Abstract
Reovirus replication occurs in the cytoplasm of infected cells and culminates in the formation of crystalline arrays of progeny virions within viral inclusions. Two viral nonstructural proteins, σNS and μNS, and structural protein σ3 form protein-RNA complexes early in reovirus infection. To better understand the minimal requirements of viral inclusion formation, we expressed σNS, μNS, and σ3 alone and in combination in the absence of viral infection. In contrast to its concentration in inclusion structures during reovirus replication, σNS expressed in cells in the absence of infection is distributed diffusely throughout the cytoplasm and does not form structures that resemble viral inclusions. Expressed σNS is functional as it complements the defect in temperature-sensitive, σNS-mutant virus tsE320. In both transfected and infected cells, μNS is found in punctate cytoplasmic structures and σ3 is distributed diffusely in the cytoplasm and the nucleus. The subcellular localization of μNS and σ3 is not altered when the proteins are expressed together or with σNS. However, when expressed with μNS, σNS colocalizes with μNS to punctate structures similar in morphology to inclusion structures observed early in viral replication. During reovirus infection, both σNS and μNS are detectable 4 h after adsorption and colocalize to punctate structures throughout the viral life cycle. In concordance with these results, σNS interacts with μNS in a yeast two-hybrid assay and by coimmunoprecipitation analysis. These data suggest that σNS and μNS are the minimal viral components required to form inclusions, which then recruit other reovirus proteins and RNA to initiate viral genome replication.
Mammalian reoviruses are members of the family Reoviridae and contain a genome of 10 segments of double-stranded RNA (dsRNA). The genome is surrounded by two protein shells termed the outer capsid and core. Following internalization by receptor-mediated endocytosis (5, 13, 73, 85) and removal of the outer capsid in the endocytic compartment (4, 18, 80, 85), the core is released into the cytoplasm (12, 41, 42, 55, 88). The core catalyzes all of the biochemical activities necessary to synthesize and release capped, message-sense, single-stranded RNA (ssRNA) copies of the 10 dsRNA gene segments (6, 14, 35, 82). These mRNAs are competent templates for translation and encode 11 proteins (14, 47, 58, 96, 97). Eight viral proteins form the virion; the remaining three proteins, σ1s, σNS, and μNS, are termed nonstructural and are found only in infected cells (9, 54, 83, 97). The viral mRNAs also serve as templates for minus-strand synthesis, resulting in nascent genomic dsRNA (52, 76). This step appears to be concomitant with assortment of the 10 viral gene segments into particles that are destined to become progeny virions (1). These particles are also transcriptionally active (62, 95, 98) and produce the majority of the viral mRNA and protein during the viral life cycle (48, 62). Outer-capsid proteins are added to progeny particles, silencing transcription and completing virion morphogenesis (3, 8). Late in infection, the majority of the cytoplasm of infected cells consists of viral inclusions replete with progeny virions (34).
Little is known about the establishment and expansion of viral inclusions. These structures have been studied by using a variety of microscopic techniques and first appear by phase-contrast microscopy as dense granules scattered throughout the cytoplasm. As infection progresses, these granules coalesce and localize about the nucleus, eventually forming perinuclear inclusions (34). Viral inclusions contain several types of filaments (78), dsRNA (81), viral proteins (34), and complete and incomplete viral particles (34). In contrast to cytoplasmic sites of replication used by several other viruses, reovirus inclusions are not thought to be associated with membranes or other cellular organelles (39, 70). A previous study implicated reovirus nonstructural proteins σNS and μNS and structural protein σ3 in early steps of genome replication by showing that these three proteins are found in the earliest detectable viral protein-RNA complexes in infected cells (1). These proteins may form the structures necessary to initiate viral inclusion formation.
Reovirus inclusions can be observed by confocal immunofluorescence microscopy to coincide with the large arrays of progeny virions visualized by electron microscopy (7). Immunofluorescence staining indicates that inclusion structures contain both structural and nonstructural proteins, the exact composition of which changes with time (7). Large cytoplasmic complexes of viral proteins also can be observed following infection of cells with some temperature-sensitive (ts) mutant viruses (7, 57). Since these infections do not yield viable progeny, these inclusion-like structures are not functional, presumably because of alterations in the activity of the mutant viral protein. A direct assay to monitor the nature and function of reovirus inclusions has not been reported.
The σNS protein is encoded by the S3 gene and consists of 366 amino acids with a molecular mass of 41 kDa (58, 63). Reovirus σNS isolated from infected cells and expressed in vitro binds ssRNA nonspecifically and forms higher-order structures (37, 38, 43, 71). Smaller complexes containing four (plus or minus two) monomers are present after treatment of either native (43) or recombinant (37) σNS with RNase A. Higher-order homo-oligomeric structures are seen when RNA is present, and these structures are most likely the protein complexes competent for binding of ssRNA in a cooperative manner (38; T.R.P., unpublished observations). During reovirus infection, σNS localizes to viral inclusions and is necessary for their formation (7).
The μNS protein is encoded by the M3 gene and consists of 766 amino acids with a molecular mass of 80 kDa (58, 63). It associates with viral mRNAs (1) and viral cores but not virions or disassembly intermediates (15). In immunofluorescence studies, μNS colocalizes with cytoskeletal elements in infected cells (61), and this interaction appears to be mediated by the μ2 protein (16, 66). Cells transfected with μNS-expressing plasmids form cytoplasmic globular structures that contain μNS (16). μNS mutants have not been reported, and its contribution to functional reovirus inclusion formation has not been defined.
The σ3 protein is encoded by the S4 gene and consists of 365 amino acids with a molecular mass of 41 kDa (58, 63). It is a component of the viral outer capsid and binds dsRNA nonspecifically (27, 43, 53, 56, 60, 74). The σ3 protein has been implicated in several processes that occur in infected cells, including blockade of the interferon-inducible dsRNA-dependent protein kinase PKR (36, 45) and inhibition of cellular protein synthesis (75, 79). However, a role for σ3 in the formation of viral protein-RNA complexes remains unclear.
To probe the steps leading to the formation of reovirus inclusions, we used confocal immunofluorescence microscopy to examine the subcellular localization of σNS in the absence of viral infection. We determined the effects of ectopic σNS expression on the production of infectious viral progeny and subcellular localization of viral proteins during infection with wild-type (wt) and ts mutant reoviruses. We examined the subcellular localization of the σNS, μNS, and σ3 proteins, individually and in combination, in the presence and absence of reovirus replication. The results suggest that σNS and μNS are the minimal components necessary to establish an intracellular site of viral replication and initiate the activities required to complete the production of progeny virions.
MATERIALS AND METHODS
Cells and viruses.
Murine L929 (L) cells were grown in either suspension or monolayer cultures in Joklik's modified Eagle's minimal essential medium (Irvine Scientific, Santa Ana, Calif.) that was supplemented to contain 5% fetal calf serum (Gibco Invitrogen Cell Culture Products, Grand Island, N.Y.), 2 mM l-glutamine, 100 U of penicillin G/ml, 100 μg of streptomycin/ml, and 250 ng of amphotericin B/ml (Irvine Scientific). Human embryonic kidney 293T cells and HeLa cells were maintained in Dulbecco's modified eagle medium (DMEM; Gibco) supplemented to contain 10% fetal calf serum (Gibco), 2 mM l-glutamine, 100 U of penicillin G/ml, 100 μg of streptomycin/ml, and 250 ng of amphotericin B/ml (Irvine Scientific). Spodoptera frugiperda (Sf21) cells were grown in Grace's medium (Gibco) supplemented to contain 10% fetal calf serum (Gibco), 100 U of penicillin G/ml, 100 μg of streptomycin/ml, and 250 ng of amphotericin B/ml (Irvine Scientific).
Reovirus strains type 1 Lang (T1L) and type 3 Dearing (T3D) are laboratory stocks. Mutant viruses tsE320 and tsH11.2 were grown from stocks originally obtained from Bernard Fields (32) and Kevin Coombs (24), respectively. Second- or third-passage L-cell lysate stocks of isolates of each strain plaque purified twice were used for subsequent studies. Virus titers were determined by plaque assay with L-cell monolayers as previously described (24).
Plasmids.
Genomic dsRNA isolated from T3D virions was used as the template for reverse transcriptase PCR amplification of the σNS open reading frame (ORF) of the S3 gene by using the following primers with 5′ SalI and 3′ SpeI restriction sites (lowercase letters) appended: 5′-gtcgacCACTATGGCTTCCTCACTCAGAG-3′ and 5′-actagtCATTACACGCGAATCGGAAA-3′. PCR products were ligated into pCRII (Invitrogen Life Technologies, Carlsbad, Calif.), and nucleotide sequences were determined by fluorescent dideoxy-chain termination with an ABI Prism 377 DNA Sequencer (Applied Biosystems, Foster City, Calif.). One nucleotide change from the reported sequence of the S3 gene (71) was found in several independent cDNA clones, but it did not result in a substitution in the deduced amino acid sequence of σNS. The S3 cDNA was subcloned into the multiple cloning site of the pTet-Splice vector (Tet-Regulated Expression System; Gibco) to generate the plasmid S3/pTet, which allows σNS expression to be regulated by the tetracycline-responsive promoter Ptet. pTet-tTAk contains the transactivator protein (tTA) under the control of Ptet. LTR-neo, a plasmid encoding the aminoglycoside O-phosphotransferase gene from Tn5 (Neor), was used as a selectable marker.
Yeast two-hybrid cloning vector pGBKT7 [GAL4(1-147) DNA-BD TRP1 Kanr c-myc epitope tag] was used for bait constructs, and pGADT7 [GAL4(768-881) DNA-AD LEU2 Ampr influenza virus hemagglutinin (HA) epitope tag] was used for prey constructs (Clontech, Palo Alto, Calif.). S3/pTet was used as the template for PCR amplification of the σNS-encoding sequence containing primer-templated 5′ EcoRI and 3′ BamHI restriction endonuclease cleavage sites (5′-GgaattcACTATGGCTTCCTCACTCAGAGCT-3′ and 5′-CGggattcTTACACGCGAATCGGAAAAACCAGCAG-3′) with Pfu DNA polymerase (Promega, Madison, Wis.). PCR products were digested with EcoRI and BamHI and ligated into pGBKT7 to generate pBS3wt. The nucleotide sequence of the insert was determined, which indicated that it correctly encoded full-length reovirus T3D σNS for fusion protein expression. The σNS-encoding region from pBS3wt was transferred to pGADT7 following digestion with EcoRI and BamHI to generate pPS3wt.
A cDNA corresponding to the T3D S4 gene (91) was used as the template for PCR amplification of the σ3-encoding sequence containing primer-templated 5′ EcoRI and 3′ BamHI restriction sites (5′-GgaattcACAATGGAGGTGTGCTTGCCCAACGGT-3′ and 5′-CGggatccTTAGCCAAGAATCATCGGATCGCCAATCAT-3′) with Pfu DNA polymerase (Promega). PCR products were digested with EcoRI and BamHI and ligated into pGBKT7 to generate pBS4wt. Confirmation of the nucleotide sequence of this insert indicated that it correctly encoded full-length reovirus T3D σ3 for fusion protein expression. The σ3-encoding region from pBS4wt was transferred to pGADT7 following digestion with EcoRI and BamHI to generate pPS4wt.
