y58, R60, S83, C93, S153, H155, and Y157. These residues are strictly conserved across all eukaryotic CDOs. S153, H155, and Y157 form a hydrogen bond network (red dashes) with a water molecule that chelates the iron center. The sulfur atom of residue C93 (yellow) forms a novel thioether bond with Y157.
A PSI-BLAST search (2) using rat CDO as the query sequence uncovers many bacterial cupin proteins of unknown function that share low similarity with rat CDO (12 to 21% identity). Many of these proteins conserve the residues in the active site pocket of CDO, but a notable difference is the lack of conservation of Cys93. In putative bacterial CDOs, this residue is highly conserved as a glycine. Some of the bacterial proteins contain nearby Cys residues and could conceivably form the thioether bond, whereas other do not. Because of this issue, combined with the fact that the overall similarity of the proteins is not much higher than that of rat CDO with functionally unrelated cupins such as cupin phosphoglucose isomerase, it is not possible to conclusively classify these proteins as bona fide CDOs based upon primary sequence data alone.
To test which proteins are functional CDO homologs, we cloned putative CDOs from Bacillus subtilis and Bacillus cereus and two homologs from Streptomyces coelicolor A3(2). These proteins were heterologously expressed, purified, and tested for CDO activity. CDO expression and activity was also evaluated in cultures of B. subtilis. These data, together with phylogenetic analyses, suggest that a subset of eubacteria is capable of cysteine sulfoxidation, a novel pathway of cysteine catabolism for prokaryotes.
MATERIALS AND METHODS
Cloning of putative CDOs.
Rattus norvegicus CDO was cloned as previously described (22). The open reading frames (ORFs) of putative CDOs NP_390992 (yubC, B. subtilis), NP_832375 (BC2617, B. cereus ATCC 14579), NP_627257 (SCO3035, S. coelicolor A3(2)), and NP_629897 (SCO5772, S. coelicolor A3(2)) were cloned and overexpressed in Escherichia coli. PCR with platinum Taq DNA polymerase high fidelity (Invitrogen) was used to generate the ORFs. Genomic DNA of each bacterial species was used as a template. For the amplification of yubC, a forward primer (5′-GTGGATCCATGGAACTGTATGAGTGTATC-3′, restriction site underlined) was designed to create a unique 5′ BamHI restriction site and reverse primer (5′-GTCTGCAGTCATGAATTTTCCAATACCTCC-3′, restriction site underlined) was designed to create a unique 3′ PstI restriction site. For the amplification of the remaining genes, primers were designed to create a 5′ BamHI restriction site and a 3′ HindIII restriction site. Those primers were (restriction sites underlined): BC2617, 5′-TCGGGATCCATGTTGACTTGTAATGGAAG-3′ (forward) and 5′-TCGAAGCTTCTATGAATTATAACTAATGTA-3′ (reverse); SCO3035, 5′-TCGGGATCCATGAACAGCGACAGCGACCTC-3′ (forward) and 5′-TCGAAGCTTTCAGCCGGTCGTCACCGCGG-3′ (reverse); and SCO5772, 5′-TCGGGATCCATGTCCCTCCCCTCCGTTTCTC-3′ (forward) and 5′-TCGAAGCTTTCACTGCCAGTCCTGCGGGCG-3′ (reverse). After PCR amplification and digestion with BamHI/PstI or BamHI/HindIII, PCR products were inserted by T4 DNA ligase (Invitrogen) into pQE30 (QIAGEN) expression vectors linearized by either BamHI/PstI or BamHI/HindIII double digestion. The ligated product was transformed into E. coli XL-1 Blue (Novagen). Plasmids generated in this strain were sequenced by the Cornell BioResource Center (Ithaca, NY). After sequence verification, plasmids were transformed into E. coli M15 (QIAGEN) for expression and overproduction of the recombinant protein.
Expression and purification of recombinant protein.
Rattus norvegicus CDO was expressed and purified as previously described (22). For expression of putative bacterial CDO protein, cells were grown in 2x Luria Broth (LB) containing 100 μg of carbenicillin/ml at 37°C with shaking at 250 rpm. For BC2617 and YubC, cells were grown until the suspension reached an A600 of ∼0.6, and then the expression of recombinant protein was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 1 mM. Cells containing these plasmid constructs were grown for an additional 3 h at 37°C and harvested by centrifugation at 6,000 × g for 15 min. Initial expression experiments with SCO3035 and SCO5772 showed that these proteins were found exclusively in inclusion bodies if induction was performed at temperatures between 22 and 37°C. Solubility was improved by growing cells at 37°C until the suspension reached an A600 of ∼0.45. Cells were then grown at room temperature until reaching an A600 of ∼0.6. IPTG was then added at a final concentration of 330 μM, and cells were grown for an additional 16 h at 8°C before being harvested by centrifugation.
