Abstract
IgG1 b12 is a broadly neutralizing antibody against human immunodeficiency virus type 1 (HIV-1). The epitope recognized by b12 overlaps the CD4 receptor-binding site (CD4bs) on gp120 and has been a target for vaccine design. Determination of the three-dimensional structure of immunoglobulin G1 (IgG1) b12 allowed modeling of the b12-gp120 interaction in which the protruding third complementarity-determining region (CDR) of the heavy chain (H3) was crucial for antibody binding. In the present study, extensive mutational analysis of the antigen-binding site of Fab b12 was carried out to investigate the validity of the model and to identify residues important for gp120 recognition and, by inference, key to the anti-HIV-1 activity of IgG1 b12. In all, 50 mutations were tested: 40 in H3, 4 each in H2 and L1, and 2 in L3. The results suggest that the interaction of gp120 with H3 of b12 is crucially dependent not only on a Trp residue at the apex of the H3 loop but also on a number of residues at the base of the loop. The arrangement of these residues, including aromatic side chains and side chains that hydrogen bond across the base of the loop, may rigidify H3 for penetration of the recessed CD4-binding cavity. The results further emphasize the importance to gp120 binding of a Tyr residue at the apex of the H2 loop that forms a second finger-like structure and a number of Arg residues in L1 that form a positively charged, shelf-like structure. In general, the data are consistent with the b12-gp120 interaction model previously proposed. At the gene level, somatic mutation is seen to be crucial for the generation of many of the structural features described. The Fab b12 mutants were also tested against the b12 epitope-mimic peptide B2.1, and the reactivity profile had many similarities but also significant differences from that observed for gp120. The paratope map of b12 may facilitate the design of molecules that are able to elicit b12-like activities.
It is of fundamental importance to the global human immunodeficiency virus type 1 (HIV-1) vaccine effort to look for potential ways in which to elicit an effective neutralizing antibody response against HIV-1 (6, 8, 10, 27, 32, 53, 68, 88, 92). The target of neutralizing antibodies against HIV-1 is the envelope spike, which consists of the surface glycoprotein gp120 and the transmembrane protein gp41. Although not formally proven, it is generally accepted that the spike is a trimer of gp120-gp41 heterodimers (12, 13, 33, 40, 60, 83, 87). One of the consequences of this quaternary arrangement is that a number of conserved epitopes that are well exposed on purified, monomeric gp120 and gp41 are buried or partially buried in the trimeric gp120-gp120, gp41-gp41, or gp120-gp41 interfaces within the native spike (29, 62, 67, 69, 86). The relative inaccessibility of conserved epitopes in the trimeric spike likely explains the paucity of neutralizing monoclonal antibodies against HIV-1 (8) as well as the low titers of isolate cross-neutralizing antibodies typically found in the serum of animals or humans immunized with soluble envelope protein (15, 20, 21, 23, 26, 45, 72, 81, 85) or even during natural infection with HIV-1 (38, 48).
Nevertheless, a few broadly neutralizing antibodies against HIV-1 have been described. Immunoglobulin G1 (IgG1) b12 binds to the CD4 receptor binding site (CD4bs) on gp120 (4, 9,11, 62), 2G12 binds to a carbohydrate-rich epitope on the silent face of gp120 (63, 70, 80), and 2F5 binds to a linear epitope close to the membrane on the ectodomain of gp41 (52, 61, 94). In addition, three novel antibodies have recently been identified as having broad neutralizing activity: Fab X5, which binds an epitope on gp120, the exposure of which is enhanced by CD4 binding (51), 4E10 (74, 94), and Z13 (94), which bind immediately C-terminal to 2F5 on gp41. These antibodies stand out among the population of known human antibodies as being relatively potent and able to neutralize a wide range of primary isolates of HIV-1 (21, 51, 94) and in combination have been shown to neutralize HIV-1 with some degree of synergy (43, 95). Moreover, IgG1 b12 has recently been shown to be effective at neutralizing primary isolates of subtype C, which is responsible for the greatest number of infections worldwide (7). Importantly, IgG1 b12 is able to completely protect macaques against vaginal challenge with the simian immunodeficiency virus-HIV hybrid SHIV162P4 (58). This study, together with other passive antibody protection studies (2, 14, 44, 46, 56, 73), establishes parameters by which antibody can mediate sterile protection against retroviral challenge and illustrates the potential of broadly neutralizing antibodies for controlling HIV-1, at least in animal models.
Of the panel of broadly neutralizing anti-HIV-1 monoclonal antibodies, IgG1 b12 is the best characterized at the molecular level. Somatic variants of b12 were available essentially since its discovery by phage display methodology, providing an early indication of residues in the antibody variable regions that influence binding activity (4, 9, 57). Later, in vitro selection experiments were performed in which a complementarity determining region (CDR) walking strategy was used to identify variants of b12 with greater affinity for monomeric gp120 and, in some cases, with an enhanced ability to neutralize HIV-1 (5, 90). Recently, the three-dimensional structure of the whole IgG1 b12 molecule was determined (66), providing a structural framework for attempts to elucidate its broadly neutralizing activity. In particular, a Trp residue displayed at the apex of a long protruding H3 loop may allow b12 to penetrate and fill the hydrophobic cavity of the CD4bs on gp120 in a way analogous to Phe43 of CD4 (66). From a docking model of b12 with the core of gp120 (39), b12 also makes contacts on the inside face of the V1/V2 loop stem of gp120 and with the D loop of gp120 by a canyon created by CDRs H3, L1, and L3 (66). This model can now be tested by additional structural and functional studies.
IgG1 b12 is also arguably the best characterized of a group of antibodies known as anti-CD4bs antibodies, which compete with CD4 and with each other in binding to gp120. Many human anti-CD4bs antibodies (besides IgG1 b12) have been described by various groups (49, 50, 75), including 15e (30), F105 (76), F91 (50), 1125H (77), 21 h (30, 75), 654-30D (41), and Fab b6 (57, 62), the last of which was isolated from the same seropositive subject from whom b12 was cloned. These anti-CD4bs antibodies often show broad reactivity with monomeric gp120s from different isolates of HIV-1 but do not, however, show the neutralizing activity of b12 (21, 22, 62). The difference has been associated with the ability of b12 but not of other anti-CD4bs antibodies to bind well to the trimeric envelope spike on the surface of virions (59). It would seem that b12 is able to bind with reasonably high affinity to both monomeric and trimeric forms of gp120, whereas the other CD4bs antibodies bind well only to the monomeric form.
Clearly, one would like to have immunogens capable of eliciting b12-like antibodies. To this end, we have been exploring the interaction of b12 with gp120 from a number of aspects. These include determination of the crystal structure of b12 (66), docking of b12 with the structure of the core of gp120 (66), and examination of the effects of mutations of gp120 residues on the b12-gp120 interaction (55). Here, we approached the problem from the point of view of the antibody by attempting to identify the key structural features of b12 required for gp120 binding through extensive mutagenesis of b12 residues. At the same time, for comparative purposes, we looked at the effects of mutations on the interaction of b12 with a peptide mimotope, B2.1, that binds b12 specifically and is being studied as a vaccine lead (93). The results provide functional data relevant to the docking model presented previously (66) and reveal specific requirements at the tip and base of the CDR H3 finger of b12 for gp120 recognition. In addition, a cluster of arginine residues in the CDR L1 region forming a shelf-like structure and a prominently displayed Tyr residue in CDR H2 are shown to be crucial. The potential implications for eliciting b12-like antibodies by vaccination are discussed in terms of the demands that this puts on the antibody repertoire.
MATERIALS AND METHODS
Mutagenesis and crude Fab preparation.
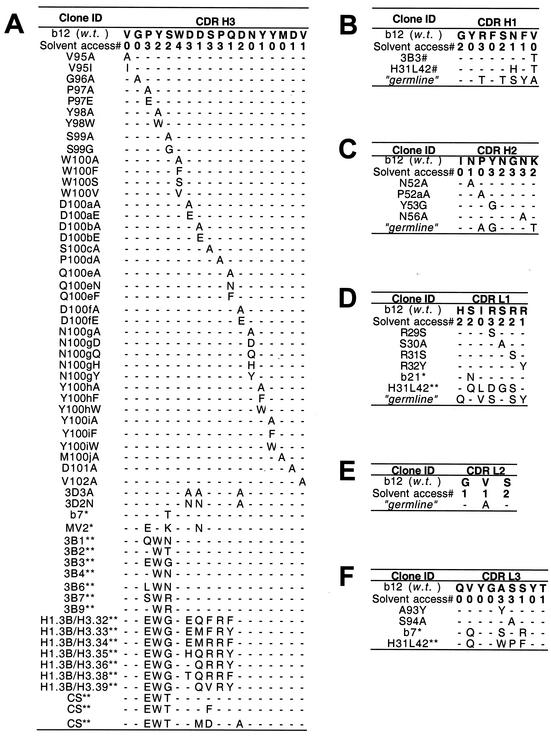
b12 Fab mutants were engineered with the QuikChange mutagenesis kit (Stratagene) according to the manufacturer's directions with pComb3H vector DNA, encoding wild-type b12 Fab, as the template. A similar approach was used to engineer the Fab b6 mutants. The sequences of the mutant clones were verified by DNA sequencing within the variable regions. A complete list of the Fab b12 mutants engineered in this study is included in Fig. 1. The CDRs were defined with IMGT delimitations (http://imgt.cnusc.fr:8104/home.html) (42) except for H3, for which conserved residues A93 and R94 were omitted, as per the Kabat and Wu definition (34).
FIG. 1.
