Abstract
Bordetella avium is a pathogen of poultry and is phylogenetically distinct from Bordetella bronchiseptica, Bordetella pertussis, and Bordetella parapertussis, which are other species in the Bordetella genus that infect mammals. In order to understand the evolutionary relatedness of Bordetella species and further the understanding of pathogenesis, we obtained the complete genome sequence of B. avium strain 197N, a pathogenic strain that has been extensively studied. With 3,732,255 base pairs of DNA and 3,417 predicted coding sequences, it has the smallest genome and gene complement of the sequenced bordetellae. In this study, the presence or absence of previously reported virulence factors from B. avium was confirmed, and the genetic bases for growth characteristics were elucidated. Over 1,100 genes present in B. avium but not in B. bronchiseptica were identified, and most were predicted to encode surface or secreted proteins that are likely to define an organism adapted to the avian rather than the mammalian respiratory tracts. These include genes coding for the synthesis of a polysaccharide capsule, hemagglutinins, a type I secretion system adjacent to two very large genes for secreted proteins, and unique genes for both lipopolysaccharide and fimbrial biogenesis. Three apparently complete prophages are also present. The BvgAS virulence regulatory system appears to have polymorphisms at a poly(C) tract that is involved in phase variation in other bordetellae. A number of putative iron-regulated outer membrane proteins were predicted from the sequence, and this regulation was confirmed experimentally for five of these.
The genus Bordetella comprises eight species: B. pertussis, B. parapertussis, B. bronchiseptica, B. avium, B. hinzii, B. holmesii, B. trematum, and B. petrii. The genus is classified as a member of the beta-proteobacteria. Bordetellae are closely related to the genera Achromobacter and Alcaligenes, which include mostly environmental bacteria with some opportunistic pathogens (reviewed in reference 112). A phylogenetic analysis based on 16S rRNA genes places Alcaligenes as the most ancient, with Achromobacter and Bordetella deriving more recently from a single node and B. petrii and B. avium being most distantly related to all other Bordetella species (112). The very close phylogenetic relationship among B. pertussis, B. parapertussis, and B. bronchiseptica has been well established (110), and it is generally accepted that B. pertussis and B. parapertussis were differentiated from B. bronchiseptica by substantial gene loss (29, 71), diverging as much as 3.5 million years ago (30). B. pertussis, B. parapertussis, and B. bronchiseptica colonize the respiratory tracts of mammals (reviewed in reference 25), but their host ranges and the severity of the diseases they cause differ. B. pertussis is restricted to the human host and is the etiological agent of an acute respiratory disease known as pertussis or whooping cough. B. parapertussis is divided into two phylogenetically distinct subspecies/populations, one containing isolates from cases of human whooping cough (B. parapertussishu) and the other containing strains isolated from sheep (B. parapertussisov) (110). B. bronchiseptica has a broad mammalian host range, causing chronic and often asymptomatic respiratory infections in a wide range of animals and occasionally humans (40).
Of the other bordetellae described, B. hinzii is usually found as a commensal of birds but has occasionally been seen as an opportunistic pathogen of immunocompromised humans (21, 52), and B. holmesii has been associated with septicemia (114) and, more recently, with respiratory illness possibly resembling pertussis (67, 116), while B. trematum has been described as an occasional pathogen of humans causing wound infections and otitis media (108). B. petrii was first isolated from a bioreactor and is not thought to be a pathogen (113).
B. avium is the causative agent of bordetellosis in avian species, which is similar to respiratory disease caused by other bordetellae (48, 53). B. avium infects many different wild and domesticated birds (47, 49, 84), but commercially raised turkeys are particularly susceptible (94). Similar to B. bronchiseptica (75, 76), B. avium survives for long periods of time in water and dilute salt solutions, which may provide a means of survival between hosts (84). B. avium has a specific tropism for ciliated respiratory tissue, a property that it shares with B. pertussis, B. parapertussis, and B. bronchiseptica (7).
Among them, B. pertussis, B. parapertussis, and B. bronchiseptica share a large complement of virulence factors which can be broadly categorized as adhesins or toxins. The adhesins include filamentous hemagglutinin (FHA), pertactin (Prn), tracheal colonization factor, serum resistance protein (BrkA), and fimbriae. The toxins include pertussis toxin (PTX), adenylate cyclase-hemolysin (CyaA), dermonecrotic toxin (DNT), and tracheal cytotoxin (TCT) (65). Of these, only FHA, fimbriae, DNT, and TCT have been identified in B. avium (36, 95). The expression of these virulence-activated genes and another set of virulence-repressed genes is regulated by a two-component master regulatory system encoded by the bvgAS locus (reviewed in reference 24), also previously identified in B. avium (95).
In this report, we present the complete genome sequence of B. avium strain 197N and a comparative analysis between this and the genomic sequences of B. bronchiseptica, B. pertussis, and B. parapertussis (71). We also present some experimental data related to phenomena suggested by the bioinformatic analysis of the genomic sequence data. Comparison of the genomes of these four bordetellae will provide insights into the virulence factors of B. avium, provide information about the mechanisms underlying the host specificities and evolution of members of the Bordetella genus, and suggest many testable hypotheses about the pathogenesis, ecology, and biology of the bordetellae.
MATERIALS AND METHODS
DNA preparation, cloning, and sequencing.
B. avium strain 197N was isolated from a diseased turkey in a commercial flock in the early 1980s in Ohio and chosen for further study on the basis of increased virulence in comparison to other isolates (36; unpublished data). DNA was extracted by the method of Marmur (63). The initial genome assembly was obtained from 63,567 paired end sequences (giving 9.33-fold coverage) derived from five genomic shotgun libraries (two in pOTW12 with insert sizes of 2 to 2.8 kb and 3 to 3.3 kb and three in pMAQ1 with insert sizes of 5.5 to 6 kb, 9 to 10 kb, and 10 to 12 kb) using dye terminator chemistry on ABI3730 automated sequencers; 1,621 paired end sequences from two pBACe3.6 libraries with insert sizes of 15 to 17 kb and 17 to 20 kb (a clone coverage of 3.96-fold) were used as a scaffold. A further 4,286 directed sequencing reads were generated during finishing.