Cell lysates were prepared as previously described (22). Lysate supernatant was filtered with a 0.2-μm-pore-size syringe filter, and the filtrate was applied to a 1-ml HisTrap HP column (Amersham Biosciences, Piscataway, NJ) to purify the His6-tagged recombinant protein. Immobilized metal affinity chromatography (IMAC) was conducted by using an AKTA system (Amersham Biosciences) at a flow rate of 1 ml/min. Increasing imidazole concentrations were created by mixing two mobile phase solutions: 20 mM Tris (pH 8.0), 5 mM imidazole, 0.1% (vol/vol) Tween, 500 mM sodium chloride (IMAC buffer A) and the same buffer with 500 mM imidazole (IMAC buffer B). After sample loading, the column was washed with 20 column volumes (CVs) of 5% buffer B. Recombinant protein was eluted over two 20-CV steps of 10 and 20% buffer B. Peak fractions were concentrated and run on sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis gels and stained with Coomassie blue to estimate protein purity and relative abundance. Protein concentrations were determined by the bicinchoninic acid assay (Pierce, Rockford, IL). Per liter of culture, the approximate yield of purified protein from the putative bacterial CDOs was 10 mg for BC2617, 12 mg for YubC, 1 mg for SCO3035, and 700 μg for SCO5772.
Induction of sporulation in B. subtilis strains.
Sporulation was induced in wild-type B. subtilis strain 168 (1A1; Bacillus Genetic Stock Center, Columbus, OH) and B. subtilis strain 168 yubC::pMUTIN4 (generously provided by Patrick Stragier, Institut de Biologie Physico-Chimique, Paris, France) by resuspension in Sterlini-Mandelstam medium (25) at 37°C. Cells were first grown overnight at 30°C in growth medium and diluted in 100 ml of growth medium to achieve an A600 of 0.1. Cultures were then grown at 37°C to an A600 of 0.55, centrifuged at 8,000 × g for 10 min, and resuspended in 100 ml of Sterlini-Mandelstam medium to start sporulation. At the indicated time points (0, 1, 5, and 10 h after induction of sporulation), aliquots of bacteria were removed for RNA extraction and CDO activity assays. Sporulation efficiency for each strain was assessed by a heat resistance assay as previously described (14). Briefly, each strain was separately inoculated into 10 ml of Difco sporulation medium and incubated at 37°C for 28 h. At this point, cultures were serially diluted in T base medium supplemented with MgSO4 (final concentration, 1 mM). An aliquot from each dilution was first plated onto Difco sporulation medium agar, and the remaining cells were heated to 80°C for 20 min. Heat challenged cells were then spread onto Difco sporulation medium agar and incubated overnight with non-heat-stressed cells at 37°C. Total CFU per ml (CFU/ml) was calculated from the number of colonies obtained from the unheated dilutions, and the number of spores per sample was calculated from the number of colonies derived from the heated dilutions.
Reverse transcription-PCR analysis of yubC expression in Bacillus subtilis.
Total RNA was isolated from aliquots of B. subtilis cultured as described above using a RNeasy Plus minikit (QIAGEN). cDNA was synthesized from 2 μg of the harvested total RNA using an AccuScript high-fidelity first-strand cDNA synthesis kit (Stratagene) and oligo(dT) primers. Amplification of the yubC mRNA transcript was done by Platinum Taq DNA Polymerase (Invitrogen) using the same set of primers used to clone yubC for recombinant protein production (see above).
Assay of CDO activity. (i) Recombinant protein.
The activity of purified recombinant protein was assayed at 37°C by a previously reported method (3). pH optima were determined by using 2-morpholinoethanesulfonic acid (MES) or Tris buffers at a final concentration of 62.5 mM. Reaction mixtures also consisted of recombinant protein (0 to 10 μg) incubated in the presence of 12.5 μM bathocuproine disulfonate, various concentrations of ferrous sulfate (0 to 400 μM), and a range of cysteine concentrations (0 to 10 mM). Bathocuproine disulfonate was added to reduce the auto-oxidation of cysteine to cystine by trace quantities of copper in the reaction mixture. For inhibition studies involving recombinant protein, three cysteine analogs (2-aminoethanethiol, 2-sulfanylethanol, and 3-sulfanylpropionic acid) were separately tested for their ability to affect activity using a cysteine concentration equivalent to the estimated Km of the queried protein and analogs at a final concentration of 1× and 10× the Km for cysteine. Reaction assays were conducted to ensure that no more than 10% of total substrate was converted to product during the incubation period (9 to 18 min). Reactions were stopped by the addition of 200 μl of 5% (wt/vol) sulfosalicylic acid. Acid supernatants were passed over a Dowex-50 column, and the eluate was used for the measurement of cysteine sulfinic acid production by high-performance liquid chromatography (HPLC) (3). HPLC data were collected and analyzed by using Empower software (Waters, Milford, MA). Km and Vmax were determined by hyperbolic regression using the equation of v = Vmax*S/[Km + S(1 + S/Ks)] and Prism 3 (GraphPad Software).
(ii) In vivo activity assays.
Aliquots of the cultures from wild-type and yubC::pMUTIN4 B. subtilis were centrifuged at 8,000 × g for 15 min. Pellets were resuspended in ice-cold 50 mM MES buffer (pH = 6.2), 150 mM NaCl, 0.1% Tween, and 1× Complete protease inhibitor cocktail (Roche). Cells were lysed by sonification and centrifuged at 16,000 × g for 20 min. Supernatant was collected and assayed for CDO activity at 37°C with 10 mM cysteine and 300 μM ferrous sulfate. The CDO assay was done as described above except that hydroxylamine hydrochloride was included in the assay mixture at a final concentration of 10 mM to prevent the dissimilation of cysteine sulfinic acid by pyridoxal-5′-phosphate-dependent enzymes present in the bacterial lysates.
