Abstract
The Mycobacterium tuberculosis protein Rv2302 (80 residues; molecular mass of 8.6 kDa) has been characterized using nuclear magnetic resonance (NMR) and circular dichroism (CD) spectroscopy. While the biochemical function of Rv2302 is still unknown, recent microarray analyses show that Rv2302 is upregulated in response to starvation and overexpression of heat shock proteins and, consequently, may play a role in the biochemical processes associated with these events. Rv2302 is a monomer in solution as shown by size exclusion chromatography and NMR spectroscopy. CD spectroscopy suggests that Rv2302 partially unfolds upon heating and that this unfolding is reversible. Using NMR-based methods, the solution structure of Rv2302 was determined. The protein contains a five-strand, antiparallel β-sheet core with one C-terminal α-helix (A61 to A75) nestled against its side. Hydrophobic interactions between residues in the α-helix and β-strands 3 and 4 hold the α-helix near the β-sheet core. The electrostatic potential on the solvent-accessible surface is primarily negative with the exception of a positive arginine pocket composed of residues R18, R70, and R74. Steady-state {1H}-15N heteronuclear nuclear Overhauser effects indicate that the protein's core is rigid on the picosecond timescale. The absence of amide cross-peaks for residues G13 to H19 in the 1H-15N heteronuclear single quantum correlation spectrum suggests that this region, a loop between β-strands 1 and 2, undergoes motion on the millisecond to microsecond timescale. Dali searches using the structure closest to the average structure do not identify any high similarities to any other known protein structure, suggesting that the structure of Rv2302 may represent a novel protein fold.
Mycobacterium tuberculosis is the etiological agent responsible for the chronic infectious disease tuberculosis. Despite the availability of effective short-term chemotherapy and extensive vaccination programs, this gram-positive tubercle bacillus still infects approximately one-third of mankind, and in 2003 it is estimated that it claimed the lives of approximately 1.7 million people (44). Indeed, there has been a recent increase in the incidence of tuberculosis in both developing and industrialized nations as new drug-resistant strains have emerged, and a deadly synergy with human immunodeficiency virus has evolved (17). The complete sequenced genome of M. tuberculosis contains approximately 4,000 genes (6). In 2000 a TB Structural Genomics Consortium was established with the goal of using the genomic information to discover and analyze the structures of the M. tuberculosis gene products (38). In addition to providing a foundation for a fundamental understanding of biology, the information and knowledge obtained from determining the structures of the proteins in the M. tuberculosis genome will enable the conception and development of new therapies and strategies to treat and control this deadly disease. In pursuit of these goals, we have used nuclear magnetic resonance (NMR)-based methods to determine the solution structure for a protein, Rv2302, that is highly conserved in the bacterium M. tuberculosis.
MATERIALS AND METHODS
Cloning, expression, and purification.
The DNA coding sequence for the Rv2302 gene was cloned into a modified pET28b vector (Novagen, Madison, WI) that provided an N-terminal 6-His tag (MGSSHHHHHHSSGLVPRGSH) upstream of the NdeI site. This DNA was then transfected into the host Escherichia coli bacterial strain BL21PRO (Clontech, Palo Alto, CA). Uniformly 15N- and 13C-15N-labeled Rv2302 was obtained by growing the transformed cells (37°C) in minimal medium (Miller) containing 15NH4Cl and d-[13C6]glucose supplemented with thiamine (1 μg/ml), Fe2Cl3 (10 μM), kanamycin (34 μg/ml), and spectinomycin (100 μg/ml). At an A600 reading of ∼0.8, protein expression was induced by making the broth concentration 1.0 μM in isopropyl-β-d-1-thiogalactopyranoside. Following 4 to 8 h of culture growth at 28°C, the cells were harvested and then frozen at −80°C. The thawed cell pellet was resuspended in 35 ml of lysis buffer (50 mM sodium phosphate, 0.3 M NaCl, 10 mM imidazole, pH 8.1), brought to 0.2 μM in phenylmethylsulfonyl fluoride, and passed three times through a French press (SLM Instruments, Rochester, NY). The cell debris was removed by centrifugation for 45 min at 24,000 × g in a JA-20 rotor (Beckman Instruments, Fullerton, CA). Following filtration through a 0.45-μm-pore-size membrane (Corning Incorporated, Corning, NY), the supernatant was loaded onto a 20 ml Ni-nitrilotriacetic acid affinity column (QIAGEN, Valencia, CA) and washed stepwise with buffer (0.3 M NaCl, 50 mM sodium phosphate, pH 8.1) containing increasing concentrations of imidazole (5 to 500 mM). The Rv2302 fraction that eluted at 250 mM imidazole (∼30 mg/liter) was dialyzed overnight at 4°C in 4 liters of thrombin cleavage buffer (150 mM NaCl, 20 mM Tris-HCl, pH 8.4). After the volume was reduced to ∼3 ml (Amicon Centriprep-3), the N-terminal polyhistidine tag was removed by overnight incubation at room temperature with ∼1 mg of thrombin (Fisher BioReagents, Fair Lawn, NJ) and 3 μl of 1.0 M CaCl2. The cleaved protein was then purified on a Superdex75 HiLoad column (Amersham Pharmacia Biotech, Piscataway, NJ) that simultaneously exchanged it into NMR buffer (100 mM KCl, 20 mM potassium phosphate, 2.0 mM dithiothreitol, pH 7.1). Using a flow rate of 1.0 ml/min, approximately 20 mg of Rv2302 eluted at 80 min, a retention time characteristic of a protein with a calculated monomeric molecular mass of 8,591 Da (Rv2302 plus the three N-terminal residues, GSH, that remain after thrombin cleavage). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed the protein to be greater than 98% pure.
