Abstract
Comparative genome hybridization of the Francisella tularensis subsp. tularensis and F. tularensis subsp. holarctica populations have shown that genome content is highly conserved, with relatively few genes in the F. tularensis subsp. tularensis genome being absent in other F. tularensis subspecies. To determine if organization of the genome differs between global populations of F. tularensis subsp. tularensis and F. tularensis subsp. holarctica, we have used paired-end sequence mapping (PESM) to identify regions of the genome where synteny is broken. The PESM approach compares the physical distances between paired-end sequencing reads of a library of a wild-type reference F. tularensis subsp. holarctica strain to the predicted lengths between the reads based on map coordinates of two different F. tularensis genome sequences. A total of 17 different continuous regions were identified in the F. tularensis subsp. holarctica genome (CRholarctica) which are noncontiguous in the F. tularensis subsp. tularensis genome. Six of the 17 different CRholarctica are positioned as adjacent pairs in the F. tularensis subsp. tularensis genome sequence but are translocated in F. tularensis subsp. holarctica, implying that their arrangements are ancestral in F. tularensis subsp. tularensis and derived in F. tularensis subsp. holarctica. PCR analysis of the CRholarctica in 88 additional F. tularensis subsp. tularensis and F. tularensis subsp. holarctica isolates showed that the arrangements of the CRholarctica are highly conserved, particularly in F. tularensis subsp. holarctica, consistent with the hypothesis that global populations of F. tularensis subsp. holarctica have recently experienced a periodic selection event or they have emerged from a recent clonal expansion. Two unique F. tularensis subsp. tularensis-like strains were also observed which likely are derived from evolutionary intermediates and may represent a new taxonomic unit.
Francisella tularensis is a nonmotile, gram-negative coccobacillus originally isolated from ground squirrels in 1911 during a plague investigation in Tulare County, CA (19). The geographic distribution of the organism spans the entire Northern Hemisphere, with only a very recent isolated recovery of the organism in the Southern Hemisphere (29, 33). The organism is a facultative intracellular pathogen and is believed to infect more animal species than any other known zoonotic pathogen (13, 15). It has been isolated from as many as 250 species of wildlife (reviewed in reference 24), including various birds, amphibians, fish, and many mammalian species. The organism can also be found in invertebrate species, including arthropod vectors such as mosquitoes and ticks (25). Human infection occurs most often through direct exposure to infected animals or by bites from infected arthropod vectors. Recently, terrestrial and aquatic life cycles have been described for F. tularensis (14, 20); protozoa, such as Acanthamoeba castellanii, may also serve as a host for maintenance of F. tularensis in the aquatic cycle (1).
The species F. tularensis is comprised of four recognized subspecies: F. tularensis subsp. tularensis (Type A), F. tularensis subsp. holarctica (Type B), F. tularensis subsp. novicida, and F. tularensis subsp. mediaasiatica. F. tularensis subsp. holarctica and F. tularensis subsp. tularensis are considered clinically significant in humans (6, 30) and have been the most studied by far. F. tularensis subsp. tularensis is believed to be more virulent in humans than F. tularensis subsp. holarctica based on epidemiological data and its higher infectivity in animals. F. tularensis subsp. tularensis and F. tularensis subsp. holarctica also show striking geographic differences in their distribution, with both F. tularensis subsp. tularensis and F. tularensis subsp. holarctica being found in North America but only F. tularensis subsp. holarctica being found in Europe and Asia (25). Populations of F. tularensis subsp. mediaasiatica may be even more geographically limited, since, as its name suggests, this subspecies has only been isolated from the Asian subcontinent (6, 27). F. tularensis subsp. novicida, which was only recently detected outside the United States (33), can be found in aquatic environments, but little is known about its ecology.
Despite the unique geographic and virulence characteristics, known genetic and phenotypic differences distinguishing F. tularensis subsp. tularensis and F. tularensis subsp. holarctica are limited. Biochemically, the two subspecies have classically been differentiated primarily on the basis of glycerol fermentation, production of citrulline ureidase, and sensitivity to erythromycin (6). High-resolution genotyping methods, such as pulsed-field gel electrophoresi (11), restriction-fragment length polymorphism (31), amplified fragment length polymorphism (11), and multilocus variable-number tandem repeat analysis (8, 9, 15), also distinguish the two subspecies genotypically and show that they are divergent but clonally related.
Given the unique geographical and virulence characteristics, there is tremendous interest in understanding the genetic basis for these characteristics. Recent comparative genome hybridization studies identified limited differences in genome content between the two subspecies but did include the pdpD region, which is associated with virulence in F. tularensis subsp. tularensis and has apparently been lost from the F. tularensis subsp. holarctica genome (4, 26). Comparative genome-sequencing efforts are also under way and promise to provide detailed information with regard to specific strains. To provide a more complete catalogue of the genomic events which arose early during divergence of the subspecies (true subspecies-specific genomic differences as opposed to strain-level differences), we have applied paired-end sequence mapping (PESM) to identify candidate regions of genomic difference and further used comparative genome PCR (CG-PCR) on a large set of strains to identify regions of genomic difference that are conserved across multiple isolates. PESM was originally developed as a simple but elegant method to identify genomic islands of Shigella dysenteriae (5). The PESM strategy measures the physical distance between paired-end reads from a clone library, specifically searching for clones whose physical distance is incongruent with the predicted distance based on available genome sequences. In our study, we constructed a library from an F. tularensis subsp. holarctica strain and compared physical distances with the F. tularensis subsp. tularensis strain SCHU S4 (referred to herein as SCHU S4) genome sequence. Cloned segments with incongruent lengths compared to the map position were further distinguished as strain specific versus potentially subspecies specific by comparison to the F. tularensis subsp. holarctica strain LVS (referred to herein as LVS) genome sequence. In instances where the length difference was conserved in the reference strain and the LVS strain, the segments were further tested among a set of strains consisting of F. tularensis subsp. holarctica and F. tularensis subsp. tularensis to confirm that the genome difference was broadly conserved across the subspecies. Using this strategy, we identified 17 regions in the genome that are continuous in 66 of 67 F. tularensis subsp. holarctica strains examined but which are discontinuous in F. tularensis subsp. tularensis strains. These continuous regions (CR), termed CRholarctica, likely arose through extensive insertion/deletion, translocation, and rearrangement events. Despite the substantial genome diversity caused by the CRholarctica, their high degree of conservation among strains of F. tularensis subsp. holarctica of distinct temporal and geographic origin implies that this subspecies has recently undergone clonal expansion and subsequent geographic spread.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Cultures for this study were propagated on chocolate agar at 37°C in 5% CO2. Glycerol fermentation was tested in a limited number of isolates, either as described previously (26) or by using Biolog (Biolog, Inc., Hayward, CA) according to the manufacturer's instructions. A summary of spatial, temporal, host, and other pertinent demographic information, as well as prior subspecies determinations of all strains and/or DNA used in this study, is listed in Table S1 in the supplemental material.
