Abstract
Salicylic acid (SAL) may impact Staphylococcus aureus virulence by activating the sigB operon (rsbU-V-W-sigB), thus leading to reductions in alpha-toxin production and decreased fibronectin binding (L. I. Kupferwasser et al., J. Clin. Investig. 112:222-233, 2003). As these prior studies were performed in strain RN6390 (an rsbU mutant) and its rsbU-repaired variant, SH1000, the current investigation was designed to determine if the SAL effect occurs via rsbU- and/or rsbV-dependent pathways in an rsbU-intact S. aureus strain (FDA486). We thus quantified the transcription from two sigB-dependent promoters (asp23 and sarA P3) in FDA486 in response to SAL exposure in vitro, using isogenic single-knockout constructs of rsbU, rsbV, or rsbW and a green fluorescent protein reporter system. SAL induced sarA P3 and asp23 promoter activities in a dose-dependent manner in the parental strain. In contrast, sigB activation by SAL was progressively more mitigated in the rsbU and rsbV mutants. As predicted, SAL caused significant reductions in both alpha-toxin production and fibrinogen and fibronectin binding in the parental strain. The extent of these reductions, compared with the parent, was reduced in the rsb mutants (rsbV > rsbU), especially at low SAL concentrations. Since generation of the free SigB protein usually requires a sequential rsbU-V-W-sigB activation cascade, the present phenotypic and genotypic data suggest key roles for both rsbU and rsbV in SAL-mediated activation of sigB in strains with a fully intact sigB operon.
The ability of Staphylococcus aureus to survive stress conditions is attributable to activation of stress-response regulatory elements, particularly sigma factor B (sigB) (10, 11). Such stress conditions include extremes of temperature, pH, and osmolarity, as well as nutrient limitations, ethanol exposures, and oxygen depletion in vitro (10, 11). The response regulator sigB can control a number of downstream gene promoters that possess a SigB recognition motif (12). Thus, during in vitro growth in routine laboratory medium, sigB activation can modulate the expression of the global regulator, sarA, by enhancing activation of the sarA P3 promoter (4, 6). We recently showed that salicylic acid (SAL), the major in vivo biometabolite of aspirin in humans, down-modulates the virulence of S. aureus in experimental infection models (e.g., infective endocarditis [IE]) by mechanisms that are largely independent of the antiplatelet effects of aspirin (17). These studies indicated that SAL was a potent activator of the sigB operon (rsbU-V-W-sigB) both in vitro and in vivo. In turn, such activation was associated with a reduction in SarA protein expression and mitigation of agr- and sarA-dependent gene expression (e.g., fnbA and hla) (3, 6). The phenotypic consequences of these genotypic effects were reduced production of alpha-toxin and reduced binding to fibronectin and fibrinogen (17). Reductions in these phenotypes have been demonstrated to be critically involved in decreased S. aureus virulence in experimental IE (17).
An interesting aspect of the above investigations was that SAL could activate sigB in strains containing either an intact or defective rsbU locus within the sigB operon, thus implicating both rsbU-dependent and rsbU-independent pathways for activation (17). The current study was designed to further examine the role of rsbU, rsbV, and rsbW in mediating the impact of SAL on sigB activation as well as upon sigB-modulated downstream virulence phenotypes. For these investigations, we utilized strategic single-knockout constructs within the sigB operon of a sigB-intact parental strain.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. FDA486 is an rsbU + S. aureus strain previously used to study expression of fibrinogen-binding proteins and sigB-dependent transcription of selected target genes (24). RN4220 is a restriction-deficient S. aureus host strain used as the initial recipient for transformation of plasmids (22). Escherichia coli XL1-Blue was used for cloning isolated DNA fragments.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description or construction | Source or reference |
|---|---|---|
| Strains | ||
| E. coli XL-1 Blue | Highly transformable strain | Stratagene |
| S. aureus | ||
| FDA486 | Wild-type strain with intact rsbU gene | 24 |
| RN4220 | Restriction-deficient derivate of 8325-4 | 22 |
| ALC2128 | rsbU mutant of FDA486 | 24 |
| ALC2129 | rsbV mutant of FDA486 | 24 |
| ALC2130 | rsbW mutant of FDA486 | This study |
| FDA486 sarA P3-gfp | FDA486 with sarA P3 promoter driving gfpuvr in pSK236 | This study |
| ALC2128 sarA P3-gfp | rsbU mutant with sarA P3 promoter driving gfpuvr in pSK236 | This study |
| ALC2129 sarA P3-gfp | rsbV mutant with sarA P3 promoter driving gfpuvr in pSK236 | This study |
| ALC2130 sarA P3-gfp | rsbW mutant with sarA P3 promoter driving gfpuvr in pSK236 | This study |
| ALC2322 | FDA486 with asp23 promoter driving gfpuvr in pSK236 | 17 |
| ALC2128 asp23-gfp | rsbU mutant with asp23 promoter driving gfpuvr in pSK236 | This study |
| ALC2129 asp23-gfp | rsbV mutant with asp23 promoter driving gfpuvr in pSK236 | This study |
| ALC2130 asp23-gfp | rsbW mutant with asp23 promoter driving gfpuvr in pSK236 | This study |
| Plasmids | ||
| pCL52.2 | Shuttle vector with temperature-sensitive origin of replication for S. aureus | 19 |
| pALC552 | pBluescript containing the ermC gene | 24 |
| pΔrsbW | This study | |
| pSK236 | Shuttle vector containing pUC19 cloned into the HindIII site of pC194 | 20 |
| pALC1484 | pSK236 containing gfpuvr | |
| pALC1672 | pSK236 containing sarA P3::gfpuvr | This study |
| pALC2201 | pSK236 containing asp23::gfpuvr | This study |
Allelic replacement in S. aureus strain FDA486.