Optical spectroscopy.
Circular dichroism (CD) data were collected on an Aviv Model 62DS spectropolarimeter calibrated with an aqueous solution of ammonium d-(+)-camphorsulfonate. The measurements were obtained on an Rv2302 sample (∼0.06 mM) in NMR buffer and in a quartz cell of 0.1 cm path length. A thermal denaturation curve for Rv2302 was obtained by recording CD spectra at intervals of 2.5°C from 5 to 80°C and plotting the ellipticity at 220 nm. Wavelength scans for Rv2302 were recorded between 200 and 250 nm at 25°C, 80°C, and 25°C (postheating). Each wavelength spectrum was the result of averaging two consecutive scans with a bandwidth of 1.0 nm and a time constant of 1.0 s. The wavelength spectra were processed by first subtracting a blank spectrum followed by baseline correction and noise reduction.
NMR spectroscopy and resonance assignments.
All NMR experiments were collected on samples with concentrations of ∼2.0 mM at 25°C using Varian 600- and 500-Inova spectrometers equipped with triple resonance probes and pulsed-field gradients. The data were processed and analyzed with Felix (MSI, San Diego, CA). All chemical shifts were referenced to DSS (2,2-dimethyl-2-silapentane-5-sulfonate; 0 ppm) using indirect methods (41).
The 13C, 1H, and 15N chemical shifts of the backbone and side chain resonances were obtained from the analysis of sensitivity-enhanced two-dimensional 1H-15N (18, 45) and 13C-1H heteronuclear single quantum correlation (HSQC) (16, 18) spectra and three-dimensional (3-D) HNCA, HN(CO)CA, CBCA(CO)NH (18, 30), HNCACB, HNCO, HCCH-total correlated spectroscopy (TOCSY) (19), HCC-TOCSY-NNH (26, 29), and CC-TOCSY-NNH (12, 25, 29) spectra. Distance restraints were obtained from a suite of multidimensional nuclear Overhauser effect spectroscopy (NOESY) experiments using a mixing time of 150 msec: 3-D 13C- and 15N-edited NOESY-HSQC (18, 33, 45) and 4-D CC-NOESY-HMQC (40). Deuterium-exchange studies were performed by lyophilizing an NMR sample and redissolving in 99.8% D2O (the 1H-15N HSQC spectrum of a lyophilized sample redissolved into 10% D2O-90% H2O was essentially identical to the 1H-15N HSQC spectrum obtained prior to lyophilization). Two-dimensional 1H-15N HSQC spectra were recorded 0.5, 2, 4, and 96 h after the exchange. Steady-state {1H}-15N heteronuclear NOE values were measured from the ratios of 1H-15N HSQC cross-peak volumes in spectra recorded in the presence (Isat) and absence (Iunsat) of three seconds of proton presaturation prior to the 15N excitation pulse (NOE = Isat/Iunsat) (9).
Structure calculations.
Structures were calculated with the CNS program (version 1.1) (2) using the experimentally derived restraints listed in Table 1. Pseudoatom corrections were added to the upper bound of the following stereochemically unassigned methylene protons and methyl groups: 1.0 Å for methylene protons, 2.0 Å for chemically equivalent aromatic protons, 1.5 Å for methyl protons, and 2.4 Å for pairs of methyl groups in Leu and Val residues. The latter pseudoatom corrections were applied only to the methyl groups for 5 out of the 11 Val residues and 1 of the 2 Leu residues because the others were stereospecifically assigned using a biosynthetically directed 13C-labeled sample and observing the carbon-carbon splitting of the Pro-R methyl group in the 13C-1H HSQC spectrum (31). Dihedral angle restraints for phi (Φ) of −57 ± 25° (α-helices) and −139 ± 40° (β-strands) were obtained through the chemical shift index analysis of the carbon and proton chemical shifts (42). One of the three Pro residues (P36) was determined to be in the cis conformation on the basis of a large difference in the Pro 13Cβ and 13Cγ chemical shift (34) and NOE pattern. Deuterium exchange experiments revealed a subset of 24 cross-peaks in the 1H-15N HSQC spectrum that still remained 30 min after the exchange. This subset of more slowly exchanging protons corresponded to amides in β-strands. When the acceptor oxygen for the slowly exchanging amide was identified from preliminary structural ensembles, hydrogen bond restraints (1.8 to 2.3 Å and 2.8 to 3.3 Å for the NH-O and N-O distances, respectively) were introduced into the structure calculations. Identical hydrogen bond restraints were added for backbone amide protons in an α-helical region (residues 65 to 75) identified in the preliminary structural ensembles (and confirmed by chemical shift index analysis). Note that the latter amides all exchanged with deuterium within 30 min, consistent with the observation that hydrogens in α-helix hydrogen bond networks generally exchange before hydrogens in β-strand hydrogen bond networks (8). When no persistent distance or dihedral angle violations were observed in the initial ensemble of calculated structures of lowest energy, dihedral angle restraints for psi (Ψ) of −47 ± 30° (α-helices) and 140 ± 40° (β-strands), based on the elements of secondary structure identified in the structural ensemble and TALOS calculations (7), were introduced into the calculations. The final set of 55 calculations, using the restraints compiled in Table 1, generated an ensemble of 25 low-energy structures (in terms of total and NOE energies). The structures in this ensemble were refined with explicit water (24) using force constants of 500 and 2,000 kcal for the NOE and dihedral restraints, respectively. This final ensemble was then used to calculate a mean structure and average root mean square deviation (RMSD) values to the mean structure. Structural quality was assessed using PROCHECK-NMR (23). Structural similarity searches were performed using the DALI server (www.ebi.ac.uk/dali/interactive.html) (13).