Subspecies-specific PCR.
To confirm the subspecies designation of all the strains in our collection, we tested them using an RD1 PCR assay previously shown to differentiate among all four F. tularensis subspecies (4). All RD1 results are included in Table S1 in the supplemental material. Primers for RD1 were as published by Broekhuijsen et al. (4). All RD1 PCRs were performed in 25-μl volumes, each containing 5 mM MgCl2 and 160 μM of each deoxynucleoside triphosphate (Idaho Technology, Salt Lake, UT), 500 nM each of forward and reverse primer (Invitrogen, Carlsbad, CA), and 2.5 U of Platinum Taq (Invitrogen). Each reaction was conducted on 1.5-μl DNA samples either prepared by a standard large-scale bacterial genomic DNA extraction protocol (34) or using Puregene DNA isolation kits (Gentra Systems, Inc., Minneapolis, MN). Thermocycling conditions were optimized and performed on a Dyad (MJ Research, Reno, NV) thermocycler according to the following cycling parameters: initial hold at 95°C for 2 min, 30 s; 30 cycles of 95°C for 30 s, 64°C for 1 min, and 72°C for 1 min; final extension at 72°C for 5 min.
Construction of λ phage library.
F. tularensis subsp. holarctica strain MS304 is a human isolate obtained in 2002 from Missouri. A library was constructed from F. tularensis strain MS304 genomic DNA by partial digestion with Sau3AI. After optimization of the partial digestion for 10- to 15-kb fragments, 7 μg of genomic DNA was digested with Sau3AI in separate reactions with 0.0625, 0.0312, and 0.0156 U per μg for 1 h at 37°C. The partially cut DNA was electrophoresed on a 0.7% agarose gel along with molecular weight markers, and the regions containing 10- to 15-kb fragments were excised from the gel. The fragments were electroeluted, pooled, and then precipitated to concentrate. Size distribution of the gel-purified fragments was confirmed by agarose gel electrophoresis of a small portion of the fragments alongside molecular weight standards. The remaining purified fragments were then ligated into Lambda DASH II BamHI (Stratagene, La Jolla, CA). The ligations were packaged using Stratagene's Gigapack III Gold Extract according to the manufacturer's recommendations. The titers of the packaged phage on Escherichia coli XL1-Blue MRA P2 host bacteria were determined (Stratagene). The titer from the packaging was approximately 5 × 106 PFU/ml. Library randomness was confirmed by restriction digest and DNA sequence analysis of inserts from 10 independent plaques. The library was then amplified once using E. coli XL1-Blue MRA P2 host, and dimethyl sulfoxide was added to 7% final concentration in the clarified supernatant. The amplified library had a titer of 2.5 × 107 PFU/ml. One-milliliter aliquots were stored at −80°C.
Direct-PCR amplification (DPA) of cloned fragments.
For high-throughput PCR amplification of individual plaques, the library was diluted and plated onto mid-logarithmic-phase E. coli XL1-Blue MRA P2 (optical density at 600 nm, ∼0.6) grown in NZYM (N-Z amine A [casein hydrolysate], yeast extract, maltose) broth with 0.2% maltose. Dilutions of the library stock were made in Lambda suspension medium buffer, and dilutions yielding approximately 120 to 150 plaques per plate were subsequently plated onto several 80-cm petri dishes using E. coli XL1-Blue MRA P2 host cells. The plates were inverted in the incubator overnight at 37°C.
To amplify plaques for insert size measurement and paired-end sequencing, plaques were chosen from dilutions giving 120 to 150 well-isolated plaques. Plates with the appropriate number of plaques were first wrapped with Parafilm and inverted at 4°C for a minimum of 4 h and a maximum of 4 days. Clearly isolated plaques were further processed by direct-PCR amplification (DPA). DPA was performed in 96-well PCR plates (Applied Biosystems, Foster City, CA) preloaded with PCR master mix. Plaques for DPA were loaded by gouging each candidate plaque with a sterile 20-μl pipette tip (such that the tip contained visible plaque material) and then mixing the tip in a single well of the PCR plate. Long-range PCR was then performed using an Ex Taq Hot Start master mix kit (TaKaRa Mirus Bio, Madison, WI) containing T3 (5′-AATTAACCCTCACTAAAGGG-3′) and T7 (5′-TAATACGACTCACTATAGGG-3′) primers. PCR mixes were prepared according to the manufacturer's recipe, with each single 25-μl reaction containing 500 nM of each primer. Thermocycling conditions were performed on a Dyad (MJ Research, Reno, NV) thermocycler according to the following cycling parameters: initial hold at 95°C for 2 min, 30 s; 36 cycles of 95°C for 50 s, 55°C for 50 s, and 72°C for 15 min; final extension at 72°C for 5 min.
Following each PCR run, clone-amplicon purification was accomplished using Montage PCRμ96 Plates (Millipore, Billerica, MA) which were processed on a SAVM 384 Vacuum Manifold (Millipore) according to the manufacturer's instructions. The final elution was performed using 30 μl of Invitrogen distilled DNase/RNase-free water, and plates were sealed with adhesive sealing lids (Bio-Rad, Hercules, CA) and stored at 4°C.
Clone-amplicon sizing experiments.
Amplicons were sized by agarose gel electrophoresis. Ethidium bromide-stained gels were analyzed using the GeneTools (Synoptics) software package. The band sizes for all clones and DNA standards were exported from GeneTools into a Microsoft Excel spreadsheet and loaded into the PESM bioinformatics pipeline (see below).
Paired-end sequencing.
DNA sequencing of the DPA products of individual clones was carried out using BigDye Terminator v3.1 Cycle Sequencing kits (Applied Biosystems) with pGEM DNA serving as the sequence reaction controls. Each sequence reaction experiment was set up in 96-well reaction plates, with each sample being divided into two reactions, one reaction with T3 primer and the other with T7 primer. Sequencing reactions were carried out on the Dyad (MJ Research) thermocycler, and the reaction plate was cleaned up using a Montage SEQ96 kit on the SAVM 384 Vacuum Manifold (Millipore). The labeled DNA was then transferred into new reaction plates and loaded onto an ABI 3100 automated capillary-electrophoresis sequencer (Applied Biosystems), which was configured with an 80-cm capillary array and loaded with performance optimized polymer 4 (POP-4) (Applied Biosystems). Control sequencing reactions were performed on the two pGEM vector reactions in each set of 94 sample reactions, and the pGEM control sequences were evaluated to ensure the sequence reaction and sequence run were successful. The ABI sequence files were imported into Sequencher V.4.0.5 (Gene Codes, Ann Arbor, MI), trimmed, merged, and output as a single FASTA file for further analysis.
Fragment length and paired-end sequence pipeline.