The construction of rsbU and rsbV mutants in FDA486 has been described in a previous study (24). We also constructed an rsbW mutant of FDA486. Briefly, a region upstream of rsbW was amplified with the primers 5′-GCTGGAATTCCGCCTGGATATATTTATC-3′ and 5′-TTCGCCCGGGGTTCCTTCATTAACATGC-3′ (EcoRI and SmaI sites, respectively, are underlined) and then digested with EcoRI and SmaI. The fragment was cloned into the EcoRI-SmaI sites of the temperature-sensitive shuttle vector pCL52.2 (24). A region downstream of rsbW was amplified with the following primers with flanking PstI and HindIII restriction sites (underlined), respectively: 5′-TAAACTGCAGGAGCAGGTGCGAAATAAT-3′ and 5′-TGCCAAGCTTTGTAATTTCTTAATTGCC-3′. The downstream fragment was digested with PstI and HindIII and cloned into the PstI-HindIII sites of pCL52.2, already containing the upstream fragment. The ermC gene was excised from pALC552 and introduced into the BamHI-SalI site of pCL52.2 (24), thus resulting in divergent transcription of the ermC gene from the rest of the sigB operon. The constructed plasmid was electroporated into S. aureus RN4220 to select for tetracycline-resistant colonies (3 μg/ml) at 32°C. The recombinant pCL52.2 was then transduced from RN4220 into strain FDA486 with phage φ11. Transductants were selected on tryptic soy agar plates containing tetracycline at 32°C. One clone containing the plasmid was grown in 0.3 GL broth (0.3 M glycerol-lactate/BHI broth) (21, 23a) with tetracycline (3 μg ml−1) and erythromycin (5 μg ml−1) at 32°C overnight, followed by 12 h of growth in medium with erythromycin at 42°C, the nonpermissive temperature for pCL52.2 replication. The culture was diluted 1:100 into fresh 0.3 GL broth and incubated at 32°C without antibiotics for 12 h; this step was repeated several times. Erythromycin-resistant but tetracycline-sensitive colonies were selected for further analysis. To validate the authenticity of the rsbW mutant, we used a primer (5′-ATGGTCTATTTCAATGGCAGTTAC-3′) corresponding to bases 331 to 335 of the ermC gene (FASTA search; GenBank accession no. Y17294) in combination with a primer downstream of the sigB gene (5′-AATATCCTTCTTTAATTCCTCAGTA-3′) to confirm replacement of rsbW with ΔrsbW::ermC by PCR. The resultant PCR fragment was sequenced with identical primers. As an additional confirmation, chromosomal DNA from parental strain FDA486 and the rsbW deletion mutant strains was digested with EcoRV and probed in Southern hybridization experiments with labeled PCR fragments of individual genes within the sigB operon. To confirm that the rsbU and rsbV mutations did not lead to polar mutations downstream in the operon, we conducted Western blot analysis of cell extracts of FDA486 and its isogenic rsbU, rsbV, and rsbW mutants with anti-SigB monoclonal antibody 1D1 as previously described (6).
Gene expression studies and assay for sigB activation by SAL.
The function of the sigB operon depends on the generation of free SigB protein, i.e., upon the release of SigB from the normally inactive RsbW-SigB complex. This release is facilitated by the competitive binding of RsbW by the dephosphorylated form of RsbV upon activation by RsbU (11, 14, 21). Since the total pool of the SigB protein is relatively constant (free plus bound SigB), the extent of the level of free SigB is typically assessed by quantifying expression of one or more sigB-dependent promoters (6, 9). For this reason, we determined the expression of the sigB-dependent sarA P3 and the alkaline shock protein (asp23) promoters by green fluorescent protein (GFP) reporter and Northern blot analyses in the presence or absence of SAL. The asp23 promoter has been previously used as a faithful surrogate for sigB activation (9, 17).
(i) GFP reporter assays.
To monitor sarA P3 promoter activation in the presence or absence of SAL, we used a promoterless red-shifted variant of the gfpuv gene (gfpuvr), as previously detailed (15). This gene was cloned downstream of the sarA P3 promoter in plasmid pALC1484, which was then electroporated into the parental strain FDA486, as well as into various rsb mutants described above. The gfpuvr fusion constructs allow detection and quantification of the upstream promoter activity by fluorometric techniques (28). All rsb mutant constructs grew at the same rate as the wild-type parental strain. Also, the rsb mutant constructs bearing the plasmid containing gfpuvr grew at similar rates in the presence of chloramphenicol (10 μg/ml). Moreover, ethidium bromide-stained gels of plasmids among the wild-type strain and rsb mutant constructs revealed similar intensities when equivalent numbers of bacterial cells were analyzed. These latter two observations underscore the equivalence of plasmid copy numbers in the parental strain and mutant constructs. To monitor sigB activation of the sarA P3 promoter (a relatively weak promoter) in the presence or absence of SAL, we utilized an FL600 microplate fluorescence reader (Bio-Tek Instrument, Winooski, VT) with 485/516 nm filters. To monitor sigB activation of the asp23 promoter (a strong promoter) in the presence or absence of SAL, we utilized a previously published, standard fluorometric assay (Turner Fluorometer; Dubuque, IA) (28). As with the sarA P3 construct above, the asp23 promoter was cloned upstream of the gfpuvr gene on plasmid pALC1484 and introduced into the FDA486 strain set as described. For the sarA P3 construct, GFP expression was quantified at 0, 2, 12, and 28 h of growth at 37°C with shaking (200 rpm). For the asp23 construct, aliquots were obtained at 0, 6, 16, and 24 h. Extensive pilot studies had indicated that these distinct sampling times provided the maximal separation in promoter expression profiles between untreated versus SAL-treated cells (data not shown). No antibiotics were included in the growth medium for gfp constructs.