TABLE 1.
Summary of the structural statistics for RV2302a
| Parameter | Value |
|---|---|
| Restraints for structure calculations | |
| Total NOEs | 882 |
| Intraresidue NOEs | 324 |
| Sequential (i, i + 1) NOEs | 235 |
| Medium-range (i, i + j; 1 < j ≤ 4) NOEs | 93 |
| Long-range (i, i + j; j > 4) NOEs | 230 |
| Phi (Φ) angle restraints | 44 |
| Psi (Ψ) angle restraints | 45 |
| Hydrogen bond restraints | 60 |
| Structure calculations | |
| No. of structures calculated | 55 |
| No. of structures used in ensemble | 25 |
| Structures with restraint violations | |
| Distance restraint violations >0.1Å | 0 |
| Dihedral restraint violations | |
| >2° | 5 |
| >5° | 0 |
| RMSD to mean (Å) | |
| Backbone N-Cα-C=O atoms | |
| Ordered residuesb | 0.57 ± 0.17 |
| All residuesc | 1.28 ± 0.17 |
| Heavy atoms | |
| Ordered residuesb | 1.12 ± 0.20 |
| All residuesc | 1.87 ± 0.20 |
| All atoms | |
| Ordered residuesb | 1.37 ± 0.20 |
| All residuesc | 2.07 ± 0.19 |
| Ramachandran plots of ordered residues | |
| (all residues) | |
| Most favored regions (%) | 84 (77) |
| Additionally allowed regions (%) | 15 (20) |
| Generously favored regions (%) | 1 (2) |
| Disallowed regions (%) | 0 (1) |
All statistics are for the 25-structure ensemble deposited in the Protein Data Bank (PDB code 2A7Y).
Residues 2 to 11, 20 to 28, 35 to 51, and 57 to 75.
Residues 1 to 80.
Protein structure accession numbers.
The atomic coordinates for the ensemble of 25 lowest energy structures for M. tuberculosis Rv2302 have been deposited in the Research Collaboratory for Structural Bioinformatics under PDB code 2A7Y. The chemical shift assignments have been deposited with the BMRB under accession number 7000.
RESULTS AND DISCUSSION
Optical spectroscopy.
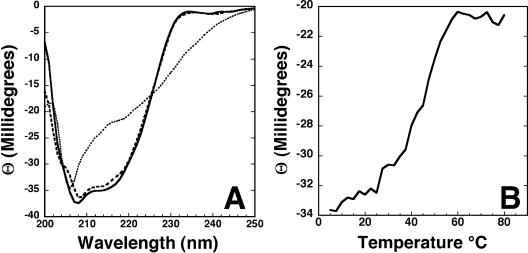
CD spectroscopy was used to characterize the secondary structure of Rv2302 over a range of temperatures (28, 35, 43). Figure 1A shows the CD spectra for Rv2302 at two temperatures. The solid line is the CD spectrum of a fresh, ∼0.06 mM sample collected at 25°C in the same buffer used to collect NMR data. The double minimum at ∼215 and ∼208 nm and projected maximum at a wavelength of <200 nm are characteristic of a structured protein with a mixture of β-sheet and α-helical content (14). The dotted line in Fig. 1A is the CD spectrum of the same sample at 80°C. Relative to the spectrum collected at 25°C, the minimum at ∼215 nm has increased and red-shifted to ∼218 nm, while there is only a slight increase and small blue shift in the minimum at ∼208 nm. These observations at 80°C indicate that Rv2302 is more unstructured at elevated temperatures. However, the absence of an extrapolated negative minimum at ∼198 nm and a positive maximum at ∼218 nm indicates that Rv2302 is not entirely random coil at 80°C and still has some β-sheet structure. When the sample is cooled to 25°C again, the CD spectrum, indicated by the dashed line in Fig. 1A, is very similar to the original CD spectrum collected prior to heating (solid line). This indicates that the temperature-induced effects to the structure of Rv2302 are largely reversible.
FIG. 1.
(A) CD spectra of Rv2302 (0.06 mM) in NMR buffer collected at 25°C (solid line), 80°C (dotted line), and 25°C postheating (dashed line). (B) CD thermal melt for Rv2302 (0.06 mM) in NMR buffer. Data points were collected at 220 nm in intervals of 2.5°C between 5 and 80°C.
To assay the thermal stability of Rv2302, the ellipticity at 220 nm was measured as a function of temperature between 5 and 80°C. Typically, a phase transition can be detected when a structured protein becomes denatured by monitoring the increase in the ellipticity at 220 nm with increasing temperature (3, 22). As shown in Fig. 1B, a gradual increase in ellipticity at 220 nm is observed when the protein is heated to ∼60°C, at which point a plateau is reached with an inflection point at approximately 45°C. Because the CD spectra in Fig. 1A suggest that at 80°C the protein is not entirely unstructured and will refold upon cooling back to 25°C, the plateau in Fig. 1B may not represent a fully unstructured protein. If this is true, then the inflection point in Fig. 1B may represent an initial stage in protein denaturation. Because β-sheets are typically more robust than α-helices (8), if the inflection point in Fig. 1B represents a first stage in the denaturation of Rv2302, then it may reflect an unraveling of the C-terminal α-helix.
Solution state NMR.