Amino acid sequences of mutants and variants of wild-type (w.t.) b12 in CDR loops H3 (A), H1 (B), H2 (C), L1 (D), L2 (E), and L3 (F). Each panel shows the solvent accessibility (Solvent access#) of the side chains of each residue, the relevant sequences of mutants of b12 engineered for this study, and natural (*) and in vitro-evolved (**) variants isolated in previous studies (references 4 and 57 and references 5 and 90, respectively). The solvent accessibilities of the side chains were determined by the NACCESS computer program (16) and are graded as follows: 0, buried (<10% exposure); 1, partially exposed (10 to 35% exposure); 2, moderately exposed (36 to 66% exposure); 3, mostly exposed (67 to 90% exposure); and 4, fully exposed (>90% exposure). H31L42 (37) is a whole-IgG version of the Fab designated h1.1 h3.33/L1.4L3.14 (90). The deduced amino acid sequences of the closest related germ line DNA are shown for comparison; the germ line sequences for the heavy and light chains are IGHV1-3*01 (DP-25; accession no. X62109) and IGKV3-20*01 (DPK22; accession no. X12686), respectively.
The preparation of crude Fab supernatants has been described previously (3). Briefly, the mutant clones, wild-type b12, and an irrelevant Fab negative control were transformed separately into Escherichia coli XL1-Blue cells (Stratagene), and single colonies were used to inoculate 10-ml cultures in SB medium containing 50 μg of carbenicillin and 10 μg of tetracycline per ml. The cultures were shaken at 300 rpm at 37°C for 6 to 8 h, then induced with 1 mM isopropylthiogalactopyranoside (IPTG), and incubated overnight at 30°C with shaking. The next day, the cultures were centrifuged at 5,000 × g for 15 min at 4°C, the pellets were resuspended in 1 ml of phosphate-buffered saline (PBS, pH 7.0), and the bacterial suspensions were subjected to four rounds of freeze-thawing. The bacterial debris was pelleted at 14 000 rpm in a microcentrifuge, and the supernatants were supplemented with bovine serum albumin (BSA) and Tween 20 (1% and 0.025% final concentrations, respectively). Duplicate or triplicate crude Fab supernatants were prepared to lessen the effect of culture-to-culture variation in Fab production, pooled, and used directly for enzyme-linked immunosorbent assays (ELISAs) as described below.
Crude Fab ELISA.
Ninety-six-well plates (one-half diameter, flat-bottomed; Costar) were coated with 50 μl of PBS containing 50 ng of goat anti-human IgG F(ab′)2 (Pierce), 75 ng of gp120JRFL (Progenics), 50 ng of oligomeric gp120IIIB (ImmunoDiagnostics, Inc.), or 7 × 109 B2.1 phage particles (the B2.1 peptide HERSYMFSDLENRCI is a disulfide-bridged, homodimeric peptide displayed as a fusion to the N terminus of pVIII on the filamentous phage [93]) and incubated overnight at 4°C. The wells were washed twice with PBS containing 0.05% Tween 20 and blocked with 3% BSA at 37°C for 1 h. The wells were washed once, and 50 μl of the bacterial supernatants containing Fab diluted in PBS containing 1% BSA and 0.025% Tween 20 was added. The plates were incubated for 2 h at 37°C, the wells were washed four times, goat anti-human Fab conjugated to alkaline phosphatase (Pierce), diluted 1:500 in PBS containing 1% BSA and 0.025% Tween 20, was added to the wells, and the plate was incubated at room temperature for 30 min. The wells were washed five times and developed by adding 50 μl of alkaline phosphatase substrate, prepared by adding one tablet of disodium p-nitrophenyl phosphate (Sigma) to 5 ml of alkaline phosphatase staining buffer (pH 9.8), according to the manufacturer's instructions.
After ≈30 min, the optical density at 405 nm was read on a microplate reader (Molecular Devices). The concentration of Fab was determined with the anti-Fab ELISA (full curve, threefold dilution series) with simple linear regression; the concentrations of Fab in the samples were usually within about twofold of that of wild-type Fab b12 except for mutants V95A, Y53G, 3D3A, and 3D3N, which were consistently 4- to 10-fold less abundant. A full threefold ELISA binding curve was also generated for groups of Fab mutants alongside wild-type b12 and a negative Fab control against gp120 or B2.1 phage. Apparent affinities were calculated as the antibody concentration at half-maximal binding. Apparent affinities as a percentage of that of wild-type Fab b12 were calculated with the formula [(apparent affinity of the wild type)/(apparent affinity of the mutant)] × 100. All samples were tested at least twice, and the mean was taken as the final reported value.
Competition ELISA with purified Fab.
Ninety-six-well plates were coated with gp120, washed, and blocked, as above. Wild-type and representative mutants of Fab b12 were purified as described previously (3) with protein G-Sepharose columns (Fast Flow; Pharmacia) and verified to be >90% pure by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The purified Fabs were added to the wells at various concentrations in the presence of a single concentration of biotinylated Fab b12 that was previously determined to generate an ELISA signal of 50 to 75% of maximal. After 2 h of incubation at 37°C, the plate was washed, and a streptavidin-horseradish peroxidase conjugate (Jackson) diluted 1:1,000 in PBS containing 1% BSA and 0.025% Tween 20 was added. After a 30-min incubation at room temperature, the plates were washed, and the tetramethylenebenzidine (TMB) substrate kit (Pierce) was used for developing, according to the manufacturer's instructions. The optical density at 450 nm was read on a microplate reader (Molecular Devices), and the results were recorded as the 50% inhibitory concentration (IC50), defined as the concentration of competing Fab that was required to reduce the maximal signal generated by biotinylated Fab b12 by 50%. All competition ELISAs were performed twice.
Synthesis of b12 CDR H3 peptide and coupling to BSA.
A peptide corresponding to CDR H3 of b12 was synthesized with the b12 crystal structure as a guide (66); the peptide has the sequence C-PGK-A93RVGPYSWDDSPQDNYYMDW103. Residues PGK were included to promote a type II β-turn (31), and a Cys residue was added to facilitate cyclization via native chemical ligation (19) and chemical coupling to a carrier molecule. The Arg 94 side chain in the third framework region (FR3) projects into solvent in the crystal structure and so was included in the peptide, as was Ala 93. The peptide was synthesized with N-tert-butoxy-carbonyl chemistry (71), purified by reverse-phase high-pressure liquid chromatography to >95% purity, and verified by mass spectrometry.
To cyclize the peptide, the backbone was linked N to C terminus by native chemical ligation (28), and the final product was purified by reverse-phase high-pressure liquid chromatography to >95% purity and verified by mass spectrometry. The free Cys of the peptide was used to cross-link the peptide to activated BSA (Pierce) by using the manufacturer's instructions. Briefly, 10 mg of sulfo-succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate-activated BSA in 1 ml of deionized H2O was mixed with 10 mg of peptide dissolved in 0.25 ml of dimethyl sulfoxide and 1.75 ml of PBS. The mixture was rocked gently for 2 h at room temperature, dialyzed extensively against PBS, and then sterilized with a 0.2-μm filter. A control peptide (GTP-binding peptide of sequence CEGNVRSRELAGHTGY; American Peptide Co.) was similarly linked to BSA. The protein concentration of each sample was determined with the Micro BCA protein assay kit (Pierce) according to the manufacturer's instructions. The conjugates were also analyzed by SDS-PAGE, and cross-linking was confirmed by an increase in the molecular weight of the activated BSA after reacting with the peptide; both the b12 CDR H3 and the control peptide caused a similar shift in molecular weight.
HIV-1 neutralization assay.
The primary isolate HIV-1JRFL was assayed for neutralization with peripheral blood mononuclear cells as target cells and detection of p24 in ELISA as a reporter assay, as described previously (94).
Nucleotide sequence accession number
The sequences of the heavy and light chains of b12 have been deposited in GenBank (accession no. AAB26315.1 and AAB26306.1, respectively).
RESULTS
Alanine-scanning mutagenesis of CDR H3 of b12.
In the mutagenesis strategy, we chose to compare binding of different mutants of b12 to gp120 with crude Fab supernatants prepared from bacterial cultures (3), which facilitated the rapid analysis of a large number of Fab mutants (Fig. 1) that otherwise would not have been feasible. Apparent affinities relative to wild-type Fab b12 were measured by ELISA, as described in Materials and Methods. As a first step in our analysis of b12, we performed a complete alanine scan of the CDR H3 loop and tested the mutant Fabs by ELISA to determine their relative binding strengths to gp120. We chose recombinant gp120 from two different strains, one from a primary isolate (gp120JRFL) and one from a T-cell-line-adapted strain (gp120IIIB), to see whether any differences in binding could be found with our mutant Fab panel. We also included in our analysis the previously described, b12-specific peptide B2.1, as displayed on filamentous phage (93). The dissociation constant (Kd) of wild-type Fab b12 against gp120LAI is ≈9 nM (62), against the B2.1 synthetic peptide is ≈2.5 μM (93), and against B2.1 phage is closer to the Kd of the b12-gp120 interaction because of the constraint that the phage coat imparts to the peptide, although a precise Kd value of b12 for the phage-associated peptide is difficult to determine (93).
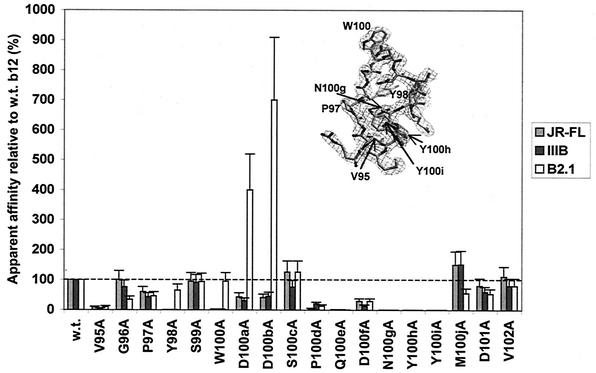
The results of the alanine scan of the H3 loop of b12 are shown in Fig. 2. Substitution of 7 out of 18 residues in H3 with Ala reduced binding of Fab b12 to both gp120JRFL and gp120IIIB by greater than 90%. The relevant mutants are V95A, Y98A, W100A, Q100eA, N100gA, Y100hA, and Y100iA. [The CDR H3 of b12 contains a 10-residue insertion. In Kabat and Wu numbering (34), these residues are designated 100A, 100B,…, 100J. For clarity, these inserted residues will be denoted with a lowercase letter after the number of the residue position so as to avoid confusion with the mutated residue (uppercase) when referring to mutations (e.g., mutation D100aA)]. The loss of antigen binding of the W100A mutant was not unexpected from the crystal structure of IgG1 b12 and a docking model of b12 and the gp120 core (66). However, the behavior of some of the other mutants was more surprising.