(ii) Northern blot hybridization.
To verify the putative importance of rsbU in activation of the sigB operon by SAL, we performed Northern blotting to monitor sarA P3 and asp23 transcriptions. RNA samples were obtained during early postexponential phase, at an optical density at 650 nm (OD650) nm of 1.7 (∼6 h of growth), using an 18-mm borosilicate glass tube in a Spectronic 20 spectrophotometer. Twenty micrograms of total cellular RNA from FDA486 and its isogenic mutants (strains carrying rsbU, rsbV and rsbW mutations) were electrophoresed through a 1.5% agarose-0.66 M formaldehyde gel in running buffer (20 mM morpholinepropanesulfonic acid, 10 mM sodium acetate, 2 mM EDTA, pH 7.0). Blotting of RNA onto Hybond N+ membranes (Amersham, Arlington Heights, IL) was performed with a Turbo-blotter alkaline transfer system (Scheicher and Schuell, Inc., Keene, NH). The intensities of the 23S and 16S rRNA bands stained by ethidium bromide were verified to be equivalent among samples prior to transfer. A [32P]dCTP-labeled sarA fragment was used to detect sarA transcript as described previously (2). We utilized specific primers (5′-TAGGTATTGGGTATATGAAAGA-3′ and 5′-TTGTCTTTCTTGGTTATTGTTT-3′) to amplify a 624-bp fragment to be used as a probe to detect asp23 expression. All the probes were radiolabeled by the random primer method (Ready-To-Go labeling kit; Amersham). The blots were hybridized under high-stringency conditions, washed, and autoradiographed as previously described (7).
Phenotypic studies. (i) Fibrinogen and fibronectin binding assays.
We have previously reported that SAL mitigates S. aureus binding to solid-phase fibrinogen and fibronectin biomatrices in vitro (17). To assess the effects of various rsb mutations on this SAL-mediated phenotype, the parent and all mutant constructs were grown to postexponential phase for maximal sigB expression (9) in the presence or absence of SAL (at 25 or 50 μg/ml). Following pelleting, washing, and bovine serum albumin blocking steps (17), 5 × 103 CFU of each construct were added to six-well polystyrene plates precoated with 50 μg/ml of either fibrinogen or fibronectin. Prior to the addition, the bacterial inoculum was briefly sonicated to ensure singlet cells and then allowed to bind to the above biomatrices for 1 h at 37°C on a rotating platform. After unbound cells were removed by three washes with phosphate-buffered saline, 2 ml of tryptic soy agar was overlaid in all wells. Plates were incubated at 37°C for 24 h, when all visible colonies were counted. Bacterial binding was quantified as the percentage of the initial inoculum bound in the presence or absence of SAL. Data were calculated as the means (± standard deviation [SD]) of three independent runs and expressed as the mean percent reductions of fibrinogen or fibronectin binding under various assay conditions.
(ii) Alpha-toxin activity in the presence of SAL.
To monitor the impact of SAL on the production of alpha-toxin in the parental strain versus various rsb mutants, we employed a well-established phenotypic assay in which the ability of alpha-toxin to lyse rabbit erythrocytes was measured (17). S. aureus strains were grown at 37°C in tryptic soy broth (control cells) or in medium containing 25 or 50 μg/ml SAL for 18 h to stationary phase on a rotary shaker. After cells were pelleted at 5,000 × g for 10 min, the number of cells in each tube was standardized by spectrophotometry, and aliquots of serial dilutions of culture supernatants were added to a 1% suspension of washed rabbit erythrocytes in 0.01 M phosphate-buffered saline (pH 7.2) containing 0.1% bovine serum albumin. Purified alpha-toxin (1 μg/ml; Toxin Technology, Sarasota, FL) was used as a positive control. Data were expressed as mean units of hemolytic activity (± SD) per ml of culture supernatant from six separate runs. The hemolytic units were defined as the reciprocal of the highest dilution of the culture supernatant causing ≥50% erythrocyte lysis as measured by optical densitometry (17).
Statistics.
Continuous data were statistically analyzed by a Kruskal-Wallis analysis of variance, with corrections for multiple comparisons where appropriate. A P value of <0.05 was considered significant.
RESULTS
The impact of rsbU, rsbV, and rsbW mutations on SigB expression.
