Abstract
The temperate double-stranded DNA bacteriophage Bam35 infects gram-positive Bacillus thuringiensis cells. Bam35 has an icosahedral protein coat surrounding the viral membrane that encloses the linear 15-kbp DNA genome. The protein coat of Bam35 uses the same assembly principle as that of PRD1, a lytic bacteriophage infecting gram-negative hosts. In this study, we dissected the process of Bam35 entry into discrete steps: receptor binding, peptidoglycan penetration, and interaction with the plasma membrane (PM). Bam35 very rapidly adsorbs to the cell surface, and N-acetyl-muramic acid is essential for Bam35 binding. Zymogram analysis demonstrated that peptidoglycan-hydrolyzing activity is associated with the Bam35 virion. We showed that the penetration of Bam35 through the PM is a divalent-cation-dependent process, whereas adsorption and peptidoglycan digestion are not.
Viruses must recognize and bind to the host cell in order to carry out the infection process. The initial recognition of the host cell, typically mediated by a cell surface receptor and leading to virus entry, is highly specific: proteins on the virion surface (receptor-binding proteins) specifically interact with molecules or molecular assemblies (receptors) exposed on the surface of a susceptible cell. Phages that infect gram-negative bacteria recognize either the polysaccharide moieties (e.g., T-even phages) or outer membrane proteins, such as porins (phages PP01 and λ) (34, 40) and transporters (T1, T5, and φ80) (23). Several filamentous single-stranded DNA and icosahedral single-stranded RNA phages of E. coli, as well as the enveloped bacteriophage φ6 of Pseudomonas syringae, adsorb to bacterial pili (38). Viruses that infect gram-positive bacteria usually attach to the cell surface polysaccharides (52). For example, phages φ29 and φ25 adsorb to the major teichoic acid in the cell wall of Bacillus subtilis (37). The host cell wall peptidoglycan moiety is the receptor for only a few bacteriophages, such as Listeria monocytogenes phage A511 and Clostridium botulinum type A 190L phage α2 (51, 57). In the cases of B. subtilis phage SPP1 (47) and Lactococcus lactis phages of the c2 group (53), the membrane proteins are necessary for irreversible adsorption.
After adsorption, virus entry involves either nucleic acid injection through a specific channel, membrane fusion, viral-capsid internalization via an intracellular vesicle, or a combination of these events (for a review, see reference 38). The entry of the tailed phages T4, T5, T7, and λ (for a review, see reference 29), as well as the filamentous phages M13, fd, and f1 (30), all infecting gram-negative hosts, has been studied most extensively. Entry of the tailed phage φ29, which infects gram-positive B. subtilis cells, is thought to be similar to that of other tailed bacteriophages and includes channel formation in the host cell envelope, followed by DNA injection (20). The membrane-containing bacteriophages PRD1 (see below), φ6 and φ13 (8, 17, 39), and most probably PM2 (26) use the viral membrane for genome delivery into the gram-negative host cytosol.
Bacteriophage Bam35 infects gram-positive Bacillus thuringiensis cells (2). It belongs to the family Tectiviridae, which includes icosahedral double-stranded DNA bacterial viruses with an internal membrane (5). Bam35 morphology closely resembles that of bacteriophage PRD1, the best-characterized member of the family Tectiviridae (2, 28, 41). Bam35 and PRD1 both have ∼15-kbp-long linear double-stranded DNA genomes with 5′-covalently linked terminal proteins (7, 49, 56) and similar genome organizations (41). Bam35, in contrast to PRD1, is a temperate phage (2) and can exist as a linear plasmid inside the host cell (19, 49). The sequence of the Bam35 genome is similar to those of the B. thuringiensis phages GIL01 and GIL16 and the linear Bacillus cereus plasmid pBClin15 (49, 55, 56). However, the Bam35 genome has no great sequence similarity to that of PRD1 (41).
Comparison of the cryoelectron microscopy-based three-dimensional image reconstruction of Bam35 with the high-resolution X-ray structure of PRD1 showed that the capsids of the two viruses are identical in size (64.5 nm between opposite faces) and have the same structures associated with assembly (1, 14, 28). Spikes protrude from the capsid at the vertices occupied by a pentameric minor capsid protein. The fold of the Bam35 major capsid protein closely resembles that of PRD1 (9, 28, 41). The virion-associated phospholipids of Bam35 and PRD1 are derived from the host plasma membrane (PM) but are incorporated selectively into the virion (27). In the case of PRD1, it was shown that during virus entry, the viral membrane undergoes structural rearrangements and transforms into a tubular structure. This tube-like structure crosses the capsid through an opening that is formed at one of the capsid vertices after receptor recognition (44). Consequently, this “tail” penetrates the cell envelope to deliver the DNA into the host cytosol (16, 22). Similar tube-like structures have been observed for Bam35 (2, 28), suggesting that the viral membrane most probably is involved in the delivery of the phage genome into the host cell. In order to overcome the barrier of the peptidoglycan (PG) layer, many bacteriophages encode PG-hydrolyzing enzymes (or endolysins) that are components of the virions, suggesting their role in virus entry (32). PRD1 contains two lytic enzymes (P7 and P15) as structural components of the virion (42, 43). The Bam35 genome harbors two open reading frames (ORFs) (26 and 30) that encode PG-hydrolyzing enzymes, as shown for a very closely related phage, GIL01 (54).
In the present study, the entry process for Bam35 was dissected into receptor binding, PG penetration, and interaction with the host PM. We showed that Bam35 very rapidly adsorbs to the cell surface. N-Acetyl-muramic acid (MurNAc) was identified as a component of the Bam35 receptor. The results of zymogram analysis demonstrated that PG-hydrolyzing activity was associated with the Bam35 virion. Furthermore, the penetration of Bam35 through the PM was shown to be a divalent-cation-dependent process, whereas adsorption and PG digestion were not.
MATERIALS AND METHODS
Bacteria and phage cultivation.
The Bacillus thuringiensis strains used in this study are listed in Table 1. B. thuringiensis cells were cultivated in Luria-Bertani broth (LB) (46). Bacteriophage Bam35 (2) was propagated on Bacillus thuringiensis serovar israelensis HER1410, as described previously (41). Briefly, to obtain Bam35 stocks, the soft agar from semiconfluent plates was collected, LB was added (3 ml/plate), and the plates were incubated for 2 h at 28°C with aeration; debris was removed by centrifugation (Sorvall SS-34 rotor; 10,000 rpm; 15 min; 5°C). Fresh Bam35 stocks were used for the adsorption experiments. For the production of phage particles, HER1410 cells were infected with a fresh Bam35 stock at a multiplicity of infection (MOI) of ∼15. After cell lysis, the particles were concentrated with polyethylene glycol 6000 and NaCl and further purified by rate-zonal centrifugation in a linear 5 to 20% (wt/vol) sucrose gradient (41). Bam35-resistant HER1410_R20 cells were obtained by isolating colonies from confluently lysed plates of B. thuringiensis HER1410 infected with Bam35 as described previously (19, 49). For DNA manipulations, E. coli K-12 strain HMS174 (13) or HMS174(DE3) (50) was used. The cells were grown in LB with appropriate antibiotics. The plasmids used in this study are listed in Table 2.
TABLE 1.