FIG. 2.
Alanine-scanning mutagenesis of CDR H3 of b12. Bars indicate the apparent affinities of Fab mutants relative to wild-type (w.t.) Fab b12 for gp120JRFL, gp120IIIB, and the B2.1 peptide. The H3 loop and corresponding electron density from the intact b12 structure (66) are shown (inset), with key residues indicated.
Substitutions in b12 that reduced gp120 binding as much as or even more than observed with the W100A mutant included V95A, Y98A, Q100eA, N100gA, Y100hA, and Y100iA. Most of these substitutions are at the base of the CDR H3 and involve side chains that are poorly exposed to solvent, as defined in Fig. 1. Solvent accessibility to side chain (Fig. 1) was not, however, predictive of the effect of a substitution on antigen binding (Fig. 2). The greatest effects of an Ala substitution on gp120 recognition occurred with residues N100g, Y100h, and Y100i at the C-terminal end of H3, which resulted in complete loss of gp120 recognition in our assay (<0.1% apparent affinity relative to wild-type b12). We confirmed these results by purifying Fab protein for these mutants and were unable to observe significant reactivity with gp120 even at 20 μg/ml (data not shown). To further explore these findings, additional substitutions were made in these positions and tested for binding to gp120 and the B2.1 peptide (see below). Significantly, there was generally good agreement in the relative binding of Ala mutants to gp120JRFL (primary isolate) and gp120IIIB (T-cell-line-adapted).
Thirteen Ala substitutions in H3 had qualitatively similar effects on the ability of b12 to bind to gp120 and B2.1, suggesting some similarity in recognition of these antigens by b12. However, the effects of other Fab substitutions such as Y98A, W100A, D100aA, and D100bA did not correlate, pointing to possible ways of improving the B2.1 peptide as a b12 epitope mimic.
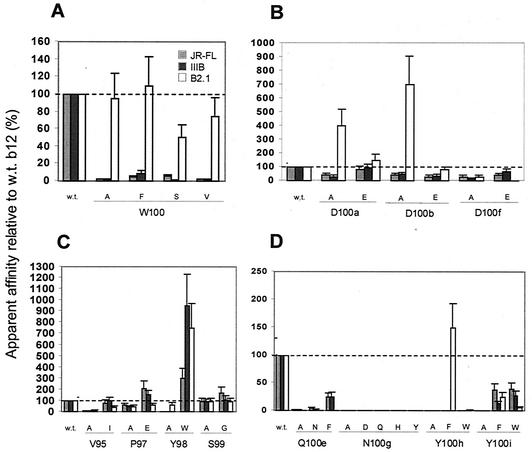
Mutations in CDR H3 W100.
A striking feature in the crystal structure of IgG1 b12 is the prominent display of the aromatic residue W100 at the apex of the long H3 loop (66). As shown in Fig. 2 and 3A, Ala substitution at W100 significantly diminished binding of Fab b12 to gp120JRFL and gp120IIIB. This result was confirmed with purified Fab: the W100A mutant inhibited biotinylated Fab-b12 with an IC50 of 29 μg/ml and 50 μg/ml for gp120JRFL and gp120IIIB, respectively, compared to an IC50 of 3 μg/ml and 1.5 μg/ml, respectively, for wild-type Fab b12 (data not shown).
FIG. 3.
Further substitution analysis of residues in the H3 loop of b12. Bars indicate the apparent affinities of Fab mutants relative to wild-type (w.t.) Fab b12 for gp120JRFL, gp120IIIB, and the B2.1 peptide. Substitutions were made to Trp100 (A), Asp residues near the crown of the H3 loop (B), selected H3 residues N-terminal to Trp100 (C), and key residues C-terminal to Trp100 (D).
In the b12-gp120 docking model, W100 was acting to fill the hydrophobic cavity of the CD4bs on gp120 in a way analogous to F43 of CD4. Thus, we also made the W100F mutant to determine whether Phe would be more or less favored than Trp at this position. Retention of an aromatic ring in the W100F mutant did promote slightly better binding than the W100A mutant to both gp120s, but the apparent affinity was ≈10% relative to that of the wild type. Two other mutants, W100S and W100V, also had considerably impaired binding to both gp120s. Taken together, these results confirm the importance of W100 for b12 by showing that Trp is preferred over four other residues, including another aromatic, Phe, at this position. In contrast to the results with the gp120s, the W100 mutants were all able to bind B2.1 at nearly wild-type levels, strongly suggesting that W100 is not involved in B2.1 recognition.
Role of Asp residues in CDR H3 of b12.
Another feature of the b12 crystal structure that initially drew our attention was a clustered grouping of acidic moieties on one face of the H3 loop (66). We wondered whether this “acidic patch” was involved in keeping the H3 loop of b12 erect via charge repulsion with a weakly acidic patch near the base of the H3 loop. The residues involved included D100a, D100b, and D100f, as well as hydroxyl groups from Ser 99, Ser 100c, and Tyr 100i (D101 was not involved in this patch and moreover was found to have little involvement in binding to gp120 and B2.1; see Fig. 2). The Ala substitutions (Fig. 2) had mostly moderate effects on binding to gp120 (≈2- to 3-fold-reduced binding relative to wild-type b12). Further mutagenesis (Fig. 3B) yielded mutants D100aE, D100bE, and D100fE, the substitutions in which either had no effect (D100aE) or again only moderate effects (D100bE and D100fE) on binding to gp120. Two additional mutants were constructed in which all three positions were changed to see if there was any cooperativity among these residues. A triple Ala mutant, dubbed 3D3A, and another triple mutant, D100aN/D100bN/D100fA, dubbed 3D2N, were still able to consistently bind to gp120JRFL and gp120IIIB, albeit at somewhat lower levels (data not shown).
The acidic patch on the paratope of b12 appears to have significance for B2.1 recognition. The Ala mutants D100aA, D100bA, and both triple mutants bound to B2.1 much better than to wild-type Fab b12. Interestingly, the D100fE mutant bound extremely poorly to B2.1 but bound gp120 almost as well as the wild type. In fact, the D100fE mutant bound more poorly to B2.1 than the “less conservative” D100fA mutant, revealing a very distinct requirement for B2.1 recognition at position 100f.
Mutagenesis in CDR H3 of b12 N-terminal to W100.
Four residues N-terminal to W100 in H3 of b12 were chosen for additional study. V95 was chosen because the Ala mutation at this position caused severe impairment in binding to gp120, and we wished to determine whether a more conservative substitution with Ile would also reduce binding. Figure 3C shows that the V95I mutant bound gp120 at nearly wild-type levels. The extra bulk of a branched aliphatic side chain at this position is probably necessary for full activity of b12. The residues P97, Y98, and S99 were also targeted for further mutagenesis, inspired by previously published variants of b12 (both naturally occurring and in vitro-enhanced variants), including 3B3, in which the sequence EWG is found in place of PYS at these positions (4, 5) (Fig. 1A). The Y98W mutant indeed bound ≈3- to 9-fold better to the gp120s than did wild-type Fab b12 (Fig. 3C). The P97E and S99G mutants bound to gp120 as well as or only slightly better than wild-type b12. Thus, the enhanced binding of 3B3 to gp120 presumably derives mostly from the preference for a W over a Y at the middle position of the EWG motif, with the flanking residues perhaps providing additional fine tuning.
In contrast to the Ala mutant, P97A, B2.1 binding was not correlated with gp120 binding for mutant P97E. Interestingly, the substitution Y98W enhanced the binding of Fab b12 to both B2.1 and gp120, whereas the S99G substitution was silent for both gp120 and B2.1.
Mutagenesis in CDR H3 of b12 C-terminal to W100.
The alanine scan (Fig. 2) showed that residues Q100e, N100g, Y100h, and Y100i in the C-terminal portion of H3 of b12 were all important for gp120 recognition and were therefore chosen for further mutational analysis. Q100eN and Q100eF mutants also bound gp120 with diminished affinity relative to wild-type Fab b12, although the gp120 binding was significantly improved for Q100eF relative to Q100eA (Fig. 3D). For verification purposes, Fab Q100eA was purified and used in a competition ELISA with biotinylated Fab b12 against gp120IIIB; the IC50 of Q100eA was ≈14 μg/ml, which is nine times greater than that of wild-type Fab b12 (IC50 ≈1.5 μg/ml) and consistent with the crude Fab ELISA. In the structure of IgG1 b12, the side chain of Q100e is only partially accessible to solvent and makes a hydrogen bond with the main chain of A93. A loss of this hydrogen bond might destabilize the interaction at the base of CDR H3 and could explain the observed reduction in binding to gp120 for the Q100e mutants.
Next, we tested more conservative substitutions at position N100g. Neither an N100gD nor an N100gQ mutant was able to bind either gp120JRFL or gp120IIIB, indicating that the removal of an amino group or the addition of a methylene group, respectively, from the side chain of Asn100g was sufficient to completely abolish gp120 recognition in our assay format (Fig. 3D). In the crystal structure of b12, the amino nitrogen on the side chain of Asn100g makes a hydrogen bond with the main-chain carbonyl of Gly96; indeed, the side chain of Asn100g is <10% exposed to solvent (Fig. 1). Again, mutations in Asn100g most likely affect the structure of the paratope of b12 due to the absence of the stabilizing hydrogen bond. Thus, not surprisingly, the substitution of Asn100g with either His or Tyr completely abolished binding to either gp120JRFL or gp120IIIB. The Tyr substitution was chosen because the JH6 family of J segments (the J-segment family used by b12 in VDJ-recombination) encodes a repeating string of Tyr residues, and thus b12-like antibodies that also use the JH6 family might encode a Tyr at this position. The complete lack of detectable binding to gp120 by any of the 100g mutants underscores the critical importance of residue N100g in the activity of b12 (see the Discussion).