Phylogenetic analyses.
Multiple sequence alignments of eukaryotic and bacterial CDO proteins were constructed by using AlignX software (Vector NTI; Invitrogen) and a blosum62mt score matrix. To create an unrooted phylogenetic tree from these data, the sequence alignment was resampled with 100 replacement replicates (bootstrapped) by using PHYLIP software version 3.65 (11). Protein distances were calculated by using the Jones-Taylor-Thornton model. Tree drawing from the distance matrix was done by using the neighbor-joining method.
RESULTS
Identification of bacterial CDOs.
To initially identify putative bacterial CDOs, a PSI-BLAST (2) search was run using the rat CDO sequence. This search resulted in 82 sequences with e-values up to 6.9. The top 43 hits were from eukaryotes and had 28% and higher overall sequence identity with rat CDO. The top bacterial hit was the YubC protein from B. subtilis (e-value = 0.05), and a total of 38 putative bacterial CDOs were found. From this PSI-BLAST search, the four proteins characterized in the present study were selected. After these bacterial CDOs were found to be active proteins, BLASTP searches of nonredundant protein databases were done using the four proteins as query sequences. Twenty-one putative bacterial homologs (e-value of ≤1.5) with good conservation of active-site residues were identified by these searches (Fig. 3). Interestingly, the BLASTP searches uncovered two proteins, ZP_00991440 from Vibrio splendidus and NP_902852 from Chromobacterium violaceum, that differed from the overwhelming majority of other putative bacterial CDOs by having conservation of the unique cysteine found in motif 1 of eukaryotic CDOs. These two proteins were not originally identified in our initial PSI-BLAST query. The searches also uncovered two cyanobacterial proteins, ZP_00112383 from Nostoc sp. strains PCC73102 and ZP_00516761 from Crocosphaera watsonii, that share some of the conserved active-site residues seen in the CDOs evaluated in the present study but that are missing other conserved residues, notably those that lie outside of cupin motifs 1 and 2. The cyanobacterial proteins, which share 21% identity with each other, are currently annotated in the NCBI database as encoding putative CDOs. Nostoc sp. strain PCC73102 was cloned from genomic DNA and tested for activity but was found to possess no detectable CDO activity (data not shown). As a consequence, these two cyanobacterial proteins were omitted from the subsequent phylogenetic analysis (discussed below).
FIG. 3.
Conservation of functional residues within eukaryotic and putative bacterial CDOs (putative bacterial CDOs tested in the present study are indicated by a box). (A) Sequence alignment showing four peptide segments (position numbers for the residue beginning each segment shown in parentheses) that contain active-site residues in eukaryotic CDO known to chelate iron (✽) or to play a role in substrate coordination and catalysis (arrows). Within cupin motif 1, most cupin proteins contain a conserved glutamate that is used for metal cofactor binding. In CDO, this residue is not conserved (▪), being replaced by a cysteine in eukaryotic CDOs and by a glycine in most prokaryotic CDOs. (B) Consensus CDO family signature of highly conserved residues was generated from an alignment of eukaryotic and bacterial CDOs. Hydrophobic residues (AILV) are indicated by “Φ.” (C) Two cyanobacterial proteins identified by BLASTP search and annotated as encoding putative CDOs in the NCBI database share some elements of the consensus CDO family signature but are missing other conserved active-site sequences. Listed e-values are from BLASTP scores using rat CDO as the query sequence. NCBI accession numbers were as follows (identification number, strain or species, accession number): 1, Rattus norvegicus, BAA11925; 2, Homo sapiens, BAA12873; 3, Xenopus tropicalis, AAH61333; 4, Histoplasma capsulatum, AAV66535; 5, Frankia sp. strain EAN1pec, ZP_00572365; 6, Janibacter sp. strain HTCC2649, ZP_00993719; 7, Mycobacterium tuberculosis, CAA17181; 8, Streptomyces avermitilis[SAV5041], NP_826218; 9, Streptomyces avermitilis[SAV2489], NP_823665; 10, Streptomyces coelicolor A3(2) [SCO3035], NP_627257; 11, Streptomyces coelicolor A3(2) [SCO5772], NP_629897; 12, Bacillus anthracis, ZP_00392898; 13, Bacillus cereus, NP_832375; 14, Bacillus subtilis, NP_390992; 15, Bacillus thuringiensis, ZP_00744219; 16, Azotobacter vinelandii, ZP_00416154; 17, Burkholderia ambifaria, ZP_00686999; 18, Burkholderia cenocepacia, ZP_00461417; 19, Myxococcus xanthus, AAF87926; 20, Polaromonas sp. strain JS666, ZP_00507221; 21, Ralstonia metallidurans (Cupriavidus metallidurans), ZP_00597470; 22, Vibrio splendidus, ZP_00991440; 23, Chromobacterium violaceum, NP_902852; 24, Nostoc PCC73102, ZP_00112383; 25, Crocosphaera watsonii, ZP_00516761.
Selection and cloning of target genes.
