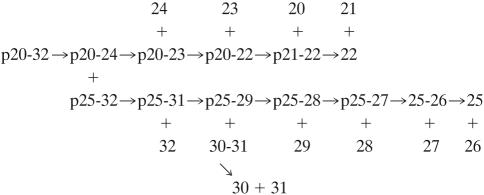Gram-negative bacteria possess a two-membrane envelope with an outer lipopolysaccharide-containing membrane that provides an effective barrier, protecting these organisms from detergents, organic solvents, drugs, and other toxic substances (24). However, the occurrence of an outer membrane poses major problems for the secretion of macromolecules (28). Consequently, gram-negative bacteria have evolved a tremendous diversity of outer membrane systems designed for the export of proteins, complex carbohydrates, nucleic acids, and lipids (4, 37).
Among the well-characterized outer membrane protein secretion systems are (i) the so-called two-partner secretion systems (transport classification [TC] 1.B.20) and (ii) the autotransporter systems (AT or AT-1; TC 1.B.12) (20, 30, 51). Following export from the cytoplasm to the periplasm via the general secretory (Sec) system, both AT and two-partner secretion system translocation domains insert into the outer membrane as β-barrel structures. They mediate export of virulence proteins or protein domains from the periplasm across the outer membrane to the extracellular medium where the exported protein or domain may either remain attached to the outer membrane or can be released in a free state (51). The exported proteins may serve as adhesins, hemolysins, proteases, cytotoxins, or mediators of intracytoplasmic actin-promoted bacterial motility (51).
Proteins of the autotransporter family possess C-terminal domains of 250 to 300 amino acyl residues that fold and insert into the outer membrane to give a β-barrel with 12 to 14 transmembrane β-strands (15, 16, 27, 29). This structure forms a pore through which the N-terminal virulence factor is presumed to be exported (13, 32). There is still some controversy as to the mechanism of protein transport (5, 6, 32, 44, 49). For example, the possible involvement of energy in the translocation process has not yet been extensively studied, and the relationship of these outer membrane translocators to mechanisms of antibiotic efflux and TonB-dependent influx, if any, has not been pursued.
A second family of autotransporters called “trimeric autotransporters,” “oligomeric coiled-coil adhesins,” or “autotransporters-2” (AT-2; TC 1.B.40) has recently been discovered (9, 17, 19, 43, 52). Among the best-characterized members of this family are the multifaceted Yersinia adhesin, YadA (2, 9, 19, 31, 36), the major adhesin of Haemophilus influenzae that allows colonization of the nasopharynx, Hia (25), and the Haemophilus “adhesin and penetration” protein, Hap (10, 11, 26, 48). These proteins define a novel family of autotransporter virulence factors. They may be able to allow translocation of their passenger domains across the outer membrane without the assistance of accessory proteins, but this postulate is still in contention.
A conserved C-terminal domain of about 70 amino acyl residues is believed to form the trimeric β-barrel that presumably allows the transport of the N-terminal “passenger” domain to the bacterial cell surface. These proteins form trimeric lollypop-like structures anchored to the outer membrane by their C-terminal autotransporter anchor domains (5, 6, 44). A superficially similar structure has been established for the outer membrane TolC protein of Escherichia coli, which has an analogous β-barrel structure. In the case of TolC, however, α-helical regions extend into the periplasm, a feature lacking in AT-2 domains (18, 22, 23). According to some investigators, the C-terminal 67- to 76-residue domains are both necessary and sufficient for translocation of the N-terminal adhesin domains (44). Each subunit AT-2 domain is believed to consist of just four transmembrane antiparallel β-strands (reviewed in reference 5). Deletion of this C-terminal domain abolishes outer membrane insertion of YadA (45), while the deletion of the linker region results in degradation of the whole protein (36). These experimental results suggest but do not establish that these C-terminal linker or outer membrane insertion regions are directly responsible for export of the passenger domain.
The few characterized protein members of the AT-2 family serve as virulence factors in animal pathogens (36). They have been termed invasins, immunoglobulin-binding proteins, serum resistance proteins, and hemagglutinins, but all appear to have adhesive properties. Because each of the few functionally characterized “passenger” domains of this class of autotransporters can function in adhesion, it is possible but not demonstrated that they are all structurally related. The characteristic feature that we will use for identification of family members, however, is the presence of the small C-terminal domain that is believed to form the outer membrane trimeric β-barrel pore.
In this minireview we present a bioinformatic analysis of the AT-2 family. We identify recognizable sequenced members of the AT-2 family and align the sequences of their autotransporter domains. The resultant multiple alignment is used to identify conserved motifs, generate a phylogenetic tree for the family, identify cluster-specific sequence characteristics, and generate average hydropathy, amphipathicity, and similarity plots that allow structural predictions. Essentially all of the AT-2 proteins analyzed here derive from α-, β-, and γ-proteobacteria and their phage, although other more distantly related members of the family are found in other gram-negative bacterial kingdoms (7). Our analyses reveal that phylogeny of the AT-2 domains does not correlate with the size of the N-terminal passenger domain. However, the passenger domains consist of homologous repeat units that are common to all members of the family. Phylogeny of the passenger domains generally follows that of the AT-2 domains. To a considerable degree, protein phylogeny follows the phylogeny of the source organisms. Our results suggest that the genes encoding these proteins have been subject to lateral transfer but that transfer occurred primarily within closely related organisms. This conclusion is substantiated by their occurrence in phage genomes (see below). We suggest that all members of the AT-2 family serve a single unifying function in cell adhesion/macromolecular recognition. This review provides the first detailed bioinformatic analysis of the AT-2 family.
ESTABLISHED PROTEIN MEMBERS OF THE AT-2 FAMILY
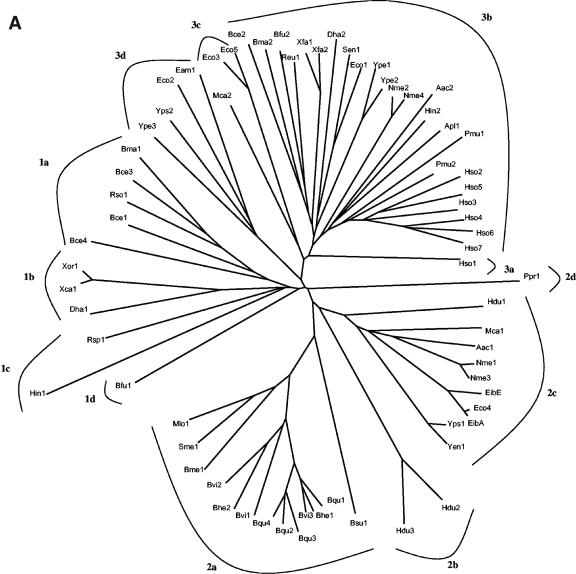
Using the PSI-BLAST search tool (1) with YadA of Yersinia enterocolitica as the query sequence and three iterations, about 140 above-threshold hits were retrieved from the NCBI database. AT-2 family members were identified on the basis of their C-terminal AT-2 domains. No homologues were identified that appeared to have the AT-2 domain anywhere other than at their extreme C termini. Redundancies, very closely related homologues, and hits that showed an insufficient degree of sequence similarity with established members of the family to establish homology (≤9 standard deviations using the GAP program [8]) were eliminated. This left 69 proteins upon which the analyses reported below were based. These proteins are presented in Table 1 while their aligned AT-2 domain sequences are shown in Fig. 1, and the phylogenetic tree based on this alignment is presented in Fig. 2A. The phylogenies of the passenger domains are presented in Fig. 2B (see below). The proteins listed in Table 1 are presented according to cluster as shown in the tree presented in Fig. 2A.
TABLE 1.