In contrast to the irreplaceability of N100g, antigen recognition was found to be at least partially maintained with conservative substitutions to Y100h and Y100i. Thus, the Y100i(F/W) mutants bound to gp120 at ≈20% to 40% wild-type levels (Fig. 3D). These results imply that a bulky aromatic at position 100i is required by Fab b12 for recognition of gp120 but that the added hydroxyl group of Tyr100h is important for full antigen binding. By contrast, only a Tyr residue appears to be sufficient for gp120 recognition at position 100h, but the Y100hF mutant was able to bind B2.1 at wild-type levels.
Aside from mutant Y100hF, all the other b12 H3 mutants (conservative mutations C-terminal to W100) had impaired binding to B2.1. Interestingly, a Phe substitution at Y100h but not at Y100i fully restored B2.1 binding. This may reflect the observation that the two Tyr residues are pointing in roughly opposite directions in the crystal structure of b12.
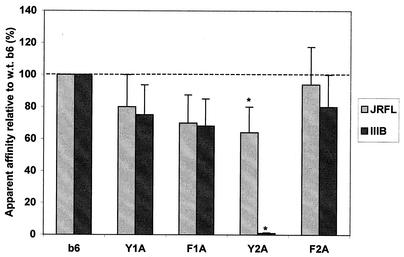
Substitution of aromatic residues in H3 loop of nonneutralizing anti-CD4bs antibody b6.
Following our analysis of the H3 loop of b12, we were particularly interested in the role of aromatics because substitution to Ala of all four aromatics in the H3 loop of b12, not only W100, caused a >95% decrease in relative binding to gp120. Thus, we chose to see if H3 loop aromatics were important in the function of another anti-CD4bs antibody, b6, which, like b12, has a long H3 loop with four aromatic residues but does not neutralize primary isolates of HIV-1 (62). The sequence of the H3 loop of b6 is given in Fig. 4, and the aromatic residues are designated Y1A, F1A, Y2A, and F2A. The results (Fig. 4) indicated that in contrast to b12, mutation to Ala of only one of the four aromatics in the H3 of b6, Y2A, led to a severe decrease in binding of Fab b6 to gp120 and only to gp120IIIB and not gp120JR-FL. In fact, the Y2A substitution decreased the half-maximal binding of purified Fab b6 to gp120IIIB from 0.003 μg/ml to 0.4 μg/ml, which was far more severe than the change in half-maximal binding to gp120JRFL (from 0.003 μg/ml to 0.007 μg/ml; data not shown). The differences in the roles of H3 aromatics between b12 and the poorly neutralizing antibody b6 are striking and are discussed below.
FIG. 4.
Effect of alanine substitutions of the four aromatic residues in the H3 loop of b6 on relative binding of Fab b6 to gp120JR-FL and gp120IIIB. Bars indicate the apparent affinities of Fab mutants relative to wild-type (w.t.) Fab b6 for gp120JR-FL and gp120IIIB. The amino acid sequence of the H3 loop of b6 is QKPRY1F1DLLSGQY2RRVAGAF2DV (the aromatic residues are in boldface and have superscript numbers to correspond with the bar graph). By ELISA, the half-maximal binding of the purified Fab of the Y2A mutant to gp120IIIB was 0.4 μg/ml, which was ≈50 times lower than the half-maximal binding of mutant Fab Y2A to gp120JRFL, 0.007 μg/ml (*); half-maximal binding of wild-type Fab b6 to both gp120IIIB and gp120JRFL was determined to be 0.003 μg/ml (data not shown).
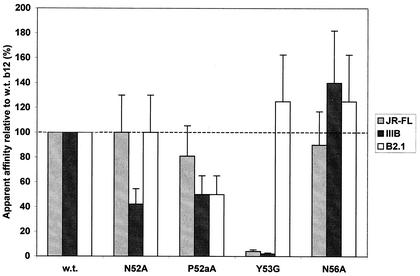
b12 H2 mutations.
In the b12-gp120 docking model (66), it was predicted that residues in H2 could contact gp120. Residue Y53 is the most prominent in H2, points directly toward gp120 in the model, and occupies a large space in the b12-gp120 interface. We suspected that residue P52a might also play a role in gp120 recognition by maintaining the H2 loop in a particular conformation rather than by making extensive contact with gp120. Interest in residues P52a and Y53 was also strong because these were nonconservatively mutated from the residue encoded by the closest germ line genes at these positions, Ala and Gly, respectively. The germ line genes closest to b12 are DPK22 and DP-25 (http://imgt.cnusc.fr:8104/ [17, 78]) for the light and heavy chains, respectively, and the residues they encode are shown for each CDR in Fig. 1. The strategy of using germ line “back mutations” for evaluating residues outside of H3, where somatic mutation is key to generating residue diversity, was adopted to determine the dependence of b12 binding on somatic mutation. Residues N52 and N56 were also in the interface between b12 and gp120 and thus were targeted for mutagenesis.
With the exception of Y53G, the H2 substitutions had only moderate to slight effects on Fab binding to gp120IIIB, gp120JRFL, and B2.1 (Fig. 5). Residue N52 was predicted to make relatively minor contact with gp120, and correspondingly the N52A substitution only moderately diminished binding against gp120IIIB. Interestingly, the N56A mutant bound gp120 at wild-type levels (Fig. 5), implying that this residue does not significantly contribute to gp120 binding. Residue N56 docks in close proximity to residue K362 of gp120; however, the side chain of N56 points away from the contact surface, potentially accounting for the absence of an effect on gp120 binding of the N56A substitution. The P52aA substitution was expected to affect H2 loop structure rather than replace a contact residue, and the effect on gp120 binding was moderate (i.e., ≈2-fold reduction in apparent affinity for gp120), indicating that P52a has a rather modest role in gp120 binding. The Y53G substitution, by contrast, greatly diminished the binding of b12 to gp120, suggesting that Y53 contributes significantly to the binding energy between b12 and gp120. Residue Y53 protrudes to form a second “finger” in the paratope of b12 and was predicted to bury into a canyon in gp120 (see Discussion). The Y53G substitution had little effect on B2.1 recognition, implying that B2.1 does not contact Y53, considering that the side chain of Y53 is mostly solvent exposed (Fig. 1). We note that attempts to purify the Y53G mutant resulted in a somewhat impure Fab preparation that bound poorly to gp120 and B2.1 (data not shown), whereas the crude Fab bound B2.1 at wild-type levels, suggesting that this mutant might be unstable to the purification conditions, which involve acid elution.
FIG. 5.
Effect of substitutions in the H2 loop of b12 on relative binding of Fab b12 to gp120JR-FL, gp120IIIB, and the B2.1 peptide. Bars indicate the apparent affinities of Fab mutants relative to wild-type (w.t.) Fab b12 against gp120JR-FL, gp120IIIB, and the B2.1 peptide. The mutations P52aA and Y53G were back mutations to the residues encoded by the closest related germ line DNA (see Fig. 1).
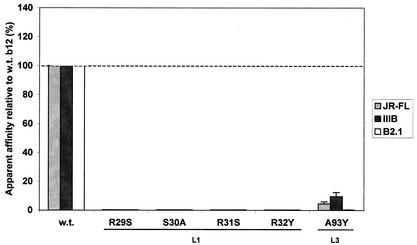
b12 L1 and L3 mutations.
Finally, the b12-gp120 docking model indicated that various residues in CDRs L1 and L3 could play a role in binding to gp120. Residues R29, R31, and R32 in L1 were chosen for substitution to S, S, and Y, respectively, because the closest germ line genes encode the substituted residues at these positions. S30 was changed simply to Ala because the closest germ line gene already encodes a Ser at this position. The results indicate that all four of these L1 mutants were essentially unable to recognize gp120 (Fig. 6) and suggest that somatic mutations found in this loop are essential to b12 specificity. Two mutations were made in L3 (A93Y and S94A), also based on predictions of their interaction with gp120. The A93Y mutant suffered a significant loss in gp120 binding (Fig. 6). Fab A93Y was purified and used in a competition ELISA with biotinylated Fab b12 against gp120IIIB; the IC50 of A93Y was ≈ 10 μg/ml, which is six to seven times greater than that of wild-type Fab b12 (IC50 ≈ 1.5 μg/ml) and consistent with the crude Fab ELISA. Unfortunately, the S94A mutant was found to be poorly produced in crude bacterial supernatants relative to the wild type (>10-fold reduction; data not shown), precluding a quantitative analysis of binding of the latter Fab mutant. Nevertheless, binding of the S94A Fab mutant to gp120 was detectable despite the low concentration of Fab in the supernatants, and we were able to conclude that the S94A mutation is at least not a complete knockout mutation (data not shown).
FIG. 6.
Effect of substitutions in the CDR loops L1 and L3 of b12 on the relative binding of Fab b12 to gp120JR-FL, gp120IIIB, and the B2.1 peptide. Bars indicate the apparent affinities of Fab mutants relative to wild-type (w.t.) Fab b12 for gp120JR-FL, gp120IIIB, and the B2.1 peptide. The mutations R29S, R31S, and R32Y were back mutations to the residues encoded by the closest related germ line DNA (see Fig. 1).
Whereas the mutations in L1 completely knocked out the binding of Fab b12 to both B2.1 and gp120 in our assay, the L3 mutation, A93Y, abolished binding to B2.1, but binding to gp120 was maintained, albeit at ≈10% of wild-type levels (Fig. 7). It is unclear whether the residues in L1 contact gp120 or if their mutation disrupts the b12 paratope. However, it appears that A93Y is important for B2.1 recognition because the paratope of the A93Y mutant was sufficiently intact to bind to gp120 with measurable affinity (Fig. 7). As for the poorly produced Fab mutant S94A, the result was similar for B2.1 as for gp120; at most, only a moderate effect on B2.1 binding was expected for this mutation (data not shown), although a quantitative analysis was not attempted.