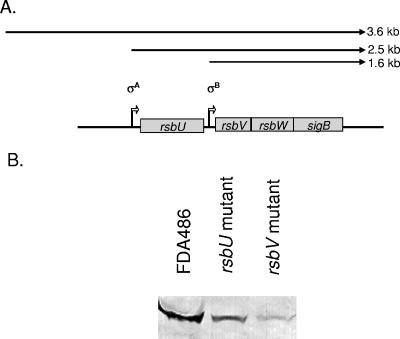
The transcription of sigB has been shown to be dependent on at least two promoters, one originating upstream of rsbU and another upstream of rsbV. A third promoter further upstream of rsbU has also been described (23) (Fig. 1A). To ensure that our mutation on rsbU and rsbV did not impact on SigB expression, we probed an immunoblot containing equivalent amounts of cell extracts from overnight cultures of the mutants with the anti-SigB monoclonal antibody 1D1. As the replacement ermC gene is transcribed divergently from the rest of the sigB operon, inactivation of rsbU and rsbV did not lead to null expression of SigB (Fig. 1B). Indeed, the SigB protein levels were clearly demonstrable, albeit at lower levels in the rsbU and rsbV mutants than in the parental strain FDA486 (24). This is consistent with the observation that rsbU and rsbV are driven by separate promoters and that activation of rsbU can occur independently of rsbV (24). The expression of SigB, encoded by the last gene in the operon, in both mutants also suggests that any potential polar effect as a result of the rsbU and rsbV mutations is probably minimal. We also examined SigB expression in the rsbW mutant. Similar to the sigB mutant, the rsbW mutant did not express any SigB as detected by immunoblotting; this finding was expected because the expression of RsbW and SigB are translationally coupled (17, 27).
FIG. 1.
The effect of rsbU and rsbV mutations on SigB expression in the FDA486 background. (A) Organization of the sigB operon in S. aureus. There are two well-described transcripts, one originating from the σA promoter upstream of rsbU and the other from the σB promoter upstream of rsbV. A third 3.6-kb transcript was recently described by Senn et al. (25). (B) Equivalent amounts of cell extracts (50 μg each) from FDA486 and its isogenic mutants were immunoblotted onto nitrocellulose. The blot was then probed with anti-SigB monoclonal antibody 1D1 at a 1:2,000 dilution. The protein band was then detected with goat anti-mouse antibody conjugated to alkaline phosphatase and developing substrate as previously described (6). We also examined expression of SigB in rsbW and sigB mutants of FDA486. In both cases, no SigB protein expression was detected. These immunoblot data have been previously published by one of our laboratories (24).
Induction of sarA P3 activity by SAL.
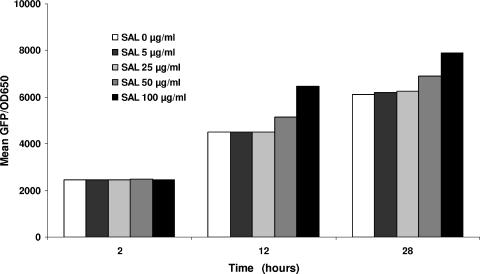
In pilot screening studies, we sought to establish a single effective SAL concentration range to utilize in the detailed phenotypic and genotypic investigations. We thus exposed the parental strain to a range of SAL concentrations (0 to 100 μg/ml) encompassing both clinically achievable human serum concentrations as well as SAL concentrations previously documented to activate sigB expression (17). Using sarA P3 promoter activation as a surrogate marker for sigB expression, the lowest SAL concentration that clearly increased sigB expression compared to untreated cells was 50 μg/ml (Fig. 2). We subsequently used the range of 25 to 50 μg/ml of SAL for the remaining phenotypic and genotypic studies. SAL-induced sarA P3 activation in this assay was apparent by 10 h of growth, reflecting both the late-logarithmic-phase maxima of this promoter as well as the time required for maturation of the GFP (13).
FIG. 2.
Effect of SAL at various drug concentrations upon sarA P3 promoter activity. Expression of gfp driven by the sarA P3 promoter was measured during the growth cycle, and fluorescence values were expressed as total GFP fluorescence/OD650 to minimize variations in fluorescence due to differing cell densities. These data represent the mean of three independent runs.
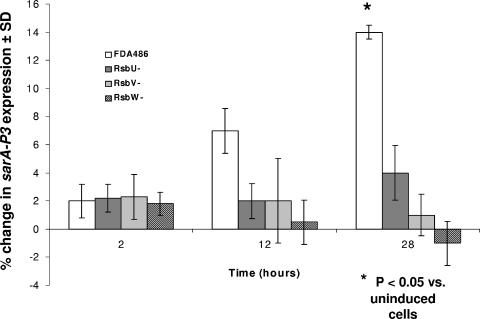
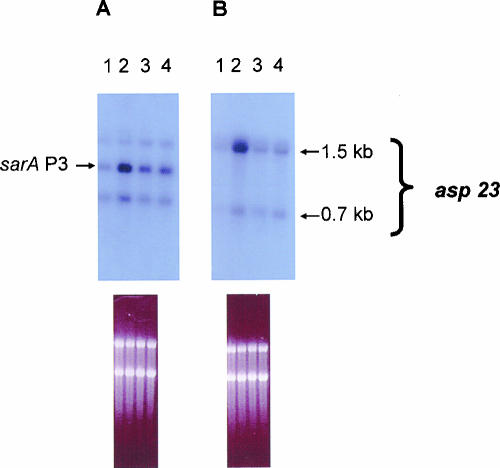
We then compared the relative, time-dependent expression of the sarA P3 promoter, using GFP reporter assays in the presence or absence of SAL (Fig. 3). Early in log-phase growth, neither the parent nor any of the rsb mutants exhibited substantial increases in sarA P3 promoter activity in the presence or absence of SAL. In contrast, at postexponential phase (12 h), the parental strain, but not the rsbU, V, or W mutants, exhibited increases in sarA P3 expression in the presence of SAL, although this difference (∼3.5-fold) did not reach statistical significance. At 28 h of growth (late stationary phase), the impact of SAL on parental strain FDA486 expression of sarA P3 was more pronounced than at 12 h, with an observed 7.5-fold increase compared to uninduced cells (P < 0.05). Interestingly, the upregulation in sarA P3 expression by SAL at 28 h was higher in the rsbU mutant than in the rsbV and rsbW mutants. This pattern of data shows that augmentation of sigB activity due to SAL (albeit relatively small) can still occur with an rsbU mutant at late stationary phase. To confirm these observations, we conducted Northern analyses with a sarA probe to ascertain sarA P3 transcription. At stationary growth phase (Fig. 4A) and also at late exponential phase (not shown), the expression level of the sarA P3 transcript was substantially increased in the parental strain with SAL exposure compared to the uninduced control (2,656 versus 1,088 densitometry units, using SigmaGel software) (Fig. 4A). Interestingly, the rsbU mutant was also able to modestly increase sarA P3 transcription in the presence of SAL (2,069 versus 1,724 densitometry units in the uninduced control). As an additional marker for sigB activation, we evaluated asp23 transcription in the parent strain FDA486 and its isogenic rsbU mutant. As shown in Fig. 4B and mirroring data in Fig. 4A, the parental strain displayed an increase in asp23 transcription with SAL exposure compared to the uninduced control, while the increase in the rsbU mutant was more modest.