T3D genomic dsRNA was used as the template for reverse transcriptase PCR amplification of the M3 gene. Nucleotide sequences corresponding to the full-length reovirus T3D μNS ORF were cloned into pGBKT7 to create pBM3wt. The nucleotide sequence of the μNS coding region of pBM3wt differs from the T3D M3 sequence reported previously (59) at three positions, each resulting in a substitution in the deduced amino acid sequence of μNS (K180E, A705V, and G707D). The μNS prey construct was made by first digesting pBM3wt with NcoI and then filling in the resulting 3′ overhang sequence with the DNA polymerase I large (Klenow) fragment (New England Biolabs, Beverly, Mass.). Linearized pBM3wt was digested with BamHI to release the μNS ORF, which was ligated into SmaI-BamHI-digested pGADT7 to generate pPM3wt.
Mammalian expression vectors encoding σNS, σ3, and μNS were constructed for transient transfections. The σNS ORF was transferred from pBS3wt to the multiple cloning site of pCMV-Script (Stratagene, La Jolla, Calif.) with the restriction sites EcoRI and SalI to generate pCMVS3wt. Similarly, the σ3 ORF was transferred from pBS4wt to pCMV-Script with the restriction sites EcoRI and SalI to generate pCMVS4wt. The μNS ORF was excised from pBM3wt by first digesting pBM3wt with NcoI and then filling in the 5′-to-3′ overhang sequence with the Klenow fragment (New England Biolabs). Linearized pBM3wt was digested with SalI to release a fragment encoding full-length μNS, which was ligated to pCMV-Script with EcoRV and SmaI sites to create pCMVM3wt.
Stable cell lines.
S3/pTet, pTet-tTak, and LTR-neo were linearized and transfected into L cells with Lipofectamine Plus (Gibco) in accordance with the manufacturer's instructions. Control cell lines were generated by transfection with nonrecombinant linearized pTet-splice, pTet-tTak, and LTR-neo. After 3 weeks of selection, cells were cloned by sorting into 96-well plates with a FACStar Plus (Becton Dickinson, San Jose, Calif.). Cloned cultures were screened for expression of σNS by confocal immunofluorescence microscopy after incubation in doxycycline-free medium for 2 days. Three cell lines expressing detectable amounts of σNS (σNS-1, -2, and -3) and two control cell lines (control-1 and -2) were maintained.
Expression and purification of recombinant σNS.
A σNS-encoding baculovirus transfer vector, pBacS3wt, was constructed by digestion of pBS3wt with EcoRI and PstI and ligation of the σNS ORF-containing fragment into these sites in pBacPAK9 (Clontech). A third-passage stock of σNS-expressing baculovirus was used to infect 1.5 × 109 Sf21 insect cells at a multiplicity of infection (MOI) of 10 PFU/cell. After incubation at 25°C for 96 h, cells were washed once with phosphate-buffered saline (PBS) and incubated on ice in 30 ml of cytoplasmic extract buffer (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride) containing Complete EDTA-free protease inhibitor cocktail (Roche Applied Science, Indianapolis, Ind.). After 15 min of incubation, 1.5 ml of 10% Igepal CA-630 (Sigma-Aldrich, Milwaukee, Wis.) was added and the mixture was vortexed. The resulting suspension was centrifuged at 8,000 × g for 10 min, and the σNS-containing supernatant was collected. The supernatant was dialyzed against 20 mM Tris-HCl-50 mM NaCl (pH 8.0), passed through a 0.22-μm-pore-size cellulose acetate filter, and loaded onto a Q Sepharose anion-exchange column (Amersham Pharmacia Biotech, Piscataway, N.J.). The column flowthrough, which contained σNS, was dialyzed against 20 mM 3-(N-morpholine)propanesulfonic acid (MOPS)-50 mM NaCl (pH 6.5) and loaded onto a Macro-Prep High S support cation-exchange column (Bio-Rad, Hercules, Calif.). The σNS-containing flowthrough was concentrated twofold with an Ultrafree-15 centrifugal filter device (Millipore, Bedford, Mass.) and passed through a 0.22-μm-pore-size cellulose acetate filter. Pure recombinant σNS was obtained from the filtered preparation with a Superdex 200 gel filtration column (Amersham Pharmacia). The σNS protein eluted from the gel filtration column as a large multimer in the void volume.
Generation of guinea pig antiserum.
σNS-specific antiserum was obtained by inoculating a guinea pig with 50 μg of purified baculovirus-expressed σNS in incomplete Freund's adjuvant, followed by 25-μg booster doses at 2, 3, and 7 weeks postinoculation (Cocalico, Reamstown, Pa.). Antiserum was obtained 4 weeks after administration of the last booster.
Antibodies.
The immunoglobulin G (IgG) fraction was purified from rabbit polyclonal σNS-specific (7), rabbit polyclonal μNS-specific (15), and guinea pig polyclonal σNS-specific antisera by protein A-Sepharose affinity chromatography (Pierce, Rockford, Ill.). Eluted IgG was dialyzed exhaustively against PBS and then concentrated to 2 mg/ml with a Centricon YM-30 centrifugal filter device (Millipore). Mouse monoclonal antibodies (MAbs) 4F2 (σ3 specific) and 8H6 (μ1/μ1C specific) (90) were purified from hybridoma supernatants (Cell Culture Center, Minneapolis, Minn.).
Immunoblotting.
Cells from each stably transfected cell line (σNS-1, -2, and -3 and control-1 and -2) were uninduced or induced to express σNS by removal of doxycycline for 1, 2, 3, or 4 days. Cells were lysed, sheared, and frozen at −70°C. Lysates were thawed on ice, and 250 μg of total cellular protein from each sample was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Trans-Blot Transfer Medium; Bio-Rad) (72). After incubation overnight with TBS-T (50 mM Tris [pH 7.5], 150 mM NaCl, 0.5% Tween 20), the blot was probed with rabbit σNS-specific antiserum at a dilution of 1:1,000, followed by donkey rabbit IgG-specific antiserum conjugated to horseradish peroxidase as the secondary antibody. The blot was incubated with peroxidase substrate (ECL; Amersham Pharmacia Biotech) and exposed to film.
293T cells (106) were plated on 100-mm-diameter tissue culture dishes overnight and either adsorbed with T3D at an MOI of 10 PFU/ml or transfected with pCMVS3wt with calcium phosphate. Infected cells were incubated at 37°C for 18 h, transfected cells were incubated at 37°C for 24 or 36 h, and both groups of cells were processed for immunoblotting with guinea pig σNS-specific antiserum.
Yeast two-hybrid direct pairwise tests.
Budding yeast two-hybrid strain AH109 (Clontech) with the HIS3, ADE2, and MEL1 reporter genes downstream of heterologous GAL4-responsive promoter elements was transformed with pBS3wt, pPS3wt, pBM3wt, pPM3wt, pBS4wt, and pPS4wt in pairwise combinations with lithium acetate. Cells were plated onto synthetic dropout medium lacking tryptophan, histidine, leucine, and adenine and supplemented with 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (Clontech) to select for interacting proteins.
In vitro coimmunoprecipitation of protein pairs.
In vitro translation of reovirus proteins was performed with the TNT T7 Quick-Coupled Transcription/Translation System (Promega) in accordance with the manufacturer's instructions. Plasmids encoding σNS, μNS, and σ3 were used as templates to generate fusion proteins with N-terminal c-Myc and HA epitope tags in the absence or presence of [35S]methionine and [35S]cysteine (Easy Tag Express 35S Protein Labeling Mix; Perkin-Elmer Life Sciences, Boston, Mass.). Plasmid pGBKT7-LAM (Clontech), which encodes human lamin C with a c-Myc epitope tag appended, was used as a control. Approximately equal amounts of translated proteins were mixed in PBS with protease inhibitors and incubated at 4°C for 120 min with gentle agitation. Either mouse anti-c-Myc MAbs or rabbit polyclonal anti-HA antibodies (Clontech) bound to protein G Sepharose (Amersham Pharmacia Biotech) were added, and the protein-antibody mixtures were incubated at 4°C for an additional 90 min. Sepharose beads were washed six times with 1% Triton X-100 in PBS, and proteins were resolved in a 10% polyacrylamide gel. Alternatively, in vitro translation mixtures were treated with RNase by the addition of affinity-purified RNase A (Ambion) to a final concentration of 100 μg/ml and incubated at room temperature for 1 h prior to electrophoresis.
Transfections.
293T cells (3 × 104) were seeded on 12-mm glass coverslips (VWR Scientific Products, Atlanta, Ga.) approximately 24 h prior to transfection. For each well of a 24-well plate, a transfection mixture containing 0.3 μg of each plasmid, 1.6 μl of 2.5 M CaCl2, 14.7 μl of sterile water, and 1.6 μl of 2× BBS (N,N-bis[2-hydroxyethyl]-2-aminoethanesulfonic acid [BES]-buffered saline, 50 mM BES [pH 6.95], 280 mM NaCl, 1.5 mM Na2HPO4) was added. After 24 h of incubation, the medium was replaced with fresh, complete DMEM. Cells were washed and fixed 36 h after transfection.
HeLa cells (3 × 104) or L cells (5 × 104) were seeded on 12-mm glass coverslips 24 h prior to transfection. Cells were transfected with 0.4 μg of each plasmid with Lipofectamine Plus Reagent (Gibco) diluted in incomplete DMEM. Cells were washed and fixed 36 h after transfection.
Immunofluorescence staining.
293T cells (3 × 104), HeLa cells, (3 × 104), or L cells (5 × 104) were seeded on 12-mm glass coverslips for 24 h prior to infection with reovirus at an MOI of 10 PFU/cell. After adsorption at 4°C for 1 h, cells were incubated at 32, 37, or 39.5°C for various intervals, washed twice with PBS, and fixed in 1:1 methanol-acetone. Fixed cells were washed two times in PBS and incubated for 15 min in PBS containing 2.5% gamma globulin-free bovine serum albumin (BSA; Sigma Aldrich). Nonspecific binding of antibody was blocked by incubation of cells for 10 min in PBS containing 1% BSA, 1% Triton X-100 (Bio-Rad), and 2% normal goat serum (Vector Laboratories, Inc., Burlingame, Calif.). Subsequent washes and dilutions were performed with this buffer unless otherwise indicated. Cells were incubated with the primary antibody for 1 h at a concentration of 10 μg/ml (MAb 4F2 or 8H6) or 2 μg/ml (IgG fraction of polyclonal σNS- or μNS-specific antiserum) and washed two times. The wash solution was replaced with PBS, and cells were incubated at 4°C for 2 to 7 days and washed once with wash solution. Cells were incubated with Alexa Fluor 488, 546, or 647 goat anti-mouse IgG, Alexa Fluor 488, 546, or 647 goat anti-rabbit IgG, and Alexa Fluor 488, 546, or 647 goat anti-guinea pig IgG (Molecular Probes, Inc., Eugene, Oreg.) at a dilution of 1:1,000 for 1 to 1.5 h. Cells were incubated with the dsDNA-specific dye TO-PRO3 (Molecular Probes) at a dilution of 1:1,000 if only two primary and secondary antibodies were used. Cells were washed twice for 15 min each time with PBS-BSA (1%)-Triton X-100 (1%) and twice for 10 min with PBS. Coverslips were either immediately mounted or left at 4°C for 1 to 2 days and then mounted on glass slides with either Aqua Poly/Mount (Polysciences, Inc., Warrington, Pa.) or ProLong Antifade (Molecular Probes). Cells were visualized with a Zeiss 40× PanFluor NA1.3 objective on a Zeiss 410 confocal fluorescence microscope (Carl Zeiss, New York, N.Y.). A differential interference contrast (DIC) image of each field of view was obtained to determine the location of cells. Coverslips containing mock-infected or -transfected cells were included in every experiment and processed in parallel with infected cells. Mock-infected or -transfected cells were examined first and used to set the background on the confocal microscope to black before obtaining images of infected or transfected cells. Images were processed and colored with Adobe Photoshop (Adobe Systems, Inc., San Jose, Calif.).