Recognized proteins of the AT-2 family
| Cluster and proteina | Organism | Database description | Size (no. of residues)b | Bacterial typec | gi |
|---|---|---|---|---|---|
| Cluster 1a | |||||
| Bce1 | Burkholderia cepacia R18194 | Autotransporter adhesin | 977 | β | 46316503 |
| Bce3 | Burkholderia cepacia R18194 | Autotransporter adhesin | 1,010 | β | 46315938 |
| Bce4 | Burkholderia cepacia R18194 | Autotransporter adhesin | 276 | β | 46322712 |
| Bma1 | Burkholderia mallei ATCC 23344 | Autotransporter adhesin | 373 | β | 53717377 |
| Rso1 | Ralstonia solanacearum | Putative hemagglutinin-related protein | 1,309 | β | 17549839 |
| Cluster 1b | |||||
| Dha1 | Desulfitobacterium hafniense | Autotransporter adhesin | 142 | Clostridia | 23115364 |
| Xca1 | Xanthomonas campestris pv. pelargonii | Unknown | 1,328 | γ | 7542317 |
| Xor1 | Xanthomonas oryzae pv. oryzae | Outer membrane protein XadA | 1,265 | γ | 9864182 |
| Cluster 1c | |||||
| Hin1 | Haemophilus influenzae R2846 | Autotransporter adhesin | 158 | γ | 42630309 |
| Rsp1 | Rhodobacter sphaeroides 2.4.1 | Large exoproteins involved in heme utilization or adhesion | 411 | α | 46192873 |
| Cluster 1d, Bfu1 | Burkholderia fungorum LB400 | Autotransporter adhesin | 3,068 | β | 48784624 |
| Cluster 2a | |||||
| Bhe1 | Bartonella henselae strain Houston-1 | Surface protein/Bartonella adhesin | 1,747 | α | 49237768 |
| Bhe2 | Bartonella henselae strain Houston-1 | Surface protein | 153 | α | 49237769 |
| Bme1 | Brucella melitensis 16M | Cell surface protein | 365 | α | 17988155 |
| Bqu1 | Bartonella quintana strain Toulouse | Surface protein/Bartonella adhesin | 1,065 | α | 49239313 |
| Bqu2 | Bartonella quintana strain Toulouse | Surface protein/Bartonella adhesin | 949 | α | 49239314 |
| Bqu3 | Bartonella quintana | VompA | 950 | α | 51949816 |
| Bqu4 | Bartonella quintana | VompC | 970 | α | 51949818 |
| Bsu1 | Brucella suis 1330 | Hypothetical protein BR1846 | 278 | α | 23502699 |
| Bvi1 | Bartonella vinsonii subsp. arupensis | BrpB | 1,760 | α | 52355211 |
| Bvi2 | Bartonella vinsonii subsp. arupensis | BrpA | 3,620 | α | 52355212 |
| Bvi3 | Bartonella vinsonii subsp. arupensis | BrpC | 1,420 | α | 52355210 |
| Mlo1 | Mesorhizobium loti MAFF303099 | Hypothetical protein mil2848 | 1,953 | α | 13472521 |
| Sme1 | Sinorhizobium meliloti 1021 | Hypothetical protein SMc01708 | 1,291 | α | 15964211 |
| Cluster 2b | |||||
| Hdu2 | Haemophilus ducreyi 35000HP | Hypothetical protein HD1920 | 296 | γ | 33152901 |
| Hdu3 | Haemophilus ducreyi | Necessary for collagen adhesion protein | 236 | γ | 45758814 |
| Cluster 2c | |||||
| Aac1 | Actinobacillus actinomycetemcomitans | Putative adhesin/invasin | 295 | γ | 19568164 |
| Eco4 | Escherichia coli | Immunoglobulin-binding protein EibF | 459 | γ | 16923467 |
| EibA | Prophage P-EibA | Immunoglobulin-binding protein EibA | 392 | dsDNA virus | 7532792 |
| EibE | Bacteriophage P-EibE | Immunoglobulin-binding protein EibE | 487 | dsDNA virus | 7523541 |
| Hdu1 | Haemophilus ducreyi 35000HP | Serum resistance protein DrsA | 257 | γ | 33151932 |
| Mca1 | Moraxella catarrhalis | Ubiquitous surface protein A2 | 686 | γ | 18568377 |
| Nme1 | Neisseria meningitidis | Putative adhesin/invasin | 405 | β | 21427129 |
| Nme3 | Neisseria meningitidis | Putative adhesin/invasin | 355 | β | 21427156 |
| Yen1 | Yersinia enterocolitica | Adhesin YadA | 454 | γ | 1955604 |
| Yps1 | Yersinia pseudotuberculosis | Adhesin YadA precursor | 434 | γ | 141104 |
| Cluster 2d, Ppr1 | Photobacterium profundum | Hypothetical protein | 288 | γ | 46917051 |
| Cluster 3a, Hso1 | Haemophilus somnus 129PT | Autotransporter adhesin | 452 | γ | 23468079 |
| Cluster 3b | |||||
| Aac2 | Actinobacillus actinomycetemcomitans | EmaA | 1,965 | γ | 33578091 |
| Apl1 | Actinobacillus pleuropneumoniae serovar 1 strain 4074 | Autotransporter adhesin | 2,600 | γ | 46143665 |
| Bce2 | Burkholderia cepacia R18194 | Autotransporter adhesin | 1,439 | β | 46313782 |
| Bfu2 | Burkholderia fungorum LB400 | Autotransporter adhesin | 770 | β | 48787852 |
| Bma2 | Burkholderia mallei ATCC 23344 | Hemagglutinin family protein | 831 | β | 53717118 |
| Dha2 | Desulfitobacterium hafniense DCB-2 | Autotransporter adhesin | 86 | Clostridia | 53684140 |
| Eco1 | Escherichia coli O157:H7 EDL933 | Putative adhesin | 1,588 | γ | 15804146 |
| Hin2 | Haemophilus influenzae | Adhesin | 1,096 | γ | 25359414 |
| Hso2 | Haemophilus somnus 2236 | Autotransporter adhesin | 2,419 | γ | 46156748 |
| Hso3 | Haemophilus somnus 2236 | Autotransporter adhesin | 2,390 | γ | 46156040 |
| Hso4 | Haemophilus somnus 129PT | Autotransporter adhesin | 611 | γ | 23467645 |
| Hso5 | Haemophilus somnus 2236 | Autotransporter adhesin | 3,391 | γ | 32030792 |
| Hso6 | Haemophilus somnus 2236 | Autotransporter adhesin | 1,550 | γ | 46156755 |
| Hso7 | Haemophilus somnus 2236 | Autotransporter adhesin | 3,674 | γ | 46156455 |
| Nme2 | Neisseria meningitidis | Adhesin | 591 | β | 15676883 |
| Nme4 | Neisseria meningitidis | NhhA outer membrane protein | 589 | β | 14578023 |
| Pmu1 | Pasteurella multocida subsp. multocida strain Pm70 | Hsf | 2,712 | γ | 15602579 |
| Pmu2 | Pasteurella multocida subsp. multocida strain Pm70 | Hsf | 1,299 | γ | 15603435 |
| Reu1 | Ralstonia eutropha JMP134 | Autotransporter adhesin | 465 | β | 53761962 |
| Sen1 | Salmonella enterica subsp. enterica serovar Typhi strain CT18 | Putative autotransporter | 1,107 | γ | 16762618 |
| Xfa1 | Xylella fastidiosa 9a5c | Surface protein | 2,059 | γ | 15838130 |
| Xfa2 | Xylella fastidiosa 9a5c | Surface protein | 1,190 | γ | 15838575 |
| Ype1 | Yersinia pestis CO92 | Putative surface protein (partial) | 658 | γ | 16121208 |
| Ype2 | Yersinia pestis KIM | Hypothetical protein y1847 | 144 | γ | 22125740 |
| Cluster 3c | |||||
| Eco3 | Escherichia coli | IHP1-like | 436 | γ | 29367636 |
| Eco5 | Escherichia coli O157:H7 EDL933 | Hypothetical protein Z0639 | 338 | γ | 15800223 |
| Mca2 | Moraxella catarrhalis | Hemagglutinin | 2,314 | γ | 22000942 |
| Cluster 3d | |||||
| Eam1 | Erwinia amylovora | Autoagglutinating adhesin | 494 | γ | 38638179 |
| Eco2 | Escherichia coli | STEC autoagglutinating adhesin | 516 | γ | 16565696 |
| Ype3 | Yersinia pestis biovar Medievalis strain 91001 | Hypothetical protein HP1206 | 364 | γ | 45441033 |
| Yps2 | Yersinia pseudotuberculosis IP 32953 | Hypothetical protein pYptb0018 | 416 | γ | 51593960 |
The cluster refers to the clustering pattern in the phylogenetic tree shown in Fig. 2A.