Bacillus thuringiensis strains used in this study
| Strain | Serovar | Relevant phenotype | Reference/sourcea |
|---|---|---|---|
| HER1410 | israelensis | Host for Bam35 | 2 |
| 4Q2 | israelensis | Inefficient host for Bam35b | BGSC |
| 4Q4 | israelensis | Bam35-type phage-producing strain | BGSC |
| 4Q5 | israelensis | Strain producing phage detectable on HER1410 | BGSC |
| 4Q7 | israelensis | Strain producing phage detectable on HER1410 | BGSC |
| 4Q8 | israelensis | Strain producing phage detectable on HER1410 | BGSC |
| 4D11 | kurstaki | Strain not sensitive for Bam35 | BGSC |
| 4D22 | kurstaki | Sensitive strain for Bam35 | BGSC |
| 4E3 | sotto | Strain not sensitive for Bam35 | BGSC |
| 4E5 | sotto | Strain not sensitive for Bam35 | BGSC |
| 4I1 | entomocidus | Inefficient host for Bam35b | BGSC |
| 4I2 | entomocidus | Strain not sensitive for Bam35 | BGSC |
| 4M1 | darmstadiensis | Strain not sensitive for Bam35 | BGSC |
| 4M2 | darmstadiensis | Strain not sensitive for Bam35 | BGSC |
| 4T1 | wuhanensis | Strain not sensitive for Bam35 | BGSC |
| HER1410_L3 | israelensis | Lysogenic derivative of HER1410 strain | 19 |
| HER1410_L5 | israelensis | Lysogenic derivative of HER1410 strain | 49 |
| HER1410_L7 | israelensis | Lysogenic derivative of HER1410 strain | 49 |
| HER1410_R19 | israelensis | Bam35-resistant derivative | 19 |
| HER1410_R20 | israelensis | Bam35-resistant derivative | This study |
BGSC, Bacillus Genetic Stock Center, Ohio State University, Columbus.
Bam35 plating efficiency was low.
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Reference/source |
|---|---|---|
| pET24 | High-level expression vector; ColE1 replicon; Kmr | Novagen |
| pJJ2 | Expression vector; ColE1 replicon; Apr | 35 |
| pMG60 | Expression vector; ColE1 replicon; Ptac Apr T7 RBS | 48 |
| pSJ1 | pET24 + nt 14023-14820 from Bam35 genome (ORF 30) | This study |
| pSJ3 | pET24 + nt 10983-11735 from Bam35 genome (ORF 26) | This study |
| pAR70 | pJJ2 + nt 12208-13122 from Bam35 genome (ORF 28) | This study |
| pAG1 | pMG60 + nt 13126-14007 from Bam35 genome (ORF 29) | This study |
Numbers (nt, nucleotides) refer to the Bam35 genomic sequence, accession number AY257527. Ap, ampicillin; Cm, chloramphenicol; Km, kanamycin.
Sacculus purification.
For zymogram analysis, PG sacculi were isolated by boiling E. coli K12 YMC or B. thuringiensis serovar kurstaki 4D22 cells (derived from stationary-phase cultures) in 4% sodium dodecyl sulfate (SDS) as described previously (24), followed by removal of PG-associated proteins with 2 M NaCl (10). The isolated sacculus preparation was resuspended in 1/100 of the original culture volume in Milli-Q water and stored at −20°C until it was used.
For the binding experiments, PG sacculi were isolated by incubating B. thuringiensis cells derived from a stationary-phase culture with 4% SDS; washing them with 3 M NaCl, followed by Milli-Q water; and freeze-drying them (11). Teichoic acids were extracted with 0.1 M NaOH under nitrogen (48 h at 37°C) as described previously (25). The sacculus preparations were washed several times with Milli-Q water to remove the NaOH and freeze-dried.
Cloning of Bam35 ORFs 28 and 29.
Standard molecular-cloning techniques were performed as described by Sambrook and Russel (46). The DNA fragment containing the coding sequence for the Bam35 gene product (gp) 28 was amplified by PCR using specific primers and was inserted under the control of a T7 promoter into plasmid pJJ2 between EcoRI-HindIII sites, resulting in plasmid pAR70. Similarly, the coding sequence for Bam35 gp29 was amplified by PCR using specific primers, and the amplicon was inserted into plasmid pMG60 between NdeI-PstI sites, resulting in plasmid pAG1. The construct for pAG1 contains the strong controllable Ptac promoter and the ribosome-binding site of the phage T7 polymerase gene.
Purification of Bam35 gp28 and gp29.
E. coli HMS174(DE3)(pAR70) or HMS174(pAG1) cells were grown in LB medium containing ampicillin (150 μg/ml) at 28°C to a density of approximately 2 × 108 CFU/ml. The cell cultures were chilled on ice, and protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. The IPTG-induced cells were incubated for an additional 12 h at 18°C. Bacteria were collected (Sorvall GS3 rotor; 5,000 rpm; 10 min; 5°C) and resuspended in 1/500 of the original culture volume of 20 mM Tris-HCl, pH 7.2. After being frozen at −80°C and thawed, the bacteria were disrupted by passage through a French pressure cell (at ∼105 MPa). The cell debris was removed by low-speed centrifugation (Sorvall SS-34 rotor; 8,000 rpm; 15 min; 5°C), and the supernatant was cleared by high-speed centrifugation (Beckman 50Ti rotor; 33,000 rpm; 2 h; 5°C). Saturated ammonium sulfate was added to the resulting supernatant to attain 35% (for gp28) or 20% (for gp29) saturation, and the mixture was kept on ice for 1 h. The precipitated proteins were collected by centrifugation (SS-34 rotor; 10,000 rpm; 30 min; 5°C).
The following purification steps were performed at 4°C. The pellet containing gp28 was resuspended in 50 mM NaCl-20 mM Tris-HCl, pH 7.2 (buffer A), and dialyzed overnight against the same buffer. After being cleared (SS-34; 10,000 rpm; 10 min), the supernatant was loaded at a flow rate of 1 ml/min onto an anion-exchange column (HiTrap HP Q-Sepharose; Amersham Biosciences) that had been equilibrated with buffer A. The proteins were eluted using a linear 0.05 to 1 M NaCl gradient buffered with 20 mM Tris-HCl, pH 7.2.
The pellet containing gp29 was resuspended in 20 mM NaCl-20 mM Na phosphate, pH 6.5 (buffer B), and loaded at a flow rate of 1 ml/min onto an anion-exchange column (HiTrap HP Q-Sepharose; Amersham Biosciences) that had been equilibrated with buffer B. The flowthrough was collected and applied to a heparin-Sepharose HP column (Amersham Biosciences) equilibrated with buffer B. The proteins were eluted using a linear 0.1 to 1 M NaCl gradient buffered with 20 mM Na phosphate, pH 6.5. Fractions containing gp28 or gp29, as determined by SDS-polyacrylamide gel electrophoresis (PAGE), were pooled and stored on ice.
Generation of polyclonal antisera against Bam35 gp28 and gp29.
To produce polyclonal antisera against gp28 and gp29, purified proteins were used as antigens to immunize rabbits. For gp28, immunizations were performed at 3-week intervals using 300 μg of protein. The antigen was emulsified in Freund's complete adjuvant for the first immunization and in Freund's incomplete adjuvant for the next two immunizations. Antiserum against gp29 was produced at Inbio Ltd., Estonia. The antisera were tested for phage neutralization ability. Approximately 500 phage particles were treated with antiserum dilutions for 30 min at 37°C and plated with HER1410 cells. To determine if the antibodies were able to precipitate the phage, 1× purified phage particles (41) were treated with antisera for 30 min at 37°C (in 10 mM K phosphate, pH 7.2) and analyzed by rate-zonal centrifugation in a 5 to 20% sucrose gradient made in the same buffer (Sorvall TH641 rotor; 24,000 rpm; 55 min; 15°C). Fractions (1 ml) were collected (BioComp fraction collector), precipitated with 10% (vol/vol) trichloroacetic acid, and analyzed by SDS-PAGE.
Virus dissociation.