RESULTS
The reovirus σNS protein is distributed diffusely in the cytoplasm when expressed in the absence of viral infection.
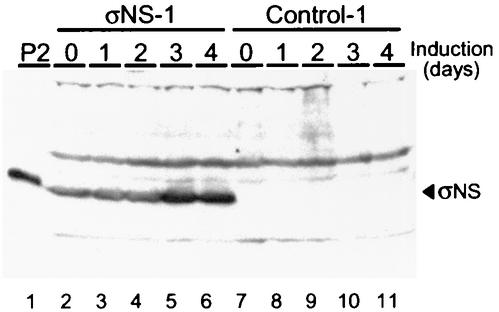
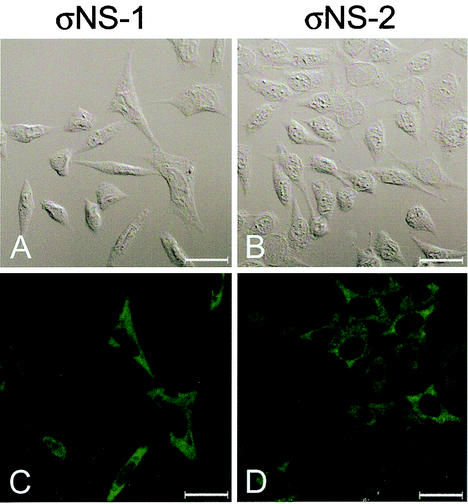
We have previously reported that functional σNS protein is required for the formation of viral inclusions and for efficient viral growth (7). To determine the subcellular localization of σNS in the absence of viral infection, L cells were engineered to express the T3D σNS protein under the control of a tetracycline-repressible promoter. Following stable transfection, three σNS-expressing cell lines (σNS-1, -2, and -3) and two control cell lines (control-1 and -2) were established and characterized. All three σNS-expressing cell lines produced a small amount of σNS in the presence of doxycycline; however, levels of σNS protein increased over time in the absence of doxycycline (Fig. 1 and data not shown). The three σNS-expressing cell lines and the two control cell lines were induced by removal of doxycycline and examined by confocal immunofluorescence microscopy (Fig. 2 and data not shown). In σNS-expressing cells, the protein was found distributed diffusely in the cytoplasm, in stark contrast to the punctate staining pattern of σNS seen in reovirus-infected cells (7). Neither of the control cell lines exhibited any σNS-specific staining. These findings indicate that the subcellular localization of σNS in reovirus-infected cells depends on viral or cellular factors acting in concert with σNS.
FIG. 1.
Protein production in stably transfected control and σNS-expressing cell lines. Whole-cell lysates were obtained from the σNS-expressing cell line σNS-1 and the control cell line control-1 after induction of σNS expression by removal of doxycycline for the times shown. Total cellular protein in each lysate was normalized, resolved by SDS-PAGE, transferred to nitrocellulose, and examined for the presence of σNS by immunoblotting. In lane 1, a second-passage (P2) lysate stock of T3D was included as a source of σNS produced during viral infection. Lanes 2 through 6 contain lysates generated from σNS-1 cells uninduced and induced for 1, 2, 3, and 4 days. Lanes 7 through 11 contain lysates generated from control-1 cells uninduced and induced for 1, 2, 3, and 4 days.
FIG. 2.
Subcellular localization of σNS protein in L cells stably transfected with a T3D S3 cDNA. σNS-1 (A and C) and σNS-2 (B and D) cells were induced by the removal of doxycycline for 2 days and then fixed. Cells were stained for σNS with σNS-specific polyclonal rabbit antiserum as the primary antibody and goat anti-rabbit Alexa Fluor 488 as the secondary antibody. A DIC image of each field was obtained (A and B). Images were obtained with a confocal microscope. The σNS protein is colored green. Images were processed with Adobe Photoshop. Bars, 25 μm.
Expressed σNS protein does not disrupt viral replication.
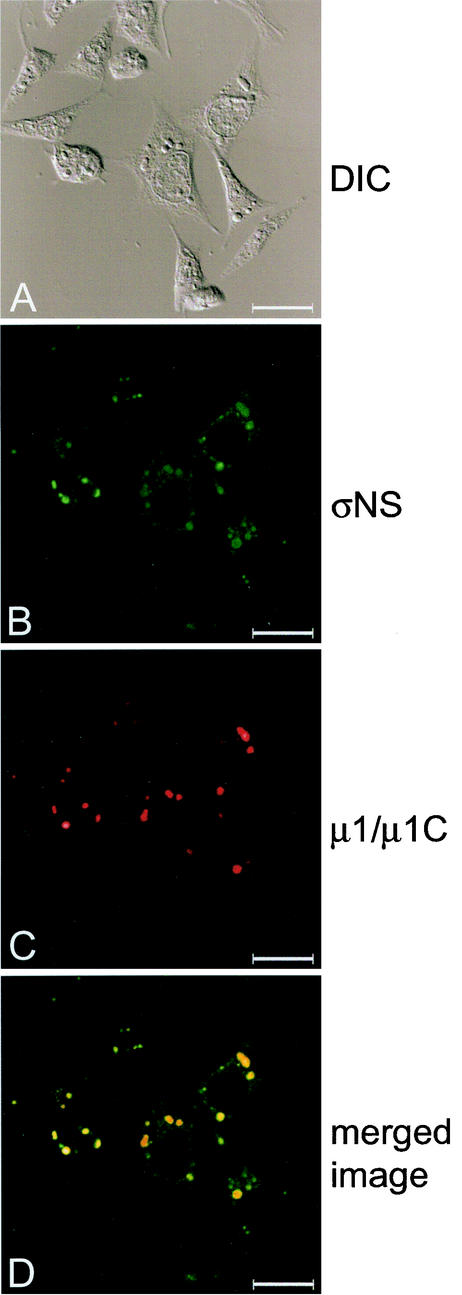
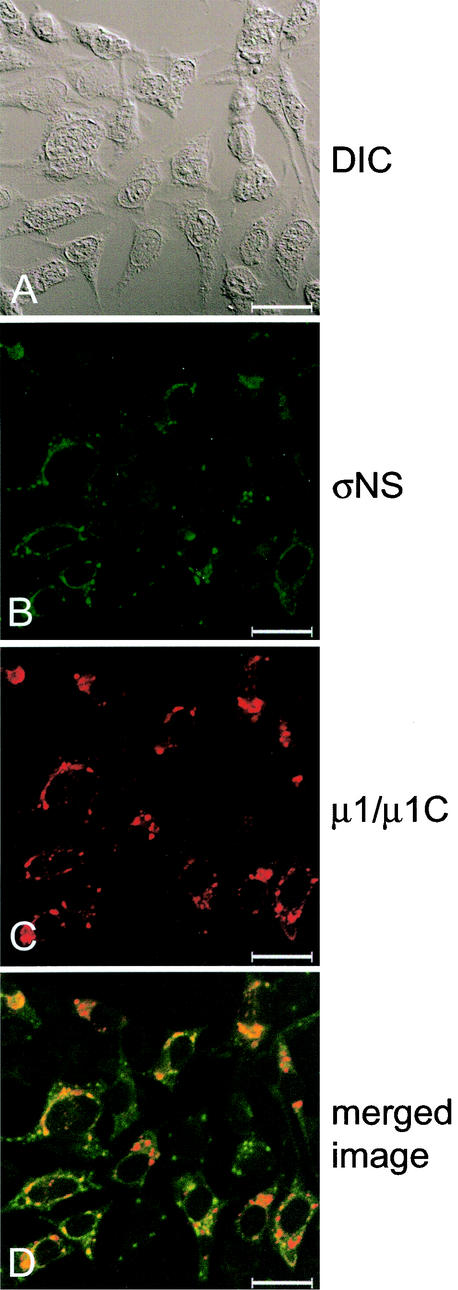
To examine whether the presence of exogenous σNS alters viral replication, we infected σNS-expressing cells with reovirus strains T1L and T3D. Control-1 and σNS-1 cells were induced by removal of doxycycline, infected with either T1L or T3D at an MOI of 10 PFU/cell, and incubated at 37°C for 24 h. Cells were then imaged with confocal immunofluorescence microscopy (Fig. 3 and data not shown). σNS-1 cells infected with either T1L or T3D were indistinguishable from control-1 cells infected with these strains, with the exception that σNS was not solely localized to viral inclusions in infected σNS-1 cells. In these cells, σNS also was dispersed to various degrees in the cytoplasm in regions between the inclusions (Fig. 3 and data not shown). Viral inclusions present in σNS-expressing cells were indistinguishable from those formed in control cells. In addition, the subcellular localization of viral outer-capsid protein μ1/μ1C, used as a marker for mature inclusions (7), was not altered by the expression of σNS prior to or during the infectious cycle (Fig. 3 and data not shown).
FIG.3.
Subcellular localization of σNS and μ1/μ1C proteins in σNS-expressing cells infected with T3D. σNS-1 cells were induced by the removal of doxycycline for 2 days and infected with T3D at an MOI of 10 PFU/cell. Following incubation at 37°C for 18 h, cells were stained for σNS with σNS-specific polyclonal guinea pig antiserum and for μ1/μ1C with μ1/μ1C-specific MAb 8H6. The secondary antibodies used were goat anti-guinea pig Alexa Fluor 488 and goat anti-mouse Alexa Fluor 546. Images were obtained with a confocal microscope. The σNS protein is colored green (B), and the μ1/μ1C protein is colored red (C). In the merged image (D), colocalization of σNS and μ1/μ1C is indicated by the color yellow. A DIC image of each field was obtained (A). Images were processed with Adobe Photoshop. Bars, 25 μm.
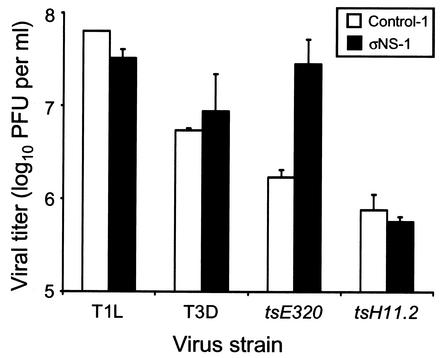
To determine whether production of infectious progeny virus was impaired by the presence of σNS prior to viral adsorption or by the increased amount of σNS during the course of viral replication, control-1 and σNS-1 cells were infected with either T1L or T3D and viral titers were determined by plaque assay after 24 h of viral growth (Fig. 4). Neither the expression of σNS prior to infection nor the presence of higher levels of σNS during the course of infection altered the yield of infectious viral progeny. These data indicate that expression of σNS during viral infection does not alter the morphology of viral replication or production of progeny virus.
FIG. 4.
Growth of wt and ts mutant viruses in control and stably transfected σNS-expressing cells. Monolayer cultures of control-1 and σNS-1 cells (5 × 105) were infected with T1L, T3D, tsE320, or tsH11.2 at an MOI of 10 PFU/cell. Cultures infected with T1L and T3D were incubated at 37°C for 24 h, and cultures infected with tsE320 and tsH11.2 were incubated at 39.5°C for 24 h. Virus titer was determined by plaque assay with L cells incubated at 37°C for T1L and T3D and at 32°C for tsE320 and tsH11.2. The results presented are the mean viral titers of at least six independent wells. Error bars indicate standard deviations of the means.
Expressed σNS protein complements ts mutant virus tsE320.