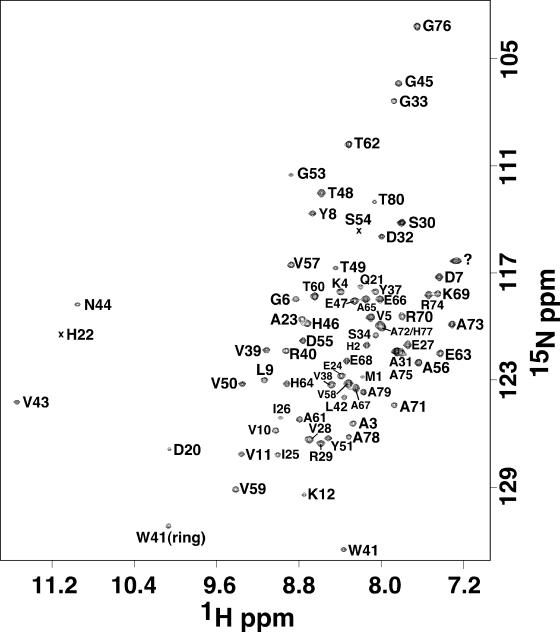
The elution time of Rv2302 on a size exclusion column was consistent with a globular 8.6-kDa protein, and this was corroborated by the 1H-15N HSQC spectrum for Rv2302 shown in Fig. 2. The line widths and chemical shift dispersion of the HSQC cross-peaks are characteristic of a folded, monomeric protein of molecular size in the 10-kDa range. Further evidence that Rv2302 behaved as a monomer in solution is the lack of intermolecular NOEs in any of the NOE experiments (3-D 13C- and 15N-edited NOESY-HSQC and 4-D CC-NOESY-HMQC). As indicated in Fig. 2, all the 1H-15N HSQC cross-peaks were unambiguously assigned except for the single cross-peak labeled with a question mark (tentatively assigned to the side chain amide of R40). Side chain and amide resonances could not be observed, or assigned, for seven residues between G13 and H19, corresponding to a loop (L1) between β-strands 1 and 2.
FIG. 2.
An 1H-15N HSQC spectrum of Rv2302 with the assigned cross-peaks labeled. Unassigned cross-peaks are identified with a question mark, and weak cross-peaks are identified with an “x.” The spectrum was collected at 25°C in NMR buffer (100 mM KCl, 20 mM potassium phosphate, 2.0 mM dithiothreitol, pH 7.1) at a 1H resonance frequency of 600 MHz.
Quality of the calculated structures.
As summarized in Table 1, a total of 882 interproton distance restraints, 60 hydrogen bond restraints, and 89 dihedral angle restraints were used in the final structure calculations. Each member of the ensemble of 25 calculated structures agrees well with the experimental data with no upper limit violation greater than 0.1 Å and no torsion angle violation greater than 5°. The quality of the structures is also reflected in good Ramachandran statistics for all the residues in the ensemble: 77% of the φ-ψ pairs for Rv2302 are found in the most favored regions, and 20% are within additionally allowed regions (calculated using PROCHECK-NMR) (23). The Ramachandran statistics improve to 84 and 15% for the φ-ψ pairs in the most favored and additionally allowed regions, respectively, when only the ordered residues in the ensemble are analyzed.
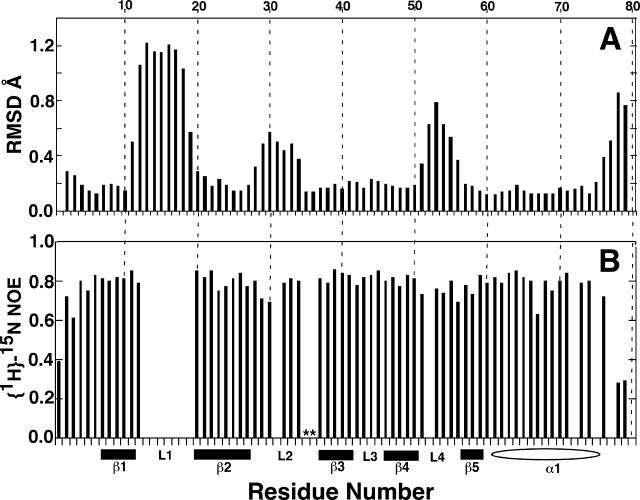
The 25 calculated structures in the ensemble converge well, as shown by the statistics in Table 1. The RMSD of the structured regions in the ensemble to the mean structure is 0.57 Å for the backbone atoms (N-Cα-C=O) and 1.12 Å for all heavy atoms. The degree and location of convergence of the 25 calculated structures in the ensemble are graphically depicted in Fig. 3A. For each residue of each structure in the ensemble, the mean pairwise RMSD to the mean structure is plotted. The values were generated by moving a window of three residues along the sequence, calculating the mean pairwise RMSD (Å) to the average structure, and plotting the value over the central residue. The figure illustrates that all five β-strands and the α-helix are well defined, with mean pairwise RMSDs per residue in the 0.2 Å range for all but one residue (V11). Two of the four loops are less ordered, with mean pairwise RMSDs per residue approaching 0.8 Å; however, the four-residue loop between β-strands 3 and 4 is well defined.
FIG. 3.
(A) Plot of the mean pairwise RMSDs to the average structure for each residue in the Rv2302 ensemble in Table 1. The data were generated by moving a window of three residues along the sequence and plotting the mean pairwise RMSD (Å) over the central residue. (B) Backbone {1H}-15N heteronuclear NOE values for Rv2302. Proline residues are indicated by asterisks. The top axis shows the residue number, and the bottom axis shows the elements of secondary structure: solid bar, β-sheet; open oval, α-helix; L, loop.
Solution structure of Rv2302.