Size of protein is given in terms of amino acyl residues.
Greek letters refer to the subcategory of the proteobacteria. dsDNA, double-stranded DNA.
FIG. 1.
Multiple alignment of the sequences of 69 putative AT-2 domains. The alignment was generated using the CLUSTAL X program (46). The positions of conserved motifs 1 and 2 are indicated above the alignment. The horizontal lines at the top left-hand side of the alignment indicate the position of the 7-residue repeat sequences, present in several homologues as illustrated in Tables 3 and 5. Residue alignment position for the AT-2 domains is indicated below the alignment. The boundaries selected between the AT-2 domains and the passenger domains were based on a multiple alignment of the intact proteins which revealed the regions of universal conservation together with previously published results (see introduction).
FIG.2.
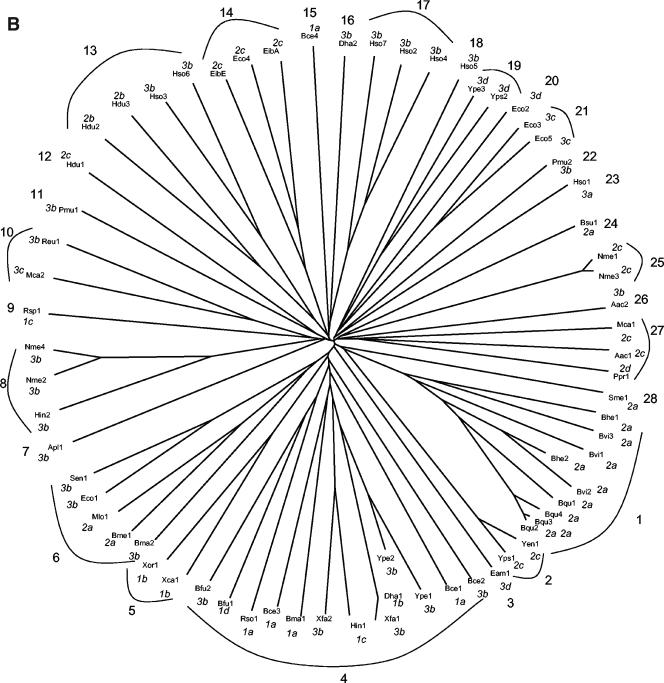
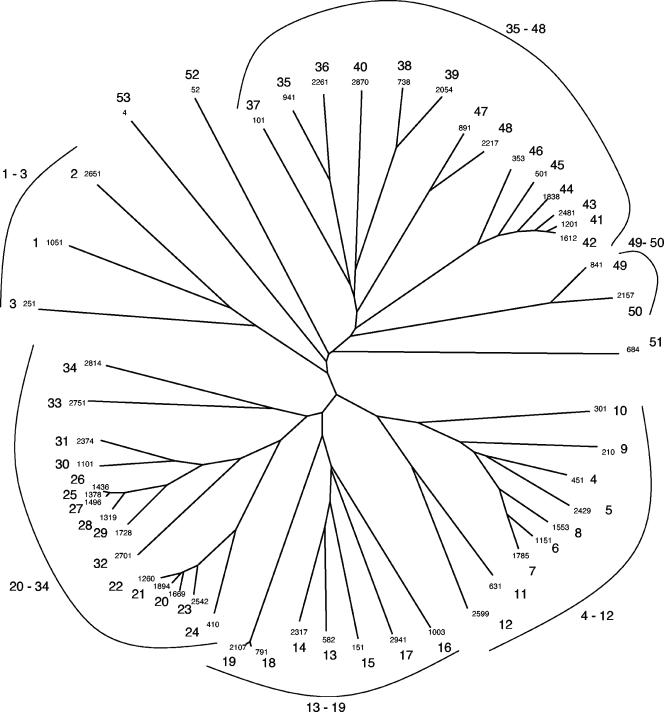
Phylogenetic trees of the C-terminal autotransporter (AT-2) domains (A) and the N-terminal passenger domains (B) of the same proteins. The clusters (1a to d, 2a to d, and 3a to d in A and 1 to 28 in B), analyzed for sequence conservation (see text), are indicated in the figure. The trees are based on the CLUSTAL X-derived multiple alignments shown in Fig. 1. The trees were drawn with the TreeView program (55).
As indicated in Table 1, the homologues exhibit tremendous variation in overall protein size (86 to 3,674 amino acyl residues). Even within a single cluster, the size variation is tremendous (Tables 1 and 2). This degree of size variation was not observed in previous studies of the AT family (51). However, this size variation is explained by the occurrence of repeat units of numbers that do not correlate with protein phylogeny (see below).
TABLE 2.
Organismal types and average sizes of the 12 phylogenetic clusters of the AT-2 family
| Cluster | Proteobacterial subcategory represented | Avg size of proteins ± SDa |
|---|---|---|
| 1a | β | 789 ± 445 |
| 1b | γ (clostridia) | 911 ± 667 |
| 1c | α, γ | 285 ± 179 |
| 1d | β | 3,068 |
| 2a | α | 1,271 ± 907 |
| 2b | γ | 266 ± 42 |
| 2c | β, γ (E. coli phage) | 422 ± 118 |
| 2d | γ | 288 |
| 3a | γ | 452 |
| 3b | β, γ (clostridia) | 1,468 ± 992 |
| 3c | γ | 1,029 ± 1,113 |
| 3d | γ | 448 ± 70 |
Average size of the proteins in a cluster is given in terms of numbers of amino acyl residues.
With the exception of four homologues, all homologues were from proteobacteria. Two close homologues are from a bacteriophage (p-EibE) and a prophage (p-EibA), both of E. coli. These two proteins are annotated as immunoglobulin binding proteins. The two small nonproteobacterial homologues (Dha2, 86 amino acyl residues, and Dha1, 142 amino acyl residues) are reported to be from Desulfitobacterium hafinense, which is a low GC-content gram-positive bacterium with no outer membrane. These proteins could not serve as autotransporters in this organism. Because the genome of D. hafinense has not been completely sequenced and is still being updated, it is possible that these sequences resulted from DNA contamination.