To determine whether σNS protein expressed in the stably transfected cell lines is functional, we infected σNS-expressing cells and control cells with σNS-mutant virus tsE320 (32) at the nonpermissive temperature. The tsE320 virus contains a lesion in the S3 gene (68) that results in a methionine-to-threonine mutation at amino acid 260 in the σNS protein (7, 92). Control-1 and σNS-1 cells were induced to express σNS by removal of doxycycline, infected with either tsE320 or tsH11.2 at an MOI of 10 PFU/cell, and incubated at the nonpermissive temperature of 39.5°C for 24 h. The ts mutant virus tsH11.2 contains a lesion in the μ2-encoding M1 gene (21, 24) and was used in this experiment as a control. Cells were examined by confocal immunofluorescence microscopy (Fig. 5), and viral titers were determined by plaque assay (Fig. 4). The σNS-1 cells infected with tsE320 at the nonpermissive temperature were morphologically indistinguishable from those infected with wt T3D. In both T3D- and tsE320-infected σNS-1 cells, σNS was observed in viral inclusions and diffusely in the cytoplasm (Fig. 5). The μ1/μ1C protein was clearly detectable in these cells (Fig. 5). However, in tsE320-infected control-1 cells, σNS was distributed throughout the cytoplasm, exhibiting a diffuse, granular staining pattern, and μ1/μ1C was not detectable (data not shown). The staining pattern of σNS and μ1/μ1C in control-1 cells infected at the nonpermissive temperature with tsE320 was identical to that following infection of untransfected L cells with tsE320 (7). With the exception of the higher levels of σNS present in σNS-1 cells, the staining patterns of σNS and μ1/μ1C were similar in σNS-1 and control-1 cells infected with tsH11.2 (data not shown). The σNS and μ1/μ1C proteins were present in punctate, cytoplasmic structures indistinguishable from the staining patterns observed previously in tsH11.2-infected L cells (7).
FIG.5.
Subcellular localization of σNS and μ1/μ1C proteins in σNS-expressing cells infected with tsE320. σNS-1 cells were induced by the removal of doxycycline for 2 days, infected with tsE320 at an MOI of 10 PFU/cell, and incubated at 39.5°C for 24 h. Cells were stained for σNS with σNS-specific polyclonal guinea pig antiserum and for μ1/μ1C with μ1/μ1C-specific MAb 8H6. The secondary antibodies used were goat anti-guinea pig Alexa Fluor 488 and goat anti-mouse Alexa Fluor 546. Images were obtained with a confocal microscope. The σNS protein is colored green (B), and the μ1/μ1C protein is colored red (C). In the merged image (D), colocalization of σNS and μ1/μ1C is indicated by the color yellow. A DIC image of each field was obtained (A). Images were processed with Adobe Photoshop. Bars, 25 μm.
The viral titer of tsE320 after growth in control-1 cells was 1.7 × 106 PFU/ml, whereas its titer after growth in σNS-1 cells was 2.8 × 107 PFU/ml. In contrast, the viral titers of tsH11.2 were low but equivalent after growth in either control-1 or σNS-1 cells (5.7 × 105 and 7.6 × 105 PFU/ml, respectively) (Fig. 4). Thus, σNS protein expressed in σNS-1 cells is functional and sufficient to rescue both the morphology of infection and the production of infectious progeny virus. This effect is specific, as expressed σNS had no effect on the production of infectious progeny by tsH11.2 at the nonpermissive temperature.
Reovirus σNS protein forms punctate structures in cells coexpressing μNS.
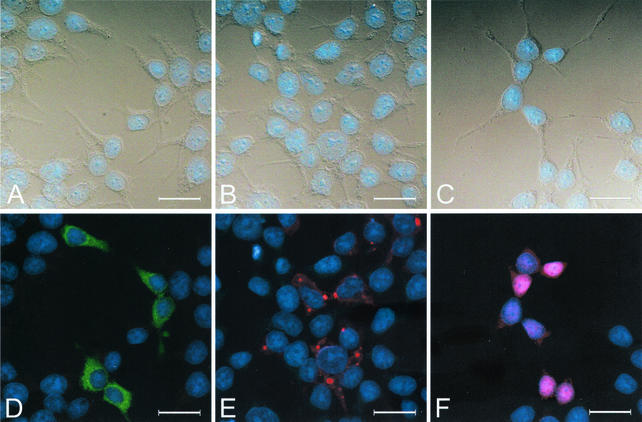
To identify viral proteins that act in concert with σNS to form viral replication structures, we engineered plasmids to express σNS, μNS, and σ3 under control of the cytomegalovirus immediate-early promoter. These viral proteins were previously identified in the earliest detectable protein-RNA complexes in reovirus-infected cells (1). 293T cells were transfected with σNS-, μNS-, or σ3-encoding plasmids singly (Fig. 6), in pairs (Fig. 7), or all together (data not shown) and imaged by confocal immunofluorescence microscopy 36 h after transfection. 293T cells were chosen for these studies because we routinely obtain transfection efficiencies of approximately 50% with these cells. Similar results were obtained in experiments with HeLa cells and L cells (data not shown). In 293T cells transfected with a σNS-expressing plasmid, σNS was distributed diffusely in the cytoplasm and did not form discrete structures (Fig. 6D). This staining pattern is identical to that observed in studies of the stably transfected σNS-expressing L-cell lines (Fig. 2). In contrast, in 293T cells transfected with a μNS-expressing plasmid, the protein localized to punctate structures within the cytoplasm. However, μNS was not present exclusively in these structures, as it was also found diffusely in the cytoplasm to various degrees (Fig. 6E). Neither σNS nor μNS was found in the nucleus. The σ3 protein showed an altogether different and more variable staining pattern. In 293T cells transfected with a σ3-expressing plasmid, the protein localized to both the cytoplasm and the nucleus and did not form discrete structures (Fig. 6F). In some cells, σ3 was distributed uniformly throughout the cytoplasm and the nucleus, whereas in others, more σ3 was detected in the nucleus than in the cytoplasm.
FIG. 6.
Subcellular localization of σNS, μNS, and σ3 in 293T cells transfected with plasmids encoding each protein. 293T cells were transfected with a plasmid encoding σNS (A and D), μNS (B and E), or σ3 (C and F) and fixed 36 h after transfection. Cells were stained for σNS with σNS-specific polyclonal guinea pig antiserum, for μNS with μNS-specific polyclonal rabbit antiserum, and for σ3 with σ3-specific MAb 4F2. The secondary antibodies used were goat anti-guinea pig Alexa Fluor 488, goat anti-rabbit Alexa Fluor 488, goat anti-mouse Alexa Fluor 488, and dsDNA-specific dye TO-PRO3. Images were obtained with a confocal microscope. The σNS protein is colored green (D), the μNS protein is colored red (E), the σ3 protein is colored red (F), and nuclei are colored blue (A to F). A DIC image of each field was obtained (A, B, and C). Images were processed with Adobe Photoshop. Bars, 25 μm.
FIG.7.
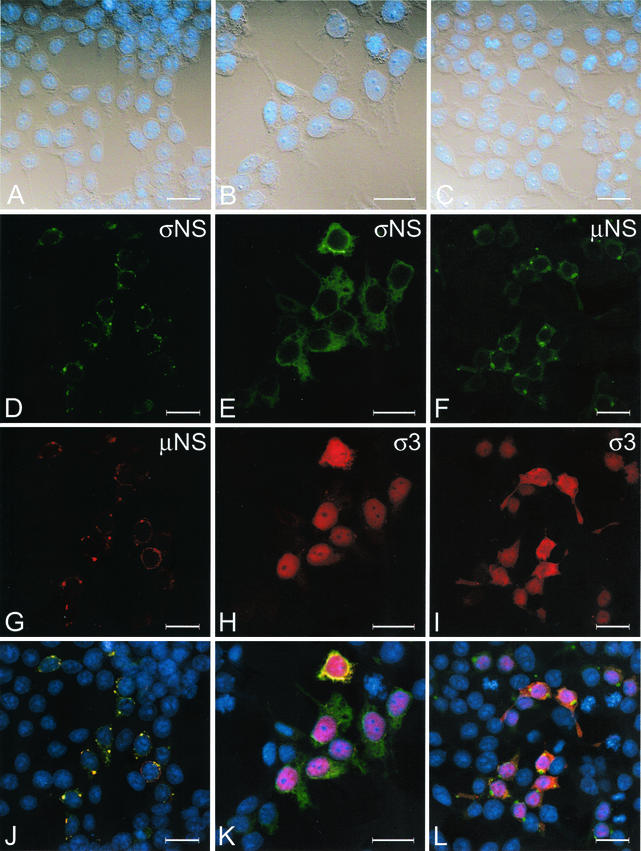
Subcellular localization of σNS, μNS, and σ3 in 293T cells transfected with plasmids encoding each protein in pairs. 293T cells were transfected with plasmids encoding σNS and μNS (A, D, G, and J), σNS and σ3 (B, E, H, and K), or μNS and σ3 (C, F, I, and L) and fixed 36 h after transfection. Cells were stained for σNS with σNS-specific polyclonal guinea pig antiserum, for μNS with μNS-specific polyclonal rabbit antiserum, and for σ3 with σ3-specific MAb 4F2. The secondary antibodies used were goat anti-guinea pig Alexa Fluor 488 and goat anti-rabbit Alexa Fluor 546, goat anti-guinea pig Alexa Fluor 546 and goat anti-mouse Alexa Fluor 488, goat anti-rabbit Alexa Fluor 488 and goat anti-mouse Alexa Fluor 546, and dsDNA-specific dye TO-PRO3. Images were obtained with a confocal microscope. The σNS protein is colored green (D, E), the μNS protein is colored green (F) or red (G), the σ3 protein is colored red (H and I), and nuclei are colored blue (A to C and J to L). In the merged images, colocalization of σNS and μNS (J), σNS and σ3 (K), or μNS and σ3 (L) is indicated by the color yellow. A DIC image of each field was obtained (A through C). Images were processed with Adobe Photoshop. Bars, 25 μm.
We next transfected 293T cells with the σNS-, μNS-, and σ3-encoding plasmids in pairs to determine whether the presence of two viral proteins altered the staining pattern of either protein when expressed alone (Fig. 7). In cells transfected with both σNS- and μNS-encoding plasmids, the σNS protein was concentrated in punctate structures and colocalized with μNS; only a small amount of σNS was distributed diffusely in the cytoplasm (Fig. 7D). The μNS protein displayed a staining pattern similar to that seen in cells transfected with μNS alone; the protein was found mostly in punctate structures, with a small amount of protein present in the cytoplasm (Fig. 7G). In cells transfected with σNS- and σ3-encoding plasmids, both proteins exhibited staining patterns indistinguishable from those seen when each protein was expressed alone (Fig. 7E and H). A similar pattern was seen in cells transfected with μNS- and σ3-encoding plasmids. The presence of μNS together with σ3 did not change the subcellular localization of either protein in comparison to the distribution of each when expressed alone (Fig. 7F and I). These findings indicate that the presence of μNS alters the subcellular localization of σNS to closely resemble the staining pattern of σNS in reovirus-infected cells (7).
To determine whether the subcellular localization of σNS, μNS, and σ3 was altered by simultaneous expression of each protein, 293T cells were transfected with all three plasmids and imaged by confocal immunofluorescence microscopy (data not shown). The σNS protein was observed in punctate structures within the cytoplasm and colocalized with μNS in these structures. The σ3 protein was present in both the cytoplasm and the nucleus, with the majority of cells showing higher levels of σ3 in the nucleus. The σNS and μNS proteins showed only limited colocalization with σ3 (data not shown). These results confirm that μNS is capable of altering the subcellular localization of σNS, whereas the subcellular localization of σ3 is not altered by coexpression of σNS, μNS, or both proteins.