The sizing and sequence files were input into a Perl-based program, referred to as the paired-end sequence mapping pipeline (PESMP), to identify coordinates of the paired-end reads from the draft F. tularensis live vaccine strain (LVS) whole-genome sequence (GenBank accession number AM233362) (21) and the completed SCHU S4 genome sequence (18) and then to compare the predicted lengths to the physical length of the cloned segment. The PESMP input files consisted of a composite FASTA file from the trimmed sequences resulting from a single 96-well plate along with a corresponding Microsoft Excel file containing the long PCR amplicon sizing data for each clone. The PESMP algorithm then uses BLASTN to localize coordinates of the reads in the respective genome sequences and outputs the coordinates, the predicted segment lengths from the two genomes based on the coordinates, and the physical measurement of the long PCR amplicon.
To identify clones with discrepancies between physical and predicted lengths between paired-end reads, the output from the PESM pipeline was loaded into an Excel spreadsheet and sorted according to the paired-end coordinates from the LVS genome sequence. Clones in which the physical size showed a >2-kb discrepancy from the predicted size of the SCHU S4 genome but not the LVS genome were chosen for further characterization. Sequence from these putative regions of genomic difference was obtained by electronic PCR, using the coordinates of the paired-end reads to extract the intervening sequence between the reads from the LVS genome sequence. These electronic clone sequences were then aligned by contig analysis using Sequencher to further delimit the physical boundaries of the CRholarctica.
Fine-structure genome mapping.
To characterize the extent and nature of the events associated with the CRholarctica, each CRholarctica was analyzed against the SCHU S4 genome sequence using BLASTN (2). Query coordinates for all segments/subsegments (shown in Table S2 of the supplemental material), except for multiple repeats of insertion (IS) elements, were used to map the location of the events in the SCHU S4 genome corresponding to the CRholarctica. CRholarctica maps were assembled using the SeqBuilder module of Lazergene V.6.0 (DNASTAR, Madison, WI) to identify corresponding gene/pseudogene content of the CRholarctica from the SCHU S4 annotation. All genes from SCHU S4 annotation found to be truncated at the flanks due to the beginning or ending of the CRholarctica clones, as well as those found internally due to rearrangements within the CRholarctica, were further analyzed using BLASTN against the LVS sequence for homology comparisons. Custom Perl scripts were used to generate graphical representations of all combined CRholarctica mapped on SCHU S4 shown in Fig. 2 to 4 and Fig. S1 to S4 in the supplemental material.
FIG. 2.
Relative positions and conservation of the CRholarctica segments in the genomes of Francisella strains. (A) The relative positioning of the CRholarctica mapped onto the SCHU S4 genome sequence. The outer scale designates coordinates of the whole-genome sequence in base pairs. The outer circle shows predicted coding regions on the plus strand color coded by role categories: violet, amino acid biosynthesis; light blue, biosynthesis of cofactors, prosthetic groups, and carriers; light green, cell envelope; red, cellular processes; brown, central intermediary metabolism; yellow, DNA metabolism; light gray, energy metabolism; magenta, fatty acid and phospholipid metabolism; pink, protein synthesis and fate; orange, purines, pyrimidines, nucleosides, and nucleotides; olive, regulatory functions and signal transduction; dark green, transcription; teal, transport and binding proteins; gray, unknown function; salmon, other categories; blue, hypothetical proteins. The second circle shows the location of all copies of ISftu1 (gray) and ISftu2 (blue) elements in the SCHU S4 genome. The fourth (innermost) circle shows the genomic location of the CRholarctica in the SCHU S4 genome sequence immediately above the corresponding CR number. The third circle shows the color-coded matching location of the CRholarctica in the LVS genome. (B) The conservation of CRholarctica within geographically and temporally distinct sets of Francisella strains. The outer scale designates coordinates in base pairs. The first circle shows predicted coding regions on the plus strand color coded by role categories: violet, amino acid biosynthesis; light blue, biosynthesis of cofactors, prosthetic groups, and carriers; light green, cell envelope; red, cellular processes; brown, central intermediary metabolism; yellow, DNA metabolism; light gray, energy metabolism; magenta, fatty acid and phospholipid metabolism; pink, protein synthesis and fate; orange, purines, pyrimidines, nucleosides, and nucleotides; olive, regulatory functions and signal transduction; dark green, transcription; teal, transport and binding proteins; gray, unknown function; salmon, other categories; blue, hypothetical proteins. The third circle depicts the genomic location of the 17 F. tularensis CRholarctica as indicated by their CRholarctica number. The second circle represents the color-coded matching location of each of the 17 F. tularensisCRholarctica distributed onto the genome of F. tularensis SCHU S4. The remaining circles display the CG-PCR results for each of the 17 loci for a panel of Francisella strains. The circles shaded in pink represent the following four strains (from the outer to inner layer): F. tularensis subsp. holarctica-japan and F. tularensis subsp. novicida strains 15482, Tu-43, and D2005067002. The circles shaded in light yellow represent the following 20 type A strains: Tu-30, SchuS4, NE-UNMC061598, NE-UNL091902, OK-00101504, OK-98041035, NC-54558-01, NC-52797-99, NC-54559-01, CDC NE 031457, UNL 072704, ATCC 6223, A88R160, 88R52, 88R144, AK-1133496, AK-1100558, AK1100559, WY-00W4114, and WY-WSVL02. The circles shaded in yellow represent the following 19 type B strains: LVS, Tu-28, Tu-29, Tu-35, Tu-42, Tu-1, Tu-2, Tu-3, Tu-4, Tu-5, Tu-6, Tu-7, Tu-8, Tu-9, Tu-10, Tu-11, Tu-12, Tu-13, and Tu-14. An additional 48 type B strains were tested that gave results identical to LVS. A negative PCR result is indicated by the absence of the CR box, and light and dark gray CR boxes correspond to PCR unique (not predicted for either SCHU S4 or LVS) for each given locus. Note that geographic locations as well as other demographic information for the strains may be obtained from Table S1 in the supplemental material.
FIG. 4.
CR13 aligned onto the circular SCHU S4 map. The top line of genes indicates the organization of the contiguous CR13 segment in the LVS genome with the coordinates of the CR indicated above. The locus tags from the SCHU S4 genome sequence are indicated below the genes, with those designated by a prefix or suffix “T” indicating genes truncated due to the beginning or ending of the cloned contig or due to altered arrangements of genomic structure within SCHU S4 compared to LVS. The relative positioning of the genes in the SCHU S4 genome is represented by rectangles on the circle and the corresponding color-coded locations in the LVS genome sequence indicated in the inner ring of the circle. Locus tag annotation is indicated to the right and left of the circle.
Comparative genome PCR (CG-PCR).