HER1410 cells were grown to 2 × 108 CFU/ml and infected with Bam35 (MOI, 15). The cell lysates were cleared, and virus particles were precipitated with polyethylene glycol 6000 and NaCl as described previously (41). The virus precipitate was dissolved in 10 mM K phosphate, pH 7.2, and incubated for 1 h at 4°C. Urea (1 M) or 0.5 M guanidine hydrochloride (final concentrations) was added, and the suspension was incubated for 40 min at 37°C. These preparations were layered on top of a linear 5 to 20% sucrose gradient in 10 mM K phosphate, pH 7.2, and centrifuged (Sorvall TH641 rotor; 24,000 rpm; 70 min; 15°C). Fractions were collected and analyzed by SDS-PAGE. Fractions containing released viral-capsid-associated proteins were pooled and dialyzed against 10 mM K phosphate, pH 7.2, overnight at 4°C; filtered (Millipore; molecular mass cutoff, 100,000 Da), and concentrated by filtration (Millipore; molecular mass cutoff, 5,000 to 10,000 Da).
Phage adsorption and infective-center (IC) formation.
Unless otherwise stated, phage adsorption tests were performed by mixing 300 to 600 phage particles with either ∼4 × 106 cells grown to a cell density of ∼2 × 108 CFU/ml or an appropriate amount of peptidoglycan sacculi isolated from B. thuringiensis cells. The mixture was incubated for 10 min at 37°C with aeration, and the cells or sacculi were removed by centrifugation (Heraeus Biofuge; 13,000 rpm for 3 min at 22°C), followed by washing with LB or buffer (100 mM Tris-HCl, pH 8). The number of nonadsorbed phage particles was determined by plating the supernatants on lawns of HER1410 cells. The number of ICs was determined by resuspending the cells in 100 mM Tris-HCl buffer, pH 8, and plating them.
To determine Bam35 adsorption kinetics, B. thuringiensis cells were grown to ∼2 × 108 CFU/ml in LB at 37°C and infected at an MOI of ∼0.1. Unless otherwise stated, samples were taken at different time points and diluted 10-fold in LB. The cells were removed as described above, and the number of nonadsorbed phage particles was determined. The adsorption rate constant was calculated as described previously (3).
For the receptor saturation assay, a constant number of HER1410 or 4D22 cells (grown to a cell density of 2 × 108 CFU/ml) was infected using MOIs between 0.1 and 1,000. After 10 min of incubation at 37°C with aeration, the cells were removed by centrifugation and washed once with LB, and the number of nonadsorbed phage particles in the supernatants was determined.
Inhibition of phage adsorption.
The abilities of purified proteins encoded by Bam35 ORFs 28 and 29, or virion-derived fractions, to interfere with the binding of phages to the cells were determined using adsorption tests. HER1410 cells grown to ∼2 × 108 CFU/ml were mixed with different amounts of purified proteins or virion-derived fractions. After 10 min of incubation at 37°C, 300 to 600 Bam35 particles were added, followed by an additional 10-min incubation at 37°C. The cells were removed by centrifugation, and nonadsorbed phage particles were determined by titration.
Zymogram analysis.
To express Bam35 gp26 and gp30 and to determine their enzymatic activities, the corresponding coding sequences were cloned into pET24 vector between BamHI and HindIII sites, resulting in plasmids pSJ1 (ORF 30) and pSJ3 (ORF 26). The inserts were sequenced (Sequencing Laboratory, Institute of Biotechnology, University of Helsinki). E. coli HMS174(DE3)(pSJ1) or HMS174(DE3)(pSJ3) cells were grown in LB medium supplemented with kanamycin (25 μg/ml) at 37°C to ∼2 × 108 CFU/ml. The cell cultures were chilled on ice, and recombinant-protein production was induced by addition of IPTG to a 1 mM final concentration. The cells were incubated for an additional 3 h at 37°C, collected (Sorvall GS3 rotor; 5,000 rpm; 10 min; 5°C), and dissolved in 20 mM Tris-HCl, pH 7.4, to obtain 1/50 of the original volume. The cells were disrupted using sonication, and the soluble and insoluble fractions were separated (Heraeus Biofuge; 13,000 rpm; 10 min; 4°C). E. coli HMS174(DE3)(pET24) cells were used as a negative control. For zymogram analysis, 14% SDS-polyacrylamide gels were used, including a 6% (vol/vol) PG sacculus preparation homogenized by sonication (36). Following electrophoresis, the gels were rinsed with Milli-Q water, transferred to a renaturation buffer (25 mM K phosphate buffer, pH 7.4, 1 mM MgCl2, 0.1% Triton X-100), and incubated overnight at 28°C. After renaturation, the gels were rinsed with Milli-Q water, stained with 0.1% methylene blue in 0.01% KOH at 28°C for 1 h, and destained with Milli-Q water.
Measurements of ion fluxes.
The concentration of tetraphenylphosphonium (TPP+) or phenyldicarbaundecaborane (PCB−) ions in the medium was monitored by selective electrodes, as described previously (15, 16, 19). Briefly, the cells were grown to ∼2 × 108 CFU/ml, collected by centrifugation, resuspended in 100 mM Tris-HCl, pH 8.0, to obtain ∼3 × 1010 CFU/ml, and kept on ice until they were used (≤4 h). The concentrated cells were added to 100 mM Tris-HCl, pH 8.0, in 5-ml thermostatic reaction vessels to obtain 109 CFU/ml. After 10 min of incubation at an appropriate temperature with or without supplements, the cells were infected with a fresh Bam35 stock at MOIs of 5 to 10. The electrodes were calibrated by adding an appropriate amount of TPP+ chloride (Sigma) or potassium salt of PCB− (synthesized and purified by A. Beganskienė, Vilnius University) at the beginning of every experiment. To determine the amount of intracellular TPP+ at the end of the experiment, the PM of the cells was permeabilized by the addition of gramicidin D (GD) (Sigma) to a final concentration of 5 μg/ml.
Analytical methods.
Protein concentrations were determined by the Bradford (12) assay using bovine serum albumin as a standard, and the protein composition was determined by SDS-PAGE (36).
RESULTS
Screening of B. thuringiensis strains and adsorption characteristics of Bam35.
Commonly available B. thuringiensis strains, as well as genuine Bam35-resistant and lysogenic HER1410 cells generated previously (19, 49) and here, were analyzed for the ability to plate Bam35, spontaneously release viruses, and bind Bam35 virions (Table 3). Plaque assays on different B. thuringiensis strains showed that Bam35 was able to propagate properly on only two strains, HER1410 and 4D22, and weakly on strains 4Q2 and 4I1. Some strains most probably carry Bam35-type prophages, because viruses able to propagate on HER1410 were detected in their culture supernatants (Table 3). Adsorption tests revealed that most of the strains studied adsorbed Bam35 particles (Table 3). The two Bam35 hosts, HER1410 and 4D22, as well as strains 4T1, HER1410_R19, and HER1410_L5, were selected for more detailed adsorption analyses. HER1410_R20 cells, which bound Bam35 virions very effectively but did not support plaque formation, were used as a control in electrochemical measurements.
TABLE 3.
Characteristics of Bacillus thuringiensis strains used in this study
| Strain | Bam35 titer (PFU/ml) | Spontaneously released phagesa (PFU/ml) | Bound phage (%) |
|---|---|---|---|
| HER1410 | 5 × 1010 | 0 | 96 |
| 4Q2b | 109 | 0 | 46 |
| 4Q4 | 0 | 106 | NAc |
| 4Q5 | 0 | 103 | 80 |
| 4Q7 | 0 | 4 × 105 | 40 |
| 4Q8 | 0 | 106 | 34 |
| 4D11 | 0 | 0 | 14 |
| 4D22 | 5 × 1010 | 0 | 60 |
| 4E3 | 0 | 0 | 40 |
| 4E5 | 0 | 0 | 50 |
| 4I1b | 109 | 0 | 60 |
| 4I2 | 0 | 0 | 50 |
| 4M1 | 0 | 0 | 45 |
| 4M2 | 0 | 0 | 40 |
| 4T1 | 0 | 0 | 20 |
| HER1410_L3 | 0 | 103 | 97 |
| HER1410_L5 | 0 | 104 | 96 |
| HER1410_L7 | 0 | 108 | NA |
| HER1410_R19 | 0 | 0 | 2 |
| HER1410_R20 | 0 | 0 | 98 |
Detected on HER1410.