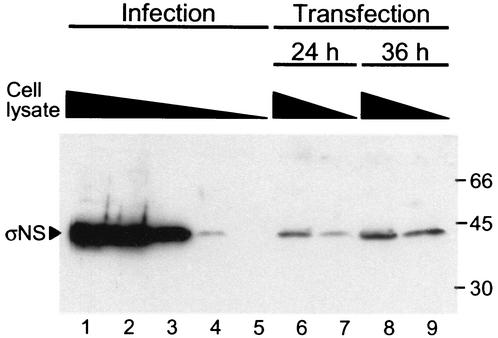
To ensure that the subcellular localization of σNS, μNS, and σ3 in the absence of viral infection was not attributable to overexpression of viral proteins during transient transfection, 293T cells were infected with T3D at an MOI of 10 PFU/cell or transfected with a plasmid encoding σNS. Infected cells were lysed at 18 h postinfection, and transfected cells were lysed at 24 and 36 h posttransfection. These times coincide with those used in the cell imaging experiments. Cell lysates were serially diluted in 10-fold increments, resolved by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with a σNS-specific antiserum (Fig. 8). The results indicate that levels of σNS following T3D infection are 100- to 1,000-fold greater than those observed after transient transfection. Thus, substantially more σNS is produced during infection than after transfection, providing confidence that the results of transient transfection are not due to overexpression of viral proteins.
FIG. 8.
Comparison of σNS levels in infected and transfected cells. 293T cells were either infected with T3D at an MOI of 10 PFU/cell or transfected with a σNS-encoding plasmid and maintained at 37°C. Infected cells were harvested 18 h postadsorption, and transfected cells were harvested at 24 and 36 h posttransfection. Cells were lysed, sheared, diluted in 10-fold increments in lysis buffer, and resolved by SDS-PAGE. Undiluted samples were normalized for protein concentration prior to dilution. Proteins were transferred to nitrocellulose and probed with guinea pig σNS-specific antiserum. Protein molecular mass standards (in kilodaltons) are shown on the right. Lanes: 1, infected and undiluted; 2, infected and diluted 1:10; 3, infected and diluted 1:100; 4, infected and diluted 1:1,000; 5, infected and diluted 1:10,000; 6, transfected for 24 h and undiluted; 7, transfected for 24 h and diluted 1:10; 8, transfected for 36 h and undiluted; 9, transfected for 36 h and diluted 1:10.
Reovirus σNS and μNS proteins interact in yeast two-hybrid and coimmunoprecipitation analyses.
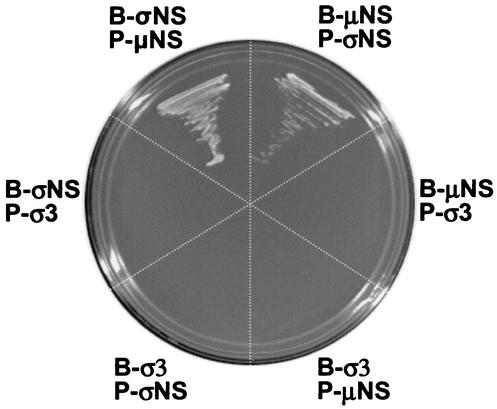
The observation that the subcellular localization of σNS is altered in cells expressing μNS suggests that these proteins directly interact. To test this hypothesis, we generated yeast two-hybrid bait and prey expression vectors for σNS, μNS, and σ3. Transformation of auxotrophic yeast strain AH109 with these expression vectors in pairwise combinations resulted in reporter gene activation only when the σNS and μNS fusion proteins were expressed together (Fig. 9). Transformation of yeast with the σ3 fusion protein in combination with either the σNS or the μNS fusion protein did not result in reporter gene activation (Fig. 9). Thus, σNS and μNS are capable of interacting when expressed as fusion proteins in yeast, and these interactions appear to be specific.
FIG. 9.
Reovirus σNS and μNS proteins interact in direct yeast two-hybrid tests. Reovirus σNS-, μNS-, and σ3-encoding bait (B) and prey (P) plasmids were used to transform yeast strain AH109 in pairwise combinations. Transformants expressing both bait and prey plasmids were selected by growth on medium lacking tryptophan and leucine. Protein interactions were evaluated by growth on medium lacking tryptophan, leucine, adenine, and histidine. Rescue of growth on selective medium was achieved by coexpression of σNS and μNS.
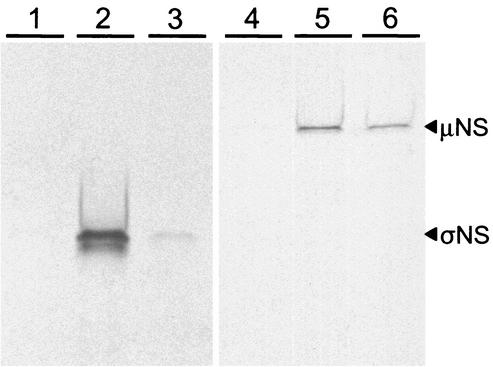
To provide further evidence that the σNS and μNS proteins physically interact, we generated epitope-tagged fusion proteins for use in coimmunoprecipitation experiments. Plasmids were engineered to express the σNS, μNS, and σ3 proteins with either a c-Myc or an HA epitope tag. Proteins were expressed by in vitro transcription and translation in either the presence or the absence of radiolabeled cysteine and methionine, incubated in different combinations, immunoprecipitated with epitope-specific antibodies, and then subjected to SDS-PAGE. Immunoprecipitation of c-Myc-μNS by anti-c-Myc antibody resulted in coimmunoprecipitation of radiolabeled HA-σNS (Fig. 10, lane 3). Similarly, immunoprecipitation of c-Myc-σNS by anti-c-Myc antibody resulted in coimmunoprecipitation of radiolabeled HA-μNS (Fig. 10, lane 5). Immunoprecipitation of control protein c-Myc-lamin C did not result in coimmunoprecipitation of either HA-σNS or HA-μNS (Fig. 10, lanes 1 and 4, respectively). Immunoprecipitation of c-Myc-σNS by anti-c-Myc antibody resulted in coimmunoprecipitation of radiolabeled HA-σNS (Fig. 10, lane 2), consistent with previous observations (37). Similarly, immunoprecipitation of c-Myc-μNS by anti-c-Myc antibody resulted in coimmunoprecipitation of radiolabeled HA-μNS (Fig. 10, lane 6), consistent with previous observations (15). Treatment of the in vitro transcription and translation reactions with RNase A prior to electrophoresis did not alter the capacity of σNS and μNS to coimmunoprecipitate (data not shown). These findings indicate that the σNS and μNS proteins are capable of both homologous and heterologous interactions in the absence of other proteins or RNA.
FIG. 10.
Coimmunoprecipitation of reovirus σNS and μNS proteins. Radiolabeled HA-σNS or HA-μNS was incubated in pairwise combinations with unlabeled c-Myc-lamin C, c-Myc-σNS, or c-Myc-μNS. Anti-c-Myc antibody was used for immunoprecipitation, and binding was analyzed by SDS-PAGE and autoradiography. Lanes: 1, c-Myc-lamin C and HA-σNS; 2, c-Myc-σNS and HA-σNS; 3, c-Myc-μNS and HA-σNS; 4, c-Myc-lamin C and HA-μNS; 5, c-Myc-σNS and HA-μNS; 6, c-Myc-μNS and HA-μNS.
Nonstructural proteins σNS and μNS form structures in reovirus-infected cells that morphologically resemble early viral inclusions.
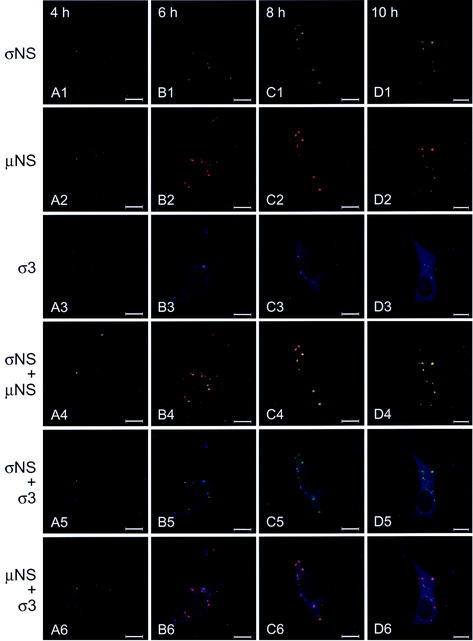
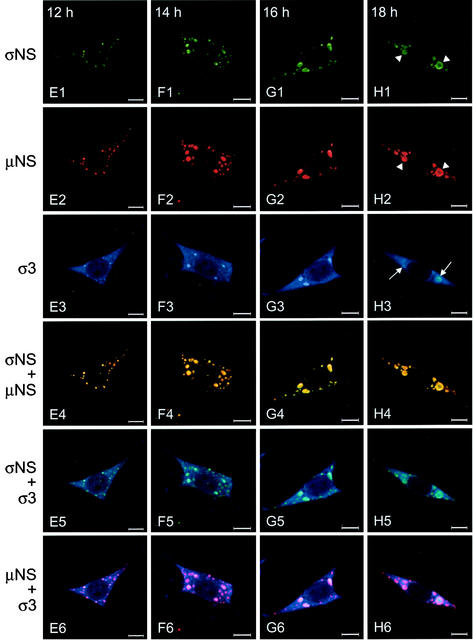
To determine whether σNS and μNS interact during a productive reovirus infection, cells were infected with T3D and imaged by confocal immunofluorescence microscopy at 2-h intervals with antibodies specific for σNS, μNS, and σ3 (Fig. 11). Each of these proteins was detectable by 4 h postinfection. The σNS and μNS proteins colocalized in punctate structures throughout the cytoplasm. At early times of infection, the inclusion structures were numerous and small, but as infection progressed, these structures decreased in number and increased in size. The σNS protein was found exclusively in these structures, whereas μNS also was observed elsewhere in the cytoplasm. In contrast, σ3 was distributed both diffusely and in punctate structures early in infection, but it did not colocalize with either σNS or μNS. Between 10 and 12 h postinfection, σ3 was detectable in the nucleus and also was found in several of the larger inclusion structures along with σNS and μNS. By 18 h postinfection, at which time infectious viral progeny can be detected (28, 91; data not shown), most of the σNS and μNS protein was observed in large viral inclusions. The σ3 protein also was present in these structures, as well as in the cytoplasm and the nuclei of infected cells. The σNS and μNS proteins were often found at the periphery of the inclusions at these late time points (Fig. 11H1 and H2, arrowheads), consistent with the observation that mature inclusions are replete with progeny virions, which do not contain nonstructural proteins (7). At these late time points, the σ3 protein continued to stain the interior of the inclusions (Fig. 11H3, arrows), commensurate with its role as a structural component of the viral outer capsid. These data indicate that both σNS and μNS are present in the earliest detectable viral structures in reovirus-infected cells. These results, together with the analysis of the subcellular localization of the σNS and μNS proteins following transient transfection, suggest that σNS and μNS initiate the formation of viral inclusions during reovirus infection.
FIG.11.