Confirmation of the CRholarctica was conducted by PCR analysis on multiple isolates of F. tularensis subsp. holarctica and F. tularensis subsp. tularensis. For each CRholarctica, a three-primer nested-PCR assay was designed based on the relative coordinates of the corresponding junctions of the CRholarctica maps. Primers for all assays were designed using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The assays were designed by first identifying a primer common to both LVS and SCHU S4, either forward or reverse (designated C-F or C-R, respectively), immediately adjacent to a breakpoint in the SCHU S4 genome sequence where synteny of the adjacent segment changed or was translocated, leaving an SCHU S4-specific region for an SCHU S4-specific (S) primer, and which likewise left a target for an LVS-specific (L) primer in the adjacent-contiguous LVS sequence. The design of all assays was such to produce “A”-type bands across all 17 CRholarctica for SCHU S4 and “B”-type bands across all 17 CRholarctica for LVS. Since either intact or truncated IS elements or their corresponding repeated elements were present near all breakpoints, care was taken to avoid placement of primers within IS elements (see Table S3 in the supplemental material for all primer coordinates, sequences, and expected amplicon sizes). All PCR assays were conventional by design and were conducted on 1.5 μl of the DNA samples used for RD1 PCR. All CG-PCRs were performed in 25-μl volumes, each containing 5 mM MgCl2 and 160 μM of each deoxynucleoside triphosphate (Idaho Technology, Salt Lake, UT), 500 nM each of common primer, LVS-specific primer, and SCHU S4-specific primer (Invitrogen, Carlsbad, CA), and 2.5 U of Platinum Taq (Invitrogen). Thermocycling conditions were optimized and performed on a Dyad (MJ Research, Reno, NV) thermocycler according to the following cycling parameters: initial hold at 95°C for 2 min, 30 s; 32 cycles of 95°C for 30 s, 60°C for 1 min, and 72°C for 1 min; final extension at 72°C. Each assay was first tested against SCHU S4 and LVS and then against a panel of 91 additional global Francisella strains representing both subspecies as well as F. tularensis subsp. novicida and a unique F. tularensis subsp. holarctica strain from Japan (23, 27, 31), tentatively called F. tularensis subsp. japonica (15) but for our studies referred to as F. tularensis subsp. holarctica-japan (see Table S1 in the supplemental material for panel composition). The amplicons were resolved by electrophoresis on 0.8% to 1% agarose gels and visualized by staining with ethidium bromide. A 100-bp PCR molecular ruler ranging from 100 bp to 3 kb (Bio-Rad) was used for size determinations.
RESULTS
Paired-end sequencing.
A total of 752 plaques were picked from the F. tularensis subsp. holarctica MS304 library and subjected to DPA, with 551 of the DPA yielding amplicons of >8 kb in length that were of sufficient quality and quantity for size determination and DNA sequence analysis (DPA success rate of 73.3%). The mean amplicon/insert size from the 551 successful DPA reactions was 14,174 bp, which corresponds to approximately 7.8 Mb of coverage, or an estimated 4.1 times the coverage of the 1.89-Mb F. tularensis subsp. tularensis genome (18).
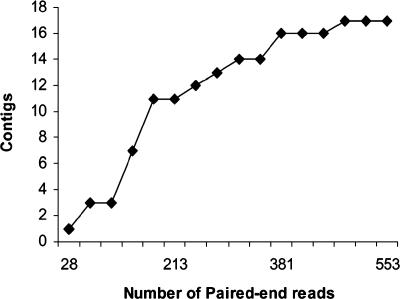
Of the 551 clones with quality paired-end reads, 66 clones had physical lengths that were not congruent with the distance between the paired-end reads relative to the SCHU S4 genome but were congruent with distances predicted from the LVS genome. These clones were therefore considered candidates for subspecies-specific genomic events, because the length differences were conserved in temporally and geographically distinct F. tularensis subsp. holarctica strains (LVS and MS304). Alignment of the sequences between the paired-end reads from the LVS genome grouped the 66 cloned segments into 17 different contiguous regions. These 17 contiguous regions (CRholarctica) are therefore defined as different regions where contiguous stretches of the F. tularensis subsp. holarctica strain LVS and MS304 genomes are otherwise dispersed or noncontiguous in the F. tularensis subsp. tularensis SCHU S4 genome sequence. As shown in Fig. 1, plotting of the cumulative number of CRholarctica identified versus the total number of clones sequenced showed that the number of new CRholarctica began to decrease sharply after 250 clones were sequenced. Of the last 301 clones sequenced, only three new CRholarctica were identified, suggesting that the library was nearly saturated.
FIG. 1.
Saturation of the PESM library. The cumulative number of contigs is plotted on the y axis as a function of the number of paired-end reads from the PESM library.
Fine-structure analysis of CRholarctica.
Fine-structure mapping and annotation of the CRholarctica was next conducted by alignment of the CRholarctica contigs from the strain LVS genome sequence with the SCHU S4 genome sequence. The corresponding locations of the aligned regions are shown in Fig. 2A, with the relative positions of the color-coded CRholarctica plotted onto the circular map of the LVS sequence (innermost ring) and SCHU S4 (next ring). The combined sequenced regions represented by the 17 CR correspond to nearly 230 genes/pseudogenes and over 30 IS elements, mainly a combination of ISftu1 and ISftu2 elements. Sixteen of the rearrangements and translocations are juxtaposed to IS elements in both the LVS and SCHU S4 sequences with the only exception occurring in CR6 (where only the SCHU S4 sequence revealed an ISftu2 element), suggesting that many of the events were likely mediated by these elements. As is evident from the illustration, these events resulted in remarkably large changes in the location of the CRholarctica between SCHU S4 and LVS. Equally intriguing, however, is that despite these massive changes in synteny, nearly all of the genes within the CRholarctica are present in both the LVS and SCHU S4 genomes, implying that if the events are indeed mediated by ISftu elements, the mechanisms have high fidelity and/or that selective pressure for maintenance of content is high.
Conservation of the CRholarctica in F. tularensis subsp. holarctica strains of wide geographic origin.
Conservation of the CRholarctica in the MS304 reference strain, which was isolated in 2002 and is temporally and geographically distinct from the LVS strain isolated in 1941, leads to the simple hypothesis that these CR likely arose early during divergence of F. tularensis subsp. tularensis and F. tularensis subsp. holarctica and therefore should be conserved across most F. tularensis subsp. holarctica strains. To confirm this, comparative genome PCR (CG-PCR) assays were developed for each CRholarctica using nested primer sets at the junctions of the CR (Table S3 in the supplemental material). The different CG-PCRs for all 17 CRholarctica were then run on a panel of DNA samples from SCHU S4 and LVS, as well as from 19 different F. tularensis subsp. tularensis strains, 67 F. tularensis subsp. holarctica strains, 3 F. tularensis subsp. novicida strains, and a single strain each of F. tularensis subsp. holarctica-japan and F. philomiragia. The results for all 17 CG-PCR panels are illustrated in Fig. 2B. The CRholarctica are represented by the same color scheme as that in Fig. 2A and uses the same coordinates. The innermost rings depict the presence of the CRholarctica as detected by CG-PCR in F. tularensis subsp. holarctica strains, with 18 of the rings illustrating individual strains and the first ring representing 48 different F. tularensis subsp. holarctica strains that also share the same CG-PCR profile as LVS and MS304. Remarkably, only a single F. tularensis subsp. holarctica strain produced an unexpected amplicon, with this single deviation occurring in F. tularensis subsp. holarctica strain Tu-42 (15th ring from the center), which produced a F. tularensis subsp. tularensis A-type band at CR2 and did not produce a detectable CG-PCR product from CR17. Thus, the CRholarctica identified through the PESM pipeline are indeed highly conserved among geographically and temporally distinct isolates of F. tularensis subsp. holarctica and therefore likely arose early during divergence of F. tularensis subsp. holarctica.