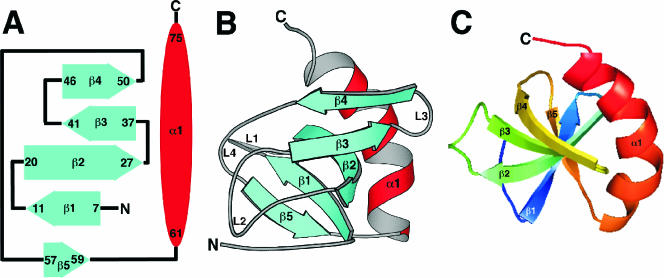
The CD spectrum of Rv2302 contained features characteristic of a protein with a mixture of α-helical and β-sheet secondary structure, and this observation is corroborated by the NMR-based structure determined for Rv2302 shown schematically in Fig. 4A and with Molscript (21) in Fig. 4B. The core of the structure is a five-strand antiparallel β-sheet with a conspicuous β-sheet twist (5) that is more clearly illustrated in Fig. 4C. The length and relative orientation of the five β-strands are illustrated in the secondary structure diagram in Fig. 4A. The absence of 1H-15N HSQC cross-peaks for most of the residues between β-strands 1 and 2 (L1) suggests that this region is not rigidly structured in solution and qualitatively implies that these residues are undergoing conformational exchange on an intermediate timescale (millisecond to microsecond) (4). The regions between the other β-strands in the sheet are more ordered relative to L1, as suggested by the lower pairwise RMSDs of these loop residues in the ensemble structures to the mean structure (Fig. 3A). Indeed, Fig. 3A indicates that the loop between β-strands 3 and 4 is very well defined. Nestled against the backside of the β-strand core is a 15-residue α-helix. A hydrophobic core consisting of the side chains of residues A72 and A75 in the α-helix and the side chains of residues V50 and T48 (methyl group) in β-strand 4 and W41 and V39 in β-strand 3 appears responsible for holding the α-helix near the β-sheet. Indeed, a number of long-range NOEs were observed between the side chains of these residues. While the α-helix is drawn continuously between residues 61 and 75 in Fig. 4B, there is actually a slight bend in the helix that is more evident in the view shown in Fig. 4C.
FIG. 4.
(A) Secondary structure diagram of Rv2302. The α-helices are drawn as red ovals and the β-strands as solid blue arrows with the residue number of the beginning and the end of each element shown. (B) Molscript ribbon representation of the average structure of the ensemble of calculated structures for Rv2302 produced using MOLMOL (20). The β-strands are shown in blue, and the α-helices are red. (C) View of the protein looking directly down upon the β-sheet core highlights the twist in the β-sheet. The protein, drawn using PyMOL, is rainbow colored (ROYGBIV [red, orange, yellow, green, blue, indigo, violet]), starting from the C terminus.
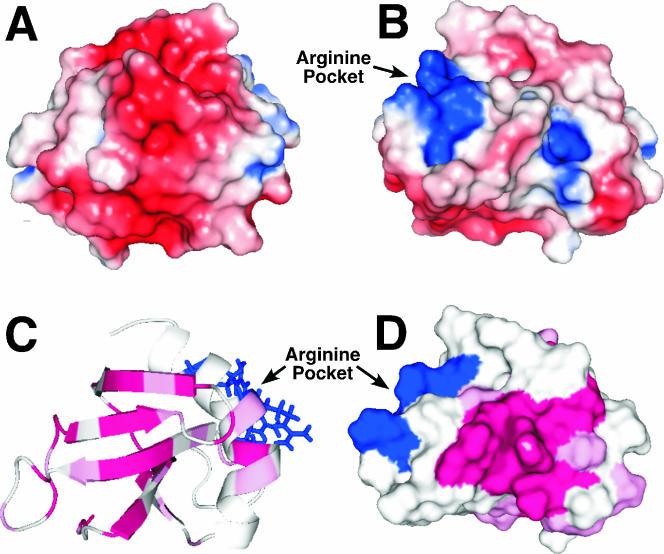
The electrostatic nature of the protein's surface often plays a role in biochemical functions (4, 10). The electrostatic potentials at the solvent-accessible surface was calculated for Rv2302 using PyMOL (DeLano Scientific, San Carlos, CA) to determine if Rv2302 has any clustering of charges on its solvent-accessible surface that may provide a continuous interface for electrostatic associations. Figure 5A and B illustrate this potential on the solvent-accessible surface in two protein orientations that differ by ∼180°. One face is highlighted by a negatively charged (red) surface, and the other face is dominated by two positively charged (blue) pockets. The largest pocket of positive charge is composed of three arginine residues, R18, R70, and R74 (Fig. 5B). While R70 and R74 are part of an α-helix, R18 is in the unstructured loop between β-strands 1 and 2 that the 1H-15N HSQC suggests has motion on the intermediate NMR timescale. Because intermediate timescale motion is often associated with activity at, or near, an active site (15), R18 may have a mechanistic role in the biochemical function of Rv2302.
FIG. 5.
(A and B) PyMOL-generated maps of the electrostatic potentials at the solvent-accessible surface of Rv2302 showing views of the protein that differ by an ∼180° rotation of the molecule about the vertical axis. Positively charged surfaces are shown in blue, and negatively charged surfaces are red. Three positively charged arginine residues, R18, R70, and R74, are clustered together to form an arginine pocket. (C) PyMOL-generated ribbon representation of the Rv2302 structure. The regions with the highest ConSurf (11) scores from Fig. 6 are shown in magenta (9) and pink (8), and the three side chains that comprise the arginine pocket are shown in blue. (D) PyMOL-generated surface representation of panel C rotated approximately 180° highlights a conserved surface-exposed region composed of loops 12 and 14.
Steady-state {1H}-15N heteronuclear NOEs.