From the list of putative CDOs from our initial PSI-BLAST search, we selected for further characterization four proteins (PSI-BLAST e-values indicated in parentheses): YubC from B. subtilis (0.05), BC2617 from B. cereus (0.83), and two divergent paralogs (pairwise identity = 25%) SCO3035 (0.059) and SCO5772 (0.45) from S. coelicolor A3(2). These four proteins were selected in part on the basis of their low overall sequence similarity to one another (<26%) but mainly because of their differing potentials for the formation of an internal thioether bond analogous to that found in eukaryotic CDO. The YubC protein contains a cysteine residue within motif 1, one residue downstream of its relative position in eukaryotic CDO, and could conceivably form an internal thioether bond. BC2617 and SCO3035 contain cysteine residues, but all lie well outside of motif 1 and thus would not be predicted to form an internal thioether. SCO5772 contains no cysteine residues at all and definitely could not form an internal thioether bond. All four proteins have been previously annotated as hypothetical cupin proteins of unknown function or putative cysteine dioxygenase-like proteins.
The ORFs of the four bacterial genes were cloned in frame into the pQE30 expression vector to yield N-terminal His6-tagged protein products. The recombinant proteins were purified to apparent homogeneity by a single chromatographic step on a nickel-Sepharose column. During purification of three of the bacterial proteins (all except SCO5772), two peaks of highly purified recombinant protein were found to elute: one at 10% (peak 1) and the second at 20% buffer B (peak 2). By SDS-PAGE, the two peaks both appeared >95% pure and were indistinguishable. However, kinetic data showed that peak 1 had a substantially lower Km for cysteine and a significantly higher Vmax compared to peak 2, so the parameters of that more active fraction are reported. The elution of heterogeneous populations of recombinant CDO when using a step gradient elution profile has been reported previously for eukaryotic CDO (22). The phenomenon is presumed to be due to differences in protein folding or posttranslational modification, some variants of which show reduced catalytic efficiency.
Kinetic properties.
Upon purification, all of the recombinant proteins were catalytically inactive. Activity could be restored, however, upon the addition of exogenous ferrous iron, with maximal activation seen at a ferrous iron concentration of 300 μM. Other transition metals (Co2+, Mn2+, Ni2+, and Zn2+) failed to restore significant activity to the purified proteins. These data are consistent with previous reports of a strict requirement of ferrous iron for CDO catalytic activity (6, 22).
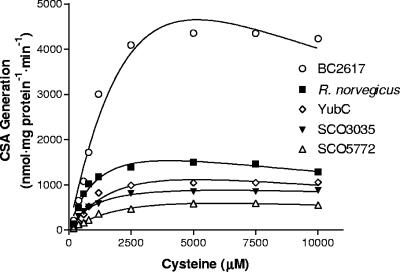
The pH optima of the bacterial CDOs were nearly identical to that seen for rat CDO (Table 1) . The reaction velocity as a function of cysteine concentration showed various degrees of substrate inhibition, with Ks values ranging from 4.6 to 32 mM (Fig. 4). The relative catalytic efficiencies, as assessed by kcat/Km values, varied by less than an order of magnitude with the relative order being BC2617/R. norvegicus CDO > SCO3035 > YubC/SCO5772 (Table 1).
TABLE 1.
Comparison of molecular properties and kinetic parameters of purified eukaryotic and bacterial CDOs
| CDO construct | % Sequence identity with rat CDOa | pH optimum | Cysteine Km (mM) | Sp act (μmol mg−1 min−1) | kcat (s−1) | kcat /Km (mM−1 s−1) |
|---|---|---|---|---|---|---|
| R. norvegicus | 100 | 6.1b | 1.5b | 1.4b | 0.62 | 0.4 |
| BC2617 | 13.0 | 6.1 | 5.7 | 4.4 | 2.0 | 0.4 |
| YubC | 18.7 | 6.2 | 3.0 | 1.0 | 0.39 | 0.1 |
| SCO3035 | 20.6 | 6.0 | 1.2 | 0.8 | 0.33 | 0.3 |
| SCO5772 | 16.4 | 6.1 | 3.8 | 0.7 | 0.30 | 0.1 |
FIG. 4.
Effect of cysteine concentration on cysteine sulfinic acid (CSA) generation. Activity assays were conducted in MES buffer at the pH optimum for each enzyme. Ferrous iron was included at a concentration of 300 μM. The datum points represent the average of a set of duplicate assays.
Substrate specificity and inhibition profile.
Rat CDO exhibits a high degree of specificity for its reaction—structurally related thiols are unable to serve as substrates or as competitive inhibitors of the reaction (31). It is generally seen that the degree of functional variation and divergence of reaction specificity increases significantly for proteins sharing less than 30% identity (28). Because all of the bacterial CDOs fall well below this level, the specificity of bacterial CDOs for cysteine was probed by the ability of cysteine analogs to inhibit the formation of cysteine sulfinic acid. As shown in Table 2, these analogs produced only a modest inhibition of activity, even when present in the reaction mixture at a concentration 10-fold higher than the concentration of cysteine. Of the analogs tested, 2-aminoethanethiol produced the highest degree of inhibition. It does not appear to be a substrate for dioxygenation, however, since no hypotaurine—the expected product—could be detected in assays where recombinant protein was incubated with 5 mM 2-aminoethanethiol and ferrous iron (data not shown).
TABLE 2.