SEVEN-RESIDUE REPEAT SEQUENCES IN THE LINKER REGIONS OF AT-2 PROTEINS AND OTHER PROTEINS
Several of the AT-2 proteins listed in Table 1 exhibit a demonstrable 7-amino-acyl repeat element between the passenger domains and the putative transmembrane regions of the AT-2 domains (i.e., in the linker regions). For many of these homologues, two, three, or more repeat elements could be identified at the N-terminal end of the AT-2 domain, often extending into the part of the protein referred to as the passenger domain (Fig. 1). In AT-2-like proteins retrieved in BLAST searches, this 7-amino-acyl repeat occurred as many as 18 times. Twelve repeats are sufficient to create a domain the length of the linker plus the AT-2 domain. An example of this is the Apl2 protein of Actinobacillus pleuropneumonia with a size of 195 amino acyl residues. The repeat elements, encompassing all but the last 12 residues of this protein, are presented in Table 3, where 12 tandem repeat elements are shown. The consensus for this repeat element is (D/E)(Q/N)(R/K)(F/I)(Q/D)(Q/K)(V/L), where the two most prevalent residues at each position are indicated in parentheses. The presence of this repeat sequence can be easily seen, for example, for Yps1 and Yen1, both of which show extensive similarity to the consensus sequence (Fig. 1). It is possible that the AT-2 domains have evolved from a primordial gene like that encoding the Apl protein, derived from an internally repeated 21-bp genetic element. These repeat sequences of several AT-2 proteins occur in the linker regions connecting the passenger domains to the AT-2 domains. Thus, AT-2 domains may have either evolved from a sequence like that shown in Apl2, as illustrated in Table 3, or they could have evolved independently of this repeat sequence and become associated with it as a result of gene fusion events.
TABLE 3.
The 7-residue repeat element comprising the C-terminal region of the Apl2 protein from A. pleuropneumoniae (gi 32035081)a
| Repeat no. | Residue position | Repeat sequence | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 100 | E | G | K | F | F | N | I |
| 2 | 107 | D | K | K | F | E | Q | V |
| 3 | 114 | D | L | R | F | Q | Q | I |
| 4 | 121 | D | Q | R | F | Q | Q | V |
| 5 | 128 | D | Q | R | F | Q | Q | V |
| 6 | 135 | D | Q | R | F | Q | Q | V |
| 7 | 142 | E | D | K | I | H | K | L |
| 8 | 149 | D | I | R | I | G | K | V |
| 9 | 156 | E | S | R | L | D | V | V |
| 10 | 163 | E | E | K | I | D | V | L |
| 11 | 170 | N | N | K | F | D | K | L |
| 12 | 177 | D | N | K | F | D | K | M |
| Consensus sequenceb | D | Q | R | F | Q | Q | V | |
| E | N | K | I | D | K | L | ||
This region, exhibiting 12 repeat units in Apl2, is about the same length as a typical AT-2 domain. Corresponding repeat units can be detected in the N-terminal regions of many AT-2 domains as revealed in Fig. 1 (horizontal lines at top left of the figure; see legend). The numbers of the 12 C-terminal 7-residue repeats in Apl2 are presented in column 1, while the residue position at the beginning of each repeat is indicated in column 2.
The two dominant residues at each position are presented in the consensus sequences.
PHYLOGENETIC CLUSTERING OF AT-2 DOMAINS ACCORDING TO ORGANISMAL TYPE
All of the proteins in Table 1 exhibit sequence similarity in their AT-2 domains. The phylogenetic tree for these domains, shown in Fig. 2A, reveals clustering according to organismal type (Table 1). Thus, cluster 1a contains only β-proteobacterial proteins; cluster 2a contains only α-proteobacterial proteins; and clusters 2b, 2d, and 3a contain only γ-proteobacterial proteins. Moreover, clusters 1b, 2c, and 3b contain only β- and γ-proteobacterial proteins with the exception of the two E. coli phage proteins and the two putative desulfitobacterial proteins, Dha1 and Dha2. Finally, cluster 1c contains only α- and γ-proteobacterial proteins. Thus, to some extent, clustering reflects the organismal type from which these proteins derive. This observation suggests that horizontal transfer of genetic material encoding AT-2 proteins has been restricted largely to organisms within any one of the proteobacterial subdivisions (see Conclusions and Perspectives).
AT-2 DOMAIN STRUCTURAL PREDICTIONS
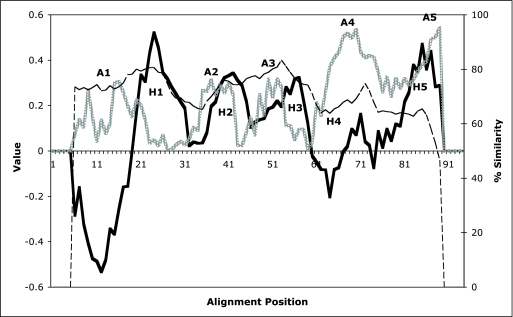
The average hydropathy, amphipathicity, and similarity plots, based on the Fig. 1 multiple alignment and obtained using the AveHas program (53), are shown in Fig. 3. There are five peaks of hydrophobicity (H1 to H5), and with the angle set at 180°, as is appropriate for a β-strand, there are five peaks of amphipathicity (A1 to A5). The average similarity plot (Fig. 3, dashed line) follows the average amphipathicity plot (dotted line) more closely than it follows the average hydrophobicity plot (solid line).
FIG. 3.
Average hydropathy, amphipathicity, and similarity plots for the AT-2 domains of the 69 AT-2 proteins included in this study. The plots were generated with the AveHas program (53). H1 to H5, five peaks of hydrophobicity; A1 to A5, five peaks of amphipathicity when the angle is set at 180° as is appropriate for a β-strand. Hydropathy, heavy solid line; amphipathicity, light dotted line; similarity, thin dashed line.
The first hydrophobic peak (H1) does not show amphipathic character, and the first amphipathic peak (A1) is not appreciably hydrophobic. These regions may not form transmembrane β-strands. However, H2 overlaps and follows A2, H3 overlaps and follows A3, H4 overlaps and slightly follows A4, and H5 overlaps and precedes H5. Established transmembrane β-strands in outer membrane porins often show overlapping but noncoincident peaks of hydrophobicity and amphipathicity (54). There are four overlapping peaks of amphipathicity and hydrophobicity that therefore serve as excellent candidates for transmembrane, pore-forming β-strands. Each of these overlapping regions is about 7 to 10 amino acyl residues long, as expected for a transmembrane β-strand. We therefore predict that these four strands form a small transmembrane β-sheet. This β-sheet presumably forms the homotrimeric pore through which the passenger domain passes (see introduction).
CONSERVED MOTIFS
As shown in Fig. 3, the most conserved regions of the alignment coincide with hydrophobic peak H1 and amphipathic peak A3. These include the two most conserved motifs among AT-2 domains. These two consensus motifs were AGIASALALA (motif 1; alignment positions 18 to 27) and SAVAIGV (motif 2; alignment positions 51 to 57). Although the majority of the proteins exhibit these conserved residues, no residue position is fully conserved, and the variation at any one position is usually considerable. The best-conserved residue is G56 which is conserved in all but one of the proteins (Hin1), where a V can be found (Fig. 1 and Table 4). Examination of the data in Table 4 reveals that at almost all conserved positions in motif 1, exceptional nonconserved residues can be hydrophilic, hydrophobic, or semipolar. Only at alignment position 21 is the residue always semipolar. This fact suggests that there is not an absolute requirement for residue type at most of the positions in putative hydrophobic peak 1 (Fig. 3).
TABLE 4.