Unlike the 67 F. tularensis subsp. holarctica strains, the 19 F. tularensis subsp. tularensis strains, shown in the second set of yellow-shaded rings in Fig. 2B, displayed significantly more heterogeneity in CRholarctica. As can be seen from the number of gray rectangles (indicating unique CG-PCR products) and the absence of rectangles (indicating no CG-PCR product), several F. tularensis subsp. tularensis strains had alterations in CR1, CR8, CR10, and CR16, while others failed to give PCR products at several CR. Based on the combinations of reaction products observed from the different CR, at least four different subgroups of the F. tularensis subspecies can be resolved. SCHU S4 and nine other F. tularensis subsp. tularensis strains comprise one subgroup, producing A-type bands across all 17 CR. A second subgroup is represented by A88R160, 88R52, 88R144, AK-1133496, AK-1100558, and AK-1100559, with each of these strains sharing an amplicon from CR10 nested-PCR that was unique in size (denoted by gray rectangles at the CR10 position). All six of these strains were isolated from rabbits or hares, with the three AK strains derived from Alaska in 2003 and 2004 and the three others isolated from the contiguous United States. The ATCC 6223 strain was originally isolated from a human patient in 1920 and has since lost its virulence (15). A third subgroup is represented by the single strain OK-98041035, which matches the SCHU S4 subgroup except that it failed to produce a CR14 PCR amplicon. The fourth subgroup comprises a very unique set of two isolates, strains WY-00W4114 and WY-WSVL02. The results from these strains are shown on the innermost two rings of the F. tularensis subsp. tularensis strains. Both produced an A-type band at CR3, CR4, CR5, CR6, CR8, CR11, CR15, and CR17, were negative at CR1 and CR13, and produced B-type bands at CR7, CR9, CR12, and CR14. Unlike any other strains, they also produced unique bands at CR16 and CR10. These two strains were also distinguishable from one another in that WY-00W4114 produced a B-type band at CR2, whereas WY-WSVL02 was negative (yellow). These two strains were also only weakly positive for glycerol fermentation (26). Collectively, the genetic and biochemical data strongly suggest these two strains represent a new taxonomic unit. If indeed this is a new taxon, then the population is likely to be virulent, since one of the isolates was obtained from a human clinical sample (26). Importantly, all four of the different groups of F. tularensis subsp. holarctica strains share the subspecies-specific RD1 region in common, implying that, though divergent, they are taxonomically true F. tularensis subsp. tularensis derivatives.
As would be expected, the F. tularensis subsp. novicida, F. tularensis subsp. holarctica-japan, and F. philomiragia strains (pink-shaded ring in Fig. 2B labeled “other”) showed heterogenous CG-PCR results. F. philomiragia was negative across each of the 17 CR (not shown in the figure). The three F. tularensis subsp. novicida strains (inner three rings of the pink-shaded region) were negative for CR1, CR4, CR14, and CR16, and they produced a uniquely sized amplicon from CR3, CR5, CR7, CR8, CR9, CR10, CR12, CR13, and CR17. All three strains produced a “B”-type allele across CR6 and CR15, and they all produced an “A”-type allele across CR11. CR2 differentiated between the Tu-43 strain, which produced a unique amplicon, while the other two F. tularensis subsp. novicida strains (from American Type Culture Collection and USAMRIID) were both negative. Consistent with its classification as a separate subspecies (15), the single F. tularensis subsp. holarctica-japan strain (outermost ring in the pink-shaded region) was also distinct from all other strains in this study; it produced a “B”-type allele across CR1, CR2, CR3, CR5, CR6, CR7, CR9, CR10, CR11, CR12, CR15, CR13, and CR16, an “A”-type allele across CR17, and was negative across CR4, CR8, and CR14.
Genes affected by rearrangements.
As shown in Fig. 2A, the distribution of the CR1 to CR17 segments around the LVS genome and the relative positions of the corresponding regions in the SCHU S4 genome have some remarkable characteristics, indicative of potential effects of selection and/or bias in the biochemical basis of the events. First, the CR in the LVS genome show some positional bias, with 15 of the 17 CRholarctica being present in roughly one-half of the genome, from 1.2 Mb to 0.45 Mb. Second, there are three notable instances where segments from different CRholarctica in LVS are positioned adjacent to one another in the SCHU S4 genome. Specifically, segments from CR1 and CR16 are adjacent in the SCHU S4 genome but are dispersed in the LVS genome, as are segments from CR4 and CR10 and segments from CR13 and CR15. These events are illustrated in detail in Fig. 3. The juxtapositioning of these segments in F. tularensis subsp. tularensis suggests that their clustering in F. tularensis subsp. tularensis was the ancestral state, while their dispersal in F. tularensis subsp. holarctica is a derived state. Further support for this hypothesis can be seen in Fig. 3C, where both CR13 and CR15 contain genes that are involved in glycerol fermentation, and it seems likely that their ancestral condition would have been functionally clustered.
FIG. 3.
Juxtapositioning of CRholarctica segments in the F. tularensis subsp. tularensis SCHU S4 genome sequence. Shown are the CRholarctica segments from CR1 and CR16, CR4 and CR10, and CR13 and CR15 in their relative organizations in the SCHU S4 genome sequence and the LVS genome sequence. (A) CR1-CR16; (B) CR4-CR10; (C) CR13-CR15. Corresponding genes from the SCHU S4 and LVS genome sequence are indicated by the same color with the locus tag identifier number listed below the genes. The relative positioning and organization of corresponding genes between the two genomes is highlighted by the peach coloring, with inversions reflected by the hourglass shape of the coloring. (C) Putative orthologues of glpK (glycerol kinase), glpF (glycerol uptake facilitator), and glpA (glycerol-3-phosphate dehydrogenase) are indicated above or below the corresponding gene.