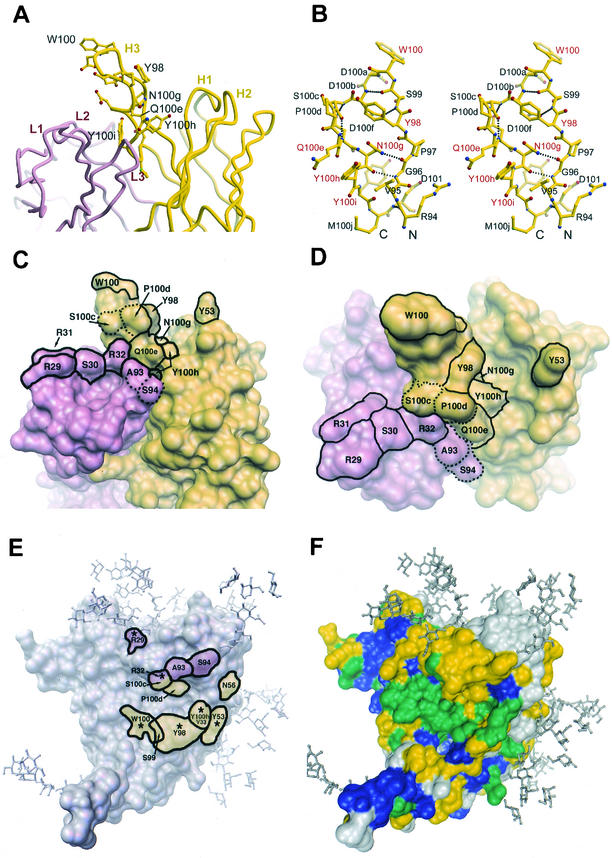
FIG.7.
Molecular features of the paratope of b12. (A) Tube diagram (36) of the combining site of Fab b12, showing the position of the protruding H3 loop relative to the other CDRs. Residues in H3 for which replacement by Ala resulted in a ≥95% decrease in apparent affinity to gp120 relative to wild-type (w.t.) b12 are labeled. (B) Stereo diagram of a ball-and-stick representation (24, 36) of the H3 loop of b12. The key residues that were labeled in A are labeled in red. (C and D) Molecular surface rendering (64, 65) of the b12 paratope (C, side view; D, top view). The light chain is colored pink, and the heavy chain is colored yellow. Residues that upon substitution caused a ≥95% decrease in apparent affinity to gp120 relative to wild-type b12 are indicated (solid outline). Note that residue Y100i is buried. Residues S100c, P100d, A93, and S94 (dotted outline) are shown for facile comparison with panel E. The b12 structure is taken from Saphire et al. (66). (E) Crystal structure of the gp120 core (39) with the residues of b12 that are predicted from the docking model (66) to be in close proximity to the outlined region on gp120. Thus, the labeled residues are those of b12, not gp120. Putative footprints in pink and yellow are from light- and heavy-chain residues, respectively. Asterisks (*) indicate the predicted contact residues that were also found to be critical for gp120 recognition by mutational analysis in this study, as defined in panels C and D. Note: a crystal structure of b12 in complex with core gp120 is as yet unavailable. (F) Sequence conservation map of core gp120 as defined by Kwong et al. (39). Residues in blue are conserved among all primate retroviruses, residues in green are conserved among all HIV-1 isolates, residues in yellow are moderately conserved among all HIV-1 isolates, and residues in grey are variable. Carbohydrate has been modeled onto the core structures (39) of gp120.
Neutralization of HIV-1 by synthetic CDR H3 b12 peptide.
We recently reported the neutralization of the T-cell-line-adapted strains HIV-1MN and HIV-1IIIB by a conjugate of BSA and a synthetic peptide corresponding to the H3 loop of b12 (66). This neutralizing activity against HIV-1MN and HIV-1IIIB was specific because a conjugate of BSA and an irrelevant peptide did not neutralize virus (66). We present here some additional observations. Whereas neutralization of HIV-1 was found for HIV-1MN and HIV-1IIIB in H9 cells at ≤1 mg of conjugate per ml (IC75 ≈ 0.5 mg/ml for each [66]), no neutralization at 4 mg of the peptide-BSA conjugate per ml was observed for the primary isolate HIV-1JRFL in a peripheral blood mononuclear cell assay (data not shown). It should be noted that, in our hands, neutralization (IC90) of HIV-1JRFL occurs at ≈0.8 μg of IgG1 b12 per ml (94), so HIV-1JRFL is as sensitive to neutralization by IgG1 b12 as HIV-1IIIB (IC90 ≈ 0.5 μg/ml). Thus, although T-cell-line-adapted viruses were neutralized at high concentrations of the b12 H3 peptide conjugate, the primary isolate, HIV-1JRFL was not neutralized by the same conjugate.
We also wished to determine whether a direct interaction between the b12 H3 peptide and recombinant gp120 could be established. By direct ELISA, no specific interaction between the b12 H3 peptide or the b12 H3 peptide-BSA conjugate and gp120 was detected by immobilizing either the peptide or recombinant gp120 (JRFL and IIIB) and then probing with the partnering molecule (data not shown). Similarly, by competition ELISA, high concentrations of b12 H3 peptide (0.5 mg/ml) or b12 H3 peptide-BSA conjugate (4 mg/ml) did not inhibit the ELISA signal generated by Fab b12 against immobilized recombinant gp120 (JRFL and IIIB; data not shown). Thus, it appears that, although the b12 H3 peptide-BSA conjugate neutralizes the T-cell-line-adapted viruses HIV-1MN and HIV-1IIIB, we could not demonstrate a specific interaction between the conjugate and recombinant gp120, at least by direct and competition ELISAs, which may not detect interactions with high micromolar to millimolar dissociation constants. These results may be explained at least in part by differences in the conformation of gp120 as it exists in the trimeric envelope spike, as probed in neutralization assays versus recombinant, plate-immobilized gp120, as probed by ELISA.
DISCUSSION
In the current study, we examined the antigen binding site of b12 with site-directed mutagenesis to create 50 mutations involving 27 different residues in four different CDRs of b12. We found that the use of crude Fab supernatants was reproducible and efficient, allowing the simultaneous analysis of a large number of Fab mutants against a panel of antigens. This type of mutational analysis can easily be adapted for other Fabs or single-chain Fvs that are amenable to production in E. coli, and we are currently using this approach for other antibodies against HIV-1. One potential drawback of the analysis is that if a substitution leads to diminished antigen binding, it is not known whether it is due to a direct effect on affinity for antigen or an effect on Fab stability or folding. However, we targeted residues in the CDR loops, most of which were at least partially exposed to solvent. For buried residues such as V95, N100g, and Y100i, it is possible that Ala substitution might partially unfold these mutants. Nevertheless, of the mutants that were chosen to be purified, W100A, Q100eA, N100gA, Y100hA, A93Y, and Y53G, most gave favorable yields of Fab and produced a single 50-kDa band by SDS-PAGE, suggesting that the Fabs were largely intact and not degraded. Moreover, the purified Fabs generally showed activities against gp120 very similar to those of the crude Fabs.
A very strong correlation was found for binding of the mutants to both gp120IIIB and gp120JR-FL. This correlation is consistent with a common binding mechanism of b12 to both the T-cell-line-adapted and primary isolate gp120 and is perhaps not surprising for an antibody with such broad reactivity to diverse HIV-1 envelopes (11, 54, 79). W100 was found to be important, as its substitution generally decreased binding of b12 to gp120. This result is also consistent with a prior experiment in which a portion of the gene segment encoding the H3 loop of b12, including W100, was randomized and incorporated into a phage display library and the library was affinity selected against gp120. A Trp residue at position 100 was absolutely conserved in all clones selected (5). These data caused us to speculate that perhaps a Trp residue (b12) might be superior to a Phe (CD4) in filling the hydrophobic pocket in gp120 that is important for CD4 receptor engagement (Phe43 is used to fulfill this role in CD4 [39]). However, an F43W mutant of CD4 did not show enhanced binding to gp120 but rather bound more poorly (i.e., the F43W mutant bound to gp120 at ≈9% of wild-type CD4 levels, and F43Y bound gp120 at ≈40% of wild-type CD4 levels; Raymond Sweet, personal communication). In addition to W100, the three remaining aromatics in the H3 loop of b12 appeared to be important for gp120 recognition; mutation of any of four aromatics (Y98, W100, Y100h, and Y100i) to Ala resulted in ≥95% decrease in relative binding to gp120.
By contrast, mutation to Ala of only one of four aromatics (i.e., residue Y2, as defined in Fig. 4) in the H3 loop of the poorly neutralizing anti-CD4bs antibody b6 resulted in a ≥95% decrease in relative binding to gp120 and then only to one of the two strains, IIIB (Fig. 4). In fact, for the Y2A mutant of Fab b6, a much greater binding differential was found between gp120 strains than for any of the 50 b12 mutants (Fig. 4). This result highlights a striking difference in the way that the aromatics of b12 and b6 are used to bind gp120, at least with respect to their H3 loops, even though the footprints of b6 and b12 on gp120 appear to be relatively similar (55). The H3 loop of an antibody is usually crucial in determining its specificity (82, 89). It may be that W100 (b12) and Y2 (b6), each of which is located near the middle of their long H3 loops, probe different regions within the CD4bs of gp120. Broadly neutralizing molecules such as b12 and CD4 might be able to occupy the conserved hydrophobic pocket of gp120, whereas b6 may not possess this ability. We speculate that the b6 H3 loop lacks rigidity and might lie across the CD4bs of gp120 rather than inserting into it like b12. Given that the Y2A mutation of b6 leads to significant strain preference (Fig. 4) but W100A of b12 does not, the Y2 residue could be interacting with or proximal to a region of gp120 that is relatively variable.