Residue composition of the two most conserved motifs in proteins of the AT-2 family
| Motif no.a | Residue position | Amino acid residues and their frequency |
|---|---|---|
| 1 | 18 | A29 Q13 G9 R4 S4 K3 N2 M2 T1 V1 |
| 19 | G46 A15 M3 N2 I1 V1 F1 | |
| 20 | I25 A15 V9 T7 L6 S3 M2 G1 K1 | |
| 21b | A64 S4 P1 | |
| 22 | S17 G14 A12 I11 N5 Q5 E2 M2 V1 | |
| 23 | A37 G12 S7 Q5 T4 M3 E1 | |
| 24 | L17 M15 A14 N7 I5 T4 S3 V2 G1 H1 | |
| 25b | A63 N2 S1 Q1 G1 V1 | |
| 26 | L19 M16 V14 A11 T6 I2 Q1 | |
| 27 | A31 S25 T8 N3 G1 L1 | |
| 2 | 51 | S32 Q11 T7 G7 N4 A2 K2 Y1 H1 R1 E1 |
| 52 | A58 G7 S4 | |
| 53 | V21 L21 F12 I9 Y2 M2 T1 K1 | |
| 54b | A61 S6 G1 P1 | |
| 55 | I19 V19 L16 F9 A5 S1 | |
| 56b | G68 V1 | |
| 57 | V19 Y16 A14 T5 G5 S4 I3 L1 M1 |
The positions of these two motifs in the multiple alignment can be found in Fig. 1.
The two most highly conserved residues in each motif.
In contrast to conserved motif 1, conserved motif 2 has a characteristic residue type at each position. Thus, at alignment position 51, all residues are semipolar or hydrophilic. At position 52, all residues are semipolar. At position 53, all residues but one are hydrophobic. At position 54, all residues are semipolar, and at positions 55 to 57, no residue is strongly hydrophilic. Motif 2, therefore, has the highest degree of conservation in terms of the residue types found at the various aligned positions. This suggests that motif 1 in hydrophobic region H1 may have evolved to serve dissimilar functions within the differing AT-2 domains, while motif 2, in putative transmembrane β-strand 2, serves a single function, common to all family members.
PHYLOGENY OF THE PASSENGER DOMAINS OF AT-2 PROTEINS
The phylogenetic tree of the passenger domains (Fig. 2B) was significantly different from that of the AT-2 domains (Fig. 2A). Cluster 1a, 1b, and 1d proteins in Fig. 2A can be found in clusters 4 and 5 in Fig. 2B, while cluster 1c proteins are found in clusters 4 and 9 in Fig. 2B (see Table S1 in the supplemental material [http://biology.ucsd.edu/∼msaier/supmat/AT2]). Thus, cluster 1 proteins in Fig. 2A are found almost exclusively in clusters 4 and 5 in Fig. 2B. Cluster 2 proteins in Fig. 2A are distributed between 10 clusters in Fig. 2B with no member in clusters 4, 5, and 9. Further, cluster 3 proteins in Fig. 2A are distributed between 16 clusters in Fig. 2B, but only 1 of these 16 clusters overlaps with the cluster 1 proteins of Fig. 2A, and only 2 of the 16 clusters shown in Fig. 2B overlap with cluster 2 proteins of Fig. 2A. It is therefore clear that while the phylogenetic trees of the passenger domains reflect a greater degree of sequence divergence than that of the AT-2 domains, there is rough segregation of the passenger domains according to the phylogenetic groupings of the AT-2 domains. Further, whenever two proteins are phylogenetically closely related, the phylogenetic positions of the passenger domains correlate well with those of the AT-2 domains. Because of (i) the greater variation in size, (ii) the presence of multiple repeat units, and (iii) the greater sequence divergence of the passenger domains relative to the AT-2 domains, the tree shown in Fig. 2A is expected to show greater accuracy than the tree in Fig. 2B. We therefore suggest that while shuffling of the passenger domains relative to the AT-2 domains may have occurred throughout evolution of these proteins, such shuffling was a relatively rare event.
LARGE INTERNAL REPEAT SEQUENCES IN THE PASSENGER DOMAINS OF AT-2 PROTEINS
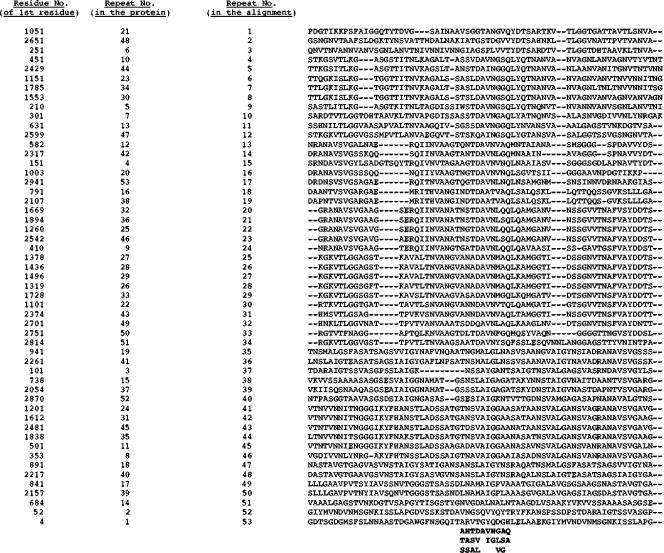
Examination of the passenger domains revealed that these consist primarily of large repeat units of about 70 residues (60 to 80 residues for individual large repeat units). The larger proteins contain greater numbers of repeat units than the smaller proteins, and for each protein examined in detail, most of the passenger domains consist of these types of repeat units. For example, 53 repeat units were identified in the 3,068-residue protein Bfu1 of Burkholderia fungorum. These were multiply aligned as shown in Fig. 4. The alignment revealed that the best-conserved region is in the centers of these repeat units where the residue consensus motif for a 10-residue sequence is (A/T/S)(N/A/S)(T/S/A)(D/V/L)A(V/I)(N/G)(G/L/V)(A/S/G)(Q/A) (Fig. 4, bolded residues under the alignment).
FIG. 4.
Multiple alignment of 53 repeat units in the passenger domain of Bfu1 of B. fungorum LB400 (gi 48784624) of 3,068 amino acyl residues. The average size of the repeat units is 63 ± 4 residues. The position of the repeat is indicated by the residue number of the first residue in the repeat unit (column 1). These repeats are numbered according to position in the protein (column 2) and according to position in the multiple alignment (column 3).
Phylogenetic clustering of these repeat units is shown in Fig. 5. It can be seen that these repeat units show striking clustering patterns where some of the repeats are extremely similar in sequence while others show relatively little sequence similarity compared with the other repeats. For example, repeats 25 and 26 in the alignment are identical to each other, while repeat 27 differs from these two at only one position (a T for an S substitution at their C termini). Further, repeat 28 differs from these at only three positions near the N termini of these repeat units. These four repeat units have the order in the Bfu1 protein of repeat units 27, 28, 29, and 26 (Fig. 4 and 5). Thus, these four identical or extremely similar repeat units occur in the protein in tandem. These elements undoubtedly arose by very recent tandem duplication events.
FIG. 5.
Phylogenetic tree of the 60-residue repeat units in the Bfu1 protein of B. fungorum. The numbers of the repeats indicate the positions in the multiple alignment shown in Fig. 4. The residue numbers in the protein of the first residue in each repeat unit (Fig. 4) is provided in small print.
Another example of similar, tandem repeat units can be seen for repeats 49, 50, and 51 in the protein. Repeat units 33 and 34 in the alignment correspond to repeat units 50 and 51 in the Bfu1 protein, while repeat unit 32 in the alignment is repeat unit 49 in the protein. They are thus adjacent to each other in the protein. Repeats 33 and 34 in the alignment (Fig. 4) differ from each other at 36 positions although they cluster loosely together on the tree (Fig. 5). The adjacent branch 32 is further from 33 and 34 but is nevertheless within the same major cluster. These repeats probably arose by late duplication events. Further, repeat units 25 to 32 probably arose as a result of more recent duplication events. If so, repeats 30 and 31 may also have arisen from the immediate precursor of 32, even though they are distant from 32 in terms of their positions in the protein (Fig. 4 and 5).