Further bioinformatics analysis of the junction of the juxtaposed CR4-CR10 segment (Fig. 3B) revealed the complete deletion of a gene of unknown function (FTT1308c) from F. tularensis subsp. holarctica. In addition, nine genes were found which are disrupted as a consequence of the rearrangements or translocation of genome segments in the CR. The intact versions of these genes in the SCHU S4 genome encode proteins with significant similarity to oligopeptide transporters (oppD and oppF), a ribosome modification gene (rimK), an acetyltransferase (FTT0177c), and genes of unknown function (corresponding to FTT0898c, FTT1122, FTT0921, and FTT1311). Figure 4 illustrates the region of CR13 in SCHU S4 containing the intact oppD and oppF genes, which are truncated in the LVS genome. The aceF gene, encoding the E2 domain of pyruvate dehydrogenase, lies near the junction of CR3 (along with aceE and lpd) and carries a 300-base in-frame deletion (Fig. S1 in the supplemental material). The deletion corresponds to loss of a repeated biotin-binding repeat region, leaving F. tularensis subsp. holarctica strains with two biotin-binding domains while the F. tularensis subsp. tularensis strains contain three. Whether the deletion occurred during the translocation event and whether it affects function of the pyruvate dehydrogenase complex is not clear. The AceF orthologues from several pathogenic species and genera, including Vibrio, Yersinia, Shigella, and Escherichia coli, do carry three domains. It has, however, been shown that deletion of two of the three domains of AceF in E. coli has little affect on function (7). It is worth noting that the aceF truncation itself was also identified by comparative genome hybridization studies (4, 26, 30), but the translocation event corresponding to CR3 was not detected, underscoring the importance of using multiple approaches for comparative genome analyses.
In addition to direct disruption as a consequence of translocation, genes near the junctions of the translocation events could also be subject to control by unique regulatory machinery. In this light, it is interesting to note that some of the genes within the CR and near the junctions could have functions related to physiology and virulence of F. tularensis. Two different genes encoding pilin subunits (pilE homologues) of a type IV pilus are present within the CR2 and CR10, and fine-structure genome comparisons of these CRholarctica mapped onto SCHU S4 are shown in Fig. S2 and Fig. S3, respectively, in the supplemental material. The gene encoding the pilin subunit pilE5 (FTT0230c) (12) is embedded within CR2. Another member of the pilE family is also present in CR10, and the region upstream is disrupted by IS elements. These IS elements are also associated with disruption and duplication of the FTT1311 gene in LVS compared to a single, intact copy in SCHU S4.
Because glycerol fermentation has classically served as a biochemical marker distinguishing F. tularensis subsp. tularensis and F. tularensis subsp. holarctica, we also scanned the CR for genes associated with glycerol metabolism. Several genes encoding enzymes associated with glycerol fermentation were found within CR11, CR13, and CR15. Fine-structure genome comparisons of each of these CRholarctica between SCHU S4 and LVS are shown in Fig. 3C and Fig. S4 in the supplemental material. There is no difference in the content of genes within CR11, CR13, and CR15 between the two subspecies; however, it is possible that their unique organization contributes to the different glycerol fermentation phenotypes.
DISCUSSION
Whole-genome sequencing has provided an outstanding resource for comparative genome studies, allowing high-resolution snapshots of the genetic diversity found within a given species. One of the drawbacks of comparative genome sequencing, however, is that only limited numbers of strains or taxa can reasonably be compared, making it difficult to distinguish between strain-level genomic differences and true lineage- or population-specific sets of genes. Comparative genome hybridization using DNA microarrays circumvents this problem to some degree by providing a single platform for comparison of multiple strains or taxa. On the other hand, the array approach is limited to assessing the diversity in genetic content that is represented on the array.
PESM was originally developed as a means to identify unique genomic islands (5). In our application, we scaled PESM for comparative genome studies. Given at least one reference genome sequence, PESM provides an economical means to identify candidate regions of genomic difference, and these regions can be further examined in larger strain sets by nested CG-PCR. The PESM library used in our study carried modest-sized fragments of the genome (averaging 14 kb), such that coverage could be obtained with a reasonable amount of sequencing without severely limiting the ability to measure physical size. PESM libraries, however, can be made using different insert sizes in different types of vectors, such that coverage per clone can be increased with larger segments while resolution can be increased by sequencing a larger number of segments from small-fragment libraries.
In addition to economy, the PESM approach allows any strain to be used as a source of the library, thereby allowing the user to choose the best taxonomic unit as a reference. This is particularly important when multiple subpopulations of a species may display unique characteristics that are of interest. Indeed, although we have used PESM in a binary comparison (F. tularensis subsp. holarctica compared to F. tularensis subsp. tularensis), it is possible to scaffold multiple libraries into the same PESM pipeline using only a single reference genome upon which to align the data.
Genome diversity in F. tularensis.
Populations of highly virulent bacteria display a wide spectrum with respect to genetic diversity. On the one hand, populations of species such as Bacillus anthracis display very little diversity, and only a limited number of clones appear to be spread worldwide (16). These clones can only be differentiated by examining variation in tandem repeats, which are some of the most rapidly evolving loci in the genome (17). With the availability of multiple genome sequences, single-nucleotide polymorphisms will soon complement or displace the approaches based on multilocus variable-number tandem repeat analysis. At the other end of the spectrum are subpopulations of E. coli O157:H7 which, despite the presence of highly clonal signatures in their genomic backbone, display substantial genomic diversity that is largely phage mediated and detectable by a relatively low-resolution method, such as pulsed-field gel electrophoresis (3, 22, 28).
Based on the data described in this study, we believe that F. tularensis may represent an intriguing model of genome evolution. Previous studies of genetic diversity in F. tularensis detected only limited diversity (6, 32). The four F. tularensis subspecies are known to share 98% identity in their 16S rRNA, show very similar biochemical profiles, and have quite similar antigenic compositions (6, 10). Only very high-resolution methods can provide any phylogenetic signal that reasonably correlates with biochemical and virulence characteristics.
Despite the apparently limited degree of genetic diversity, the F. tularensis subspecies display distinct geographic distribution and virulence characteristics. Thus, it was initially believed that although limited, the diversity in genomic content would parallel phylogeographic and epidemiologic characteristics and provide clues to the genetic basis for these traits. With the exception of differences in numbers of pilin-like genes encoded by the pilE-like loci (12) and the loss of the pdpD locus within the F. tularensis pathogenicity island (21), no other obvious candidate virulence genes (18, 24) emerged from comparative genome hybridization studies (4, 26, 30), and >99% of the genetic material present in the more virulent F. tularensis subsp. tularensis can also be found in the F. tularensis subsp. holarctica genome.
In the present study, we now show that despite the high degree of genetic conservation, the genome organization of F. tularensis subsp. tularensis and F. tularensis subsp. holarctica is vastly different. At least 17 substantial genomic events have occurred during divergence of these two subspecies and have been preserved among multiple strains of each population. The events correspond to extensive translocations and rearrangements, many if not all of which were mediated by movement of IS elements. As shown in Fig. 2, the IS elements are plentiful in the SCHU S4 genome, with 50 different copies of ISftu1 and 16 copies of ISftu2 being distributed around the genome (18). Given the large number of these elements, it is therefore not surprising to find them at or near the junctions of all 17 CR. IS elements were also found abutting subspecies-specific regions of genomic difference observed in comparative genome hybridization studies (4, 26), and our data here further reinforce the thinking that IS elements are the primary means through which this genome diversifies, be it through transposition or through homologous recombination at the numerous IS element sites.