Subcellular localization of σNS, μNS, and σ3 proteins in L cells at different times postinfection with T3D. L cells were infected with T3D at an MOI of 10 PFU/cell and incubated at 37°C for the times shown. Cells were stained for σNS with σNS-specific polyclonal guinea pig antiserum, for μNS with μNS-specific polyclonal rabbit antiserum, and for σ3 with σ3-specific MAb 4F2. The secondary antibodies used were goat anti-guinea pig Alexa Fluor 546, goat anti-rabbit Alexa Fluor 488, and goat anti-mouse Alexa Fluor 647. Images were obtained with a confocal microscope. The σNS protein is colored green (A1 to H1), the μNS protein is colored red (A2 to H2), and the σ3 protein is colored blue (A3 to H3). In the merged images, colocalization of σNS and μNS is indicated by the color yellow (A4 to H4), colocalization of σNS and σ3 is indicated by the color blue-green (A5 to H5), and colocalization of μNS and σ3 is indicated by the color purple (A6 to H6). Arrowheads indicate the presence of σNS (H1) and μNS (H2) at the periphery of mature viral inclusions; arrows (H3) indicate the presence of σ3 in the interior of mature viral inclusions. Images were processed with Adobe Photoshop. Bars, 10 μm.
DISCUSSION
Little is known about the steps leading to the formation of inclusion structures in which reovirus replication and morphogenesis take place in infected cells. We previously demonstrated that these structures form only when functional σNS is present (7). To further examine the role of σNS in early steps of viral inclusion formation, we sought to determine the subcellular localization of σNS when it is expressed in cells in the absence of viral infection. In these cells, σNS is distributed diffusely throughout the cytoplasm and does not form inclusion-like structures. Expressed σNS rescues infection by σNS-mutant virus tsE320 at the nonpermissive temperature but does not alter infection with wt reovirus. Only when σNS is expressed with μNS does σNS localize to punctate structures in the cytoplasm that resemble viral inclusions seen during reovirus infection. Nonstructural proteins σNS and μNS directly interact, leading to the hypothesis that the capacity of μNS to redirect the subcellular localization of σNS is mediated by protein-protein interactions.
Early studies of reovirus-infected cells by electron microscopy described crystalline arrays of progeny virions termed viral inclusions (26, 34, 39, 70, 78, 84). Reovirus-infected cells examined by phase-contrast and immunofluorescence microscopy exhibit large phase-dense and viral protein-containing structures, respectively, that correspond to the viral inclusions seen by electron microscopy (7, 34, 70, 78). Immunofluorescence microscopy has become a valuable tool with which to probe early steps in the formation of viral inclusions. This technique facilitates the identification of specific viral proteins in inclusion structures. Functional viral inclusions lead to the production of infectious progeny, and thus, inclusion function can only be tested indirectly by assays of viral infectivity. A method by which to directly determine whether a viral inclusion structure is replication competent has not been developed. Therefore, we used both techniques in this study.
Expressed σNS is diffuse in the cytoplasm.
In both stably and transiently transfected cells, expressed σNS is distributed diffusely throughout the cytoplasm. Therefore, σNS is necessary for the formation of functional viral inclusions, but it is not capable of establishing inclusions in the absence of other viral proteins. Interestingly, lesions in the σNS and μ2 proteins in ts mutant viruses tsE320 and tsH11.2, respectively, lead to altered subcellular localization of the mutant protein at the nonpermissive temperature, indicating a link between the function and localization of these proteins (7). Many cellular proteins are functional only when localized to a particular site within the cell, and the movement of these proteins to the appropriate intracellular site can serve to regulate protein function. Our data support the idea that reovirus proteins involved in replication are active only within functional centers characterized by a particular location and protein composition. The function of σNS appears to be regulated by its subcellular localization in that σNS expressed alone exhibits a subcellular localization markedly different from that following viral infection.
Both tsE320 and tsH11.2 are classified as dsRNA-negative viruses, and neither produces dsRNA or inclusions of viral progeny at the nonpermissive temperature (7, 21, 24, 25, 32). These viruses differ in that tsH11.2 generates ssRNA approaching wt levels early in infection (21), whereas tsE320 produces less ssRNA (25). Previous reports indicate that the two viruses generate similar amounts of viral protein (21, 22, 33); however, confocal imaging data indicate that tsE320 produces less protein than tsH11.2 (7; data not shown), which is consistent with the levels of ssRNA produced by each virus (21, 25) Cells infected with tsH11.2 at the nonpermissive temperature produce structures resembling viral inclusions, but progeny virions are not detected (7). In contrast, tsE320-infected cells do not produce inclusion structures detectable by either confocal or electron microscopy (7). These findings suggest that, in addition to a role in inclusion formation, σNS may be involved in viral mRNA or protein synthesis.
Expressed σNS rescues tsE320 infection but does not alter infection with wt reovirus.
It was previously thought that the mutant form of σNS produced in tsE320-infected cells does not act as a dominant-negative inhibitor since coinfection with other ts mutants or wt reovirus is able to rescue the defect in σNS (46). Our data support this hypothesis by demonstrating that expressed σNS is capable of complementing the mutation specifically in tsE320 without altering the growth of tsH11.2 or wt reovirus. These results parallel those reported previously indicating that tsH11.2 can be complemented by expression of wt μ2 protein (94). Expressed σNS is sufficient to form punctate structures in tsE320-infected cells and restore viral titers to wt levels, indicating that the protein is functional in both its subcellular localization and its role in viral replication. Our data also indicate that the presence of expressed σNS prior to infection with wt reovirus, as well as higher levels of σNS during infection, does not alter yields of viral progeny. Therefore, the correlation between the capacity of σNS to function in viral replication and its subcellular localization strengthens the hypothesis that σNS is spatially regulated and functions only when properly localized through interactions with viral proteins or RNA.
Expression of μNS is sufficient to redirect the subcellular localization of σNS.
The staining patterns seen in cells transfected with σNS, μNS, and σ3 are distinct. The subcellular localization of μNS in transfected cells is similar to that observed in T3D-infected cells. The μNS protein forms cytoplasmic punctate structures in both settings, as reported previously (16). In transfected cells, more σ3 is present in the nucleus, but it is also found in the cytoplasm. The overall pattern of σ3 distribution is the same in reovirus-infected cells; however, σ3 is present at higher levels in the cytoplasm during infection. These findings are similar to those of a previous study (93). The staining pattern of σNS is distinctly different in transfected cells and infected cells, diffuse within the cytoplasm or concentrated in punctate structures, respectively. By expressing these proteins in pairwise combinations, we found that only when μNS is expressed with σNS does the staining pattern change: expression of μNS mediates a striking redistribution of σNS to punctate structures.
Since σNS and μNS are viral nonstructural proteins that associate with ssRNA and have been found at sites of viral replication or in complex with viral replication intermediates (1, 43, 61, 62, 66, 78), it is likely that both are involved in the earliest steps of reovirus genome replication. It is possible that either or both proteins (i) participate in the formation of viral inclusions, (ii) recruit or transport viral mRNA or proteins to inclusion structures, or (iii) enhance viral translation. Since it has been hypothesized that assortment and perhaps packaging of the reovirus genome occur prior to dsRNA synthesis (1), it is also possible that σNS or μNS acts to chaperone viral mRNA in these critical steps, perhaps within inclusions.
Reovirus strains exhibit differences in viral inclusion morphology (16, 57, 66). Strain T1L forms filamentous inclusions, whereas T3D forms punctate or globular inclusions. This difference in inclusion morphology segregates with the M1 gene (66), which encodes viral structural protein μ2, a component of the viral core (23). The T1L μ2 protein associates with microtubules, but T3D μ2 does not (16, 66). These findings suggest that μNS, perhaps in concert with μ2, selects intracellular sites for viral inclusion formation and may form the scaffolding on which inclusions are built (16, 66). Since the subcellular localization of σNS is altered in the presence of μNS, it is most likely that σNS functions subsequent to μNS localization in infected cells. Studies of tsE320 indicate that at least one function of σNS is executed prior to dsRNA synthesis (32); however, it is possible that σNS and μNS have additional roles during virion morphogenesis (15).
Nonstructural proteins σNS and μNS directly interact.
We found that σNS and μNS directly interact by using both yeast two-hybrid assays and coimmunoprecipitation studies. These experiments also indicate that each protein is capable of self-association, consistent with previous reports (37, 38, 43, 59, 71), and concur with a previous study indicating that μNS can be immunoprecipitated with σNS-specific MAbs (51). Because of differences in σNS localization in the presence and absence of μNS, the direct protein-protein interaction between σNS and μNS is likely to be a critical early step in the establishment of an intracellular site for viral genome replication and virion morphogenesis. Our biochemical data also correlate with the staining patterns seen in reovirus-infected cells, in which σNS and μNS colocalize in punctate structures soon after virus adsorption and are maintained in these structures throughout the infectious cycle. The σ3 protein does not colocalize with σNS and μNS until later in infection, perhaps as outer capsid proteins are assembled onto viral cores to form mature viral progeny. While interactions of σ3 with σNS or μNS during viral replication cannot be excluded by our studies, it is interesting that pairwise yeast two-hybrid tests do not demonstrate interactions between σ3 and either σNS or μNS.
Comparison to other members of the family Reoviridae.
Mammalian reoviruses make up one genus of the family Reoviridae and have a number of features in common with other viruses in this family. Rotavirus, another genus of the family Reoviridae, encodes a nonstructural protein, NSP2, with properties similar to those of σNS. NSP2 is a highly conserved basic protein with a molecular mass of 35 kDa. NSP2 also forms higher-order homo-oligomers (77), binds ssRNA nonspecifically (50), and accumulates in inclusions (67, 86). Like σNS, NSP2 has helix-destabilizing activity not dependent on Mg2+ or a nucleoside triphosphate (NTP) energy source (38, 87). However, NSP2 has several properties that have not been attributed to σNS, including an Mg2+-dependent NTPase and related autokinase activity (86). NSP2 also associates with the rotavirus RNA-dependent RNA polymerase (20, 49) and partially replicated RNA (2). Therefore, NSP2 has been implicated in early steps of rotavirus replication, including mRNA packaging or minus-strand synthesis. A ts rotavirus with a lesion in the NSP2-encoding gene does not synthesize the viral genome at the nonpermissive temperature, but it does produce empty particles (19, 69). These findings are in contrast to the absence of particles following infection with σNS-mutant virus tsE320 (7), suggesting that the functions of NSP2 and σNS differ in important respects. Interestingly, NSP2 expressed in cells is diffuse, but when expressed with another rotavirus nonstructural protein, NSP5, it colocalizes with NSP5 in punctate structures (31).
Bluetongue virus (BTV), the prototype member of the Orbivirus genus of the family Reoviridae, encodes NS2, a protein that is most likely a σNS homolog. NS2 has a molecular mass of 41 kDa, binds ssRNA in a nonspecific manner, exists as a homo-oligomer (89), and accumulates in viral inclusion bodies (17, 30). However, NS2 is functionally more similar to rotavirus NSP2 than reovirus σNS in that NS2 has the capacity to hydrolyze NTPs and can undergo phosphorylation (29), properties also exhibited by NSP2 (44, 87).
Although reovirus σNS, rotavirus NSP2, and BTV NS2 are similar in several respects, NSP2 and NS2 mediate enzymatic functions not attributable to σNS, such as NTP hydrolysis and kinase activity (38). However, it seems likely that these members of the family Reoviridae all require the biochemical activities attributed to NSP2 and NS2, but these necessary functions may be accomplished by other proteins in the mammalian reoviruses. For example, reovirus μ2 has NTPase activity and is hypothesized to be a component of the polymerase complex (65). Also, structural protein λ1 has NTPase and helicase activities (10, 11, 40, 64). The μNS protein has no obvious homologs in other members of the family Reoviridae, which raises the possibility that it may serve a functional role analogous to that of rotavirus NSP2 or BTV NS2.