Sequence analysis and annotation.
The sequence was assembled, finished, and annotated as described previously (71), using Artemis (88) to collate data and facilitate annotation. The DNA and predicted protein sequences of B. avium were compared to genomic sequences of the other three Bordetella species using the Artemis Comparison Tool (15). Orthologous gene sets were calculated by reciprocal best-match FASTA comparisons. Pseudogenes had one or more mutations that would prevent translation; each of the inactivating mutations was checked against the original sequencing data.
Polymorphism sequence analysis.
For identifying polymorphisms in bvgS, the following primers were used: 5′GCACGCCCATCTATGCCATCAC3′ and 5′TGCAGCAGATGTTCGGCATCC3′. Amplification of the 301-base-pair fragment spanning the poly(C) region was accomplished utilizing GC-cDNA polymerase from Clontech. Amplicons were sequenced by MWG Biotechnologies.
Construction of mutants.
Internal fragments of the target genes (bfrH, bfrB, and bfeA) were amplified from B. avium genomic DNA using PCR and cloned into the HinDIII and BamHI sites of pFUS2 (6). Gene disruptions were performed by conjugation using B. avium strain KO1, a bhuR mutant of wild-type strain 4169rif (69), to generate KO1bfrH, KO1bfrB, and KO1bfeA. Transconjugants were selected on brain heart infusion (BHI) agar containing gentamicin.
Outer membrane protein isolation.
A modification of the protocol of Leyh and Griffith (59) was used to purify B. avium outer membrane proteins (OMPs) from Fe-replete and Fe-stressed cultures and was described elsewhere (69). Fe-stressed conditions were achieved by adding enough ethylene-di-o-hydroxyphenylacetic acid (EDDHA) to reduce the cell density (measured by the optical density at 600 nm) of an 18-h culture (cultured at 37°C and 300 rpm) to 50 to 60% of the growth of duplicate culture grown under Fe-replete conditions. OMPs were harvested from B. avium cultured at 37°C to stationary phase in 200 ml of BHI (Difco Laboratories, Detroit, MI) broth containing either 36 μM FeSO4 (Fe replete) or 50 μM EDDHA (Sigma Biochemicals, St. Louis, Mo.) (Fe stressed). The total protein concentration of the OMP suspensions was determined with a Bio-Rad protein assay using bovine serum albumin as the standard. OMPs were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (56) and visualized by staining with colloidal Coomassie brilliant blue (70).
LPS preparation and analysis.
For lipopolysaccharide (LPS) analysis and immunological staining, bacteria were prepared as described previously (81). Briefly, approximately 1 × 109 bacteria were pelleted from a 1-ml suspension and resuspended in 500 μl of phosphate-buffered saline. Next, 250 μl of LPS buffer I (0.1875 M Tris-HCl, pH 6.8, 6% [wt/vol] glycerol) was added, and the mixture was heated in a boiling water bath for 5 min. Then, 10 μl of the cell lysate was added to 35 μl of LPS buffer II (0.0624 M Tris-HCl, pH 6.8, 0.1% SDS, 10% [wt/vol] glycerol, 0.1% [wt/vol] bromophenol blue) along with 10 μl of proteinase K (25 mg/ml). The sample was incubated at 55°C for 12 to 16 h. Samples were heated in a boiling water bath for 5 min prior to SDS-PAGE. Twenty microliters of sample was loaded onto SDS-PAGE gels. LPS samples were analyzed by SDS-PAGE in the Tricine-buffered gel system originally described by Schagger and von Jagow (89), with the modifications described by Lesse and colleagues (58). Silver staining was performed according to the method of Tsai and Frasch (105). Western blot analysis was performed as previously described (3) using the monoclonal antibodies BL-2 (band A LPS specific) and BL-8 (band B LPS specific) (64).
Nucleotide sequence accession number.
The genome sequence has been submitted to EMBL with the accession number AM167904.
RESULTS AND DISCUSSION
Genome structure.
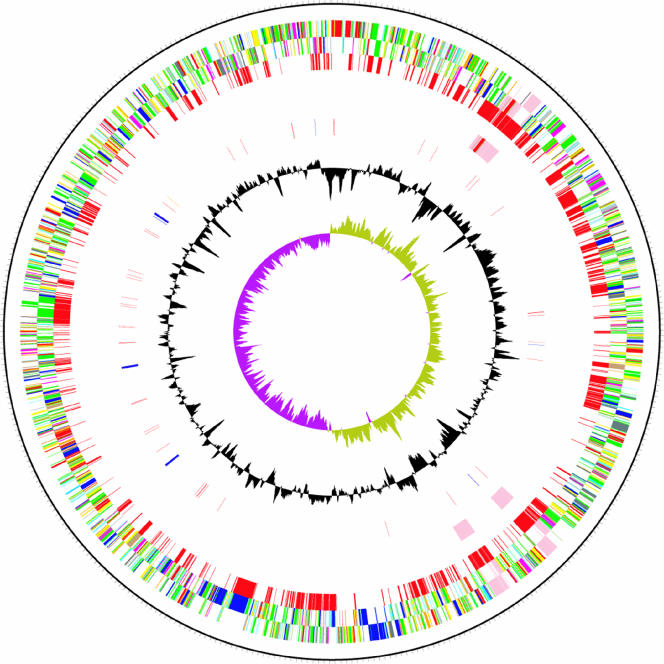
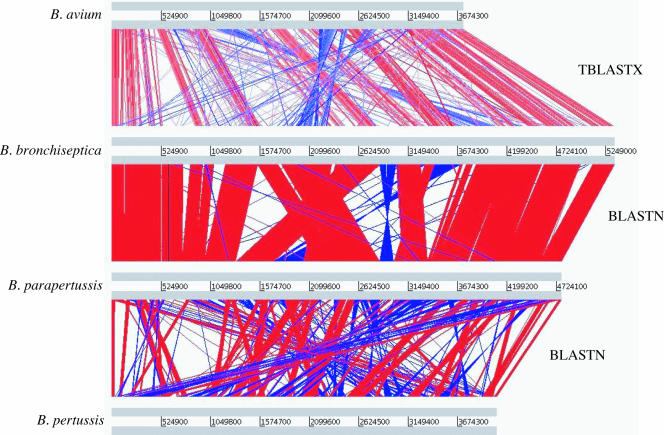
The general features of the B. avium genome are shown in Table 1 and Fig. 1. The B. avium genome comprises 3,732,255 base pairs, which is 1.6 Mb smaller than that of B. bronchiseptica. The B. avium genome has limited synteny with the other Bordetella genomes, and the overall similarity at the DNA (97%) and protein (75% average identity between orthologues) levels is lower than among B. bronchiseptica, B. pertussis, and B. parapertussis, which show approximately 99% identity at the DNA level and 99.1 to 99.5% average protein identity between orthologues (Fig. 2).