Recently, Zhu et al. showed that an engineered molecule (MBri) containing the V1/V2 loop and a portion of the bridging sheet of gp120 (i.e., β2, β3, β20, and β21) was able to bind to b12 (91). These authors showed that a V3 loop peptide is able to partially inhibit the binding of b12 to MBri and attributed this effect to a physical interaction between the V1/V2 loop and V3 loop. One might speculate that b12 is able to bind to gp120 in spite of an interaction between variable loops on the native trimer, whereas other anti-CD4bs antibodies, such as b6, cannot. In this vein, we have observed that most anti-CD4bs antibodies but not b12 inhibit the binding of a novel loop-dependent antibody to gp120 (MBZ; Robert Kelleher, Richard Jensen, Aran Labrijn, Meng Wang, Gerald Quinnan, Paul W. H. I. Parren, and Dennis R. Burton, submitted for publication), suggesting that the variable loops affect other anti-CD4bs antibodies in a manner different from how they affect b12.
Further comparative studies, both structural and functional, between b12 and other nonneutralizing anti-CD4bs antibodies like b6 could help elucidate the conformational differences between monomeric gp120 and trimeric gp120 on the HIV-1 envelope spike. In terms of vaccine design, these types of analyses should be extremely helpful to the design of improved gp120 constructs that would elicit b12-like antibodies by maximizing the exposure and immunogenicity of the b12 epitope while limiting the antibody response against overlapping epitopes targeted by b6 and other poorly neutralizing anti-CD4bs antibodies.
A molecular (ribbon) model of the Fab of b12 (Fig. 7A) illustrates the relative orientations of the CDRs and the prominent H3 loop. Figure 7B details the contour of the H3 loop and how the H3 polypeptide extends down on either side from W100 in an extended β-ladder with a distinct twist. The β-ladder is roughly 4 Å in width and contains five hydrogen bonds between strands; most notably, the main-chain carbonyl of G96 hydrogen bonds with the amino group of the side chain of N100g. Our data strongly suggest that this stabilizing hydrogen bond is crucial for the interaction of b12 with both gp120 and B2.1, since mutation of N100g to A, D, Q, H, or Y completely abolishes b12 binding to gp120 and B2.1. Another potentially loop-stabilizing hydrogen bond exists between Q100e and A93, and we found that changing Q100e to Ala, Asn, or Phe resulted in diminished binding to gp120. We note that in a recent and related study by McHugh et al. (47), it was shown by mutational analysis of a single chain Fv fragment corresponding to a b12 variant, 3B3-PE, that gp120 recognition was enhanced ≈2-fold by a Q100eY mutation. Thus, it appears that an aromatic residue at Q100e is compatible with strong gp120 binding in certain contexts.
The last few C-terminal residues of the H3 loop of b12 are encoded by DNA contributed by the joining region, which, in the case of b12, belongs to the family JH6 (4). The JH6 segment of DNA can potentially contribute up to six consecutive Tyr residues. We considered the possibility that this portion of the H3 loop of b12 may be a part of a more conserved and general structural motif, since a single segment of DNA encodes it as a group. We performed a database search (1) on the motif NYYMDV and found only three exact matches: one is an antihapten human heavy chain, VH-37 (accession no. S46393 [25]), and two are highly homologous antibodies against rhesus D blood group antigen (accession no. Y08177 and Y08186; S. M. Miescher, personal communication). Unfortunately, no structure for these antibodies is currently available to compare with that of IgG1 b12.
The strong dependence of b12 on a CDR of the light chain is consistent with a previous chain-shuffling experiment in which the heavy chain of b12 exclusively paired with the same light chain following affinity selection on gp120 (4). Significantly, of the six non-H3 mutations that were found to decrease gp120 binding by ≥90%, four arose via somatic mutation, and three of these were in L1. [The four substitutions, G53Y(H2), S29R, S31R, and Y32R, (L1) were determined to have arisen via somatic mutation on the basis of sequence alignment (http://imgt.cnusc.fr:8104/home.html) with known human germ line genes (see Fig. 1), and their positions in the paratope of b12 can be seen in Fig. 7C and D.] Although germ line antibodies can neutralize some viruses (35), given our results, it is very unlikely that a nonsomatically mutated version of b12 would have any HIV-1-neutralizing activity.
In a prior experiment, the gene segment encoding six residues in L1 (including R29, R31, and R32) was randomized and incorporated into a phage display library, and the library was affinity selected against gp120. Although there was positive selection for Arg residues in the enriched phage pools, the highest-affinity variant differed from b12 at every targeted position but R32, which was absolutely conserved in all the clones sequenced (90). The L1 sequence of the highest-affinity Fab is identical to that of H31L42, its whole-IgG counterpart (37) (Fig. 1). Note, however, that the multiple substitutions in CDR L1 of H31L42 occurred in the background of additional substitutions in H3 and H1, and the effects of most of these substitutions on affinity for gp120 were nonadditive (90).
A footprint of the putative contact residues of b12 on the deglycosylated core of gp120 (39), according to our docking model (66), is shown in Fig. 7E. The b12 residues whose substitution diminished gp120 binding by ≥95% relative to the wild type are indicated by asterisk. A useful guide in orienting the two molecules is the insertion of the “fingers” H3 (W100) and H2 (Y53) of b12 into the CD4 hydrophobic pocket and into a gap between T373 and N386 in gp120, respectively. The diminished gp120 binding of mutants W100A, Y98A, and Y53G are supportive of and supplement the model.
We postulate that W100 and Y53 contribute to the binding energy of b12 to gp120 largely by burying into a hydrophobic pocket and canyon, respectively, on either side of a ridge including S365 and D368 on gp120. Y98 of b12 is also very close to the S365-D368 ridge, which has been shown to be important for b12 binding by mutational analysis (55). Residues R29 and R32 in L1 are in close proximity to residues N276/K282 and N280/A281 in the D-loop of gp120, respectively. A recent mutational analysis of gp120 shows that substitutions N276A, K282A, and N280A slightly enhance, diminish, and have no effect on the binding of b12 to gp120, respectively (55).
From the crystal structure of b12, we calculated that the side chain of R32 was only partially accessible to solvent (Fig. 1), suggesting that the observed effect of the R32Y substitution may be due more to changes in local paratope structure than to direct contact with gp120. By contrast, the side chain of R29 is mostly accessible to solvent, suggesting that this residue might contact gp120. Alternatively, long-range electrostatic effects due to the cluster of basic residues in L1 might play a significant role in the observed effects caused by replacing any one of these b12 residues. N56 of b12 docks in close proximity to K362 of gp120; however, the N56 side chain points away from the hydrophobic canyon into which Y53 is situated, potentially explaining the absence of an effect on gp120 binding of substituting this residue. We would also add the caveat that some Ala substitutions can be energetically neutral in receptor-ligand interactions, despite replacing contact residues, as found for example in the Fab D1.3-hen egg white lysozyme complex (18). Exactly how the b12 residues discussed above spatially relate to residues in gp120 will require determination of a crystal structure of b12 in complex with gp120.
Despite the extreme differences between gp120 (≈120 kDa, heavily glycosylated protein) and the B2.1 peptide (dimer ≈ 4.3 kDa, nonglycosylated peptide), there are many similarities in the effects of b12 mutations on its binding to these antigens: 36 mutations had qualitatively similar effects, whereas 14 had different effects. A recently solved crystal structure of Fab b12 in complex with the B2.1 peptide should help in identifying which of these residues represent differential contacts for B2.1 and gp120 (E. O. Saphire, M. Montero, A. Menendez, M. B. Irving, M. B. Zwick, P. W. H. I. Parren, D. R. Burton, J. K. Scott, and I. A. Wilson, submitted for publication). The D100fE mutation, in particular, is interesting because it is conservative yet it severely impairs b12 binding to B2.1, whereas gp120 binding is almost unchanged. In addition, mutations D100aA, D100bA, 3D3A, and 3D2N all enhance b12 binding to B2.1 but reduce binding to gp120, suggesting that the “acidic patch” on the H3 loop (66) is more favorable for binding to gp120 than to B2.1. It is also noteworthy that binding of the W100 mutants to the B2.1 peptide was the same as wild-type b12, which may be expected since B2.1 does not have a deep hydrophobic cavity like gp120.
IgG1 b12 is one of a very few antibodies that exhibit potent cross-isolate anti-HIV-1 neutralizing activity (7, 8, 11, 21, 37, 79). It is therefore instructive to gather structural and functional information with respect to the neutralizing activity of IgG1 b12 in the hope of learning how to reproducibly elicit b12-like antibodies. From the antibody perspective, a question that surfaces in regard to this goal is how close to b12 does an antibody need to be in order to have the same ability to neutralize HIV-1. Two possibilities may exist: b12 is a unique specificity that cannot be reproduced with antibodies with low sequence homology to b12, or the specificity of b12 may be reproduced by a wide spectrum of nonhomologous antibodies.
Obviously, the former possibility would present a much greater challenge to template-driven vaccine design in that only anti-CD4bs antibodies that use the same germ line genes as b12 and have the correct critical residues in the CDRs would be broadly HIV-1 neutralizing. Some of these crucial residues are encoded by non-germ line DNA sequences. However, one of the very hallmarks of the humoral immune response is its ability to devise novel solutions to biomolecular recognition with molecules that have low sequence homology (i.e., the variable regions of antibodies) yet can still recognize the same epitope. Such plasticity in antigen recognition has been observed in other ligand-receptor systems (84). Clearly, additional broadly neutralizing anti-CD4bs antibodies are sorely needed in order to determine whether or not the key molecular features of b12, such as the long and rigid CDR H3 finger, the positively charged shelf-like structure in L1, and the high level of somatic mutation in b12, are required of other anti-CD4bs antibodies in order to be as broadly neutralizing against HIV-1.