Bam35 formed small and hazy plaques on these strains.
NA, information is not available because of high background of spontaneously released viruses.
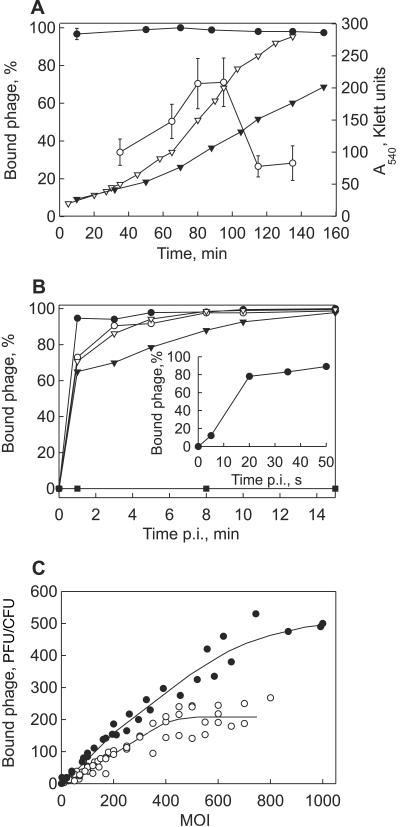
Bam35 adsorption efficiency dependence on the host cell densities was studied using HER1410 and 4D22 cells. HER1410 cells taken from early, middle, or late logarithmic growth phase all adsorbed approximately 96% of virus particles. In the case of 4D22, mid-log-phase cells bound Bam35 particles more effectively than the cells from early or late log phase (Fig. 1A).
FIG. 1.
Bam35 adsorption characteristics. (A) Efficiency of Bam35 adsorption to HER1410 (closed circles) and 4D22 (open circles) cells at different cell densities (MOI ∼10−4). The bars represent standard errors (of at least three experiments). The closed triangles and open triangles show HER1410 and 4D22 growth curves, respectively. (B) Kinetics of Bam35 adsorption to B. thuringiensis strains HER1410 (closed circles), 4D22 (open circles), 4I1 (closed triangles), 4Q2 (open triangles), and HER1410_R19 (closed squares). The inset depicts the kinetics of Bam35 adsorption to HER1410 during the first minute of infection. (C) Receptor saturation by Bam35 particles on HER1410 (closed circles) and 4D22 (open circles) cells. The experiments were performed as described in Materials and Methods.
Bam35 adsorption kinetics was studied using several B. thuringiensis strains. In all cases, the cells bound more than 60% of virus particles during the first minute after phage addition (Fig. 1B). The exception was Bam35-resistant HER1410_R19 cells, which showed no adsorption. Bam35 adsorption onto HER1410 cells revealed an adsorption rate constant of 2.3 × 10−8 ml/min for samples collected 20 s after phage addition (Fig. 1B, inset).
To determine how many virus particles could attach to a host cell, the dependence of Bam35 binding on the MOI was examined (Fig. 1C). HER1410 cells bound ∼500 Bam35 particles per cell, with signs of saturation at an MOI of ∼600. Receptor saturation of 4D22 cells was achieved at ∼200 virions bound per cell when an MOI of ∼400 was used.
Identification of the Bam35 receptor.
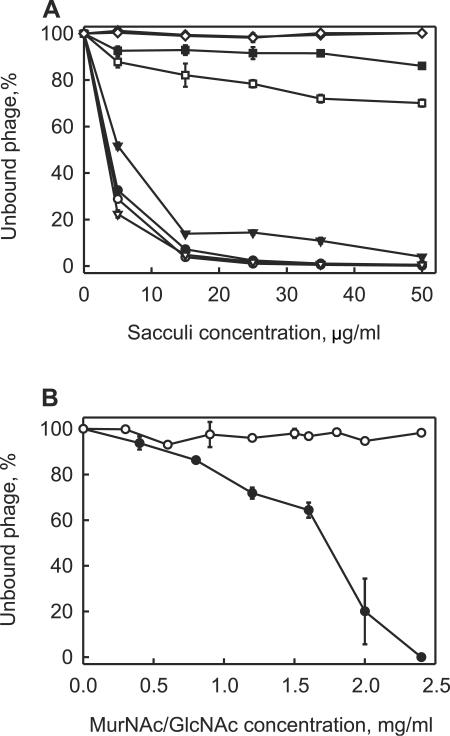
Bam35 adsorption characteristics suggested that a major repeated constituent of the gram-positive cell wall functions as a receptor. We isolated PG sacculi from different B. thuringiensis strains and tested whether they bound Bam35 particles (Fig. 2A). Sacculi isolated from the wild-type HER1410 cells very effectively eliminated plaque formation. In that assay, the lysogenic derivative HER1410_L5 sacculi behaved like those of nonlysogenic cells (HER1410), indicating unchanged cell wall properties (data not shown). Sacculi from 4D22 adsorbed phage particles less effectively than the sacculi from HER1410 cells. It is worth noting that intact 4D22 cells, when used instead of the sacculi, also bound Bam35 virions less effectively (Table 3). In agreement with the results of the previous experiment (Fig. 1B), sacculi of Bam35-resistant HER1410_R19 cells did not reduce plaque formation. PG sacculi derived from B. thuringiensis serovar wuhanensis 4T1 showed weak binding of virus particles. The isolated gram-positive sacculi have several major components: PG, teichoic acids (TA), and teichuronic acids (45). The teichoic acids were removed, and the PG sacculi without TA were tested for their ability to bind virus. In the case of HER1410, these sacculi bound virions with the same efficiency as the sacculi with TA. By comparison, 4D22 and 4T1 sacculi bound Bam35 particles more efficiently after the removal of TA (Fig. 2A), thereby excluding TA as the Bam35 receptor. Gram-negative cells do not have any teichoic or teichuronic acids in their cell walls (45). However, Bam35 particles adsorbed to the PG sacculi isolated from E. coli K-12 (data not shown). These observations indicated that the PG moiety of the sacculus is likely the receptor. Consequently, the PG constituents, MurNAc and N-acetyl-glucosamine (GlcNAc), were tested for the ability to inhibit plaque formation. MurNAc inactivated the virus, whereas GlcNAc at the same or higher concentrations did not (Fig. 2C). These results led to the conclusion that PG is used for Bam35 binding and that MurNAc is involved in the process.
FIG. 2.
Bam35 adsorption to the PG sacculi or PG constituents. (A) Bam35 adsorption to PG sacculi (closed symbols) or PG sacculi lacking teichoic acid (open symbols) from B. thuringiensis strains HER1410 (circles), 4T1 (squares), 4D22 (triangles), and HER1410_R19 (diamonds). (B) Bam35 inactivation by MurNAc (closed circles) and GlcNAc (open circles). Approximately 300 Bam35 particles were incubated with different concentrations of MurNAc or GlcNAc (15 min at 37°C), and titers were determined on HER1410 lawns. The bars represent standard errors (of at least three experiments). The experiments were performed as described in Materials and Methods.
Putative Bam35 receptor-binding proteins.