FIG. 3.
Effect of SAL (50 μg/ml) on sarA P3 promoter activation in the FDA486 parental strains versus the corresponding rsbU, rsbV, and rsbW mutants. The percent differences were calculated by the following formula: [(fluorescence with SAL/fluorescence without SAL) − 1.0] × 100. Results are the means (± SD) of triplicates from a representative experiment that was repeated thee times.
FIG. 4.
Transcription of the sarA P3 (A) and asp23 (B) promoter in response to SAL (50 μg/ml) by Northern blotting of S. aureus FDA486 (parental) and its corresponding rsbU deletion mutants at the postexponential phase of bacterial growth. Lane 1, wild-type strain FDA486; lane 2, wild-type strain FDA486 with SAL; lane 3, rsbU mutant; lane 4, rsbU mutant with SAL. The figures underneath the blots indicate equivalent loading as reflected by similar ethidium bromide staining of the 16S and 23S rRNA bands.
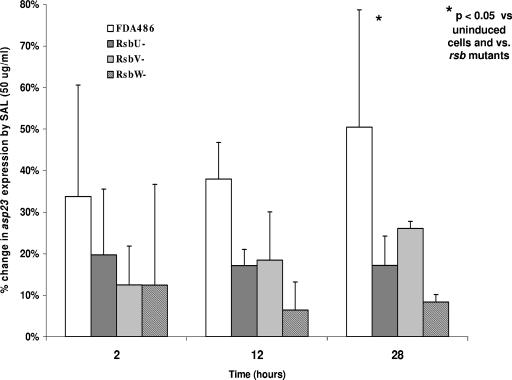
We next compared the relative, time-dependent expression from the asp23 promoter, using GFP as a reporter, in the presence or absence of SAL (Fig. 5). Similar to the data for sarA P3, the parental strain exhibited a substantial increase in asp23 expression in the presence of SAL versus uninduced control over the 28-h growth cycle (range, mean percentage change of 1 to 28% at 25 μg/ml versus uninduced controls [data not shown] and 36 to 66% at 50 μg/ml). These differences reached statistical significance at the 28-h time point of SAL exposure (P < 0.05 versus both uninduced parental cells and induced rsb mutant cells). In contrast, asp23 expression in the three rsb mutants was only modestly induced by SAL at the same drug concentrations. However, these differences (versus uninduced cells) did not reach statistical significance for the mutants.
FIG. 5.
Effect of SAL (50 μg/ml) on asp23 promoter activation in the FDA486 parental strain versus the corresponding rsbU, rsbV, and rsbW mutants. The percent differences were calculated by the following formula: [(fluorescence with SAL/fluorescence without SAL) − 1.0] × 100. Results are the means (± SD) of triplicates from a representative experiment that was repeated thee times.
Phenotypic assays. (i) Fibrinogen and fibronectin binding.
In previous studies, we have shown that SAL can reduce the fibrinogen- and fibronectin-binding capacity of wild-type S. aureus cells (17). Interestingly, baseline fibrinogen binding, as a reflection of intrinsic bacterial adhesion capacity, was similar between untreated parental cells and the three rsb mutants (range, 4.2 to 4.9% of the initial inoculum) (Table 2). Parallel data were also observed for fibronectin binding, showing similar binding among all constructs (range, 3.8 to 4.8% of initial inoculum) (Table 2). Upon SAL exposure, the parental cells showed substantial reductions in binding to fibrinogen in a dose-dependent manner, with more reduction at 50 μg/ml (P < 0.05 versus control cells) than at 25 μg/ml. Similarly, the SAL-mediated reduction in fibronectin binding for parental cells was greater at 50 μg/ml SAL than the untreated control (P < 0.05) (Table 2). For both assays, the reductions in ligand-binding capacity seen in the parental strain were blunted in all rsb mutants. Of note, there was somewhat greater dampening of the SAL impact in the rsbV (versus uninduced cells) compared to the rsbU mutant at 25 μg/ml SAL (but not at 50 μg/ml), although this did not reach statistical significance.