TABLE 1.
Comparison of size, composition, and coding sequences between B. avium and the previously sequenced bordetellae
| Characteristic | Value for
|
|||
|---|---|---|---|---|
| B. avium | B. bronchiseptica | B. parapertussis | B. pertussis | |
| Size (bp) | 3,732,255 | 5,339,179 | 4,773,551 | 4,086,186 |
| G+C content (%) | 61.58 | 68.08 | 68.10 | 67.72 |
| No. of coding sequences | 3,417 | 5,011 | 4,404 | 3,816 |
| Coding percentage | 88.6 | 91.9 | 86.6 | 82.9 |
| Average gene size (bp) | 972 | 982 | 987 | 978 |
| No. of pseudogenes | 68 | 18 | 220 | 358 |
| No. of rRNA operons | 3 | 3 | 3 | 3 |
| No. of tRNA operons | 61 | 55 | 53 | 51 |
FIG. 1.
Circular representations of the genome of B. avium. The circles represent, from the outside in: circles 1 and 2, all genes (transcribed clockwise and counterclockwise); 3, B. avium unique genes; 4, bacteriophage genes; 5, RNA genes (blue, rRNAs; red, tRNAs; and green, stable RNAs); 6, G+C content (plotted using a 10-kb window); and 7, GC deviation ([G−C]/[G+C] plotted using a 10-kb window; khaki indicates values of >1, and purple indicates values of <1). Color coding for genes is as follows: dark blue, pathogenicity/adaptation; black, energy metabolism; red, information transfer; dark green, surface associated; cyan, degradation of large molecules; magenta, degradation of small molecules; yellow, central/intermediary metabolism; pale green, unknown; pale blue, regulators; orange, conserved hypothetical; brown, pseudogenes; pink, phage and insertion sequence elements; and gray, miscellaneous.
FIG. 2.
Linear genomic comparison of B. avium, B. bronchiseptica, B. pertussis, and B. parapertussis. The gray bars represent the forward and reverse DNA strands. The red and blue lines between the genomes represent protein similarity (TBLASTX) between B. avium and B. bronchiseptica or DNA-DNA similarities (BLASTN matches) between B. bronchiseptica and B. pertussis or B. parapertussis (red lines represent direct matches, while blue lines represent inverted matches).
Gene complements.
We have chosen to focus this analysis primarily on a comparison between B. avium and B. bronchiseptica. Previous comparative genomics and microarray analyses revealed that B. pertussis and B. parapertussis are recent derivatives of B. bronchiseptica that have evolved primarily by genome reduction; excluding insertion sequence elements, there are no genes unique to B. parapertussis and only 11 genes unique to B. pertussis relative to B. bronchiseptica (29, 71). In contrast, there are 602 genes, including 277 coding sequences (CDSs) on prophage or plasmid-like regions, which are unique to B. bronchiseptica relative to both B. pertussis and B. parapertussis. Thus, among the four sequenced Bordetella species, B. bronchiseptica has the largest gene set and has not undergone significant reductive evolution. In addition, we have previously shown that the large majority of genes unique to B. bronchiseptica are unlikely to have been acquired recently but are more likely to appear unique due to their having been deleted from B. pertussis and/or B. parapertussis (71). B. avium is similar in niche to B. bronchiseptica, infecting a wide variety of hosts, and similarly has fewer transposons and pseudogenes than B. pertussis or B. parapertussis; therefore, the B. avium-B. bronchiseptica comparison is likely to be the most informative and constitutes the bulk of the analysis described below.
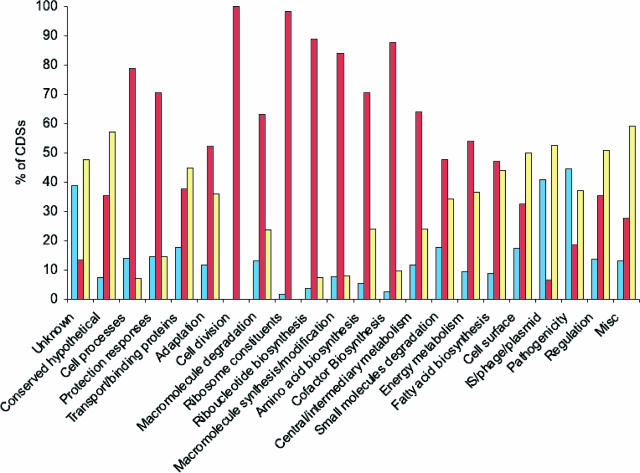
To distinguish CDSs orthologous between B. avium and B. bronchiseptica or unique to each (which may include paralogous genes), reciprocal FASTA analysis followed by manual curation was performed. B. avium and B. bronchiseptica share only 2,380 orthologous CDSs (Fig. 3). These shared CDSs encode primarily core/essential functions, whereas the species-specific CDSs are involved mainly in accessory functions (Fig. 4), which is likely to reflect the difference in lifestyle and pathogenicity of these two organisms (for example, infection of avian versus mammalian respiratory epithelia). The genome of B. avium contains 1,109 CDSs (31.5%) that do not have orthologues in B. bronchiseptica (Fig. 1 and 3). B. bronchiseptica, with nearly 46% more coding regions than B. avium, has 2,628 CDSs (52.5%) that are absent from B. avium (Fig. 3). These figures are larger than the variations observed within other species (e.g., Escherichia coli O157:H7 contains 1,387 genes [26%] not present in E. coli K-12, which has 528 [12%] unique genes [73]) and roughly comparable to those identified from other congeneric species comparisons (e.g., Bacteroides fragilis versus Bacteroides thetaiotaomicron, where 1,941 [44.3%] and 2,337 [48.9%] of the genes are unique, respectively [16]).