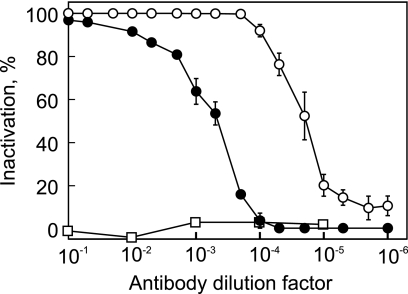
The Bam35 virion has spikes protruding from the capsid vertices (2, 28). Such spikes are considered to be responsible for host cell recognition and binding (21). Amino acid sequence similarities to PRD1 and the results of threading of all predicted Bam35 gene product amino acid sequences onto all available PRD1 X-ray structures suggested two Bam35 proteins, gp28 and gp29, as possible candidates for the Bam35 spike complex (28, 41). These proteins were expressed in E. coli and purified to near homogeneity (not shown). The purified proteins were used in the Bam35 adsorption inhibition assay. However, neither gp28 nor gp29 had any significant effect on the adsorption of Bam35 to HER1410 when the cells were preincubated with either of the purified proteins (not shown). Due to these negative results, a method to dissociate the viral capsid was developed (see Materials and Methods). Released and partially purified capsid proteins (containing mostly gp28) did not compete with Bam35 virions in the adsorption assay (not shown). Polyclonal antibodies against gp28 and gp29, however, inactivated the virions (Fig. 3) and caused virion aggregation, indicating that both of the proteins reside on the virion surface.
FIG. 3.
Inactivation of Bam35 virions by polyclonal antibodies against gp28 (closed circles) and gp29 (open circles). Preimmune serum was used as a control (open squares). The bars represent standard errors (of at least three experiments).
Endolysins of Bam35.
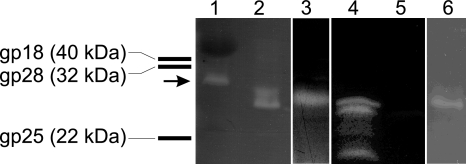
To determine if Bam35 virions contain the PG-hydrolyzing enzymes as structural components, purified Bam35 particles were analyzed using zymography (Fig. 4). Purified bacteriophage PRD1 virions were used as a control; the clear band visible in the zymogram resulted from the ∼27-kDa lytic enzyme, P7 (42). Bam35 structural-protein species yielded a clear zone with an upward smear at the position corresponding to a molecular mass of 20 to 30 kDa, indicating lytic activity associated with the virus particle (Fig. 4). ORF 26, which encodes a protein with a deduced molecular mass of ∼26.5 kDa, and ORF 30, which encodes a protein of ∼30.8 kDa, were cloned; the gene products were expressed in E. coli; and the crude cell extracts were analyzed in a zymogram assay (Fig. 4). The cell extracts of both clones showed PG degradation zones (using E. coli-derived PG) close to the migrating position of the Bam35-derived zone and were absent in the negative control (vector plasmid without the insert). We tested numerous gel systems with no success in order to make a clearer distinction between the two zones. We can conclude that gp26 clearly caused the lower major band in the zymograms, as this protein was also identified in the virion by N-terminal amino acid sequencing (41). Bam35 endolysin gp26 was not stable and degraded in the crude cell extract, causing smaller clear bands visible in the zymogram (Fig. 4). We observed here that recombinant protein gp26 showed strong lytic activity on the PG derived from E. coli whereas gp30 was more active on PG of B. thuringiensis serovar kurstaki 4D22 (data not shown). It was shown previously that these enzymes possess different PG-hydrolyzing activities, depending on the PG source (54). Bam35-associated lytic activity was also observed when the renaturation buffer contained 1 mM EDTA and no Mg2+ ions, suggesting that divalent cations are not needed for endolysin activity (Fig. 4).
FIG. 4.
Zymogram analysis of purified Bam35 virions and cell extracts with expressed Bam35 proteins gp26 and gp30 using E. coli-derived PG. Zymogram renaturation, except for lane 6, was performed as described in Materials and Methods. In the case of lane 6, the renaturation buffer contained 1 mM EDTA and no Mg2+. Lane 1, purified PRD1 particles (the arrow marks the migration position of PRD1 endolysin P7); lanes 2 and 6, purified Bam35 particles; lanes 3 to 5, crude cell extracts of E. coli HMS174(DE3)(pSJ1) containing gp30, HMS174(DE3)(pSJ3) containing gp26, and HMS174(DE3)(pET24) containing the vector without insert, respectively. The bars on the left represent the migration positions of some Bam35 proteins in the zymogram.
Bam35-induced effects on the PM.
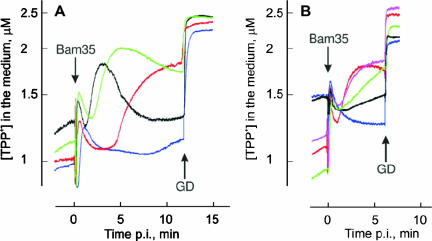
We measured TPP+ ion fluxes across the host PM to monitor temporal-permeability-related changes in the membrane voltage (Δψ; negative inside) during Bam35 entry. Bam35 infection induced PM depolarization, resulting in an efflux of the cell-accumulated TPP+, which at 37°C started 1.5 to 2 min postinfection (p.i.). This was followed by PM repolarization, a decrease in the external concentration of TPP+, starting 5 to 7 min p.i. (Fig. 5A) (19). Experiments with Bam35-resistant but virus-adsorbing cells of strain HER1410_R20 showed that adsorption alone did not cause PM depolarization (data not shown). Accordingly, by monitoring TPP+ fluxes, it is possible to detect when Bam35 reaches the cell PM during the entry process.
FIG. 5.
Effects of temperature and osmotic pressure on Bam35-induced depolarization of HER1410 cells. After 10 min of incubation in 100 mM Tris-HCl, pH 8.0, containing 3 μM TPP+, the cells were infected with Bam35 using an MOI of ∼10. GD was added to the final concentration of 5 μg/ml. (A) Temperature effects (37°C, black; 32°C, green; 27°C, red; 21°C, blue). (B) Effects of osmotic pressure (95 mM sucrose, red; 190 mM sucrose, pink; 375 mM sucrose, green; 500 mM sucrose, black; 750 mM sucrose, blue). The experiments were performed at 37°C.
At temperatures lower than 37°C, the initiation of phage-induced PM depolarization was delayed and the kinetics of depolarization was slower (Fig. 5A). At 21°C, PM depolarization started ∼7 to 8 min p.i., although the efficiency of adsorption was not greatly affected, and at 1 min p.i., the number of cell-bound viruses was similar to that at 37°C (not shown). However, we were not able to determine which of the processes was more temperature sensitive, PG hydrolysis or interaction with the PM.
Extracellular osmotic pressure plays an important role in bacteriophage entry (33). An increase in the osmotic pressure of the medium by addition of sucrose decreased the amplitude of Bam35-induced PM depolarization (Fig. 5B). At sucrose concentrations over 350 mM, depolarization and IC formation were prevented (Table 4 and Fig. 5B), although the cells still accumulated large amounts of TPP+.
TABLE 4.