Acknowledgments
We thank Ray Sweet for sharing mutagenesis data on CD4, Robert Kelleher and Nienke van Houten for excellent technical assistance, and Liang Yan for assistance with peptide synthesis. We also thank Bill Olson and Paul Maddon (Progenics, Tarrytown, N.Y.) for the kind gift of gp120JRFL.
We acknowledge support from the Elizabeth Glaser Pediatric AIDS Foundation and the Natural Sciences and Engineering Research Council of Canada (M.B.Z.); MH62261 (P.E.D.); AI49111, AI49808, and MRC HOP14562 (J.K.S.); GM46192 (I.A.W.); AI40377 (P.W.H.I.P.); and AI33292 (D.R.B.). E.O.S. is a fellow of the Universitywide AIDS Research Program. D.R.B. and I.A.W. are supported by IAVI through the Neutralizing Antibody Consortium.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. [DOI] [PubMed] [Google Scholar]
- 3.Barbas, C. F., III, D. R. Burton, J. K. Scott, and G. J. Silverman. 2001. Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 4.Barbas, C. F., III, T. A. Collet, W. Amberg, P. Roben, J. M. Binley, D. Hoekstra, D. Cababa, T. M. Jones, R. A. Williamson, G. R. Pilkington, N. L. Haigwood, E. Cabezas, A. C. Satterthwait, I. Sanz, and D. R. Burton. 1993. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J. Mol. Biol. 230:812-823. [DOI] [PubMed] [Google Scholar]
- 5.Barbas, C. F., III, D. Hu, N. Dunlop, L. Sawyers, D. Cababa, R. M. Hendry, P. L. Nara, and D. R. Burton. 1994. In vitro evolution of a neutralizing human antibody to HIV-1 to enhance affinity and broaden stain cross-reactivity. Proc. Natl. Acad. Sci. USA 91:3809-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bures, R., L. Morris, C. Williamson, G. Ramjee, M. Deers, S. A. Fiscus, S. Abdool-Karim, and D. C. Montefiori. 2002. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J. Virol. 76:2233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton, D. R. 1997. A vaccine for HIV type 1: the antibody perspective. Proc. Natl. Acad. Sci. USA 94:10018-10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burton, D. R., C. F. Barbas III, M. A. A. Persson, S. Koenig, R. M. Chanock, and R. A. Lerner. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA 88:10134-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton, D. R., and J. P. Moore. 1998. Why do we not have an HIV vaccine and how can we make one? Nat. Med. 4:495-498. [DOI] [PubMed] [Google Scholar]
- 11.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. H. I. Parren, L. S. W. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, M. Lamacchia, E. Garratty, E. R. Stiehm, Y. J. Bryson, Y. Cao, J. P. Moore, D. D. Ho, and C. F. Barbas. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 12.Caffrey, M., M. L. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, D. G. Covell, A. M. Gronenborn, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17:4572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 14.Conley, A. J., J. A. Kessler II, L. J. Boots, P. M. McKenna, W. A. Schleif, E. A. Emini, G. E. Mark III, H. Katinger, E. K. Cobb, S. M. Lunceford, S. R. Rouse, and K. K. Murthy. 1996. The consequence of passive administration of an anti-human immunodeficiency virus type 1 neutralizing monoclonal antibody before challenge of chimpanzees with a primary virus isolate. J. Virol. 70:6751-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connor, R. I., D. C. Montefiori, J. M. Binley, J. P. Moore, S. Bonhoeffer, A. Gettie, E. A. Fenamore, K. E. Sheridan, D. D. Ho, P. J. Dailey, and P. A. Marx. 1998. Temporal analyses of virus replication, immune responses, and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J. Virol. 72:7501-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cormier, E. G., and T. Dragic. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox, J. P., I. M. Tomlinson, and G. Winter. 1994. A directory of human germ-line V kappa segments reveals a strong bias in their usage. Eur. J. Immunol. 24:827-836. [DOI] [PubMed] [Google Scholar]
- 18.Dall'Acqua, W., E. R. Goldman, W. Lin, C. Teng, D. Tsuchiya, H. Li, X. Ysern, B. C. Braden, Y. Li, S. J. Smith-Gill, and R. A. Mariuzza. 1998. A mutational analysis of binding interactions in an antigen-antibody protein-protein complex. Biochemistry 37:7981-7991. [DOI] [PubMed] [Google Scholar]
- 19.Dawson, P. E., T. W. Muir, I. Clark-Lewis, and S. B. Kent. 1994. Synthesis of proteins by native chemical ligation. Science 266:776-779. [DOI] [PubMed] [Google Scholar]
- 20.D'Souza, M. P., S. J. Geyer, C. V. Hanson, R. M. Hendry, and G. Milman. 1994. Evaluation of monoclonal antibodies to HIV-1 envelope by neutralization and binding assays: an international collaboration. AIDS 8:169-181. [PubMed] [Google Scholar]
- 21.D'Souza, M. P., D. Livnat, J. A. Bradac, S. Bridges, the AIDS Clinical Trials Group Antibody Selection Working Group, and Collaborating Investigators. 1997. Evaluation of monoclonal antibodies to HIV-1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 22.D'Souza, M. P., G. Milman, J. A. Bradac, D. McPhee, C. V. Hanson, R. M. Hendry, and Collaborating Investigators. 1995. Neutralisation of primary HIV-1 isolates by anti-envelope monoclonal antibodies. AIDS 9:867-874. [DOI] [PubMed] [Google Scholar]
- 23.Earl, P. L., C. C. Broder, R. W. Doms, and B. Moss. 1997. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J. Virol. 71:2674-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esnouf, R. M. 1999. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 55:938-940. [DOI] [PubMed] [Google Scholar]
- 25.Figini, M., J. D. Marks, G. Winter, and A. D. Griffiths. 1994. In vitro assembly of repertoires of antibody chains on the surface of phage by renaturation. J. Mol. Biol. 239:68-78. [DOI] [PubMed] [Google Scholar]
- 26.Graham, B. S., M. J. McElrath, R. I. Connor, D. H. Schwartz, G. J. Gorse, M. C. Keefer, M. J. Mulligan, T. J. Matthews, S. M. Wolinsky, D. C. Montefiori, S. H. Vermund, J. S. Lambert, L. Corey, R. B. Belshe, R. Dolin, P. F. Wright, B. T. Korber, M. C. Wolff, P. E. Fast, the AIDS Vaccine Evaluation Group, and the Correlates of HIV Immune Protection Group. 1998. Analysis of intercurrent human immunodeficiency virus type 1 infections in phase I and II trials of current AIDS vaccines. J. Infect. Dis. 177:310-319. [DOI] [PubMed] [Google Scholar]
- 27.Grundner, C., T. Mirzabekov, J. Sodroski, and R. Wyatt. 2002. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J. Virol. 76:3511-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hackeng, T. M., J. H. Griffin, and P. E. Dawson. 1999. Protein synthesis by native chemical ligation: expanded scope by with straightforward methodology. Proc. Natl. Acad. Sci. USA 96:10068-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helseth, E., U. Olshevsky, C. Furman, and J. Sodroski. 1991. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J. Virol. 65:2119-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho, D. D., J. A. McKeating, X. L. Li, T. Moudgil, E. S. Daar, N. C. Sun, and J. E. Robinson. 1991. Conformational epitope on gp120 important in CD4 binding and human immunodeficiency virus type 1 neutralization identified by a human monoclonal antibody. J. Virol. 65:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutchinson, E. G., and J. M. Thornton. 1994. A revised set of potentials for beta-turn formation in proteins. Protein Sci. 3:2207-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, R. P., and R. C. Desrosiers. 1998. Protective immunity induced by live attenuated simian immunodeficiency virus. Curr. Opin. Immunol. 10:436-443. [DOI] [PubMed] [Google Scholar]
- 33.Joshi, S. B., R. E. Dutch, and R. A. Lamb. 1998. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology 248:20-34. [DOI] [PubMed] [Google Scholar]
- 34.Kabat, E. A., T. T. Wu, H. M. Perry, K. S. Gottesman, and C. Foeller. 1991. Sequences of proteins of immunological interest. Department of Health and Human Services, Washington, D.C.