TABLE 2.
Adhesion of S. aureus strain FDA486 and corresponding rsbU, rsbV, and rsbW mutants to immobilized fibrinogen and fibronectin in the presence or absence of SAL
| Strain or mutant | Fibrinogen binding
|
Fibronectin binding
|
||||
|---|---|---|---|---|---|---|
| Controla | SAL (%)b
|
Controla | SAL (%)b
|
|||
| 25 μg/ml | 50 μg/ml | 25 μg/ml | 50 μg/ml | |||
| FDA486 | 4.2 ± 1.1 | −25.40 | −47.10c | 3.8 ± 1.1 | −20.90 | −45.50c |
| rsbU mutant | 4.5 ± 1.2 | −14.20 | −23.30 | 4.8 ± 1.3 | −15.40 | −24.30 |
| rsbV mutant | 4.5 ± 1.1 | −8.90 | −22.10 | 4.8 ± 1.0 | −13.90 | −25.30 |
| rsbW mutant | 4.9 ± 0.8 | −11.80 | −21.10 | 4.5 ± 1.4 | −8.30 | −13.10 |
Percent binding of initial inoculum to ligand in the absence of SAL.
Mean percent reduction in binding comparing SAL-treated versus untreated (control) S. aureus cells. These data are derived from the means of three independent runs and are expressed as mean percent reductions of fibrinogen or fibronectin binding under various assay conditions.
P < 0.05 versus untreated control cells.
(ii) Alpha-toxin assays.
In an earlier study (17), we found that preexposure of wild-type S. aureus cells to SAL reduced their capacity to express alpha-toxin, presumably by up-regulating sigB activity. To investigate the role of specific genes within the sigB operon in mediating this effect, rsb mutants were assayed for hemolytic activity (attributable to alpha-toxin) following SAL exposures. As expected, supernatants from stationary-phase parental cells treated with SAL exhibited a significantly diminished capacity to lyse erythrocytes, compared to supernatants from untreated cells, in a dose-dependent manner (∼35 to 50% reductions; P < 0.05 versus untreated control cells) (Table 3). Of interest, the stationary-phase supernatants from all untreated rsb mutant cells exhibited higher baseline hemolytic activity than parental cells, with the rsbV and rsbW mutants showing a more prominent increase than the rsbU mutant. Remarkably, the capacity of SAL to reduce hemolytic activity, as seen in parental cells, was blunted in the rsbU mutant. The rsbV and rsbW mutants also displayed minimal decreases in hemolytic titer in the presence of 25 or 50 μg/ml of SAL compared with nontreated controls. For the rsbU mutant, the hemolytic titer at 50 μg/ml of SAL was equal to the parental strain without SAL exposure.
TABLE 3.
α-Toxin hemolytic activity in S. aureus strain FDA486 and its isogenic mutants in the presence or absence of SAL
| SAL concn (μg/ml) | Hemolytic activity (mean ± SD)a
|
|||
|---|---|---|---|---|
| FDA486 | rsbU mutant | rsbV mutant | rsbW mutant | |
| Control | 658 ± 38 | 825 ± 69 | 983 ± 104 | 992 ± 38 |
| 25 | 383 ± 52b | 700 ± 71 | 933 ± 58 | 917 ± 41 |
| 50 | 333 ± 41b | 658 ± 31 | 867 ± 29 | 883 ± 26 |
Results are expressed in hemolytic units. See text for details.
P < 0.05 versus untreated control cells.
DISCUSSION
The sigB operon of S. aureus represents a global regulatory system that enables the organism to deal with environmental stresses. However, it differs from the well-characterized sigB operon of Bacillus subtilis in that it is smaller (four genes versus eight genes) and lacks distinct environmental and energy-sensing modules (27) (Fig. 1A). The sigB operon of S. aureus consists of a putative sensor (rsbU) which responds to a broad range of microenvironmental cues via autophosphorylation to activate the next gene in this operon (rsbV). Phosphorylated RsbU acts as a phosphatase (or an anti-anti-sigma factor) to dephosphorylate RsbV. Dephosphorylated RsbV binds competitively to the anti-sigma factor, RsbW, thus displacing the normally inhibitory RsbW from the RsbW-SigB complex (21). The collective result of this activation cascade is the release of free SigB to activate genes with a SigB recognition motif within their promoter region (e.g., sarA P3 and asp23) (3) via recruitment of RNA polymerase.
Likely related to its role in countering environmental stresses, the sigB operon has been shown to be intimately involved in biofilm formation (1), as well as in the regulation of virulence factors. As an example, the activation of the sarA P3 promoter impacts the expression of sarA-dependent structural genes, including the genes for alpha-toxin (hla), V8 protease (sspA), and fibronectin-binding proteins (e.g., fnbA) (3). SigB may also repress expression of the two-component regulatory system, saeRS, which itself positively regulates hla and fnbA expression (26). Importantly, during in vitro growth, sigB is activated at early stationary growth phase, corresponding to its activation of the sarA P3 promoter. Since SarA production is the net result of activation of the three sarA promoters (sarA P1, P2, and P3), sarA P3 activation normally leads to enhanced SarA production. However, Karlsson et al. (16) have shown recent data suggesting that clinical S. aureus strains differ substantially in intrinsic “tone levels” of sigB and SigB-dependent gene expression. This concept was exemplified by the strain-to-strain variability in the production of V8 protease (a SigB-SarA repressible event) (16). Similarly, we have shown that sigB can be exogenously stimulated by SAL to a high “tone level,” resulting in augmented transcription from the sarA P3 promoter. Contrary to growth phase-related effects on SarA expression (i.e., upregulation), hyperactivation of sigB due to exogenous SAL leads to reductions in net sarA activation (manifested by both reduced SarA protein levels and enhanced V8 protease and lipase production) (16, 17). The mechanism(s) by which excess sarA P3 activation by SAL mitigates overall sarA expression is not understood but may involve promoter occlusion of the proximal, but more prominent, sarA P1 promoter. Alternatively, a direct effect of SAL on the sarA promoter complex cannot be ruled out.