Effects of divalent cations and osmotic pressure on Bam35 viability, adsorption, and infectivitya
| Supplementb | Bam35 viabilityc (%) | Adsorption (%) | IC (%) |
|---|---|---|---|
| 500 mM sucrose | 70 ± 3 | 85 ± 5 b.i. | 1 ± 1 b.i. |
| 94 ± 1 p.i. | 22 ± 8 p.i. | ||
| 2 mM EGTA | 52 ± 4 | 99 ± 1 b.i. | 2 ± 1 b.i. |
| 91 ± 2 p.i. | 59 ± 8 p.i. | ||
| 2 mM EDTA | 64 ± 5 | 99 ± 1 b.i. | 1 ± 1 b.i. |
| 92 ± 2 p.i. | 50 ± 9 p.i. | ||
| 2 mM EGTA/4 mM Ca2+ | 124 ± 2 | 97 ± 1 b.i. | 237 ± 17 b.i. |
| 2 mM EGTA/4 mM Mg2+ | 130 ± 3 | 64 ± 2 b.i. | 102 ± 12 b.i. |
| 2 mM EDTA/4 mM Ca2+ | 127 ± 10 | 96 ± 2 b.i. | 145 ± 5 b.i. |
| 2 mM EDTA/4 mM Mg2+ | 130 ± 6 | 56 ± 8 b.i. | 114 ± 6 b.i. |
Measurements were performed using 100 mM Tris-HCl, pH 8, at 37°C. The given value is the mean ± standard error (of at least three experiments); the level of Bam35 viability or infective-center formation without supplement addition was set as 100%; b.i., supplement added before infection; p.i., supplement added 5 min postinfection.
In the case of two supplements, the first was added 5 min prior to the addition of the second.
Bam35 titer after 10-min incubation.
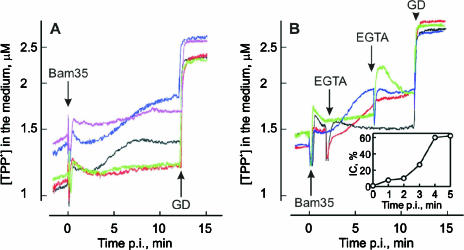
Divalent cations play an important role in bacteriophage entry (29, 47). A divalent cation chelator, EGTA or EDTA, at 2 mM concentration decreased the phage titer by ∼50% (Table 4). However, it was possible to totally restore phage viability by the addition of higher concentrations of Ca2+ or Mg2+ (Table 4). EGTA or EDTA (2 mM final concentration) added to the cell suspension before infection prevented Bam35-induced PM depolarization (Fig. 6A) and inhibited formation of the ICs (Table 4). Addition of excessive concentrations of Ca2+ to EGTA- or EDTA-containing cell suspensions effectively reversed the effects of the chelators on IC formation (Table 4). However, the depolarization caused by infection was not observed in the presence of Ca2+ (Fig. 6A). Conversely, addition of Mg2+ to EGTA-containing medium restored both IC formation and the depolarization caused by the entering phage (Table 4 and Fig. 6A). These results indicate that entry does not require Ca2+ addition to the medium, but the removal of cell surface- and/or virion-bound Ca2+ ions blocks Bam35 entry. As Mg2+ can effectively replace Ca2+, Bam35 entry is a divalent-cation-dependent process.
FIG. 6.
Effects of divalent cations on the Bam35-induced depolarization of HER1410 cells. Prior to infection, the cells were incubated for 10 min at 37°C in 100 mM Tris-HCl, pH 8.0, containing 3 μM TPP+ (A, black curve) and, in addition, 2 mM EGTA (A, green), 2 mM EDTA (A, red), 2 mM EGTA and 4 mM CaCl2 (A, pink), 2 mM EGTA and 4 mM MgCl2 (A, blue), 2 mM CaCl2 (B, black and green), or 2 mM MgCl2 (B, red and blue) was added. The cells were infected using an MOI of ∼5. (B) EGTA was added at different time points, as indicated, to obtain a 4 mM final concentration. GD was added to a final concentration of 5 μg/ml. The inset in panel B shows IC formation after the addition of EGTA (2 mM final concentration) at different time points. IC formation without EGTA was set as 100%; time zero corresponds to the addition of EGTA before infection.
To further dissect the cation-associated processes, the chelators were introduced at different time points. Both EGTA (Fig. 6B, inset) and EDTA (data not shown) reduced IC formation very effectively when added either prior to infection or during the first 2 minutes p.i. but only moderately when added 4 min p.i. or later. This positions the divalent-cation-dependent stage between 1.5 and 2 min p.i., corresponding to the time point when PM depolarization starts (Fig. 6A).
Ca2+ ions induce Bam35 entry without induction of depolarization (see above). To gain further information about this, EGTA was added to Mg2+- or Ca2+-containing medium before the phage-induced depolarization in Mg2+-containing medium started (1.75 min p.i.) or later, when the depolarization in the presence of Mg2+ reached its maximal level (7.25 min p.i.) (Fig. 6B). The early addition of Ca2+-depleating amounts of EGTA had no great effect on the accumulation of TPP+ (Fig. 6B), but late addition induced a strong reversible leakage of TPP+, indicating depolarization and repolarization of the PM (Fig. 6B). The amplitude of the depolarization increased in a time-dependent manner if EGTA was added between these two time points (not shown). Addition of EGTA to Mg2+-containing medium did not greatly change the course of Bam35-induced depolarization (Fig. 6B). In the absence of divalent cations, EGTA and EDTA induced additional reversible leakage of TPP+ from infected cells when added ∼7 min p.i. (data not shown). These results indicate that Ca2+ ions prevent Bam35-induced depolarization, e.g., by affecting the ion permeability of infected cells. Comparison of the Bam35-induced depolarization-blocking effects of alkaline earth metals revealed the sequence Ca2+>Sr2+>Ba2+>Mg2+ (data not shown).
Structural changes in the bacterial PM during virus entry can be detected by monitoring the binding of PCB− ions to membranes of the infected cells (15). The channel-forming phages T4 and λ (15), as well as PM2 (26), induce strong binding of PCB− to infected bacteria. Bam35, like phages PRD1 (16) and φ13 (17), cause only weak PCB− binding to the host membrane (results not shown), indicating that the PM structure is not severely disturbed by the entering virus.
DISCUSSION
In this study, we dissected the Bam35 entry process into three distinct steps: (i) receptor binding, (ii) PG digestion, and (iii) interaction with the PM. Bam35 very rapidly adsorbed to the cell surface and reached up to 500 particles per HER1410 cell (Fig. 1). Using an effective virion diameter of 100 nm, it could be calculated that close to one-half of the cell surface was covered by viral particles. This result is consistent with previously published electron microscopy data demonstrating closely packed virions attached to the cell (41).
Virus binding experiments with the PG constituents allowed the identification of MurNAc as a component of the Bam35 receptor. It should be noted, however, that when using MurNAc, the concentration needed to compete with phage adsorption was ∼3 orders of magnitude higher than when using sacculi (Fig. 2). This observation indicated that some components in addition to MurNAc, such as the variable peptide linkages, might be involved in Bam35 adsorption. Most likely, these PG components were altered in HER1410_R19 cells, as the sacculi from these cells did not bind Bam35 at all (Fig. 2A). In contrast to Bam35, which uses the PG sacculus, most of the bacteriophages infecting gram-positive bacteria use saccharide components of teichoic or teichuronic acid for irreversible adsorption (52).
Bam35 aggregation and neutralization assays using polyclonal antibodies showed that gp28 and gp29 reside on the virion surface (Fig. 3). However, the recombinant proteins or capsid proteins dissociated from the virion did not compete for receptor binding, leaving the identity of the receptor-binding protein obscure. It might be that the folding or the multimericity of the separated proteins was not correct and prevented their biological activity.
Lytic activities of recombinant endolysins from phage GIL01 (a close relative of Bam35) have been demonstrated (54). Here, we showed that Bam35 possesses PG-hydrolyzing activity associated with the virus particle. Although it was difficult to resolve the identities of the PG-hydrolyzing enzymes in the Bam35 particle, we consider that gp26 is responsible for the major lytic activity of the virion. Due to the strong signal and double-band character of the virion-associated lytic activity in zymograms (Fig. 4), we suggest that both PG-hydrolyzing enzymes encoded by the Bam35 genome, gp26 and gp30, are associated with the virion.