Results presented in this report indicate that σNS and μNS interact to facilitate early steps in reovirus genome replication that are dependent on the proper subcellular localization of these proteins. Once these nascent replication structures are established, subsequent steps in RNA synthesis and virion morphogenesis can occur, perhaps through the recruitment of viral mRNA, proteins, or cellular components. Our ongoing work will focus on mechanisms by which these viral proteins facilitate the establishment and maturation of viral inclusions that culminate in the formation of progeny virions.
Acknowledgments
We express our appreciation to Mark Denison, Erik Prentice, and Sam Wells for essential discussions and to Jim Chappell for review of the manuscript. We are grateful to Howard Price and Brenda Starks-Stanton for excellent technical assistance. We thank Paul Spearman for HEK 293T cells and Teresa Broering and Max Nibert for rabbit polyclonal anti-μNS antiserum.
This work was supported by Public Health Service award T32 GM08554 (M.M.B.) from the National Institute of General Medical Sciences, the Howard Hughes Medical Institute (T.R.P.), Public Health Service award AI32539 from the National Institute of Allergy and Infectious Diseases, and the Elizabeth B. Lamb Center for Pediatric Research. Additional support was provided by Public Health Service awards CA68485 and DK20593 for the Vanderbilt DNA Sequencing Shared Resource and Vanderbilt Cell Imaging Core.
REFERENCES
- 1.Antczak, J. B., and W. K. Joklik. 1992. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal antibodies directed against reovirus proteins. Virology 187:760-776. [DOI] [PubMed]
- 2.Aponte, C., D. Poncet, and J. Cohen. 1996. Recovery and characterization of a replicase complex in rotavirus-infected cells by using a monoclonal antibody against NSP2. J. Virol. 70:985-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astell, C., S. C. Silverstein, D. H. Levin, and G. Acs. 1972. Regulation of the reovirus RNA transcriptase by a viral capsomere protein. Virology 48:648-654. [DOI] [PubMed] [Google Scholar]
- 4.Baer, G. S., and T. S. Dermody. 1997. Mutations in reovirus outer-capsid protein σ3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J. Virol. 71:4921-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baer, G. S., D. H. Ebert, C. J. Chung, A. H. Erickson, and T. S. Dermody. 1999. Mutant cells selected during persistent reovirus infection do not express mature cathepsin L and do not support reovirus disassembly. J. Virol. 73:9532-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee, A. K., and A. J. Shatkin. 1970. Transcription in vitro by reovirus-associated ribonucleic acid-dependent polymerase. J. Virol. 6:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker, M. M., M. I. Goral, P. R. Hazelton, G. S. Baer, S. E. Rodgers, E. G. Brown, K. M. Coombs, and T. S. Dermody. 2001. Reovirus σNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions. J. Virol. 75:1459-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellamy, A. R., J. L. Nichols, and W. K. Joklik. 1972. Nucleotide sequences of reovirus oligonucleotides: evidence for abortive RNA synthesis during virus maturation. Nature 238:49-51. [DOI] [PubMed] [Google Scholar]
- 9.Belli, B. A., and C. E. Samuel. 1991. Biosynthesis of reovirus-specified polypeptides: expression of reovirus S1 encoded σ1NS protein in transfected and infected cells as measured with serotype specific polyclonal antibody. Virology 185:698-709. [DOI] [PubMed] [Google Scholar]
- 10.Bisaillon, M., and G. Lemay. 1997. Characterization of the reovirus lambda1 protein RNA 5′-triphosphatase activity. J. Biol. Chem. 272:29954-29957. [DOI] [PubMed] [Google Scholar]
- 11.Bisaillon, M., and G. Lemay. 1997. Molecular dissection of the reovirus λ1 protein nucleic acids binding site. Virus Res. 51:231-237. [DOI] [PubMed] [Google Scholar]
- 12.Borsa, J., B. D. Morash, M. D. Sargent, T. P. Copps, P. A. Lievaart, and J. G. Szekely. 1979. Two modes of entry of reovirus particles into L cells. J. Gen. Virol. 45:161-170. [DOI] [PubMed] [Google Scholar]
- 13.Borsa, J., M. D. Sargent, P. A. Lievaart, and T. P. Copps. 1981. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology 111:191-200. [DOI] [PubMed] [Google Scholar]
- 14.Both, G. W., S. Lavi, and A. J. Shatkin. 1975. Synthesis of all the gene products of the reovirus genome in vivo and in vitro. Cell 4:173-180. [DOI] [PubMed] [Google Scholar]
- 15.Broering, T. J., A. M. McCutcheon, V. E. Centonze, and M. L. Nibert. 2000. Reovirus nonstructural protein μNS binds to core particles but does not inhibit their transcription and capping activities. J. Virol. 74:5516-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broering, T. J., J. S. Parker, P. L. Joyce, J. Kim, and M. L. Nibert. 2002. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J. Virol. 76:8285-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brookes, S. M., A. D. Hyatt, and B. T. Eaton. 1993. Characterization of virus inclusion bodies in bluetongue virus-infected cells. J. Gen. Virol. 74:525-530. [DOI] [PubMed] [Google Scholar]
- 18.Chang, C. T., and H. J. Zweerink. 1971. Fate of parental reovirus in infected cell. Virology 46:544-555. [DOI] [PubMed] [Google Scholar]
- 19.Chen, D., J. L. Gombold, and R. F. Ramig. 1990. Intracellular RNA synthesis directed by temperature-sensitive mutants of simian rotavirus SA11. Virology 178:143-151. [DOI] [PubMed] [Google Scholar]
- 20.Chen, D., and J. T. Patton. 1998. Rotavirus RNA replication requires a single-stranded 3′ end for efficient minus-strand synthesis. J. Virol. 72:7387-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coombs, K. M. 1996. Identification and characterization of a double-stranded RNA− reovirus temperature-sensitive mutant defective in minor core protein μ2. J. Virol. 70:4237-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coombs, K. M. 1998. Temperature-sensitive mutants of reovirus. Curr. Top. Microbiol. Immunol. 233:69-107. [DOI] [PubMed] [Google Scholar]
- 23.Coombs, K. M., B. N. Fields, and S. C. Harrison. 1990. Crystallization of the reovirus type 3 Dearing core: crystal packing is determined by the λ2 protein. J. Mol. Biol. 215:1-5. [DOI] [PubMed] [Google Scholar]
- 24.Coombs, K. M., S.-C. Mak, and L. D. Petrycky-Cox. 1994. Studies of the major reovirus core protein σ2: reversion of the assembly-defective mutant tsC447 is an intragenic process and involves back mutation of Asp-383 to Asn. J. Virol. 68:177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cross, R. K., and B. N. Fields. 1972. Temperature-sensitive mutants of reovirus type 3: studies on the synthesis of viral RNA. J. Virol. 50:799-809. [DOI] [PubMed] [Google Scholar]
- 26.Dales, S. 1963. Association between the spindle apparatus and reovirus. Proc. Natl. Acad. Sci. USA 50:268-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denzler, K. L., and B. L. Jacobs. 1994. Site-directed mutagenic analysis of reovirus σ3 protein binding to dsRNA. Virology 204:190-199. [DOI] [PubMed] [Google Scholar]
- 28.Dermody, T. S., M. L. Nibert, J. D. Wetzel, X. Tong, and B. N. Fields. 1993. Cells and viruses with mutations affecting viral entry are selected during persistent infections of L cells with mammalian reoviruses. J. Virol. 67:2055-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devaney, M. A., J. Kendall, and M. J. Grubman. 1988. Characterization of a nonstructural phosphoprotein of two orbiviruses. Virus Res. 11:151-164. [DOI] [PubMed] [Google Scholar]
- 30.Eaton, B. T., A. D. Hyatt, and S. M. Brookes. 1990. The replication of bluetongue virus. Curr. Top. Microbiol. Immunol. 162:89-118. [DOI] [PubMed] [Google Scholar]
- 31.Fabbretti, E., I. Afrikanova, F. Vascotto, and O. R. Burrone. 1999. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80:333-339. [DOI] [PubMed] [Google Scholar]
- 32.Fields, B. N., and W. K. Joklik. 1969. Isolation and preliminary genetic and biochemical characterization of temperature-sensitive mutants of reovirus. Virology 37:335-342. [DOI] [PubMed] [Google Scholar]
- 33.Fields, B. N., R. Laskov, and M. D. Scharff. 1972. Temperature-sensitive mutants of reovirus type 3: studies on the synthesis of peptides. Virology 50:209-215. [DOI] [PubMed] [Google Scholar]
- 34.Fields, B. N., C. S. Raine, and S. G. Baum. 1971. Temperature-sensitive mutants of reovirus type 3: defects in viral maturation as studied by immunofluorescence and electron microscopy. Virology 43:569-578. [DOI] [PubMed] [Google Scholar]
- 35.Furuichi, Y., M. Morgan, S. Muthukrishnan, and A. J. Shatkin. 1975. Reovirus messenger RNA contains a methylated blocked 5′-terminal structure M7G(5′)ppp(5′)GmpCp-. Proc. Natl. Acad. Sci. USA 72:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giantini, M., and A. J. Shatkin. 1989. Stimulation of chloramphenicol acetyltransferase mRNA translation by reovirus capsid polypeptide σ3 in cotransfected COS cells. J. Virol. 63:2415-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillian, A. L., and M. L. Nibert. 1998. Amino terminus of reovirus nonstructural protein σNS is important for ssRNA binding and nucleoprotein complex formation. Virology 240:1-11. [DOI] [PubMed] [Google Scholar]
- 38.Gillian, A. L., S. C. Schmechel, J. Livny, L. A. Schiff, and M. L. Nibert. 2000. Reovirus protein σNS binds in multiple copies to single-stranded RNA and shares properties with single-stranded DNA binding proteins. J. Virol. 74:5939-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomatos, P. J., I. Tamm, S. Dales, and R. M. Franklin. 1962. Reovirus type 3: physical characteristics and interactions with L cells. Virology 17:441-454. [DOI] [PubMed] [Google Scholar]
- 40.Harrison, S. J., D. L. Farsetta, J. Kim, S. Noble, T. J. Broering, and M. L. Nibert. 1999. Mammalian reovirus L3 gene sequences and evidence for a distinct amino-terminal region of the λ1 protein. Virology 258:54-64. [DOI] [PubMed] [Google Scholar]
- 41.Hooper, J. W., and B. N. Fields. 1996. Monoclonal antibodies to reovirus σ1 and μ1 proteins inhibit chromium release from mouse L cells. J. Virol. 70:672-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hooper, J. W., and B. N. Fields. 1996. Role of the μ1 protein in reovirus stability and capacity to cause chromium release from host cells. J. Virol. 70:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huismans, H., and W. K. Joklik. 1976. Reovirus-coded polypeptides in infected cells: isolation of two native monomeric polypeptides with high affinity for single-stranded and double-stranded RNA, respectively. Virology 70:411-424. [DOI] [PubMed] [Google Scholar]
- 44.Huismans, H., A. A. van Dijk, and A. R. Bauskin. 1987. In vitro phosphorylation and purification of a nonstructural protein of bluetongue virus with affinity for single-stranded RNA. J. Virol. 61:3589-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imani, F., and B. L. Jacobs. 1988. Inhibitory activity for the interferon-induced protein kinase is associated with the reovirus serotype 1 σ3 protein. Proc. Natl. Acad. Sci. USA 85:7887-7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito, Y., and W. K. Joklik. 1972. Temperature-sensitive mutants of reovirus. I. Patterns of gene expression by mutants of groups C, D, and E. Virology 50:189-201. [DOI] [PubMed] [Google Scholar]