We previously demonstrated that the major biometabolite of aspirin, SAL, exerts potent antivirulence effects in vitro and in vivo against a number of well-characterized S. aureus strains, including RN6390, SH1000, ISP479, COL, and Newman (17). These antivirulence effects include reduction in binding to a variety of matrix ligands involved in tissue colonization by S. aureus (i.e., fibrinogen, fibronectin, and fibrin), reduction in binding to endothelial cells and platelets (18), and reduction in alpha-toxin production (17). All these phenotypic traits have been linked to the virulence of S. aureus in endovascular and other infection models (5). These in vitro phenotypic effects were mirrored in vivo in experimental endocarditis (IE) models as exemplified by the reduced capacity of SAL-treated S. aureus cells to bind to sterile aortic valve vegetations in vivo (17). Treatment of animals with established S. aureus IE by aspirin or SAL also mitigated virulence, as manifested by reductions in bacterial densities in cardiac vegetations and kidneys, decreases in vegetation size and weight, and prevention of embolic renal infarcts (17, 18). Importantly, the fact that SAL (which is devoid of antiplatelet activities) demonstrated antivirulence properties virtually identical to those of aspirin argued against the idea that the antiplatelet property of aspirin is the principal abating factor and, instead, indicated the possibility that an antibacterial pathway is at work. This hypothesis was validated by our genetic analyses in vitro and in vivo, clearly showing that activation of sigB is a critical event in initiating the antivirulence properties of aspirin and SAL (17). This mechanism then leads to down-modulation of global regulons downstream of sigB (e.g., sarA and agr), as well as of key structural genes involved in matrix ligand binding and alpha-toxin production (3). Of note, a recent investigation by Entenza et al. (8) has confirmed that sigB-hyperexpressing strains of S. aureus exhibit reduced virulence during well-established stages of experimental endocarditis compared to wild-type strains.
Despite the unambiguous role of sigB activation by aspirin or SAL in antivirulence properties, the contribution of each gene within the sigB operon (i.e., rsbU, rsbV, or rsbW) to this impact is not known. As sigB activation by aspirin or SAL occurred in strains RN6390 and ISP479 (rsbU-deficient lineage strains of 8325-4) (14) as well as in the rsbU-intact strains SH1000, COL, and Newman, this suggested that activation of sigB by these compounds could proceed via both rsbU-dependent and rsbU-independent pathways.
The current study was designed to establish the relative roles of individual genes within the sigB operon in mediating the in vitro activation by SAL and in impacting two representative phenotypes (ligand binding and alpha-toxin production). Several interesting findings emerged from this investigation. (i) In parental strain FDA486 (with an intact sigB operon), deletion of rsbU eliminated a major portion of the capability of the strain to respond to SAL, corresponding to a lesser capacity to activate sigB in the rsbU mutant. This relationship was evidenced by a much lower level of sigB-dependent promoter activation (e.g., sarA P3) in the rsbU mutant as confirmed by Northern blotting and transcriptional fusions. (ii) The influences of SAL on sigB activation were concentration dependent, and the differences between untreated cells and SAL-treated cells were greatest during stationary phases of growth (when sigB expression is maximal). (iii) Our data suggested that both rsbU and rsbV can be targets for SAL. This notion was supported by a hierarchy in the reduction of ligand-binding capacity between the parent and these two latter mutants at 25 μg/ml of SAL (Table 2). This hierarchy was recapitulated in the reduction in hemolytic titers (Table 3) at both SAL concentrations (i.e., parent > rsbU mutant > rsbV mutant). Based on our previous studies, we recognized that even in the rsbU mutant (the putative stress-sensing locus within sigB), sigB could still be partially activated by energy-dependent stresses (24). This observation and the data from the present study underscore the notion that sigB can be activated by rsbU-dependent and rsbV-dependent pathways. (iv) SigB is normally a repressor of alpha-toxin gene (hla) expression; thus, deletion of genes within the sigB operon normally results in alpha-toxin hyperexpression (6, 24), as confirmed in the rsbU and rsbV mutants in the current investigation. Because rsbW and sigB are translationally coupled (21), the rsbW mutant behaves essentially like a sigB mutant. Whether SigB represses alpha-toxin production in the presence of SAL via inhibition of sarA, agr, and/or sae remains to be defined. (v) As noted above, SAL exposure in parental strain FDA486 resulted in a reduction in ligand-binding phenotypes; this effect was blunted in the rsbU mutant and more so in the rsbV and rsbW mutants at 25 μg/ml. The basis for the disappearance of this differential effect between rsbU and rsbV mutants at 50 μg/ml of SAL is not immediately evident. It is plausible that SAL at higher concentrations may affect the baseline phosphorylation of RsbV.