Percent inhibition of activity by l-cysteine analogsa
| CDO construzcts | % Inhibition by cysteine analog
|
|||||
|---|---|---|---|---|---|---|
| 2-Amino-ethanethiol
|
2-Sulfanyl-ethanol
|
3-Sulfanyl-propionic acid
|
||||
| 1 × Km | 10 × Km | 1 × Km | 10 × Km | 1 × Km | 10 × Km | |
| R. norvegicus | 8.7 | 30 | 5.9 | 13 | 4.9 | 26 |
| BC2617 | −2.5 | 20 | 4.8 | 11 | 3.2 | 12 |
| YubC | 12 | 19 | −0.7 | 6.6 | 4.7 | 11 |
| SCO3035 | 7.5 | 27 | 8.6 | 15 | 2.9 | 11 |
| SCO5772 | 0.9 | 28 | 2.6 | 13 | 5.6 | 16 |
Analogs were tested at two different concentrations: 1× and 10× the Km for cysteine. These competition assays were done with a cysteine concentration equivalent to the Km as determined (Table 1) for each respective homolog.
CDO expression in B. subtilis.
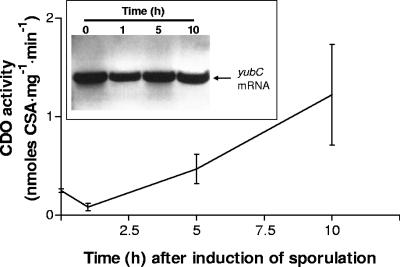
A report using microarray analyses has recently described yubC expression as being significantly upregulated in B. subtilis during sporulation (24). To verify that CDO is indeed expressed and is functionally active in a living bacterium, we decided to evaluate CDO gene expression and catalytic activity in wild-type B. subtilis and a mutant B. subtilis strain (yubC::pMUTIN4) containing an insertional mutation that inactivates expression of the yubC locus as previously described (29). As shown in Fig. 5, yubC mRNA was expressed during vegetative growth (0-h time point) and at least 10 h after the induction of sporulation in wild-type B. subtilis. CDO activity was also detectable in wild-type B. subtilis during vegetative growth and increased to ∼350% 10 h after sporulation induction. Neither yubC gene expression nor CDO catalytic activity could be detected in the yubC::pMUTIN4 strain. Tests for sporulation revealed that both strains were capable of forming spores, although with slightly different efficiencies. Wild-type B. subtilis exhibited a sporulation efficiency of 88% (total cell titer of 7.3 × 108 CFU/ml; spore titer of 6.4 × 108 CFU/ml), whereas the mutant strain had a reduced sporulation efficiency of 50% (total cell titer of 4.6 × 108 CFU/ml; spore titer of 2.3 × 108 CFU/ml).
FIG. 5.
CDO expression in sporulating and nonsporulating wild-type B. subtilis cultures. CDO activity was assayed from culture aliquots taken 0, 1, 5, and 10 h after induction of sporulation via the Sterlini-Mandelstam method (25). The data are averages ± the standard deviations of three independent experiments. (Inset) RT-PCR analysis of yubC gene expression 0, 1, 5, and 10 h after induction of sporulation.
Phylogenetic analyses.
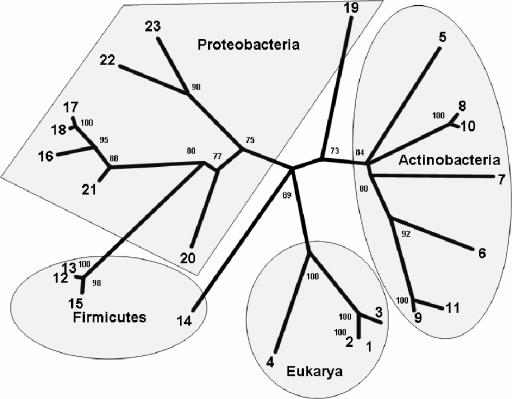
To better understand the phylogenetic relationship between eukaryotic and eubacterial CDOs, phylogenetic analyses were conducted on CDO homologs from 4 representative eukaryotic species and 19 bacterial species (Fig. 3). The topology of the phylogram, which is supported by good bootstrapping values, shows roughly four separate CDO clusters or subfamilies separated by relatively large sequence differences (Fig. 6). The eukaryotic CDOs make up one of these subfamilies. The remaining three subfamilies are composed of bacterial CDOs and are roughly divided by phyla into actinobacteria, proteobacteria, and firmicutes clusters, with the firmicutes branching from the same lineage that gives rise to the proteobacteria. The two exceptions to this categorization are the YubC CDO from B. subtilis, which is found to be more closely related to the eukaryotic CDO lineage, and the putative CDO from M. xanthus, a deltaproteobacterium, which is found here to be more closely related to the CDOs of actinobacteria than other proteobacteria.
FIG. 6.
Phylogenetic relationship of proteins belonging to the CDO family. In the unrooted dendrogram shown, branch lengths are proportional to the level of sequence divergence, and the numbers at the branch nodes of the consensus tree are bootstrapping values indicating the number of times the partition of the species into the two sets that are separated by that branch occurred among the 100 replicate trees. The orientation of some branches was rotated about their nodes to highlight the natural grouping of CDOs by phyla and to better show which species, such B. subtilis and M. xanthus, clearly lie outside of these natural groupings. The species identification numbers correspond to the numbers found in the multiple sequence alignment in Fig. 3.