While the degree of diversity in organization between the genomes of F. tularensis subsp. tularensis and F. tularensis subsp. holarctica is remarkable, perhaps equally remarkable is the degree to which the unique genome structure has been preserved across temporally and spatially distinct taxa of F. tularensis subsp. holarctica strains. This observation leads to several interesting possible hypotheses. First, it is possible that population growth is very minimal, such that little diversity has accrued. However, because F. tularensis is free-living and is also capable of infecting many different mammalian hosts, slow population turnover in the environment would seem to be an unlikely explanation. A second explanation is that the IS elements move only at a very low frequency, thus generating diversity only on a very slow timescale. In this instance, the divergence would have been quite ancestral given the degree of diversity that has accrued. With the number of ISftu1 and ISftu2 elements in the genome, this explanation also seems unlikely. More plausibly, we believe that the existing F. tularensis subsp. holarctica populations are quite homogenous, because they share very recent common ancestry. This hypothesis would imply that the populations have recently been through periodic selection or they arose from recent emergence, expansion, and geographic spread of a successful clone. If true, the clonal expansion hypothesis would also beg the question of why the recently emerged F. tularensis subsp. holarctica population can be found in North America and Eurasia, whereas the older F. tularensis subsp. tularensis populations seem to be confined to North America.
F. tularensis subsp. holarctica is likely a derived state.
Given the high degree of virulence that is displayed by F. tularensis subsp. tularensis, it has been speculated that it represents the ancestral state while the less virulent subspecies are derived states (30) that are more adept at infecting hosts without killing. Evolutionary analyses of variable-number tandem repeate loci also suggest that F. tularensis subsp. tularensis is likely more similar to the common ancestor (8, 9, 15). With respect to genome organization, our data also support this hypothesis, showing that organization of different CRholarctica appear to be a derived state, arising by dissociation of genomic units through translocation events occurring in an immediate ancestor of the F. tularensis subsp. holarctica populations. At least three genomic segments were found to be single contigs in the F. tularensis subsp. tularensis genome but are dispersed into six different CR in F. tularensis subsp. holarctica (Fig. 3). Moreover, some genes at the junctions of these events are disrupted or even deleted in F. tularensis subsp. holarctica, whereas the respective genes are present with no remnants of gene fragments in F. tularensis subsp. tularensis. Furthermore, the translocation events in CR13-CR15 have separated functionally associated genes that are putatively involved with glycerol fermentation in F. tularensis subsp. tularensis, again their scattering in F. tularensis subsp. holarctica being consistent with a derived condition (Fig. 3). We also note that three additional CR in F. tularensis subsp. tularensis (CR3-CR9, CR4-CR8, and CR9-CR11) are adjacent but not contiguous, whereas they are highly dispersed in F. tularensis subsp. holarctica (Fig. 2A). Therefore, several lines of evidence are beginning to mount in favor of the hypothesis that F. tularensis subsp. tularensis is more similar to the ancestral state, while populations of F. tularensis subsp. holarctica are derived states. If this is true, then analysis of genomic content and organization between the different subspecies should provide insights not only into virulence characteristics but also into selective pressures that have led to emergence and geographical spread of F. tularensis subsp. holarctica populations.
Although we believe these hypotheses are compelling, they should be tempered by the fact that little is known about the true biology of the francisellae. The fact that F. novicida can be found free living in water underscores the fact that although much emphasis is placed on studying species with the most impact on human health, the evolutionary and phylogenetic relationships of F. tularensis subsp. holarctica and F. tularensis subsp. tularensis are best understood within the context of the true ecology and biology of the francisellae. Therefore, our interpretation of recent evolutionary events within the F. tularensis subspecies will likely be more informed when we have a better understanding of the biology of the francisellae.
Two U.S. strains may represent a unique taxonomic unit within Francisella.
Although our search was primarily focused on identifying population-specific regions of genomic difference, the genome organization observed in F. tularensis subsp. tularensis strains WY-00W4114 and WY-WSLVL02 is very intriguing. Their pattern of genome organization is clearly distinct from the F. tularensis subsp. tularensis and F. tularensis subsp. holarctica populations, sharing CR “alleles” at some loci with F. tularensis subsp. tularensis strains, CR “alleles” at other loci with F. tularensis subsp. holarctica strains, and unique alleles at other loci (Fig. 2B). The fact that some CR alleles are shared with both subspecies suggests three different possibilities. First, the strains could have descended from a recombination event in which DNA was acquired from one subspecies and recombined into another. Given that the distribution of the CRholarctica “alleles” present in these strains is highly dispersed, multiple such recombination events would have to be invoked to account for the genomic organization. A second explanation is that the CRholarctica in these strains are homoplasious, arising through independent events within a separate lineage of descent from the common ancestor. This explanation is also unlikely, in that multiple such events must be invoked, each of which occurred similarly in at least two different lineages. Lastly, it is possible that these strains descended from an evolutionary intermediate, and thus only some of the CRholarctica have occurred and are preserved. This explanation invokes the fewest number of events and requires that the events only occur in a single lineage. Thus, we favor the evolutionary intermediate explanation. Regardless of the explanation for the events, the unique combination of CR in these strains implies that populations represented by these strains should have distinct taxonomic status (subspecies). To test the evolutionary intermediate model, further analyses on additional isolates should be performed.
Supplementary Material
Acknowledgments
We thank John McGraw and Mark Chrustowski for coordination of the AFIP Genomics Laboratory and assistance with DNA sequence analysis. We also thank Steve Francesconi, Bob Burgess, Wendall Thomas, Pete Iwen, Paul Fey, and Bob Wickert for cultivation of strains and preparation of DNAs for the Francisella panel. We are grateful to Patrick Chain and Emilio Garcia from Lawrence Livermore National Laboratories for their contribution of the LVS whole-genome sequence.
This work was supported by 1R21AI057755-01A1 to A.K.B. from the National Institutes of Health.
The views expressed in this article are those of the authors and do not reflect the official policy or position of the U.S. Air Force, Department of Defense, or government.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
This paper is a contribution of the University of Nebraska Agricultural Research Division, Lincoln (journal series no. 15265).
REFERENCES
- 1.Abd, H., T. Johansson, I. Golovliov, G. Sandstrom, and M. Forsman. 2003. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Arbeit, R. D., M. Arthur, R. Dunn, C. Kim, R. K. Selander, and R. Goldstein. 1990. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field electrophoresis to molecular epidemiology. J. Infect. Dis. 161:230-235. [DOI] [PubMed] [Google Scholar]
- 4.Broekhuijsen, M., P. Larsson, A. Johansson, M. Bystrom, U. Eriksson, E. Larsson, R. G. Prior, A. Sjostedt, R. W. Titball, and M. Forsman. 2003. Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. tularensis. J. Clin. Microbiol. 41:2924-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bumbaugh, A. C., R. F. Mangold, A. E. Plovanich-Jones, and T. S. Whittam. 2003. Genomic variation in Shigella dysenteriae type 1 using a new approach called paired-end sequence mapping, p. 622. In Proceedings of the 103rd American Society for Microbiology General Meeting. ASM Press, Washington, D.C.