FIG. 3.
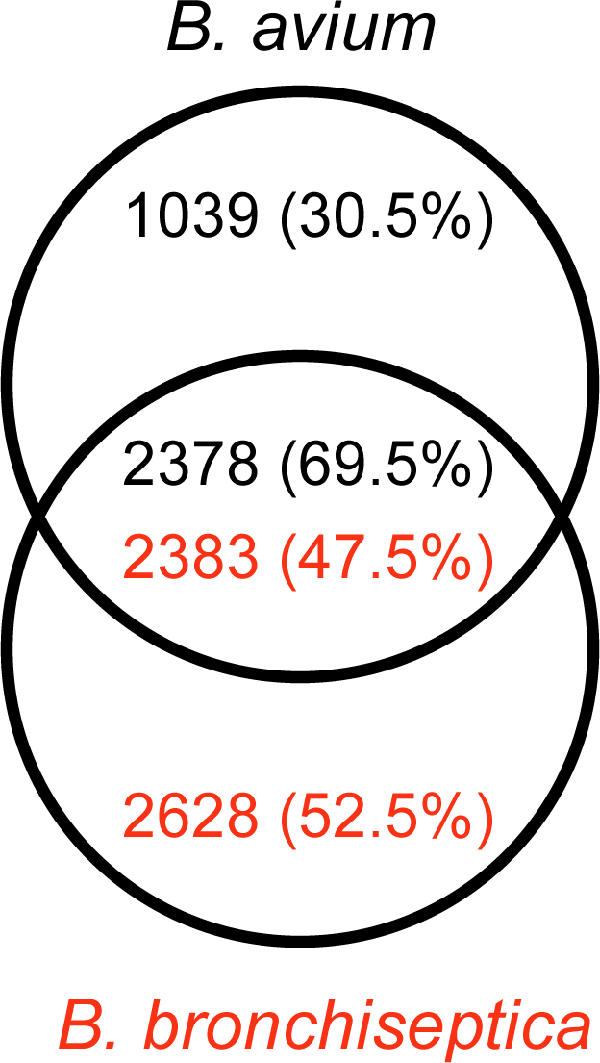
Venn diagram showing gene complements of B. avium and B. bronchiseptica. Shown are the number of B. avium-unique CDSs (top), the number of B. bronchiseptica-unique CDSs (bottom), and the number of CDSs from one organism that have orthologues in the other (middle). Numbers in parentheses show percentages of total CDSs. Orthologous genes were identified by reciprocal best-match FASTA comparison (see Materials and Methods).
FIG. 4.
Representation of the distribution, by functional categories, of the CDSs unique to B. avium (blue), unique to B. bronchiseptica (yellow), and shared between the two organisms (purple). Figures are expressed as a percentage of the total number of CDSs in each functional category.
The B. avium-specific genes, described in more detail below, include a unique fimbrial gene cluster, two hemagglutinins and their cognate secretion accessory proteins, a type II secretion system, an LPS biosynthesis locus (BAV0081-BAV0089), two separate loci (BAV1243-BAV1257 and BAV3077-BAV3087) potentially involved in the biosynthesis of an LPS O antigen, an operon that appears to encode enzymes for the biosynthesis of a cellulose-like polysaccharide (BAV2635-BAV2653), and three prophages. B. bronchiseptica-specific genes include a seven-gene cluster encoding O antigen biosynthesis, a type IV pilus locus, four prophages that are also not present in B. pertussis or B. parapertussis, the pertussis toxin locus, and the adenylate cyclase toxin gene.
There are 68 pseudogenes in the B. avium genome, more than in B. bronchiseptica (18) but substantially fewer than in B. parapertussis (220) or B. pertussis (358). B. pertussis and B. parapertussis are clonal and of recent descent. The presence of relatively few pseudogenes suggests that B. avium has not undergone a recent evolutionary bottleneck and may be older than B. pertussis and B. parapertussis. This correlates with the fact that B. avium more closely resembles B. bronchiseptica in host range and ability to survive in multiple niches (84).
Host interaction/virulence.
The overall comparison between B. avium and B. bronchiseptica shows a very large degree of variation in genes for surface structures, both proteins and polysaccharides. It is likely that these differences are central to their different specificities for avian and mammalian tracheae. These components include agglutinins/adhesins, LPSs, capsules and extracellular polymers, fimbriae and pili, autotransporters, large secreted proteins, secretion systems, and toxins and are discussed in detail in the following sections.
Agglutinins/adhesins.