DISCUSSION
CDO activity has been thought to be limited to the metazoan and fungal kingdoms, which comprise organisms that are occasionally forced to dispose of high intracellular cysteine burdens as a consequence of diet and environment. However, the four bacterial homologs evaluated in the present study possess catalytic activity and kinetic properties very similar to those of rat CDO and, despite their low sequence identity to rat CDO, the bacterial CDOs show a high degree of specificity for cysteine. These results strongly suggest that cysteine is also the authentic physiological substrate for these bacterial enzymes and, thus, that CDO is also present in eubacteria.
Based on the alignments of prokaryotic and eukaryotic CDOs, we created a phylogenetic topology of the CDO family. Within bacteria, CDO appears to be limited to the Actinobacteria, Firmicutes, and Proteobacteria. Two potential cyanobacterial homologs were identified in our BLAST searches, but the lack of conservation of some active-site residues and lack of CDO activity for the Nostoc sp. strain PCC73102 protein suggests that these proteins are not CDOs. Two proteins that did not naturally group with other members of their phyla were YubC from B. subtilis and the CDO from M. xanthus. The reason for this outgrouping could be a horizontal gene transfer event, i.e., a eukaryotic CDO gene may have been incorporated by B. subtilis and an actinomycete CDO gene may have been incorporated by M. xanthus at some point during the course of these respective species' evolution. At least for the case of B. subtilis, the unusually close relationship of B. subtilis and eukaryotic CDO seen here has been documented for other cupin proteins; in terms of both the total number of cupins encoded and their specific domain composition, B. subtilis is closer to eukaryotes than any other microbe for which whole genomic sequence data are available (10).
The physiological role of CDO in eubacteria is not clear. A common element among many of the bacteria having CDOs is that they undergo a complex life cycle involving morphological changes. Moreover, many, including the Bacillus and Streptomyces species, are spore formers. Concordant with a previous report (24), we have seen that CDO is expressed in the vegetative state and undergoes a significant increase in activity after the initiation of sporulation. It is possible that, in this context, CDO may help to reduce intracellular free cysteine levels and promote an environment conducive to the formation of disulfide bonds in the coat proteins of the nascent prespore within the mother cell. However, CDO does not appear to be absolutely necessary in this process since obliteration of its expression in the yubC::pMUTIN4 mutant B. subtilis did not appear to affect its capacity to form spores, although it did reduce the overall sporulation efficiency. Similarly, disruption of the expression of SCO5772 in S. coelicolor A3(2) by an insertional mutagenesis event was shown to produce a significant delay in the development of aerial mycelia in this species but did not completely inhibit sporulation (12). Nevertheless, given our finding in the present study that S. coelicolor A3(2) contains two functional CDO paralogs (SCO3035 and SCO5772), presumably due to the duplication of chromosomal DNA during the evolution of this genus (5), it would be interesting to see what phenotype results from complete ablation of expression of the two homologs in this organism. Interestingly, there is evidence from lower eukaryotes showing that CDO and its ability to reduce intracellular cysteine levels may play an important role in the progression of morphological differentiation. In Candida albicans, CDO expression is upregulated in the transition from the W-cell to O-cell phenotype (30). In Histoplasma capsulatum, CDO has been shown to be an important participant in the transition from the mycelial phase to yeast phase (16, 20). Collectively, these data suggest that, in addition to the important role of biosynthesis in regulating steady-state cysteine levels in organisms such as B. subtilis (1, 13), degradation of cysteine in these bacteria that have CDO may also facilitate bacterial life cycle progression and this possibility deserves further study. We should state, however, that not all bacteria with multistage life cycles have CDO. Caulobacter crescentus, for instance, is a proteobacterium that cycles between swarmer and stalk-cell forms (19). A search of its genome did not reveal any close CDO homologs.
The presence of CDO in bacteria also raises questions about the fate of its product, cysteine sulfinic acid. In mammals, cysteine sulfinic acid is either decarboxylated to hypotaurine and then spontaneously oxidized to taurine or dissimilated by aspartate aminotransferase into pyruvate and sulfite (which is subsequently oxidized to sulfate by sulfite oxidase). There have been no reports of cysteine sulfinic acid decarboxylase activity in bacteria, and thus it seems unlikely that hypotaurine/taurine would be synthesized, although many species of bacteria are capable of using these sulfonates as sources of carbon, nitrogen, or sulfur (7). Like mammals, however, bacteria do possess aspartate aminotransferase and would be able to break down cysteine sulfinic acid to pyruvate and sulfite. Some bacterial NifS proteins, which initiate Fe-S cluster protein assembly by generating persulfide through cysteine desulfuration, are also capable of dissimilating cysteine sulfinic acid through desulfination (15, 18). Indeed, the best substrate for one such NifS-like protein from Escherichia coli (a bacterium that does not have a CDO homolog in its genome) was cysteine sulfinic acid (18). The presence of NifS-like proteins in bacilli, streptomyces, azotobacteria, and mycobacteria suggests that cysteine sulfinic acid generated by CDO could be used by these proteins, although the direct incorporation of its product into iron-sulfur proteins seems unlikely given that the product would not be reduced sulfur. A final alternative pathway for cysteine sulfinic acid metabolism in bacteria would be spontaneous oxidation to cysteate, whereupon it would be broken down to sulfite/sulfate, pyruvate, and ammonium ions (8). The metabolic fate of cysteine in eubacteria containing CDO, such as the highly abundant soil saprophyte Streptomyces, has important implications for models of the geochemical sulfur cycle. The direct oxidation of cysteine by saprophytic aerobic bacteria could be an alternate pathway for the generation of inorganic sulfate from decaying organic matter and deserves further study to verify whether quantitatively significant amounts of sulfate are generated by these organisms.