- 6.Chu, M. C., and R. S. Weyant. 2003. Francisella and Brucella, p. 20. In P. R. Murray, E. J. Barron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 7.Cronan, J. E., Jr. 2002. Interchangeable enzyme modules. Functional replacement of the essential linker of the biotinylated subunit of acetyl-CoA carboxylase with a linker from the lipoylated subunit of pyruvate dehydrogenase. J. Biol. Chem. 277:22520-22527. [DOI] [PubMed] [Google Scholar]
- 8.Farlow, J., K. L. Smith, J. Wong, M. Abrams, M. Lytle, and P. Keim. 2001. Francisella tularensis strain typing using multiple-locus, variable-number tandem repeat analysis. J. Clin. Microbiol. 39:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farlow, J., D. M. Wagner, M. Dukerich, M. Stanley, M. Chu, K. Kubota, J. Petersen, and P. Keim. 2005. Francisella tularensis in the United States. Emerg. Infect. Dis. 11:1835-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsman, M., G. Sandstrom, and A. Sjostedt. 1994. Analysis of 16S ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int. J. Syst. Bacteriol. 44:38-46. [DOI] [PubMed] [Google Scholar]
- 11.Garcia Del Blanco, N., M. E. Dobson, A. I. Vela, V. A. De La Puente, C. B. Gutierrez, T. L. Hadfield, P. Kuhnert, J. Frey, L. Dominguez, and E. F. Rodriguez Ferri. 2002. Genotyping of Francisella tularensis strains by pulsed-field gel electrophoresis, amplified fragment length polymorphism fingerprinting, and 16S rRNA gene sequencing. J. Clin. Microbiol. 40:2964-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gil, H., J. L. Benach, and D. G. Thanassi. 2004. Presence of pili on the surface of Francisella tularensis. Infect. Immun. 72:3042-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopla, C. E., and A. K. Hopla. 1994. Tularemia, 2nd ed. CRC Press, Boca Raton, FL.
- 14.Jellison, W. L. 1974. Tularemia in North America, 1930-1974. Ph.D. dissertation. University of Montana, Missoula.
- 15.Johansson, A., J. Farlow, P. Larsson, M. Dukerich, E. Chambers, M. Bystrom, J. Fox, M. Chu, M. Forsman, A. Sjostedt, and P. Keim. 2004. Worldwide genetic relationships among Francisella tularensis isolates determined by multiple-locus variable-number tandem repeat analysis. J. Bacteriol. 186:5808-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keim, P., M. N. Van Ert, T. Pearson, A. J. Vogler, L. Y. Huynh, and D. M. Wagner. 2004. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect. Genet. Evol. 4:205-213. [DOI] [PubMed] [Google Scholar]
- 18.Larsson, P., P. C. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Halltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjostedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37:153-159. [DOI] [PubMed] [Google Scholar]
- 19.McCoy, G. W., and C. W. Chapin. 1912. Further observations on a plague-like disease of rodents with a preliminary note on the causative agent, Bacterium tularense. J. Infect. Dis. 10:61-72. [Google Scholar]
- 20.Morner, T. 1992. The ecology of tularaemia. Rev. Sci. Technol. 11:1123-1130. [PubMed] [Google Scholar]
- 21.Nano, F. E., N. Zhang, S. C. Cowley, K. E. Klose, K. K. Cheung, M. J. Roberts, J. S. Ludu, G. W. Letendre, A. I. Meierovics, G. Stephens, and K. L. Elkins. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 186:6430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsufjev, N. G., and I. S. Meshcheryakova. 1982. Infraspecific taxonomy of tularemia agent Francisella tularensis McCoy et Chapin. J. Hyg. Epidemiol. Microbiol. Immunol. 26:291-299. [PubMed] [Google Scholar]
- 24.Oyston, P. C., A. Sjostedt, and R. W. Titball. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2:967-978. [DOI] [PubMed] [Google Scholar]
- 25.Petersen, J. M., and M. E. Schriefer. 2005. Tularemia: emergence/re-emergence. Vet. Res. 36:455-467. [DOI] [PubMed] [Google Scholar]
- 26.Samrakandi, M. M., C. Zhang, M. Zhang, J. Nietfeldt, J. Kim, P. C. Iwen, M. E. Olson, P. D. Fey, G. E. Duhamel, S. H. Hinrichs, J. D. Cirillo, and A. K. Benson. 2004. Genome diversity among regional populations of Francisella tularensis subspecies tularensis and Francisella tularensis subspecies holarctica isolated from the US. FEMS Microbiol. Lett. 237:9-17. [DOI] [PubMed] [Google Scholar]
- 27.Sandstrom, G., A. Sjostedt, M. Forsman, N. V. Pavlovich, and B. N. Mishankin. 1992. Characterization and classification of strains of Francisella tularensis isolated in the central Asian focus of the Soviet Union and in Japan. J. Clin. Microbiol. 30:172-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott, L., P. McGee, D. Minihan, J. J. Sheridan, B. Earley, and N. Leonard. 2006. The characterisation of E. coli O157:H7 isolates from cattle faeces and feedlot environment using PFGE. Vet. Microbiol. 114:331-336. [DOI] [PubMed] [Google Scholar]
- 29.Sjöstedt, A. 2003. Family XVII, Francisellaceae. Genus I, Francisella, p. 111-113. In G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer-Verlag, New York, N.Y. [Google Scholar]
- 30.Svensson, K., P. Larsson, D. Johansson, M. Bystrom, M. Forsman, and A. Johansson. 2005. Evolution of subspecies of Francisella tularensis. J. Bacteriol. 187:3903-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas, R., A. Johansson, B. Neeson, K. Isherwood, A. Sjostedt, J. Ellis, and R. W. Titball. 2003. Discrimination of human pathogenic subspecies of Francisella tularensis by using restriction fragment length polymorphism. J. Clin. Microbiol. 41:50-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Titball, R. W., A. Johansson, and M. Forsman. 2003. Will the enigma of Francisella tularensis virulence soon be solved? Trends Microbiol. 11:118-123. [DOI] [PubMed] [Google Scholar]
- 33.Whipp, M. J., J. M. Davis, G. Lum, J. de Boer, Y. Zhou, S. W. Bearden, J. M. Petersen, M. C. Chu, and G. Hogg. 2003. Characterization of a novicida-like subspecies of Francisella tularensis isolated in Australia. J. Med. Microbiol. 52:839-842. [DOI] [PubMed] [Google Scholar]
- 34.Wilson, K. 1997. Preparation of genomic DNA from bacteria, p. 2.4.1-2.4.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, Chicago, Ill. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.