These two examples represent the only cases where the closest homologues in the protein are adjacent to each other on the tree. In all other cases, phylogenetically close homologues are distant from each other in the protein. For example, repeats 20 to 24 in the alignment shown in Fig. 4 are phylogenetically close (Fig. 5), but they represent repeats 32, 36, 25, 46, and 9, respectively, in the protein. Assuming that these sequence-similar repeats arose recently, we must conclude that they arose either by tandem duplications followed by shuffling or by a copy process, possibly involving polymerase hopping from one repeat unit in the DNA to another nontandem repeat. Such an event could have resulted from DNA looping during replication or from an event involving RNA polymerase and reverse transcriptase. Although the analysis shown in Fig. 5 suggests a mechanism of the latter type, we know of no experimental evidence supporting such a postulate. The proposed pathway for generation of all repeats in Bfu1 (assuming uniform rates of sequence divergence) is shown in Fig. S1 in the supplemental material (http://biology.ucsd.edu/∼msaier/supmat/AT2). Repeats 20 to 34 occur on one primary branch of the phylogenetic tree (Fig. 5). The original precursor repeat unit (p) first duplicated and then diverged to give the precursors of repeats 33 and 34 (p33-34) and of repeats 20 to 32 (p20-32). The former primordial unit then duplicated a second time to give repeats 33 and 34. The precursor of repeats 20 to 32 (p20-32) underwent up to eight successive duplication events as follows:
REPEAT UNITS IDENTIFIED IN THE Yen1 PROTEIN
To exemplify the occurrence of repeat units of differing lengths in the AT-2 linker and passenger domains, we analyzed the 454-residue Yen1 protein in detail. The C-terminal 75 residues in Yen1 comprise the AT-2 domain. The linker region of 21 residues consists of three 7-residue repeat units (R71 to R73) (Table 5). The first 7-residue repeat unit (R71, beginning at position 365) is less similar in sequence to the other two repeat units (R72 and R73 at positions 372 and 379, respectively) than these latter two sequences are to each other (Table 5).
TABLE 5.
Repeat units of 7, 14, and 60 residues identified in the linker and passenger domains of Y. enterocolitica protein Yen1
| Repeat designation | Initial residue position | Sequence with similaritya |
|---|---|---|
| 7-residue repeats | ||
| R71 | 365 | DHKFRQL |
| R72 | 372 | DNRLDKL |
| R73 | 379 | DTRVDKG |
| 14-residue repeats | ||
| R141 | 258 | KSAETLENARKEAF |
| R142 | 273 | QSKDVLNMAKAHSN |
| R143 | 288 | VARTTLETAEEHAN |
| R144 | 303 | VARTTLETAEEHAN |
| R145 | 319 | KSAEALASANVYAD |
| R146 | 333 | KSSHTLKTANSYTD |
| 60-residue repeats | ||
| R601 | 3 | KDFKISVSAALISALFSSPYAFADDYNGIPNLTAVQISPNADPALGQEYPVRPPVPGAGG |
| R602 | 63 | LNCSAKGIHS-IAIGATAEAAKGAAVA-VGAGSIATGVN--SVAIGPLSKALGDSAVTYG |
| R603 | 120 | IGARASTSDTGVAVGFNSKADAKNSVA-IGHSSHVAANHGYSIAIGDRSKTDRENSVSIG |
| R604 | 190 | ESLNRQLTHLAAGTKDTDAVNVAQLKKEIEKTQENTNKKSAELLAKPNAYADNKSSSVLG |
For each group of repeats, relative similarity is indicated as follows: boldface, identity; italics, close similarity; underlining, more distant similarity (see GAP program [8]).
Upstream of the 7-residue repeats can be found at least six 14-residue repeats (R141-R146) (Table 5). These 14-residue repeats could, of course, have arisen by sequence divergence of a duplicated 7-residue repeat. The similarities of these consecutive 14-residue repeat sequences are apparent, but the degrees of identity observed for these repeats differ substantially. Thus, repeats R143 and R144 are identical in all but one position (13 of 14 positions). Repeats R145 and R146, as well as repeats R141 and R145, exhibit 7 of 14 identities (50% identity). The other repeat unit comparisons reveal lower degrees of identity but still enough to suggest homology.
Upstream of the 14-residue repeats are the apparent ∼60 residue repeats (Table 5). Repeats R602 and R603 show the greatest percent identity (16 out of 60, or 27% identity). Next, R601 and R603 exhibit 8 out of 60 identities (13.5% identity), while R602 and R604 exhibit 7 out of 40 identities (18% identity). All of the AT-2 protein passenger domains proved to be homologous in the regions exhibiting the 60-residue repeat units. They differed with respect to degrees of sequence similarity and numbers of repeat units. However, the results obtained explain why all of these proteins are homologous and why proteins of very different sizes cluster together on the phylogenetic tree (Fig. 2B).
CONCLUSIONS AND PERSPECTIVES
In this minireview, we summarize the available experimental evidence and report bioinformatic analyses of the newly discovered AT-2 proteins, believed to form trimeric structures in the outer membranes of gram-negative bacteria. These trimers are thought to form 12-β-strand transmembrane pores that allow export of the N-terminal passenger domains from the periplasm to the external milieu (see introduction). Our analyses have led to several important evolutionary conclusions or suggestions. (i) AT-2 domains are found in proteobacteria of the α-, β-, and γ-subdivisions and their phage although sequence-divergent members of the family are found in other gram-negative bacterial kingdoms (7). (ii) Two homologues found outside of these bacterial subkingdoms were from a low GC-content gram-positive bacterium with an incompletely sequenced genome. We suggest that these two sequences resulted from DNA contamination. (iii) Several paralogues can be present in a single organism; for example, Haemophilus somnus 2336 has five paralogues of similar AT-2 domain sequence, while Burkholderia cepacia R18194 has four AT-2 domain paralogues, three of which are similar in sequence. (iv) AT-2 sequence similarity does not imply similarly sized passenger domains, as phylogeny of the AT-2 domains does not correlate well with protein size. (v) Although there is a poor correlation between position in the AT-2 domain tree and protein size, there is a reasonably good correlation between AT-2 protein domain phylogeny and the source organismal type (with a few potential exceptions). (vi) Linker domains appear to consist of 7-residue repeats. (vii) Adjacent to these are 14-residue repeats that may have arisen by sequence divergence of duplicated 7-residue repeats (8). Finally, most of the passenger domains consist of ∼60-residue repeats of variable numbers.
Points iii to v above imply that the shuffling of AT-2 domains relative to their passenger domains and/or the modification of passenger domain size during recent evolution has occurred repeatedly, even though horizontal transfer of these proteins across bacterial phylogenetic groupings has been relatively rare. It also appears that recent AT-2 domain-encoding gene duplication events have given rise to most of the paralogues in organisms such as H. somnus and B. cepacia. A recent increase or decrease in the numbers of ∼60-residue repeat units in the passenger domains is largely responsible for the size variations observed for close homologues.
Sequence analyses led to a very tentative but plausible suggestion that AT-2 domains may have evolved from domains that arose by repeated duplication of a genetic element of 21 nucleotides, encoding a 7-amino-acyl residue peptide. This peptide had the probable sequence of (D/E)(Q/N)(R/K)(F/I)(Q/D)(Q/K)(V/L). This is a strongly hydrophilic heptapeptide with only two hydrophobic residue positions. This repeat unit could be identified in the N-terminal “linker” regions of several AT-2 domains. This hydrophilic “linker” connects the AT-2 domain with the passenger domain. Surprisingly, it could be found throughout most of the C-terminal regions of other proteins that exhibit certain characteristics of AT-2 proteins and that were retrieved with PSI-BLAST iterations (Table 3). It is clear that if this repeated heptapeptide provided the basis for formation of the AT-2 domain, extensive sequence divergence had to have occurred in order to form the more hydrophobic, strongly amphipathic, β-structured AT-2 domains that are thought to mediate pore formation.