FHA, encoded by the fhaB gene, is the major adhesin and hemagglutinin in B. pertussis, B. parapertussis, and B. bronchiseptica (54, 87). In B. bronchiseptica and B. pertussis, FHA is produced in the form of a preprotein, FhaB, which is exported to the cell surface via a two-partner secretion system, mediated by the secretory protein FhaC (50). The C-terminal region of FhaB is removed by a specific, anchored, subtilisin autotransporter, SphB1, to yield the mature protein FHA, which is partitioned between the cell surface and the extracellular milieu (26). B. avium has orthologues of fhaB and fhaC (BAV1966 and BAV1961, respectively) in exactly the same arrangement as B. bronchiseptica, with fhaC located as the fifth gene in the fimABCD operon (see “Fimbriae” below). FhaB of B. avium exhibits a low level (29% identity) of sequence similarity to and is shorter than (2,621 amino acids [aa]) that of B. bronchiseptica (3,652 aa). Furthermore, B. avium fhaB and fhaC insertion mutants retain hemagglutination ability but are avirulent in the turkey poult model (95). B. bronchiseptica has two additional FHA-like proteins, FhaL and FhaS, orthologues of which are also encoded in the B. avium genome (BAV2159 and BAV1551). In addition, the genome of B. avium contains six novel genes encoding FHA-like proteins (BAV0121, BAV0712, BAV0937, BAV1217, BAV2819, and BAV2825), of which two (BAV0937 and BAV1217) are pseudogenes.
Among the novel genes are two, BAV2824 (hagA) and BAV2825 (hagB), that encode the hemagglutination phenotype of B. avium (102); a mutation in either gene abolishes hemagglutination (unpublished data). The paired loci are similar to the fhaB/fhaC pair of B. avium and the other sequenced bordetellae, suggesting that they may also form a two-partner secretion system. This gene pair is physically located 4 CDSs from a very similar pair of genes, BAV2818 and BAV2819, with predicted proteins of 96% (hagB and BAV2819) and 53% (hagA and BAV2818) identity to each other at the amino acid level. Apart from the extreme N and C termini, the FhaB-like predicted proteins from hagB and BAV2819 are identical at the amino acid level across most of their lengths. While hagA and hagB are divergently transcribed, BAV2818 and BAV2819 occur in tandem and may constitute an operon. Work to determine the role, if any, of the second set of genes in hemagglutination and virulence is ongoing.
Two additional CDSs unique to B. avium, BAV1656 and BAV0856, encode adhesin-like proteins. The protein encoded by BAV1656 is 29% identical to E. coli Tia, an outer membrane protein involved in adherence and invasion of epithelial cells (35); it is also similar to Hek, a hemagglutinin encoded in a pathogenicity island of uropathogenic E. coli (32). BAV0856 encodes a potential adhesin that is 31% identical to Bap, a Staphylococcus aureus surface protein involved in biofilm formation (28), and 24% identical to E. coli intimin (51).
LPS.
LPS plays an important role in the virulence of the bordetellae (80). In B. avium, it is required for colonization of turkeys, serum resistance, tracheal ring binding, and resistance to antimicrobial peptide-mediated killing (96). It also serves as the attachment site for the B. avium bacteriophage Ba1 (92, 96). There is very little information regarding the structure of B. avium LPS except for the characterization of the O antigen domain (see below). Thus, the genome sequence provides the first clues as to the relatedness of the LPS of B. avium and the previously sequenced bordetellae. B. avium contains orthologues of many of the LPS loci characterized in B. bronchiseptica (Table 2), which suggests that B. avium LPS is structurally similar to B. bronchiseptica LPS, although it should be noted that single amino acid differences between some lipid A biosynthesis genes can result in an altered substrate specificity that can affect the precise lipid A structure and have profound effects on endotoxin activity, for example (100). Structural characterization of B. avium LPS to address issues such as these is under way.
TABLE 2.
LPS-related loci in B. avium and B. bronchiseptica
| B. avium locus | B. bronchiseptica locus | Function in LPS expression (reference) |
|---|---|---|
| BAV1744-BAV1747 (LpxA, LpxB, and LpxD) | BB2615-BB2618 (LpxA, LpxB, and LpxD) | Lipid A biosynthesis |
| BAV2873 (LpxC) | BB4192 (LpxC) | Lipid A biosynthesis |
| BAV2565 (LpxH) | BB1732 (LpxH) | Lipid A biosynthesis |
| BAV2100 (LpxK) | BB2008 (LpxK) | Lipid A biosynthesis |
| BAV2864 (PagP) | BB4181 (PagP) | Lipid A modification (74, 82) |
| BAV3272-BAV3275 | BB4814-BB4817 | Core biosynthesis |
| BAV2229-BAV2238 | BB3390-BB3400 | Core biosynthesis |
| BAV0510-BAV0518 | BB0875-BB0883 | Core biosynthesis |
| BAV0090-BAV0098 | BB0145-BB0155 (Wlb) | Band A trisaccharide |
Genome sequence comparisons identified several notable differences between B. avium and B. bronchiseptica LPS biosynthesis genes, including the O antigen and terminal trisaccharide.
The O antigen is the distal domain of the LPS molecule relative to the bacterial cell surface. In the B. avium type strain, it has been determined as a polymer of 1-4-linked 2-acetamidino-3-[3-hydroxybutanamido]-2,3-dideoxy-β-d-glucopyranosyluronic acid (57). The O antigens of B. bronchiseptica and B. parapertussis are a polymer of 2,3-diacetamido-2,3-dideoxy-α-l-galactopyranosyluronic acid (2,3diNAcGalA) (31), and in B. hinzii the O antigen is composed of trisaccharide repeating units of both the gluco and galacto isomers of the same uronic acid (8, 111). Thus, all of these O antigens are comprised of similar 2,3-dideoxy-hexuronic acids. This suggests that the genetics of O antigen expression in these bordetellae are similar. In B. bronchiseptica and B. parapertussis, the locus responsible for O antigen expression, wbm, contains 24 genes, including those encoding enzymes that are presumably involved in modification of the O antigen sugars (e.g., epimerase/dehyratases) and an ABC transport system similar to other O antigen export systems (79). At an equivalent location in B. avium, a putative polysaccharide biosynthesis locus is present. However, despite the structural similarity between the B. avium and B. bronchiseptica/B. parapertussis O antigens, the B. avium locus displays no similarities to wbm. It comprises only nine genes, including those encoding an O antigen translocase (BAV0081), a possible polysaccharide repeat unit exporter/flippase (BAV0084), a glycosyl transferase (BAV0086), an amidotransferase (BAV0087), and an acyltransferase (BAV0089), but no obvious transport system. While it remains to be confirmed that this locus is involved in B. avium O antigen expression, the difference between this locus and B. bronchiseptica/B. parapertussis wbm may suggest that it is involved in modifications of 2,3diNAcGlcA (for example, the unusual 3-hydroxybutanamido and the 2-acetamidino substitutions) and that the genes responsible for expression of the O antigen polymer backbone are elsewhere in the chromosome. However, we have not identified any obvious wbm homologues in the B. avium genome, and thus it must be concluded that B. avium utilizes a novel biosynthesis pathway for expression of its O antigen.