PRD1 entry does not cause PM depolarization, whereas Bam35 entry does (16, 19). However, both PRD1 and Bam35 entries induce only a weak accumulation of PCB−, indicating that the PM structure remains practically intact. We propose that a reversible opening of specific channels is responsible for the depolarization of the PM during entry. On the other hand, when the Ca2+ concentration in the infection medium was increased, IC formation was not altered, but no depolarization was observed (Table 4 and Fig. 6A), showing that phage DNA entry and depolarization of the PM are coupled but separate processes. These observations are consistent with a model in which ion fluxes leading to the depolarization of the PM utilize pathways different than the one used for phage DNA entry. Permeability of the channels responsible for depolarization during the Bam35 entry process can be controlled in two ways: (i) the phage-induced PM depolarization is blocked in the presence of increased Ca2+ concentrations (>2 mM) (Fig. 6A) and (ii) after depolarization, the channels close in the absence of extracellular Ca2+ (Fig. 6B), leading to repolarization of the PM.
The 7.3-Å-resolution structure of the Bam35 virion revealed large membrane protein complexes that are not present in PRD1 (28). It is possible that these complexes are the ion channels causing depolarization during virus interaction with the host PM, making a connection from the external milieu to the cytosol via the cell-associated phage particle. It has been shown that several viruses, such as influenza A, B, and C viruses; human immunodeficiency virus type 1; and Paramecium bursaria chlorella virus type 1 (PBCV-1), contain ion-permeable channels in their membranes (18).
PBCV-1 induces depolarization of the infected host cells, and the depolarization is associated with the K+ channels present in the viral membrane (31). Bam35 also induces the leakage of K+ from infected cells (19). Ba2+ ions are able to block PBCV-1-formed channels (31), whereas Ca2+ ions are better blockers in the case of Bam35. It has been suggested (31) that the viral channels cause the reduction of host cell internal pressure and allow viral DNA to be delivered into the cell. However, blocking of the Bam35-induced channels by Ca2+ ions did not lead to the prevention of phage DNA entry under the conditions studied.
We observed that Bam35 penetration differs considerably from that of PRD1, although their virion architectures are very similar and both viruses form a tail-like structure during genomic-DNA release. This result is in accordance with the idea that the entry-related viral structures and functions evolve rapidly but the architectural principles of capsid shell-associated structures (the viral innate “self”) are highly conserved (4, 6).
Acknowledgments
Silja T. Jaatinen is acknowledged for the construction of plasmids pSJ1 and pSJ3. The technical assistance of Anna Rantala is gratefully acknowledged.
This work was supported by the Academy of Finland (grant 1201964 to J.K.H.B.) and the Finnish Center of Excellence Program (2006-2011) of the Academy of Finland (grants 1213467 and 1213992 to D.H.B.).
REFERENCES
- 1.Abrescia, N. G., J. J. Cockburn, J. M. Grimes, G. C. Sutton, J. M. Diprose, S. J. Butcher, S. D. Fuller, C. San Martin, R. M. Burnett, D. I. Stuart, D. H. Bamford, and J. K. Bamford. 2004. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 432:68-74. [DOI] [PubMed] [Google Scholar]
- 2.Ackermann, H. W., R. Roy, M. Martin, M. R. Murthy, and W. A. Smirnoff. 1978. Partial characterization of a cubic Bacillus phage. Can. J. Microbiol. 24:986-993. [DOI] [PubMed] [Google Scholar]
- 3.Adams, M. H. 1959. Bacteriophages. Interscience Publishers, Inc., New York, N.Y.
- 4.Bamford, D. H. 2003. Do viruses form lineages across different domains of life? Res. Microbiol. 154:231-236. [DOI] [PubMed] [Google Scholar]
- 5.Bamford, D. H., and H.-W. Ackermann. 2000. Family Tectiviridae, p. 111-116. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Classification and nomenclature of viruses. Academic Press, San Diego, Calif.
- 6.Bamford, D. H., R. M. Burnett, and D. I. Stuart. 2002. Evolution of viral structure. Theor. Popul. Biol. 61:461-470. [DOI] [PubMed] [Google Scholar]
- 7.Bamford, D. H., and L. Mindich. 1984. Characterization of the DNA-protein complex at the termini of the bacteriophage PRD1 genome. J. Virol. 50:309-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bamford, D. H., M. Romantschuk, and P. J. Somerharju. 1987. Membrane fusion in prokaryotes: bacteriophage φ6 membrane fuses with the Pseudomonas syringae outer membrane. EMBO J. 6:1467-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16:673-685. [DOI] [PubMed] [Google Scholar]
- 10.Bernadsky, G., T. J. Beveridge, and A. J. Clarke. 1994. Analysis of the sodium dodecyl sulfate-stable peptidoglycan autolysins of select gram-negative pathogens by using renaturing polyacrylamide gel electrophoresis. J. Bacteriol. 176:5225-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beveridge, T. J., C. W. Forsberg, and R. J. Doyle. 1982. Major sites of metal binding in Bacillus licheniformis walls. J. Bacteriol. 150:1438-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 13.Campbell, J. L., C. C. Richardson, and F. W. Studier. 1978. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc. Natl. Acad. Sci. USA 75:2276-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cockburn, J. J., N. G. Abrescia, J. M. Grimes, G. C. Sutton, J. M. Diprose, J. M. Benevides, G. J. Thomas, Jr., J. K. Bamford, D. H. Bamford, and D. I. Stuart. 2004. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature 432:122-125. [DOI] [PubMed] [Google Scholar]
- 15.Daugelavicius, R., E. Bakienė, J. Berzinskienė, and D. H. Bamford. 2000. Use of lipophilic anions for estimation of biomass and cell viability. Biotechnol. Bioeng. 71:208-216. [DOI] [PubMed] [Google Scholar]
- 16.Daugelavicius, R., J. K. Bamford, and D. H. Bamford. 1997. Changes in host cell energetics in response to bacteriophage PRD1 DNA entry. J. Bacteriol. 179:5203-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daugelavicius, R., V. Cvirkaitė, A. Gaidelytė, E. Bakienė, R. Gabrėnaitė-Verkhovskaya, and D. H. Bamford. 2005. Penetration of enveloped double-stranded RNA bacteriophages φ3 and φ6 into Pseudomonas syringae cells. J. Virol. 79:5017-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer, W. B., and M. S. Sansom. 2002. Viral ion channels: structure and function. Biochim. Biophys. Acta 1561:27-45. [DOI] [PubMed] [Google Scholar]
- 19.Gaidelytė, A., S. T. Jaatinen, R. Daugelavicius, J. K. Bamford, and D. H. Bamford. 2005. The linear double-stranded DNA of phage Bam35 enters lysogenic host cells, but the late phage functions are suppressed. J. Bacteriol. 187:3521-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González-Huici, V., M. Salas, and J. M. Hermoso. 2004. The push-pull mechanism of bacteriophage Φ29 DNA injection. Mol. Microbiol. 52:529-540. [DOI] [PubMed] [Google Scholar]
- 21.Grahn, A. M., J. Caldentey, J. K. Bamford, and D. H. Bamford. 1999. Stable packaging of phage PRD1 DNA requires adsorption protein P2, which binds to the IncP plasmid-encoded conjugative transfer complex. J. Bacteriol. 181:6689-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grahn, A. M., R. Daugelavicius, and D. H. Bamford. 2002. Sequential model of phage PRD1 DNA delivery: active involvement of the viral membrane. Mol. Microbiol. 46:1199-1209. [DOI] [PubMed] [Google Scholar]