We identified two particularly well-conserved sequence motifs in the AT-2 domain that must be of structural and functional significance. One proved to be in the N-terminal region of the AT-2 domain in a strongly hydrophobic region (Fig. 3, peak H1), while the other was in a strongly amphipathic region in putative transmembrane β-strand 2 (Fig. 3, peak A3). The former proved to be more hydrophobic than the latter. Most interestingly, motif 1 exhibited AT-2 domain-specific residue-type differences that were lacking in motif 2. Motif 2 exhibited conservation in the different clusters typically characteristic of the entire AT-2 family. Since only in motif 1 was there a suggestion of residue (and hence functional) specialization and since full residue conservation was not observed at any one position, we suggest that the pores formed from AT-2 domains are fairly flexible and nonspecific, accommodating a range of passenger proteins. It is possible, however, that substrate protein selectivity is a function performed by motif 1.
The proposed mechanism of membrane transport by proteins like YadA, Hia, and Hap is by no means established. The notion that 12-stranded β-barrels form export portals is in doubt. For example, in the crystal structure of the 12-stranded β-barrel from the E. coli outer membrane phospholipase A2, the ribbon diagram shows the existence of a pore formed by the barrel, but the space-filling form indicates that this channel is too small to permit export of a polypeptide in either α or β form (21, 33, 41, 42). The limitations of biochemistry to physiological theories are important to note in order to stimulate discussion of the overall validity of the proposed translocation model. A crucial point in this respect is the proposed multimeric structure of AT-2 C domains. The conclusion that AT-2 proteins are homotrimers should be evaluated carefully in view of the potential inability of a 12-stranded β-barrel to transport polypeptide strands. In this regard, however, it is also important to note that transmembrane channels can be flexible, opening and closing in response to conformational changes that alter the angle of the polypeptide relative to the plane of the membrane (35).
Outer membrane porins with 8 transmembrane β-strands (TβSs) (OmpA of E. coli, TC 1.B.6 [12, 34]), 10 probable TβSs (TP0453 of Treponema pallidum, TC 1.B.45 [14]), 12 TβSs (Tsx of E. coli, TC 1.B.10 [50]; NalP of Neisseria meningitidis, TC 1.B.12 [32]; TolC of E. coli, TC 1.B.11 [23]), and 14 TβSs (FadL of E. coli, TC 1.B.9 [47]) have been identified and have been shown to have porin activities in spite of their small pore sizes. Quite conceivably, pore activity is transient, being induced by specific conditions such as substrate binding or response to osmotic conditions (3, 35).
The analyses reported in this minireview make several predictions concerning the structures, functions, and evolutionary origins of a novel family of autotransporter proteins. A four-transmembrane strand β-sheet possibly serves as the pore-forming element, and oligomerization is likely to be required for function, as is the case for all well-characterized channel-forming peptides (38-40). The functional significance of conserved motifs 1 and 2 has not been investigated. The fact that all passenger domains are homologous, consisting of large repeats of various numbers, suggests a unified general function in adhesion/macromolecular recognition. Further studies will be required to understand the structure-function relationships of these interesting virulence-related proteins.
ADDENDUM IN PROOF
After the completion of this work, the complete genome sequence of D. hafniense Y51 has become available (H. Nonaka et al., J. Bacteriol. 188:2262-2274, 2006). The two sequences, Dha7 and Dha2, that we suspected to be contaminants are not in the completed sequence.
Acknowledgments
This work was supported by NIH grant GM64368 and GM077402 from the National Institute of General Medical Sciences.
We thank Mary Beth Hiller for her assistance in the preparation of the manuscript.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biedzka-Sarek, M., R. Venho, and M. Skurnik. 2005. Role of YadA, Ail, and lipopolysaccharide in serum resistance of Yersinia enterocolitica serotype O:3. Infect. Immun. 73:2232-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bostina, M., B. Mohsin, W. Kuhlbrandt, and I. Collinson. 2005. Atomic model of the E. coli membrane-bound protein translocation complex SecYEG. J. Mol. Biol. 352:1035-1043. [DOI] [PubMed] [Google Scholar]
- 4.Busch, W. and M. H. Saier, Jr. 2002. The transporter classification (TC) system. 2002. CRC Crit. Rev. Biochem. Mol. Biol. 37:287-337. [DOI] [PubMed] [Google Scholar]
- 5.Cotter, S. E., N. K. Surana, and J. W. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13:199-205. [DOI] [PubMed] [Google Scholar]
- 6.Cotter, S. E., H. J. Yeo, T. Juehne, and J. W. St. Geme III. 2005. Architecture and adhesive activity of the Haemophilus influenzae Hsf adhesin. J. Bacteriol. 187:4656-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desvaux, M., A. Khan, S. A. Beatson, A. Scott-Tucker, and I. R. Henderson. 2005. Protein secretion systems in Fusobacterium nucleatum: genomic identification of type 4 piliation and complete type V pathways brings new insight into mechanisms of pathogenesis. Biochim. Biophys. Acta 1713:92-112. [DOI] [PubMed] [Google Scholar]
- 8.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Tahir, Y., and M. Skurnik. 2001. YadA, the multifaceted Yersinia adhesin. Int. J. Med. Microbiol. 291:209-218. [DOI] [PubMed] [Google Scholar]
- 10.Fink, D. L., A. Z. Buscher, B. Green, P. Fernsten, and J. W. St. Geme III. 2003. The Haemophilus influenzae Hap autotransporter mediates microcolony formation and adherence to epithelial cells and extracellular matrix via binding regions in the C-terminal end of the passenger domain. Cell Microbiol. 5:175-186. [DOI] [PubMed] [Google Scholar]
- 11.Fink, D. L., and J. W. St. Geme III. 2003. Chromosomal expression of the Haemophilus influenzae Hap autotransporter allows fine-tuned regulation of adhesive potential via inhibition of intermolecular autoproteolysis. J. Bacteriol. 185:1608-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gribun, A., D. J. Katcoff, G. Hershkovits, I. Pechatnikov, and Y. Nitzan. 2004. Cloning and characterization of the gene encoding for OMP-PD porin: the major Photobacterium damsela outer membrane protein. Curr. Microbiol. 48:167-174. [DOI] [PubMed] [Google Scholar]
- 13.Guyer, D. M., I. R. Henderson, J. P. Nataro, and H. L. T. Mobley. 2000. Identification of Sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol. Microbiol. 38:53-66. [DOI] [PubMed] [Google Scholar]
- 14.Hazlett, K. R., D. L. Cox, M. Decaffmeyer, M. P. Bennett, D. C. Desrosiers, C. J. La Vake, M. E. La Vake, K. W. Bourell, E. J. Robinson, R. Brasseur, and J. D. Radolf. 2005. TP0453, a concealed outer membrane protein of Treponema pallidum, enhances membrane permeability. J. Bacteriol. 187:6499-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson, I. R., R. Cappello, and J. P. Nataro. 2000. Autotransporter proteins, evolution and redefining protein secretion. Trends Microbiol. 8:529-532. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins, M. K., J. Eswaran, P. Edwards, G. F. Schertler, C. Hughes, and V. Koronakis. 2004. Structure of the ligand-blocked periplasmic entrance of the bacterial multidrug efflux protein TolC. J. Mol. Biol. 342:697-702. [DOI] [PubMed] [Google Scholar]