- 47.Jacobs, B. L., and C. E. Samuel. 1985. Biosynthesis of reovirus-specified polypeptides: the reovirus S1 mRNA encodes two primary translation products. Virology 143:63-74. [DOI] [PubMed] [Google Scholar]
- 48.Joklik, W. K. 1981. Structure and function of the reovirus genome. Microbiol. Rev. 45:483-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kattoura, M. D., X. Chen, and J. T. Patton. 1994. The rotavirus RNA-binding protein NS35 (NSP2) forms 10S multimers and interacts with the viral RNA polymerase. Virology 202:803-813. [DOI] [PubMed] [Google Scholar]
- 50.Kattoura, M. D., L. L. Clapp, and J. T. Patton. 1992. The rotavirus nonstructural protein, NS35, possesses RNA-binding activity in vitro and in vivo. Virology 191:698-708. [DOI] [PubMed] [Google Scholar]
- 51.Lee, P. W. K., E. C. Hayes, and W. K. Joklik. 1981. Characterization of anti-reovirus immunoglobulins secreted by cloned hybridoma cell lines. Virology 108:134-146. [DOI] [PubMed] [Google Scholar]
- 52.Li, J. K.-K., P. P. Scheible, J. D. Keene, and W. K. Joklik. 1980. The plus strand of reovirus gene S2 is identical with its in vitro transcript. Virology 105:282-286. [DOI] [PubMed] [Google Scholar]
- 53.Liemann, S., K. Chandran, T. S. Baker, M. L. Nibert, and S. C. Harrison. 2002. Structure of the reovirus membrane-penetration protein, μ1, in a complex with is protector protein, σ3. Cell 108:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loh, P. C., and A. J. Shatkin. 1968. Structural proteins of reovirus. J. Virol. 2:1353-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lucia-Jandris, P., J. W. Hooper, and B. N. Fields. 1993. Reovirus M2 gene is associated with chromium release from mouse L cells. J. Virol. 67:5339-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mabrouk, T., C. Danis, and G. Lemay. 1995. Two basic motifs of reovirus σ3 protein are involved in double-stranded RNA binding. Biochem. Cell Biol. 73:137-145. [DOI] [PubMed] [Google Scholar]
- 57.Mbisa, J. L., M. M. Becker, S. Zou, T. S. Dermody, and E. G. Brown. 2000. Reovirus μ2 protein determines strain-specific differences in the rate of viral inclusion formation in L929 cells. Virology 272:16-26. [DOI] [PubMed] [Google Scholar]
- 58.McCrae, M. A., and W. K. Joklik. 1978. The nature of the polypeptide encoded by each of the ten double-stranded RNA segments of reovirus type 3. Virology 89:578-593. [DOI] [PubMed] [Google Scholar]
- 59.McCutcheon, A. M., T. J. Broering, and M. L. Nibert. 1999. Mammalian reovirus M3 gene sequences and conservation of coiled-coil motifs near the carboxyl terminus of the μNS protein. Virology 264:16-24. [DOI] [PubMed] [Google Scholar]
- 60.Miller, J. E., and C. E. Samuel. 1992. Proteolytic cleavage of the reovirus σ3 protein results in enhanced double-stranded RNA-binding activity: identification of a repeated basic amino acid motif within the C-terminal binding region. J. Virol. 66:5347-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mora, M., K. Partin, M. Bhatia, J. Partin, and C. Carter. 1987. Association of reovirus proteins with the structural matrix of infected cells. Virology 159:265-277. [DOI] [PubMed] [Google Scholar]
- 62.Morgan, E. M., and H. J. Zweerink. 1975. Characterization of transcriptase and replicase particles isolated from reovirus infected cells. Virology 68:455-466. [DOI] [PubMed] [Google Scholar]
- 63.Mustoe, T. A., R. F. Ramig, A. H. Sharpe, and B. N. Fields. 1978. Genetics of reovirus: identification of the dsRNA segments encoding the polypeptides of the μ and σ size classes. Virology 89:594-604. [DOI] [PubMed] [Google Scholar]
- 64.Noble, S., and M. L. Nibert. 1997. Characterization of an ATPase activity in reovirus cores and its genetic association with core-shell protein λ1. J. Virol. 71:2182-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Noble, S., and M. L. Nibert. 1997. Core protein μ2 is a second determinant of nucleoside triphosphatase activities by reovirus cores. J. Virol. 71:7728-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parker, J. S., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petrie, B. L., H. B. Greenberg, D. Y. Graham, and M. K. Estes. 1984. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res. 1:133-152. [DOI] [PubMed] [Google Scholar]
- 68.Ramig, R. F., T. A. Mustoe, A. H. Sharpe, and B. N. Fields. 1978. A genetic map of reovirus. II. Assignment of the double-stranded RNA-negative mutant groups C, D, and E to genome segments. Virology 85:531-534. [DOI] [PubMed] [Google Scholar]
- 69.Ramig, R. F., and B. L. Petrie. 1984. Characterization of temperature-sensitive mutants of simian rotavirus SA11: protein synthesis and morphogenesis. J. Virol. 49:665-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rhim, J. S., L. E. Jordan, and H. D. Mayor. 1962. Cytochemical, fluorescent-antibody and electron microscopic studies on the growth of reovirus (ECHO 10) in tissue culture. Virology 17:342-355. [DOI] [PubMed] [Google Scholar]
- 71.Richardson, M. A., and Y. Furuichi. 1985. Synthesis in Escherichia coli of the reovirus nonstructural protein σNS. J. Virol. 56:527-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodgers, S. E., J. L. Connolly, J. D. Chappell, and T. S. Dermody. 1998. Reovirus growth in cell culture does not require the full complement of viral proteins: identification of a σ1s-null mutant. J. Virol. 72:8597-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rubin, D. H., D. B. Weiner, C. Dworkin, M. I. Greene, G. G. Maul, and W. V. Williams. 1992. Receptor utilization by reovirus type 3: distinct binding sites on thymoma and fibroblast cell lines result in differential compartmentalization of virions. Microb. Pathog. 12:351-365. [DOI] [PubMed] [Google Scholar]
- 74.Schiff, L. A., M. L. Nibert, M. S. Co, E. G. Brown, and B. N. Fields. 1988. Distinct binding sites for zinc and double-stranded RNA in the reovirus outer capsid protein σ3. Mol. Cell. Biol. 8:273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmechel, S., M. Chute, P. Skinner, R. Anderson, and L. Schiff. 1997. Preferential translation of reovirus mRNA by a σ3-dependent mechanism. Virology 232:62-73. [DOI] [PubMed] [Google Scholar]
- 76.Schonberg, M., S. C. Silverstein, D. H. Levin, and G. Acs. 1971. Asynchronous synthesis of the complementary strands of the reovirus genome. Proc. Natl. Acad. Sci. USA 68:505-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schuck, P., Z. Taraporewala, P. McPhie, and J. T. Patton. 2001. Rotavirus nonstructural protein NSP2 self-assembles into octamers that undergo ligand-induced conformational changes. J. Biol. Chem. 276:9679-9687. [DOI] [PubMed] [Google Scholar]
- 78.Sharpe, A. H., L. B. Chen, and B. N. Fields. 1982. The interaction of mammalian reoviruses with the cytoskeleton of monkey kidney CV-1 cells. Virology 120:399-411. [DOI] [PubMed] [Google Scholar]
- 79.Sharpe, A. H., and B. N. Fields. 1982. Reovirus inhibition of cellular RNA and protein synthesis: role of the S4 gene. Virology 122:381-391. [DOI] [PubMed] [Google Scholar]
- 80.Silverstein, S. C., C. Astell, D. H. Levin, M. Schonberg, and G. Acs. 1972. The mechanism of reovirus uncoating and gene activation in vivo. Virology 47:797-806. [DOI] [PubMed] [Google Scholar]
- 81.Silverstein, S. C., and P. H. Schur. 1970. Immunofluorescent localization of double-stranded RNA in reovirus-infected cells. Virology 41:564-566. [DOI] [PubMed] [Google Scholar]
- 82.Skehel, J. J., and W. K. Joklik. 1969. Studies on the in vitro transcription of reovirus RNA catalyzed by reovirus cores. Virology 39:822-831. [DOI] [PubMed] [Google Scholar]
- 83.Smith, R. E., H. J. Zweerink, and W. K. Joklik. 1969. Polypeptide components of virions, top component and cores of reovirus type 3. Virology 39:791-810. [DOI] [PubMed] [Google Scholar]
- 84.Spendlove, R. S., E. H. Lennette, C. O. Knight, and J. N. Chin. 1963. Development of viral antigens and infectious virus on HeLa cells infected with reovirus. J. Immunol. 90:548-553. [PubMed] [Google Scholar]
- 85.Sturzenbecker, L. J., M. L. Nibert, D. B. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taraporewala, Z., D. Chen, and J. T. Patton. 1999. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J. Virol. 73:9934-9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taraporewala, Z. F., and J. T. Patton. 2001. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J. Virol. 75:4519-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tosteson, M. T., M. L. Nibert, and B. N. Fields. 1993. Ion channels induced in lipid bilayers by subvirion particles of the nonenveloped mammalian reoviruses. Proc. Natl. Acad. Sci. USA 90:10549-10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Uitenweerde, J. M., J. Theron, M. A. Stoltz, and H. Huismans. 1995. The multimeric nonstructural NS2 proteins of bluetongue virus, African horsesickness virus, and epizootic hemorrhagic disease virus differ in their single-stranded RNA-binding ability. Virology 209:624-632. [DOI] [PubMed] [Google Scholar]
- 90.Virgin, H. W., IV, M. A. Mann, B. N. Fields, and K. L. Tyler. 1991. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J. Virol. 65:6772-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wetzel, J. D., G. J. Wilson, G. S. Baer, L. R. Dunnigan, J. P. Wright, D. S. H. Tang, and T. S. Dermody. 1997. Reovirus variants selected during persistent infections of L cells contain mutations in the viral S1 and S4 genes and are altered in viral disassembly. J. Virol. 71:1362-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wiener, J. R., and W. K. Joklik. 1987. Comparison of the reovirus serotype 1, 2, and 3 S3 genome segments encoding the nonstructural protein σNS. Virology 161:332-339. [DOI] [PubMed] [Google Scholar]
- 93.Yue, Z., and A. J. Shatkin. 1996. Regulated, stable expression and nuclear presence of retrovirus double-stranded RNA-binding protein σ3 in HeLa cells. J. Virol. 70:3497-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zou, S., and E. G. Brown. 1996. Stable expression of the reovirus μ2 protein in mouse L cells complements the growth of a reovirus ts mutant with a defect in its M1 gene. Virology 217:42-48. [DOI] [PubMed] [Google Scholar]
- 95.Zweerink, H. J. 1974. Multiple forms of ss-dsRNA polymerase activity in reovirus-infected cells. Nature 247:313-315. [DOI] [PubMed] [Google Scholar]
- 96.Zweerink, H. J., and W. K. Joklik. 1970. Studies on the intracellular synthesis of reovirus-specified proteins. Virology 41:501-518. [DOI] [PubMed] [Google Scholar]
- 97.Zweerink, H. J., M. J. McDowell, and W. K. Joklik. 1971. Essential and non-essential non-capsid reovirus proteins. Virology 45:716-723. [DOI] [PubMed] [Google Scholar]
- 98.Zweerink, H. J., E. M. Morgan, and J. S. Skyler. 1976. Reovirus morphogenesis: characterization of subviral particles in infected cells. Virology 73:442-453. [DOI] [PubMed] [Google Scholar]