A sequence alignment of both eukaryotic and bacterial CDOs yielded a fingerprint consensus sequence for CDO that, in addition to the two cupin motif segments, includes key residues in segments just upstream of the first cupin motif (the equivalents of rat CDO Tyr58 and Arg60) and just downstream of the second cupin motif (the equivalents of rat CDO Ser153, His155, and Tyr157) (Fig. 3). One of the major questions resolved here is whether the Cys-Tyr cross-link seen in eukaryotic CDO is crucial for activity. Since this residue is in almost all cases substituted by a Gly in bacterial CDOs, many of the bacterial CDOs, such as BC2617, SCO3035, and SCO5772, would not be capable of forming this Cys-Tyr bond because of the lack of a nearby cysteine residue. Because they are nonetheless capable of sulfoxidation, we conclude that the ability to form an internal thioether bond is not necessary for CDO catalysis, although it could certainly still influence activity in some way that is accomplished differently in enzymes lacking the Cys. The presence of the Cys in the Vibrio splendidus and Chromobacterium violaceum CDOs (Fig. 3), which are otherwise not particularly similar to eukaryotic CDOs, suggests that the Cys-Tyr linkage may have occurred independently in this lineage.
The perfect conservation of the equivalents of Ser153 (as Ser or Thr), His155, and Tyr157 (rat CDO numbering) supports the hypothesis that they form a catalytic triad important for supporting the role of Tyr157 as a catalytic acid/base (23). In contrast, two active-site residues—rat Tyr58 and Arg60—that are conserved in eukaryotic CDOs have some variation among the bacterial CDOs. The acceptable substitution of Trp for Tyr58 in some bacterial CDOs argues against the phenolic hydroxyl playing a key role in catalysis, and the acceptable substitution of Gln for Arg60 argues that the NE atom (conserved between Arg and Gln) may be the part of Arg60 that plays an important role in substrate binding. We note, however, that during evolution, residues that are crucial to folding and/or catalysis need not be linearly preserved in the sequence. Indeed, these residues can be repositioned elsewhere within the primary sequence and nevertheless end up in the appropriate structural position and carry out the same biological/chemical role (28). Because of this, the conservative fingerprint analysis used to identify putative CDOs in bacteria may have missed some distantly related functional homologs in bacteria.
While the present study was under review, the Joint Center for Structural Genomics released coordinates for the structure of a bacterial CDO: Ralstonia eutropha (Cupriavidus necator) JMP134 (PDB 2GM6). The overall structure of the protein is very similar to rat CDO (root mean square deviation of 2.2 Å) despite a low sequence conservation (16% identity). Most importantly, the bacterial active site is well conserved with rat CDO despite the absence of a cysteinyl-tyrosine bond.
In summary, our data show that a subset of eubacteria contain catalytically active CDO. The presence of a functional thiol dioxygenase in bacteria poses many interesting avenues of future research regarding its specific physiological role in bacterial sulfur metabolism, as well as its role in the larger geochemical sulfur cycle.
Acknowledgments
We thank Relicardo Coloso for the HPLC analysis of hypotaurine production and Lawrence Hirschberger for technical assistance.
Support for this research was provided by a grant from the National Institutes of Health, grant PHS DK056649, awarded to M.H.S. J.E.D. was supported by a graduate student fellowship from the National Science Foundation.
REFERENCES
- 1.Albanesi, D., M. C. Mansilla, G. E. Schujman, and D. de Mendoza. 2005. Bacillus subtilis cysteine synthetase is a global regulator of the expression of genes involved in sulfur assimilation. J. Bacteriol. 187:7631-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attschul, S. F., T. C. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3384-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagley, P. J., L. L. Hirschberger, and M. H. Stipanuk. 1995. Evaluation and modification of an assay procedure for cysteine dioxygenase activity: high-performance liquid chromatography method for measurement of cysteine sulfinate and demonstration of physiological relevance of cysteine dioxygenase activity in cysteine catabolism. Anal. Biochem. 227:40-48. [DOI] [PubMed] [Google Scholar]
- 4.Bella, D. L., L. L. Hirschberger, Y. Hosokawa, and M. H. Stipanuk. 1999. Mechanisms involved in the regulation of key enzymes of cysteine metabolism in rat liver in vivo. Am. J. Physiol. 276:E326-E335. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 6.Chai, S. C., A. A. Jerkins, J. J. Banik, I. Shalev, J. L. Pinkham, P. C. Uden, and M. J. Maroney. 2005. Heterologous expression, purification, and characterization of recombinant rat cysteine dioxygenase. J. Biol. Chem. 280:9865-9869. [DOI] [PubMed] [Google Scholar]
- 7.Cook, A. M., K. Denger, and T. H. Smits. 2005. Dissimilation of C3-sulfonates. Arch. Microbiol. 185:183-190. [DOI] [PubMed] [Google Scholar]
- 8.Denger, K., T. H. Smits, and A. M. Cook. 2006. l-Cysteate sulpho-lyase, a widespread, pyridoxal 5′-phosphate-coupled desulphonative enzyme purified from Silicibacter pomeroyi DSS-3T. Biochem. J. 394:657-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominy, J. E., Jr., L. L. Hirschberger, R. M. Coloso, and M. H. Stipanuk. 2006. Regulation of cysteine dioxygenase degradation is mediated by intracellular cysteine levels and the ubiquitin-26 S proteasome system in the living rat. Biochem. J. 394:267-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunwell, J. M., S. Khuri, and P. J. Gane. 2000. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64:153-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1997. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 46:101-111. [DOI] [PubMed] [Google Scholar]
- 12.Gehring, A. M., J. R. Nodwell, S. M. Beverley, and R. Losick. 2000. Genomewide insertional mutagenesis in Streptomyces coelicolor reveals additional genes involved in morphological differentiation. Proc. Natl. Acad. Sci. USA 97:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30:737-749. [DOI] [PubMed] [Google Scholar]
- 14.Harwood, C. R., and S.M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Inc., New York, N.Y.