- 19.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacob-Dubuisson, F., R. Fernandez, and L. Coutte. 2004. Protein secretion through autotransporter and two-partner pathways. Biochim. Biophys. Acta 1694:235-257. [DOI] [PubMed] [Google Scholar]
- 21.Kingma, R. L., M. Fragiathaki, H. J. Snijder, B. W. Dijkstra, H. M. Verheij, N. Dekker, and M. R. Egmond. 2000. Unusual catalytic triad of Escherichia coli outer membrane phospholipase A. Biochemistry 39:10017-10022. [DOI] [PubMed] [Google Scholar]
- 22.Koronakis, V., J. Eswaran, and C. Hughes. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73:467-489. [DOI] [PubMed] [Google Scholar]
- 23.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, A., and H. P. Schweizer. 2005. Bacterial resistance to antibiotics: active efflux and reduced uptake. Adv. Drug Deliv. Rev. 57:1486-1513. [DOI] [PubMed] [Google Scholar]
- 25.Laarmann, S., D. Cutter, T. Juehne, S. J. Barenkamp, and J. W. St. Geme. 2002. The Haemophilus influenzae Hia autotransporter harbours two adhesive pockets that reside in the passenger domain and recognize the same host cell receptor. Mol. Microbiol. 46:731-743. [DOI] [PubMed] [Google Scholar]
- 26.Liu, D. F., K. W. Mason, M. Mastri, M. Pazirandeh, D. Cutter, D. L. Fink, J. W. St. Geme III, D. Zhu, and B. A. Green. 2004. The C-terminal fragment of the internal 110-kilodalton passenger domain of the Hap protein of nontypeable Haemophilus influenzae is a potential vaccine candidate. Infect. Immun. 72:6961-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loveless, B. J., and M. H. Saier, Jr. 1997. A novel family of autotransporting, channel-forming, bacterial virulence proteins. Mol. Membr. Biol. 14:113-123. [DOI] [PubMed] [Google Scholar]
- 28.Ma, Q., Y. Zhai, C. J. Schneider, T. M. Ramseier, and M. H. Saier, Jr. 2003. Protein secretion systems of Pseudomonas aeruginosa and P. fluorescens. Biochim. Biophys. Acta 1611:223-233. [DOI] [PubMed] [Google Scholar]
- 29.Maurer, J., J. Jose, and T. F. Meyer. 1999. Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J. Bacteriol. 181:7014-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman, C. L., and C. Stathopoulos. 2004. Autotransporter and two-partner secretion: delivery of large-size virulence factors by gram-negative bacterial pathogens. Crit. Rev. Microbiol. 30:275-286. [DOI] [PubMed] [Google Scholar]
- 31.Nummelin, H., M. C. Merckel, J. C. Leo, H. Lankinen, M. Skurnik, and A. Goldman. 2004. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. EMBO J. 23:701-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oomen, C. J., P. van Ulsen, P. van Gelder, M. Feijen, J. Tommassen, and P. Gros. 2004. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23:1257-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otto, B. R., R. Sijbrandi, J. Luirink, B. Oudega, J. G. Heddle, K. Mizutani, S. Y. Park, and J. R. Tame. 2005. Crystal structure of hemoglobin protease, a heme binding autotransporter protein from pathogenic Escherichia coli. J. Biol. Chem. 280:17339-17345. [DOI] [PubMed] [Google Scholar]
- 34.Pautsch, A., and G. E. Schulz. 2000. High-resolution structure of the OmpA membrane domain. J. Mol. Biol. 298:273-282. [DOI] [PubMed] [Google Scholar]
- 35.Pivetti, C. D., M.-R. Yen, S. Miller, W. Busch, Y.-H. Tseng, I. R. Booth, and M. H. Saier, Jr. 2003. Two families of prokaryotic mechanosensitive channel proteins. Microbiol. Mol. Biol. Rev. 67:66-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roggenkamp, A., N. Ackermann, C. A. Jacobi, K. Truelzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 185:3735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saier, M. H., Jr. 2000a. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saier, M. H., Jr. 2000b. Families of proteins forming transmembrane channels. J. Membr. Biol. 175:165-180. [DOI] [PubMed] [Google Scholar]
- 39.Saier, M. H., Jr. 2003a. Answering fundamental questions in biology with bioinformatics. ASM News 69:175-181. [Google Scholar]
- 40.Saier, M. H., Jr. 2003b. Tracing pathways of transport protein evolution. Mol. Microbiol. 48:1145-1156. [DOI] [PubMed] [Google Scholar]
- 41.Snijder, H. J., and B. W. Dijkstra. 2000. Bacterial phospholipase A: structure and function of an integral membrane phospholipase. Biochim. Biophys. Acta 1488:91-101. [DOI] [PubMed] [Google Scholar]
- 42.Snijder, H. J., I. Ubarretxena-Belandia, M. Blaauw, K. H. Kalk, H. M. Verheij, M. R. Egmond, N. Dekker, and B. W. Dijkstra. 1999. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature 401:717-721. [DOI] [PubMed] [Google Scholar]
- 43.St. Geme, J. W., III, and D. Cutter. 2000. The Haemophilus influenzae Hia adhesin is an autotransporter protein that remains uncleaved at the C terminus and fully cell associated. J. Bacteriol. 182:6005-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Surana, N. K., D. Cutter, S. J. Barenkamp, and J. W. St. Geme III. 2004. The Haemophilus influenzae Hia autotransporter contains an unusually short trimeric translocator domain. J. Biol. Chem. 279:14679-14685. [DOI] [PubMed] [Google Scholar]
- 45.Tamm, A., A. M. Tarkkanen, T. K. Korhonen, P. Kuusela, P. Toivanen, and M. Skurnik. 1993. Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol. Microbiol. 10:995-1011. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The clustal x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Berg, B., P. N. Black, W. M. Clemons, Jr., and T. A. Rapoport. 2004. Crystal structure of the long-chain fatty acid transporter FadL. Science 304:1506-1509. [DOI] [PubMed] [Google Scholar]
- 48.van Ulsen, P., L. van Alphen, C. T. Hopman, A. van der Ende, and J. Tommassen. 2001. In vivo expression of Neisseria meningitidis proteins homologous to the Haemophilus influenzae Hap and Hia autotransporters. FEMS Immunol. Med. Microbiol. 32:53-64. [DOI] [PubMed] [Google Scholar]
- 49.Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262-265. [DOI] [PubMed] [Google Scholar]
- 50.Ye, J., and B. van den Berg. 2004. Crystal structure of the bacterial nucleoside transporter Tsx. EMBO J. 23:3187-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yen, M. R., C. R. Peabody, S. M. Partovi, Y. Zhai, Y. H. Tseng, and M. H. Saier, Jr. 2002. Protein-translocating outer membrane porins of gram-negative bacteria. Biochim. Biophys. Acta 1562:6-31. [DOI] [PubMed] [Google Scholar]
- 52.Yeo, H. J., S. E. Cotter, S. Laarmann, T. Juehne, J. W. St. Geme, and G. Waksman. 2004. Structural basis for host recognition by the Haemophilus influenzae Hia autotransporter. EMBO J. 23:1245-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhai, Y., and M. H. Saier, Jr. 2001. A web-based program for the prediction of average hydropathy, average amphipathicity and average similarity of multiply aligned homologous proteins. J. Mol. Microbiol. Biotechnol. 3:285-286. [PubMed] [Google Scholar]
- 54.Zhai, Y., and M. H. Saier, Jr. 2002. The β-barrel finder (BBF) program, allowing identification of outer membrane β-barrel proteins encoded within prokaryotic genomes. Protein Sci. 11:2196-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhai, Y., J. Tchieu, and M. H. Saier, Jr. 2002. A web-based Tree View (TV) program for the visualization of phylogenetic trees. J. Mol. Microbiol. Biotechnol. 4:69-70. [PubMed] [Google Scholar]