Protein dynamics often plays a role in the binding and catalysis properties of proteins at and around active sites (15). Small, or negative, heteronuclear steady-state {1H}-15N NOE values identify regions of the protein experiencing picosecond motion (27). Figure 3B is a plot of the backbone {1H}-15N heteronuclear NOE values for Rv2302. No 1H-15N HSQC cross-peaks were observed for the backbone amides between G13 to H19, and, therefore, no values are shown for these residues. Otherwise, heteronuclear NOE values were not obtained for only five residues due to spectral overlap that prevented reliable volume measurements. Except for the N and C termini, all the heteronuclear NOE values are between 0.7 to 0.85, indicating an overall structure that is rigid and an absence of residues undergoing motion on a picosecond timescale. Consequently, the only dynamic region of Rv2302 is the part that is invisible in the 1H-15N HSQC spectra, loop L1, that undergoes motion on the millisecond to microsecond timescale.
Identification and mapping of the conserved regions of Rv2302.
A BLAST search of the Institute for Genomic Research Comprehensive Microbial Resource data bank results in only four proteins with 58% or more sequence conservation and 40% or more sequence identity with residues M1 to A71 of Rv2302. CLUSTAL W (39) sequence alignment of these four proteins relative to Rv2302 is illustrated in Fig. 6 along with the ConSurf (11) preliminary identification of the most conserved residues in this group, residues contained in β-strands 1 through 4 and loops 2 and 4. Loops 2 and 4 both contain five consecutive, highly conserved residues of consensus sequence: G(S/T)PPY and PGPD(A/S), respectively. In Fig. 5C and D the residues with the two highest ConSurf scores, 9 (magenta) and 8 (pink), are highlighted on the ribbon (5C) and surface (5D) structure of Rv2302 in orientations that differ by 180°. The conservation in the β-strands results in a highly conserved surface on one face of the β-sheet, as shown in Fig. 5C. On the opposite face (Fig. 5D), the conserved residues in loops 2 and 4 occupy a significant patch on the protein's surface. Further genomic sequencing of additional organisms and biochemical studies on Rv2302 are necessary to determine if these putative conserved regions have any role in the biochemical function of Rv2302. Interestingly, the three arginine residues that form a pocket on the surface of Rv2302 are not conserved in the aligned sequences and occupy a region adjacent to the highly conserved surfaces. Hence, if the arginine pocket has a biological function in Rv2302, it may be unique to Mycobacteria.
FIG. 6.
Sequence alignment of Rv2302 (residues M1 to A71) with four other closely related sequences using the program CLUSTAL W (39). The color scheme is as follows: hydrophobic, red; hydrophilic, green; acidic, blue; basic, magenta. Sav, Streptomyces avermitilis (Sav6750); Sco, Streptomyces coelicolor (Sco0174 and Sco1589); Tfu, Thermobifida fusca YX (Tfu1961). Residue positions with the highest ConSurf scores (11), 9 (asterisk) and 8 (caret), are indicated below the alignment. The helical and β-strand regions of Rv2302 are identified by red ovals and blue rectangles, respectively.
Insight into biological function.
To identify a possible biochemical function for Rv2302 based on its structure, the Protein Data Bank was searched for structures with similarities to Rv2302 using the Dali search engine (13). The search revealed two proteins with Z-scores of 3.2, the SH3 domain of a human obscurin fragment (1V1C) and the transcription antitermination protein nu (1M1G). All the other 30 “hits” had Z-scores between 2 and 3. The known functions of these 32 proteins varied considerably. The combination of low Z-scores and wide range of function of the proteins identified in the Dali search of the Protein Data Bank suggests that the protein fold for Rv2302 may be unique to the world of known protein structures.
Because the Dali search failed to identify a likely biochemical function for Rv2302, the program ProKnow was used to infer a possible biochemical role for the protein (32). ProKnow uses the primary amino acid sequence along with the known, or predicted (using DASEY), structure to identify and score the most likely biochemical functions of a protein (http://www.doe-mbi.ucla.edu/Services/ProKnow/biolatlas.phl). This program predicts that Rv2302 is a DNA binding protein, having a Bayesian score of 0.71, an evidence rank of 2.7, and 6 clues (Bayesian score range of 0 to 1 [best], evidence rank range of 0 to 6 [best], and clue range of 0 to 9 [best]).
Microarray studies of M. tuberculosis implicate Rv2302 in a couple of biological responses. In one study the Rv2302 gene was upregulated 4.14-, 4.84-, and 6.58-fold in response to 4, 24, and 96 h of starvation, respectively (1). Identification of such genes is important because M. tuberculosis is known to exist for long periods in a nongrowing, drug-resistant state, and proteins that are upregulated following exposure to conditions that mimic this latent state may be potential targets for new M. tuberculosis drugs. In a second study, the Rv2302 gene was upregulated 1.83-fold relative to wild-type cells in the double-deletion mutant of two heat-shock regulons, HrcA and HspR (37). Partial disruption of the regulatory circuits that affect an overexpression of heat-shock proteins in M. tuberculosis appears to play an important role in pathogenesis, impairing the ability of M. tuberculosis to establish a chronic infection (36).
Conclusions.