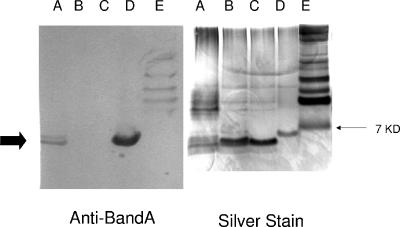
In B. pertussis, B. parapertussis, and B. bronchiseptica, the wlb gene cluster (notated wlbA to wlbL), which is immediately adjacent to the wbm locus, is involved in the addition of a trisaccharide to the distal portion of the LPS core (4), generating a structure known as band A LPS. B. avium also possesses a wlb locus at an equivalent location that is highly similar to that of B. bronchiseptica, which suggests that the two loci perform the same function; thus, it is suggested that B. avium expresses an LPS containing a band A trisaccharide structure. However, there are two differences between the B. avium and B. bronchiseptica loci, in that wlbD and wlbI are missing from B. avium. The function of WlbI is unknown, but WlbD is an epimerase that is required for expression of full-length LPS. The phenotype of a B. pertussis wlbD mutant suggests that WlbD is involved in the synthesis of the middle sugar of the trisaccharide 2,3diNAcManA (83). The absence of wlbD in B. avium suggests that any band A-like trisaccharide expressed by B. avium would comprise the gluco rather than the manno epimer of the di-N-acetylated uronic acid. However, LPS from B. avium is antigenically related to that of B. bronchiseptica, as shown by Western analysis using anti-LPS antibodies (Fig. 5). Antibodies against band A (the core plus trisaccharide) reacted with crude LPS preparations from B. avium strain 197N. In these experiments, LPS from B. avium wlbA and wlbL mutants did not react with anti-band A antibodies. Further, B. avium wlbA and wlbL mutations were complemented with the wlb region from B. bronchiseptica, as shown by normal LPS synthesis and phage Ba1 sensitivity (96).
FIG. 5.
SDS-PAGE and Western analysis of crude LPS preparations from wild-type and mutant B. avium and B. bronchiseptica using antibodies to the band A form of LPS. Lane A, B. avium wild type; lane B, B. avium wlbA mutant; lane C, B. avium wlbL mutant; lane D, B. bronchiseptica strain RB50; and lane E, molecular mass markers (the 7-kDa band is marked).
Thus, B. avium contains numerous orthologues of LPS biosynthesis genes found in B. pertussis, B. parapertussis, and B. bronchiseptica, which suggests that the LPSs share structural similarity. However, there are a number of interesting differences in B. avium LPS gene content and gene order that suggest some differences in structure and biosynthetic mechanism.
Capsular polysaccharide.
B. avium is known to produce a capsule (53), and a putative capsular polysaccharide biosynthesis locus is present in the B. avium genome (BAV2635-BAV2653). This large locus contains several genes that are predicted to encode putative sugar modification functions, including a homologue of GDP-mannose pyrophosphorylase/phosphomannose isomerase, four glycosyltransferases, an oxidoreductase, and an epimerase/dehydratase. The clustering of these genes suggests that the locus is involved in polysaccharide biosynthesis, but bioinformatic analysis does not suggest a structure. The remainder of the locus comprises genes of unknown function. There is no obvious polysaccharide export/transport apparatus encoded by this locus. This locus is different from that in the B. bronchiseptica genome (BB2918-BB2934), suggesting that B. avium and B. bronchiseptica produce different types of capsular polysaccharides. In contrast, B. pertussis and B. parapertussis appear to be devoid of capsular polysaccharide, an observation supported by the presence of pseudogenes in their capsular polysaccharide loci.
The B. avium capsular polysaccharide locus is directly adjacent to a 12-gene operon, BAV2623-BAV2634, which carries CDSs similar in both sequence and organization to those required for the biosynthesis of a cellulose-like polymer (wssA-wssJ) and which contributes to biofilm formation, niche specialization, and fitness of Pseudomonas fluorescens (97). The presence of a cellulose-like polymer has not been reported for B. avium, but if it exists, it may well suggest that B. avium is able to form biofilms.
Fimbriae.
In B. pertussis and B. bronchiseptica, fimbriae bind to sulfated sugars that are present on cell surfaces of the respiratory tract and therefore play an important role in tracheal colonization and development of a humoral immune response (38, 66). There are 11 putative fimbrial subunit genes in the B. avium genome (BAV0313, BAV0314, BAV0315, BAV0316, BAV0321, BAV0465, BAV1776, BAV1777, BAV1965, BAV2661, and BAV3039), whereas B. bronchiseptica, B. pertussis, and B. parapertussis have 6 intact genes, 3 intact genes and 1 pseudogene, and 4 intact genes and 2 pseudogenes, respectively (71).