In summary, we have confirmed that SAL exerts substantial effects on phenotypes involved in endovascular virulence via activation of the sigB operon; interruption of the sigB gene cascade by mutating loci within the operon will blunt this response. Further, the phenotypic effects of SAL appear to proceed via both rsbU-dependent and rsbV-dependent pathways. Whether SAL has direct influences upon downstream structural genes (e.g., hla) or if SAL can upregulate pathways outside of sigB to impact the above phenotypes remains to be defined.
Acknowledgments
This research was supported by grants from the National Institutes of Health to A.S.B. (AI-39108) and M.R.Y. (AI-48031) and from the American Heart Association (Western Affiliate) to A.S.B. (0150699Y).
REFERENCES
- 1.Bateman, B. Y., N. P. Donegan, T. M. Jarry, M. Palma, and A. L. Cheung. 2001. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 69:7851-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayer, M. G., J. H. Heinrichs, and A. L. Cheung. 1996. The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178:4563-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y.-Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Microbiol. Lett. 1649:1-9. [DOI] [PubMed] [Google Scholar]
- 6.Cheung, A. L., Y. T. Chien, and A. S. Bayer. 1999. Hyperproduction of alpha hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 67:1331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, A. L., and A. C. Manna. 2005. Role of the distal sarA promoters in SarA expression in Staphylococcus aureus. Infect. Immun. 73:4391-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Entenza, J. M., P. Moreillon, M. M. Senn, J. Kormanec, P. M. Dunman, B. Berger-Bachi, S. Projan, and M. Bischoff. 2005. Role of σB in the expression of Staphylococcus aureus cell wall adhesins ClfA and FnbA and contribution to infectivity in a rat model of experimental endocarditis. Infect. Immun. 73:990-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of σB-dependent transcription of sigB and asp23 in two different S. aureus strains. Mol. Gen. Genet. 261:558-566. [DOI] [PubMed] [Google Scholar]
- 10.Gertz, S., S. Engelmann, R. Schmid, K. Ziebandt, K. Tischer, C. Scharf, J. Hacker, and M. Hecker. 2000. Characterization of the σB regulon in Staphylococcus aureus. J. Bacteriol. 182:6983-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:506-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heim, R., D. C. Prasher, and R. Y. Tsien. 1994. Wavelength mutations and posttranslational autooxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA 91:12501-12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahl, B., M. Goulian, W. Van Wamel, M. Herrmann, S. Simon, G. Kaplan, G. Peters, and A. L. Cheung. 2000. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line derived from a cystic fibrosis patient. Infect. Immun. 68:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsson, A., and S. Arvidson. 2002. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor sarA. Infect. Immun. 70:4239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kupferwasser, L. I., M. R. Yeaman, C. C. Nast, D. Kupferwasser, Y. Q. Xiong, M. Palma, A. L. Cheung, and A. S. Bayer. 2003. Salicylic acid attenuates virulence in endovascular infections by targeting global regulatory pathways in Staphylococcus aureus. J. Clin. Investig. 112:222-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kupferwasser, L. I., M. R. Yeaman, S. M. Shapiro, C. C. Nast, P. M. Sullam, S. G. Filler, and A. S. Bayer. 1999. Acetylsalicylic acid reduces vegetation bacterial density, hematogenous bacterial dissemination, and frequency of embolic events in experimental Staphylococcus aureus endocarditis through antiplatelet and antibacterial effects. Circulation 99:2791-2797. [DOI] [PubMed] [Google Scholar]
- 19.Lee, C. Y. 1992. Cloning of genes affecting capsule expression in Staphylococcus aureus strain M. Mol. Microbiol. 6:1515-1522. [DOI] [PubMed] [Google Scholar]
- 20.Mahmood, R., and S. A. Khan. 1990. Role of upstream sequences in the expression of the staphylococcal enterotoxin B gene. J. Biol. Chem. 265:4652-4656. [PubMed] [Google Scholar]
- 21.Miyazaki, E., J. M. Chen, C. Ko, and W. R. Bishai. 1999. The Staphylococcus aureus rsbW (orf159) gene encodes an anti-sigma factor of SigB. J. Bacteriol. 181:2846-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novick, R. P. 1990. The staphylococcus as a molecular genetic system, p. 1-40. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 23.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 23a.Novick, R. P., and R. Brodsky. 1972. Studies on plasmid replication. Plasmid incompatibility and establishment in Staphylococcus aureus. J. Mol. Biol. 68:285-302. [DOI] [PubMed] [Google Scholar]
- 24.Palma, M., and A. L. Cheung. 2001. SigB activity in Staphylococcus aureus is controlled by RsbU and additional factors during growth. Infect. Immun. 69:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senn, M. M., P. Giachino, D. Homerova, A. Steinhuber, J. Strassner, J. Kormanec, U. Flückiger, B. Berger-Bächi, and M. Bischoff. 2005. Molecular analysis and organization of the σB operon in Staphylococcus aureus. J. Bacteriol. 187:8006-8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinhuber, A., C. Goerke, M. G. Bayer, G. Doring, and C. Wolz. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 185:6278-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu, S., H. de Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong, Y.-Q., W. Van Wamel, C. C. Nast, M. R. Yeaman, A. L. Cheung, and A. S. Bayer. 2002. Activation and transcriptional interaction between agr RNAII and RNAIII in Staphylococcus aureus in vitro and in an experimental endocarditis model. J. Infect. Dis. 186:668-677. [DOI] [PubMed] [Google Scholar]