- 15.Jaschkowitz, K., and A. Seidler. 2000. Role of a NifS-like protein from the cyanobacterium Synechocystis PCC 6803 in the maturation of FeS proteins. Biochemistry 39:3416-3423. [DOI] [PubMed] [Google Scholar]
- 16.Kumar, V., B. Maresca, M. Sacco, R. Goewert, G. S. Kobayashi, and G. Medoff. 1983. Purification and characterization of a cysteine dioxygenase from the yeast phase of Histoplasma capsulatum. Biochemistry 22:762-768. [DOI] [PubMed] [Google Scholar]
- 17.McCoy, J. G., L. J. Bailey, E. Bitto, C. A. Bingman, D. J. Aceti, B. G. Fox, and G. N. Phillips, Jr. 2006. Structure and mechanism of mouse cysteine dioxygenase. Proc. Natl. Acad. Sci. USA 103:3084-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mihara, H., T. Kurihara, T. Yoshimura, K. Soda, and N. Esaki. 1997. Cysteine sulfinate desulfinase, a NIFS-like protein of Escherichia coli with selenocysteine lyase and cysteine desulfurase activities: gene cloning, purification, and characterization of a novel pyridoxal enzyme. J. Biol. Chem. 272:22417-22424. [DOI] [PubMed] [Google Scholar]
- 19.Ryan, K. R., and L. Shapiro. 2003. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu. Rev. Biochem. 72:367-394. [DOI] [PubMed] [Google Scholar]
- 20.Sacco, M., B. Maresca, B. V. Kumar, G. S. Kobayashi, and G. Medoff. 1981. Temperature- and cyclic nucleotide-induced phase transitions of Histoplasma capsulatum. J. Bacteriol. 146:117-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekowska, A., H. F. Kung, and A. Danchin. 2000. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J. Mol. Microbiol. Biotechnol. 2:145-177. [PubMed] [Google Scholar]
- 22.Simmons, C. R., L. L. Hirschberger, M. S. Machi, and M. H. Stipanuk. 2006. Expression, purification, and kinetic characterization of recombinant rat cysteine dioxygenase, a non-heme metalloenzyme necessary for regulation of cellular cysteine levels. Protein Expr. Purif. 47:74-81. [DOI] [PubMed] [Google Scholar]
- 23.Simmons, C. R., Q. Liu, Q. Huang, Q. Hao, T. P. Begley, P. A. Karplus, and M. H. Stipanuk. Crystal structure of mammalian cysteine dioxygenase: a novel mononuclear iron center for cysteine thiol oxidation. J. Biol. Chem., in press. [DOI] [PubMed]
- 24.Steil, L., M. Serrano, A. O. Henriques, and U. Volker. 2005. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151:399-420. [DOI] [PubMed] [Google Scholar]
- 25.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stipanuk, M. H. 2004. Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Annu. Rev. Nutr. 24:539-577. [DOI] [PubMed] [Google Scholar]
- 27.Stipanuk, M. H., P. J. Bagley, R. M. Coloso, and M. F. Banks. 1992. Metabolism of cysteine to taurine by rat hepatocytes. Adv. Exp. Med. Biol. 315:413-421. [DOI] [PubMed] [Google Scholar]
- 28.Todd, A. E., C. A. Orengo, and J. M. Thornton. 2001. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 307:1113-1143. [DOI] [PubMed] [Google Scholar]
- 29.Vagner, V., Dervyn, E., and S. Dusko Ehrlich. 1998. A vector for systematic gene inactivation by Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 30.Valtavaara, M., H. Papponen, A. M. Pirttila, K. Hiltunen, H. Helander, and R. Myllyla. 1997. Cloning and characterization of a novel human lysyl hydroxylase isoform highly expressed in pancreas and muscle. J. Biol. Chem. 272:6831-6834. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi, K., Y. Hosokawa, N. Kohashi, Y. Kori, S. Sakakibara, and I. Ueda. 1978. Rat liver cysteine dioxygenase (cysteine oxidase). Further purification, characterization, and analysis of the activation and inactivation. J. Biochem. 83:479-491. [DOI] [PubMed] [Google Scholar]