Among the B. avium fimbrial genes, only one, BAV1965, is a true orthologue of a fimbrial subunit gene, fimA found in B. bronchiseptica, B. pertussis, and B. parapertussis. All the others are different in terms of both sequence similarity and chromosomal location.
In B. avium and the previously sequenced bordetellae, the fimA gene is part of an operon (fimABCD fhaC) encoding a chaperone-usher fimbrial biosynthesis system, which consists of a fimbrial subunit (FimA/BAV1965), a chaperone (FimB/BAV1964), a fimbrial usher (FimC/BAV1963), and a fimbrial adhesin (FimD/BAV1962). The location of fhaC in this fimbrial biogenesis operon is identical to the arrangement in B. pertussis and B. bronchiseptica (see “Agglutinins/adhesins” above). The fimABCD fhaC operon is functional and essential for virulence, since transposon mutants in fhaC and fimC were identified by signature tag mutagenesis (STM) using infection of turkey poults as a screening model (95). A second locus (BAV1773-BAV1777), unique to B. avium, carries two adjacent fimbrial subunits (BAV1776 and BAV1777), a chaperone (BAV1775), a fimbrial usher (BAV1774), and a fimbrial adhesin (BAV1773). Four other putative B. avium fimbrial subunit genes, BAV0313, BAV0314, BAV0315, and BAV0316, are contiguous and form a fimbrial gene cluster. Five of the B. avium fimbrial genes (BAV0313, BAV0315, BAV0316, BAV0321, and BAV2661) contain between six and nine repeats of a 7-bp sequence in their 5′ ends. Slipped-strand mispairing within these repeats may provide a mechanism for phase variation among the different fimbrial structural proteins. It is interesting that under conditions of normal laboratory growth in rich medium, such as BHI broth, negatively stained cells examined by transmission electron microscopy exhibit few if any visible fimbriae (unpublished observations). This may mean that the genes encoding the putative structures are expressed under conditions that we have not yet tested, such as within the host. Work to determine which, if any, of these many potential fimbrial genes are expressed and under what conditions is ongoing.
Type IV pilus.
The type IV pilus is involved in adherence and twitching motility and is important for virulence in some microorganisms (reviewed in reference 27). For example, Neisseria spp. require these pili for initial attachment to human epithelial cells, and type IV PAK pili mediate adhesion of Pseudomonas aeruginosa to mucosal surfaces. There are three potential type IV pilus gene clusters in the B. avium genome, two that appear intact (BAV2283-BAV2291 and BAV2357-BAV2364) and one that is probably nonfunctional due to the presence of several pseudogenes (BAV2525A-BAV2534). The same three loci are present in B. bronchiseptica, B. pertussis, and B. parapertussis, but all those in B. pertussis contain pseudogenes (71). Moreover, B. bronchiseptica has a fourth type IV pilus gene cluster (BB0776-BB0792) that is absent in B. pertussis, B. parapertussis, and B. avium.
Autotransporters.
Autotransporters are modular proteins consisting of a conserved C-terminal pore-forming domain through which the N-terminal passenger domain is exported (46). The variable passenger domains have diverse functions. Several autotransporters (BapA, BapB, BapC, BipA, BrkA, Prn, TcfA, and Vag8) from the previously sequenced bordetellae are known to be important for host interaction and virulence (46).
B. avium has seven intact autotransporter CDSs (BAV0466, BAV0811, BAV1572, BAV1641, BAV1864, BAV2846, BAV3269), while BAV1640 is a pseudogene (containing a stop codon, a frameshift, and a premature truncation) and BAV2452 is potentially phase variable, as it contains two frameshifts, one at a nine-guanine [G(9)] tract and one at a G(10). This tally compares to 21 intact autotransporters in B. bronchiseptica, of which 2 are potentially phase variable (71). Although some of the B. avium autotransporters share similarity with those present in the other bordetellae (for example, BipA; see below), they are not true orthologues. This observation suggests that these autotransporters were independently acquired by ancestral strains of B. avium and the other sequenced bordetellae or that they may be different samples from a common gene pool. One autotransporter gene, BAV2846, was previously identified by virtue of its homology to an adhesin, AIDA-I from diarrheagenic E. coli, which is involved in host cell attachment, autoaggregation, and biofilm formation (93). This gene was subsequently cloned, sequenced, expressed in E. coli, and shown to be expressed in B. avium by reverse transcription-PCR (unpublished data).
Large novel surface proteins.
The adjacent CDSs BAV1944 (4,342 aa, 447 kDa) and BAV1945 (6,460 aa, 650 kDa) are the largest in the genome. They are flanked by CDSs encoding components of a type I secretion system and potentially encode cell surface or exported proteins. Both proteins contain numerous locally repeated sequences. BAV1944 encodes a glycine- and serine-rich protein that is weakly similar to several proteins from a wide range of bacteria. A highly repetitive region occurs in the C-terminal half, with 10 occurrences of a 44-aa repeat sequence that are identical at 39 residues. These motifs are similar to hemolysin-type calcium-binding repeats (PF00353) that are conserved in proteins involved in host interactions (33). BAV1945 is most similar to a predicted bacterial immunoglobulin-like domain protein from Yersinia pseudotuberculosis (18) and to a putative cell wall-associated protein, srpA, of Streptococcus cristatus (44). These two predicted proteins, BAV1944 and BAV1945, are also weakly similar to each other and to other predicted autotransporters in the B. avium genome.
Motility.
B. bronchiseptica is motile, while B. pertussis and B. parapertussis are not; this can be ascribed to the fact that several genes encoding motility- or chemotaxis-related functions are inactivated or deleted in B. pertussis and B. parapertussis (71). B. avium appears to have a complete and intact set of motility and chemotaxis genes. With regard to its role in virulence, motility appears to be completely shut off above 40°C, approaching the body temperature of turkeys (102). Kersters et al. (53) previously reported a significantly greater number of motile rods in cultures grown at room temperature than in those grown at 35°C. Motility in B. avium has an effect on bird-to-bird transmission but has no effect on the 50% infective dose in turkeys (102). This suggests a role in environmental spread that may also be the case in B. bronchiseptica (1, 2). A B. avium bvgS mutant (95) (see below) exhibits motility identical to the wild type at 35°C and 42°C (unpublished data); thus, unlike B. bronchiseptica (2), motility of B. avium may not be regulated by the BvgAS two-component system.