Rv2302 is a small, 80-residue polypeptide with no known biological function that folds into a five-strand antiparallel β-sheet with a 15-residue C-terminal α-helix nestled onto one side. While antiparallel β-sheets are common, a Dali search of this structure versus all known structures in the Research Collaboratory for Structural Bioinformatics Protein Data Bank failed to identify any other proteins with a similar structure and/or configuration of antiparallel β-strands, indicating that the Rv2302 fold has not been observed before. CD spectroscopy suggests that Rv2302 is rather robust, being able to refold back into its native structure after being heated to 80°C. If unusual molecular dynamics, relative to the molecule as a whole, plays a role in the protein's function, then loop 1 between β-strands 1 and 2 may be important. Well-defined regions of positive and negative charges on the surface of Rv2302 may provide an interface for electrostatic associations between a substrate with a well-defined electrostatic surface. For example, ProKnow predicts that Rv2302 binds DNA, and a small positively charged region is observed on the surface of Rv2302 that may be a potential surface to bind to the negatively charged phosphodiester backbone of DNA. Microarray studies of M. tuberculosis response to starvation and overexpression of heat shock proteins show that Rv2302 is upregulated, suggesting that it may play a biochemical role in starvation survival and the heat shock response. However, further studies are necessary to uncover the biochemical function of Rv2302. If a function for Rv2302 is found, the structure presented here will contribute to a molecular understanding of its function and potentially speed up the conception and development of new therapies to fight the spread of tuberculosis around the world.
Acknowledgments
The research was performed in the Environmental Molecular Sciences Laboratory (a national scientific user facility sponsored by the Department of Energy Biological and Environmental Research) located at Pacific Northwest National Laboratory and operated for the Department of Energy by Battelle. We are grateful for the support from the NIH Protein Structure Initiative.
We thank Debnath Pal at the UCLA-DOE Institute for Genomics and Proteomics for assistance with ProKnow.
REFERENCES
- 1.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 2.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. Delano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. Warren. 1998. Crystallography and NMR system (CNS): a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 3.Buchko, G. W., N. J. Hess, V. Bandaru, S. S. Wallace, and M. A. Kennedy. 2000. Spectroscopic studies of zinc(II)- and colbalt(II)-associated Escherichia coli formamidopyrimidine-DNA glycosylase: extended X-ray absorption fine structure evidence for a metal-binding domain. Biochemistry 40:12441-12449. [DOI] [PubMed] [Google Scholar]
- 4.Buchko, G. W., K. McAteer, S. S. Wallace, and M. A. Kennedy. 2005. Solution-state NMR investigation of DNA binding interactions in Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg): a dynamic description of the DNA/protein interface. DNA Repair 4:327-339. [DOI] [PubMed] [Google Scholar]
- 5.Chothia, C. 1973. Conformation of twisted β-pleated sheets in proteins. J. Mol. Biol. 75:295-302. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Cornilescu, G., F. Delagio, and A. Bax. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13:289-301. [DOI] [PubMed] [Google Scholar]
- 8.Creighton, T. E. 1993. Proteins: structure and molecular properties. W. H. Freeman and Company, New York, N.Y.
- 9.Farrow, N. A., D. R. Muhandiram, A. U. Singer, S. M. Pascal, L. E. Kay, G. Gish, S. E. Shoelson, T. Pawson, and J. D. Forman-Kay. 1994. Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33:5984-6003. [DOI] [PubMed] [Google Scholar]
- 10.Gilboa, R., D. O. Zharkov, G. Golan, A. S. Fernandes, S. E. Gerchman, E. Matz, J. H. Kycia, A. P. Grollman, and G. Shoham. 2002. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J. Biol. Chem. 277:19811-19816. [DOI] [PubMed] [Google Scholar]
- 11.Glaser, F., T. Pupko, I. Paz, R. E. Bell, D. Bechor-Shental, E. Martz, and N. Ben-Tal. 2003. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19:163-164. [DOI] [PubMed] [Google Scholar]
- 12.Grzesiek, S., J. Anglister, and A. Bax. 1993. Correlation of backbone amide and aliphatic side-chain resonaces in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Res. B 101:114-119. [Google Scholar]
- 13.Holm, L., and C. Sander. 1998. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 26:316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holzwarth, G. M., and P. Doty. 1965. The ultra-violet circular dichroism of polypeptides. J. Am. Chem. Soc. 87:218-228. [DOI] [PubMed] [Google Scholar]
- 15.Ishima, R., and D. A. Torchia. 2000. Protein dynamics from NMR. Nat. Struct. Biol. 7:740-743. [DOI] [PubMed] [Google Scholar]
- 16.John, B. K., D. Plant, and R. E. Hurd. 1993. Improved proton-detected heteronuclear correlations using gradient-enhanced Z and ZZ filters. J. Magn. Reson. B 101:113-117. [Google Scholar]
- 17.Kamholz, S. L. 2002. Drug resistant tuberculosis. J. Assoc. Acad. Minor. Phys. 13:53-56. [PubMed] [Google Scholar]
- 18.Kay, L. E., P. Keifer, and T. Saarinen. 1992. Pure absorption gradient enhanced heteronuclear single quantum correlated spectroscopy with improved sensitivity. J. Amer. Chem. Soc. 114:10663-10665. [Google Scholar]
- 19.Kay, L. E., G. Y. Xu, A. U. Singer, D. R. Muhandiram, and J. D. Forman-Kay. 1993. A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples of proteins. J. Magn. Reson. B 101:333-337. [Google Scholar]