- 35.Kalinke, U., A. Oxenius, C. Lopez-Macias, R. M. Zinkernagel, and H. Hengartner. 2000. Virus neutralization by germ-line vs. hypermutated antibodies. Proc. Natl. Acad. Sci. USA 97:10126-10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 37.Kessler, J. A., P. M. McKenna, E. A. Emini, C. P. Chan, M. D. Patel, S. K. Gupta, G. E. Mark III, C. F. Barbas III, D. R. Burton, and A. J. Conley. 1997. Recombinant human monoclonal antibody IgG1 b12 neutralizes diverse human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retrovir. 13:575-581. [DOI] [PubMed] [Google Scholar]
- 38.Kostrikis, L. G., Y. Cao, H. Ngai, J. P. Moore, and D. D. Ho. 1996. Quantitative analysis of serum neutralization of human immunodeficiency virus type 1 from subtypes A, B, C, D, E, F, and I: lack of direct correlation between neutralization serotypes and genetic subtypes and evidence for prevalent serum-dependent infectivity enhancement. J. Virol. 70:445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laal, S., S. Burda, M. K. Gorny, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1994. Synergistic neutralization of human immunodeficiency virus type 1 by combinations of human monoclonal antibodies. J. Virol. 68:4001-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lefranc, M. P. 2001. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 29:207-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, A., H. Katinger, M. R. Posner, L. Cavacini, S. Zolla-Pazner, M. K. Gorny, J. Sodroski, T. C. Chou, T. W. Baba, and R. M. Ruprecht. 1998. Synergistic neutralization of simian-human immunodeficiency virus SHIV-vpu+ by triple and quadruple combinations of human monoclonal antibodies and high-titer anti-human immunodeficiency virus type 1 immunoglobulins. J. Virol. 72:3235-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic SHIV-89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mascola, J. R., M. K. Louder, S. R. Surman, T. C. VanCott, X. F. Yu, J. Bradac, K. R. Porter, K. E. Nelson, M. Girard, J. G. McNeil, F. E. McCutchan, D. L. Birx, and D. S. Burke. 1996. Human immunodeficiency virus type 1 neutralizing antibody serotyping with serum pools and an infectivity reduction assay. AIDS Res. Hum. Retrovir. 12:1319-1328. [DOI] [PubMed] [Google Scholar]
- 46.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 47.McHugh, L., S. Hu, B. K. Lee, K. Santora, P. E. Kennedy, E. A. Berger, I. Pastan, and D. H. Hamer. 2002. Increased affinity and stability of an anti-HIV-1 envelope immunotoxin by structure based mutagenesis. J. Biol. Chem. 277:34383-34390. [DOI] [PubMed] [Google Scholar]
- 48.Moog, C., H. J. A. Fleury, I. Pellegrin, A. Kirn, and A. M. Aubertin. 1997. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J. Virol. 71:3734-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore, J. P., Q. J. Sattentau, R. Wyatt, and J. Sodroski. 1994. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J. Virol. 68:469-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moulard, M., S. K. Phogat, Y. Shu, A. F. Labrijn, X. Xiao, J. M. Binley, M. Y. Zhang, I. A. Sidorov, C. C. Broder, J. Robinson, P. W. H. I. Parren, D. R. Burton, and D. S. Dimitrov. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. USA 99:6913-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Rüker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nunberg, J. H., K. E. Follis, M. Trahey, and R. A. LaCasse. 2000. Turning a corner on HIV neutralization? Microbes Infect. 2:213-221. [DOI] [PubMed] [Google Scholar]
- 54.Nyambi, P. N., H. A. Mbah, S. Burda, C. Williams, M. K. Gorny, A. Nadas, and S. Zolla-Pazner. 2000. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J. Virol. 74:7096-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pantophlet, R., E. O. Saphire, P. Poignard, P. W. H. I. Parren, I. A. Wilson, and D. R. Burton. 2003. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77:642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parren, P. W. H. I., H. J. Ditzel, R. J. Gulizia, J. M. Binley, C. F. Barbas III, D. R. Burton, and D. E. Mosier. 1995. Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS 9:F1-F6. [DOI] [PubMed] [Google Scholar]
- 57.Parren, P. W. H. I., P. Fisicaro, A. F. Labrijn, J. M. Binley, W. P. Yang, H. J. Ditzel, C. F. Barbas III, and D. R. Burton. 1996. In vitro antigen challenge of human antibody libraries for vaccine evaluation: the human immunodeficiency virus type I envelope. J. Virol. 70:9046-9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parren, P. W. H. I., P. A. Marx, A. J. Hessell, A. Luckay, J. Harouse, C. Cheng-Mayer, J. P. Moore, and D. R. Burton. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parren, P. W. H. I., I. Mondor, D. Naniche, H. J. Ditzel, P. J. Klasse, D. R. Burton, and Q. J. Sattentau. 1998. Neutralization of HIV-1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 72:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poignard, P., E. O. Saphire, P. W. H. I. Parren, and D. R. Burton. 2001. gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 61.Purtscher, M., A. Trkola, G. Gruber, A. Buchacher, R. Predl, F. Steindl, C. Tauer, R. Berger, N. Barrett, A. Jungbauer, and H. Katinger. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 10:1651-1658. [DOI] [PubMed] [Google Scholar]
- 62.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanner, M. F., B. S. Duncan, C. J. Carrillo, and A. J. Olson. 1999. Integrating computation and visualization for biomolecular analysis: an example with PYTHON and AVS, p. 401-412. In R. B. Altman, K. Lauderdale, A. K. Dunker, L. Hunter, and T. E. Klein (ed.), Biocomputing '99: Proceedings of the Pacific Symposium. World Scientific Press, Mauna Lani, Hawaii. [DOI] [PubMed]
- 65.Sanner, M. F., A. J. Olson, and J. C. Spehner. 1996. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38:305-320. [DOI] [PubMed] [Google Scholar]
- 66.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 67.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sattentau, Q. J., M. Moulard, B. Brivet, F. Botto, J. C. Cuillemot, I. Mondor, P. Poignard, and S. Ugolini. 1999. Antibody neutralization of HIV-1 and the potential for vaccine design. Immunol. Lett. 66:143-149. [DOI] [PubMed] [Google Scholar]
- 69.Sattentau, Q. J., S. Zolla-Pazner, and P. Poignard. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206:713-717. [DOI] [PubMed] [Google Scholar]
- 70.Scanlan, C. N., R. Pantophlet, M. R. Wormwald, E. O. Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schnolzer, M., P. Alewood, A. Jones, D. Alewood, and S. B. Kent. 1992. In situ neutralization in Boc-chemistry solid phase peptide synthesis. Rapid, high yield assembly of difficult sequences. Int. J. Peptide Protein. Res. 40:180-183. [DOI] [PubMed] [Google Scholar]
- 72.Schønning, K., A. Bolmstedt, J. Novotny, O. S. Lund, S. Olofsson, and J. E. S. Hansen. 1998. Induction of antibodies against epitopes inaccessible on the HIV type 1 envelope oligomer by immunization with recombinant monomeric glycoprotein 120. AIDS Res. Hum. Retroviruses 16:1451-1456. [DOI] [PubMed] [Google Scholar]
- 73.Shibata, R., T. Igarashi, N. Haigwood, A. Buckler-White, R. Ogert, W. Ross, R. Willey, M. W. Cho, and M. A. Martin. 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 5:204-210. [DOI] [PubMed] [Google Scholar]
- 74.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 75.Thali, M., C. Furman, D. D. Ho, J. Robinson, S. Tilley, A. Pinter, and J. Sodroski. 1992. Discontinuous, conserved neutralization epitopes overlapping the CD4 binding region of the HIV-1 gp120 envelope glycoprotein. J. Virol. 66:5635-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thali, M., U. Olshevshy, C. Furman, D. Gabuzda, M. Posner, and J. Sodroski. 1991. Characterization of a discontinuous human immunodeficiency virus type 1 gp120-epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J. Virol. 65:6188-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tilley, S. A., W. J. Honnen, M. E. Racho, M. Hilgartner, and A. Pinter. 1991. A human monoclonal antibody against the CD4-binding site of HIV-1 gp120 exhibits potent, broadly neutralizing activity. Res. Virol. 142:247-259. [DOI] [PubMed] [Google Scholar]
- 78.Tomlinson, I. M., G. Walter, J. D. Marks, M. B. Llewelyn, and G. Winter. 1992. The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J. Mol. Biol. 227:776-798. [DOI] [PubMed] [Google Scholar]
- 79.Trkola, A., A. P. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. Allaway, H. Katinger, C. F. Barbas III, D. R. Burton, D. D. Ho, and J. P. Moore. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type I. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.VanCott, T. C., J. R. Mascola, L. D. Loomis-Price, F. Sinangil, N. Zitomersky, J. McNeil, M. L. Robb, D. L. Birx, and S. Barnett. 1999. Cross-subtype neutralizing antibodies induced in baboons by a subtype E gp120 immunogen based on an R5 primary HIV-1 envelope. J. Virol. 73:4640-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.VanDyk, L., and K. Meek. 1992. Assembly of IgH CDR3: mechanism, regulation, and influence on antibody diversity. Int. Rev. Immunol. 8:123-133. [DOI] [PubMed] [Google Scholar]
- 83.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 84.Wilson, I. A., and L. K. Jolliffe. 1999. The structure, organization, activation and plasticity of the erythropoietin receptor. Curr. Opin. Struct. Biol. 9:696-704. [DOI] [PubMed] [Google Scholar]
- 85.Wrin, T., L. Crawford, L. Sawyer, P. Weber, H. W. Sheppard, and C. V. Hanson. 1994. Neutralizing antibody responses to autologous and heterologous isolates of human immunodeficiency virus. J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 7:211-219. [PubMed] [Google Scholar]
- 86.Wyatt, R., E. Desjardin, U. Olshevsky, C. Nixon, J. Binley, V. Olshevsky, and J. Sodroski. 1997. Analysis of the interaction of the human immunodeficiency virus type 1 grp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J. Virol. 71:9722-9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 88.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 89.Xu, J. L., and M. M. Davis. 2000. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 13:37-45. [DOI] [PubMed] [Google Scholar]
- 90.Yang, W. P., K. Green, S. Pinz-Sweeney, A. T. Briones, D. R. Burton, and C. F. Barbas III. 1995. CDR walking mutagenesis for the affinity maturation of a potent human anti-HIV-1 antibody into the picomolar range. J. Mol. Biol. 254:392-403. [DOI] [PubMed] [Google Scholar]
- 91.Zhu, C. B., L. Zhu, S. Holz-Smith, T. J. Matthews, and C. H. Chen. 2001. The role of the third beta strand in gp120 conformation and neutralization sensitivity of the HIV-1 primary isolate DH012. Proc. Natl. Acad. Sci. USA 98:15227-15232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zolla-Pazner, S., M. K. Gomy, and P. N. Nyambi. 1999. The implications of antigenic diversity for vaccine development. Immunol. Lett. 66:159-164. [DOI] [PubMed] [Google Scholar]
- 93.Zwick, M. B., L. L. C. Bonnycastle, A. Menendez, M. B. Irving, C. F. Barbas III, P. W. H. I. Parren, D. R. Burton, and J. K. Scott. 2001. Identification and characterization of a peptide that specifically binds the human, broadly neutralizing anti-human immunodeficiency virus type 1 antibody b12. J. Virol. 75:6692-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. H. I. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zwick, M. B., M. Wang, P. Poignard, G. Stiegler, H. Katinger, D. R. Burton, and P. W. H. I. Parren. 2001. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J. Virol. 75:12198-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]