- 23.Hantke, K., and V. Braun. 1978. Functional interaction of the tonA/tonB receptor system in Escherichia coli. J. Bacteriol. 135:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoyle, B. D., and T. J. Beveridge. 1984. Metal binding by the peptidoglycan sacculus of Escherichia coli K-12. Can. J. Microbiol. 30:204-211. [DOI] [PubMed] [Google Scholar]
- 25.Hughes, R. C., and P. J. Tanner. 1968. The action of dilute alkali on some bacterial cell walls. Biochem. Biophys. Res. Commun. 33:22-28. [DOI] [PubMed] [Google Scholar]
- 26.Kivelä, H. M., R. Daugelavicius, R. H. Hankkio, J. K. Bamford, and D. H. Bamford. 2004. Penetration of membrane-containing double-stranded-DNA bacteriophage PM2 into Pseudoalteromonas hosts. J. Bacteriol. 186:5342-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laurinavicius, S., R. Käkelä, P. Somerharju, and D. H. Bamford. 2004. Phospholipid molecular species profiles of tectiviruses infecting Gram-negative and Gram-positive hosts. Virology 322:328-336. [DOI] [PubMed] [Google Scholar]
- 28.Laurinmäki, P. A., J. T. Huiskonen, D. H. Bamford, and S. J. Butcher. 2005. Membrane proteins modulate the bilayer curvature in the bacterial virus Bam35. Structure 13:1819-1828. [DOI] [PubMed] [Google Scholar]
- 29.Letellier, L., P. Boulanger, M. de Frutos, and P. Jacquot. 2003. Channeling phage DNA through membranes: from in vivo to in vitro. Res. Microbiol. 154:283-287. [DOI] [PubMed] [Google Scholar]
- 30.Lubkowski, J., F. Hennecke, A. Plückthun, and A. Wlodawer. 1999. Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA. Structure Fold Des. 7:711-722. [DOI] [PubMed] [Google Scholar]
- 31.Mehmel, M., M. Rothermel, T. Meckel, J. L. Van Etten, A. Moroni, and G. Thiel. 2003. Possible function for virus encoded K+ channel Kcv in the replication of chlorella virus PBCV-1. FEBS Lett. 552:7-11. [DOI] [PubMed] [Google Scholar]
- 32.Moak, M., and I. J. Molineux. 2004. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 51:1169-1183. [DOI] [PubMed] [Google Scholar]
- 33.Molineux, I. J. 2006. Fifty-three years since Hershey and Chase; much ado about pressure but which pressure is it? Virology 344:221-229. [DOI] [PubMed] [Google Scholar]
- 34.Morita, M., Y. Tanji, K. Mizoguchi, T. Akitsu, N. Kijima, and H. Unno. 2002. Characterization of a virulent bacteriophage specific for Escherichia coli O157:H7 and analysis of its cellular receptor and tail fiber genes. FEMS Microbiol. Lett. 211:77-83. [DOI] [PubMed] [Google Scholar]
- 35.Ojala, P. M., J. T. Juuti, and D. H. Bamford. 1993. Protein P4 of double-stranded RNA bacteriophage φ6 is accessible on the nucleocapsid surface: epitope mapping and orientation of the protein. J. Virol. 67:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olkkonen, V. M., and D. H. Bamford. 1989. Quantitation of the adsorption and penetration stages of bacteriophage φ6 infection. Virology 171:229-238. [DOI] [PubMed] [Google Scholar]
- 37.Pooley, H. M., D. Paschoud, and D. Karamata. 1987. The gtaB marker in Bacillus subtilis 168 is associated with a deficiency in UDPglucose pyrophosphorylase. J. Gen. Microbiol. 133:3481-3493. [DOI] [PubMed] [Google Scholar]
- 38.Poranen, M. M., R. Daugelavicius, and D. H. Bamford. 2002. Common principles in viral entry. Annu. Rev. Microbiol. 56:521-538. [DOI] [PubMed] [Google Scholar]
- 39.Poranen, M. M., R. Daugelavicius, P. M. Ojala, M. W. Hess, and D. H. Bamford. 1999. A novel virus-host cell membrane interaction. Membrane voltage-dependent endocytic-like entry of bacteriophage φ6 nucleocapsid. J. Cell Biol. 147:671-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Randall-Hazelbauer, L., and M. Schwartz. 1973. Isolation of the bacteriophage lambda receptor from Eschericia coli. J. Bacteriol. 116:1436-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravantti, J. J., A. Gaidelyte, D. H. Bamford, and J. K. Bamford. 2003. Comparative analysis of bacterial viruses Bam35, infecting a gram-positive host, and PRD1, infecting gram-negative hosts, demonstrates a viral lineage. Virology 313:401-414. [DOI] [PubMed] [Google Scholar]
- 42.Rydman, P. S., and D. H. Bamford. 2000. Bacteriophage PRD1 DNA entry uses a viral membrane-associated transglycosylase activity. Mol. Microbiol. 37:356-363. [DOI] [PubMed] [Google Scholar]
- 43.Rydman, P. S., and D. H. Bamford. 2002. The lytic enzyme of bacteriophage PRD1 is associated with the viral membrane. J. Bacteriol. 184:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rydman, P. S., J. Caldentey, S. J. Butcher, S. D. Fuller, T. Rutten, and D. H. Bamford. 1999. Bacteriophage PRD1 contains a labile receptor-binding structure at each vertex. J. Mol. Biol. 291:575-587. [DOI] [PubMed] [Google Scholar]
- 45.Salton, M. R. J. 1994. The bacterial cell envelope—a historical perspective, p. 1-22. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier Science B.V., Amsterdam, The Netherlands.
- 46.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.São-José, C., C. Baptista, and M. A. Santos. 2004. Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J. Bacteriol. 186:8337-8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strömsten, N. J., D. H. Bamford, and J. K. Bamford. 2005. In vitro DNA packaging of PRD1: a common mechanism for internal-membrane viruses. J. Mol. Biol. 348:617-629. [DOI] [PubMed] [Google Scholar]
- 49.Strömsten, N. J., S. D. Benson, R. M. Burnett, D. H. Bamford, and J. K. Bamford. 2003. The Bacillus thuringiensis linear double-stranded DNA phage Bam35, which is highly similar to the Bacillus cereus linear plasmid pBClin15, has a prophage state. J. Bacteriol. 185:6985-6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNApolymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 51.Takumi, K., A. Takeoka, T. Kinouchi, and T. Kawata. 1985. Solubilization and partial properties of receptor substance for bacteriophage α2 induced from Clostridium botulinum type A 190L. Microbiol. Immunol. 29:1185-1195. [DOI] [PubMed] [Google Scholar]
- 52.Valyasevi, R., W. E. Sandine, and B. L. Geller. 1990. The bacteriophage kh receptor of Lactococcus lactis subsp. cremoris KH is the rhamnose of the extracellular wall polysaccharide. Appl. Environ Microbiol. 56:1882-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valyasevi, R., W. E. Sandine, and B. L. Geller. 1991. A membrane protein is required for bacteriophage c2 infection of Lactococcus lactis subsp. lactis C2. J. Bacteriol. 173:6095-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verheust, C., N. Fornelos, and J. Mahillon. 2004. The Bacillus thuringiensis phage GIL01 encodes two enzymes with peptidoglycan hydrolase activity. FEMS Microbiol. Lett. 237:289-295. [DOI] [PubMed] [Google Scholar]
- 55.Verheust, C., N. Fornelos, and J. Mahillon. 2005. GIL16, a new gram-positive tectiviral phage related to the Bacillus thuringiensis GIL01 and the Bacillus cereus pBClin15 elements. J. Bacteriol. 187:1966-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verheust, C., G. Jensen, and J. Mahillon. 2003. pGIL01, a linear tectiviral plasmid prophage originating from Bacillus thuringiensis serovar israelensis. Microbiology 149:2083-2092. [DOI] [PubMed] [Google Scholar]
- 57.Wendlinger, G., M. J. Loessner, and S. Scherer. 1996. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology 142:985-992. [DOI] [PubMed] [Google Scholar]