- 20.Koradi, R., M. Billeter, and K. Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics 14:51-55. [DOI] [PubMed] [Google Scholar]
- 21.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structure. J. Appl. Cryst. 24:946-950. [Google Scholar]
- 22.Kwok, S. C., and R. S. Hodges. 2003. Clustering of large hydrophobes in the hydrophobic core of two-stranded α-helical coiled-coils controls protein folding and stability. J. Biol. Chem. 278:35248-35254. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski, R. A., J. A. C. Rullmann, M. W. MacArthur, R. Kaptein, and J. M. Thornton. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477-486. [DOI] [PubMed] [Google Scholar]
- 24.Linge, J. P., and M. Nilges. 1999. Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. J. Biomol. NMR 13:51-59. [DOI] [PubMed] [Google Scholar]
- 25.Logan, T. M., E. T. Olejniczak, R. X. Xu, and S. W. Fesik. 1993. A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J. Biomol. NMR 3:225-231. [DOI] [PubMed] [Google Scholar]
- 26.Lyons, B. A., and G. T. Montelione. 1993. An HCCNH triple-resonance experiment using 13C isotropic mixing for correlating backbone amide and side-chain aliphatic resonances in isotopically enriched proteins. J. Magn. Res. B 101:206-209. [Google Scholar]
- 27.Mandel, A. M., M. Akke, and A. G. Palmer III. 1995. Backbone dynamics of Escherichia coli ribonuclease H1-correlations with structure and function in an active enzyme. J. Mol. Biol. 246:144-163. [DOI] [PubMed] [Google Scholar]
- 28.Manning, C. M., M. Illangaseke, and R. W. Woody. 1988. Circular dichroism studies of distorted α-helices, twisted β-sheets and β-turns. Biophys. Chem. 31:77-86. [DOI] [PubMed] [Google Scholar]
- 29.Montelione, G. T., B. A. Lyons, S. D. Emerson, and M. Tashiro. 1992. An efficient triple resonance experiment using carbon-13 isotropic mixing for determining sequence-specific resonance assignments of isotopically enriched proteins. J. Am. Chem. Soc. 114:10974-10975. [Google Scholar]
- 30.Muhandiram, D. R., and L. E. Kay. 1994. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. B 103:203-216. [Google Scholar]
- 31.Neri, D., T. Szyperski, G. Otting, H. Senn, and K. Wütrich. 1989. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional carbon-13 labeling. Biochemistry 28:7510-7516. [DOI] [PubMed] [Google Scholar]
- 32.Pal, D., and D. Eisenberg. 2005. Inference of protein function from protein structure. Structure 13:121-130. [DOI] [PubMed] [Google Scholar]
- 33.Pascal, S. M., D. R. Muhandiram, T. Yamazaki, J. D. Forman-Kay, and L. E. Kay. 1994. Simultaneous acquisition of 15N- and 13C-edited NOE spectra of proteins dissolved in H2O. J. Magn. Res. B 103:197-201. [Google Scholar]
- 34.Schubert, M., D. Labudde, H. Oschkinat, and P. Schmieder. 2002. A software tool for the prediction of Xaa-Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J. Biomol. NMR 24:149-154. [DOI] [PubMed] [Google Scholar]
- 35.Smith, J. A., and L. G. Pease. 1980. Reverse turns in peptides and proteins. CRC Crit. Rev. Biochem. 8:315-399. [DOI] [PubMed] [Google Scholar]
- 36.Stewart, G. R., V. A. Snewsin, G. Walzl, T. Hussell, P. Tormay, P. O'Gaora, M. Goyal, J. C. Betts, I. N. Brown, and D. B. Young. 2001. Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection. Nat. Med. 7:732-737. [DOI] [PubMed] [Google Scholar]
- 37.Stewart, G. R., L. Wernisch, R. Stabler, J. A. Mangan, J. Hinds, K. G. Laing, D. B. Young, and P. D. Butcher. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148:3129-3138. [DOI] [PubMed] [Google Scholar]
- 38.Terwilliger, T. C., M. S. Park, G. S. Waldo, J. Berendzen, L.-W. Hung, C.-Y. Kim, C. V. Smith, J. C. Sacchettini, M. Bellinzoni, R. Bossi, E. De Rossi, A. Mattevi, A. Milano, G. Riccardi, M. Rizzi, M. M. Roberts, A. R. Coker, G. Fossati, P. Mascagni, A. R. M. Coates, S. P. Wood, C. W. Goulding, M. I. Apostol, D. H. Anderson, H. S. Gill, D. S. Eisenberg, B. Taneja, S. Mande, E. Pohl, V. Lamzin, P. Tucker, M. Wilmanns, C. Colovos, W. Meyer-Klaucke, A. W. Munro, K. J. McLean, K. R. Marshall, D. Leys, J. K. Yang, H.-J. Yoon, B. I. Lee, J. E. Kwak, B. W. Han, J. Y. Lee, S.-H. Baek, S. W. Suh, M. M. Komen, V. L. Arcus, E. N. Baker, J. S. Lott, W. Jacobs, Jr., T. Albers, and B. Rupp. 2003. The TB structural genomics consortium: a resource for Mycobacterium tuberculosis biology. Tuberculosis 8:223-249. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vuister, G. W., G. M. Clore, A. M. Gronenborn, R. Powers, D. S. Garrett, R. Tschudin, and A. Bax. 1993. Increased resolution and improved spectral quality in 4-dimensional 13C/13C-separated HMQC-NOESY-HMQC spectra using pulsed-field gradients. J. Magn. Res. B 101:201-213. [Google Scholar]
- 41.Wishart, D. S., C. G. Bigam, J. Yao, F. Abildgaard, H. J. Dyson, E. Oldfield, J. L. Markley, and B. D. Sykes. 1995. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6:135-140. [DOI] [PubMed] [Google Scholar]
- 42.Wishart, D. S., and B. D. Sykes. 1994. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical shift data. J. Biomol. NMR 4:171-180. [DOI] [PubMed] [Google Scholar]
- 43.Woody, R. W. 1974. Studies of theoretical circular dichroism of polypeptides: Contributions of β-turns. John Wiley & Sons, New York, N.Y.
- 44.World Health Organization. 2005. Tuberculosis fact sheet no. 104. World Health Organization, Geneva, Switzerland.
- 45.Zhang, O., L. E. Kay, J. P. Olivier, and J. D. Forman-Kay. 1994. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed-field gradient NMR techniques. J. Biomol. NMR 4:845-858. [DOI] [PubMed] [Google Scholar]