Toxins.
As expected from previous reports (36), the two gene clusters encoding components of the pertussis toxin (PTX) and adenylate cyclase (cya) and their respective type IV and type I secretion systems are not present in the B. avium genome. Adenylate cyclase is produced by all species of Bordetella that are pathogens of mammals. On the other hand, PTX is produced by the human pathogen B. pertussis but not B. bronchiseptica and B. parapertussis, in spite of the presence of all the genes encoding PTX components. Thus, it appears that these two toxins are not required for infection of birds, and hence, upon its adaptation to these hosts, B. avium may have lost the genes encoding PTX and Cya. Alternatively, these toxin genes could have been acquired by the common ancestor of the mammalian species of Bordetella since diverging from the ancestor of B. avium.
The gene encoding another Bordetella toxin, dermonecrotic toxin (dnt), is present in the B. avium genome but at a location different from those of the previously sequenced species. The genes flanking the dnt gene in B. avium and the sequenced bordetellae are different. Although they are 41% identical, they may have been independently acquired and hence not true orthologues. DNT production is controlled by the presence of nicotinic acid or magnesium sulfate (36), implicating the bvgAS system (see below) in its expression control; however, bvgA and bvgS insertion mutants still produce DNT (unpublished observations). In contrast, a transposon insertion that abolished the production of DNT in strain WBA16 (102) was mapped in BAV0989, which encodes a LysR-family regulator (unpublished data). There are 54 LysR-type transcriptional regulators in the B. avium genome, many of which have orthologues in the previously sequenced bordetellae; interestingly, BAV0989 does not have an orthologue in those species. The DNT-negative strain is avirulent in the turkey poult model (102). These data indicate that global regulation of virulence genes in B. avium may be different from the other bordetellae.
Tracheal cytotoxin (TCT) is a glycopeptide cell wall fragment that is released in large quantities by B. pertussis in particular, to a lesser extent by B. bronchiseptica and B. parapertussis, and in small but unquantified amounts by B. avium (36). This molecule is cytotoxic in cell cultures via induction of nitric oxide biosynthesis pathways (39, 45). AmpG permease (ampG) is required in E. coli (17) and presumably other gram-negative bacteria for recycling of these cell wall anhydromuropeptides; this gene is present in the previously sequenced bordetellae. A gene similar to ampG occurs in B. avium, but in B. pertussis, this gene has an insertion sequence immediately upstream that may affect transcription (71), offering a potential explanation for the copious quantities of TCT produced in culture supernatants of B. pertussis. Whether TCT released by B. avium is significant in the pathogenesis of bordetellosis is unknown.
BvgAS system.
The two-component regulatory system BvgAS is the master expression regulator of many virulence factors and is the basis of phase variation in Bordetella mammalian pathogens (25). In response to environmental signals, the inner membrane sensor histidine kinase, BvgS, initiates a complex phosphorelay involving three of its domains, the transmitter, receiver, and C terminus, before phosphorylating the response regulator, BvgA, which then acts as a transcriptional regulator of a number of genes. Translational frameshifting at a poly(C) site near the 3′ end of bvgS, between codons for two of the phosphorelay domains, is responsible for phase variation via slip-strand mispairing in B. pertussis (98). Phase variation has been reported for B. avium strain 197 (37), its derivative strain 197N (the sequenced strain), and the type strain, ATCC 25086 (unpublished data). Interestingly, the B. avium phase variants have been reported to revert to the wild-type phenotype upon passage through the natural host, turkey (37). The molecular basis for phase variation in B. avium is unknown.
Unlike in the previously reported Bordetella genomes, the bvgAS genes in B. avium are not linked to the fhaB-fimABCD-fhaC gene cluster (this report and reference 95). Compared to those of B. bronchiseptica, the B. avium bvgAS genes vary in similarity in different regions of the predicted proteins (95). However, in the predicted BvgA protein, the phosphorylation site (D54) and helix-turn-helix motif are both retained, and in the predicted B. avium BvgS protein, the conserved phosphorelay sites in the putative transmitter (H729), receiver (D1023), and C terminus (H1172) domains (25) are also conserved.
The B. avium bvgS gene in the sequenced strain 197N has a frameshift within a homopolymeric tract of 8 cytosine residues in a location similar to the tract in B. pertussis described previously (98). This tract occurs between the predicted second and third phosphorylation sites described above. The C(8) tract appears in all clones of this region that were sequenced, and it predicts an earlier termination of the protein compared to other sequenced bordetellae. The presence of this bvgS variant in virulent strain 197N might lead to the conclusion that the B. avium bvgS gene is a pseudogene and that the bvgAS system is not involved in virulence in B. avium. However, Spears et al. (95) showed that bvgS was essential for virulence by using STM of strain 197N. To address this conundrum, we analyzed the poly(C) tract from chromosomal amplicons or cloned fragments and made the following observations (Table 3). (i) All PCR products from a number of different B. avium strain 197N derivatives that had been constructed and stored over the last 10 years had identical C(8) tracts, as in the sequenced strain. This group included the STM-derived strain PAS356, a nonvirulent mutant with a transposon insertion in bvgS (95), thus indicating that the shorter predicted BvgS protein in strain 197N is functional. To test the possibility that in vivo growth could select or enrich for bvgS mutants with an altered poly(C) tract (e.g., of 7 or 4 residues), we inoculated 1-week-old turkey poults with 197N (102) and sequenced bacterial DNA directly from tracheal swabs of infected turkeys taken 2 weeks after inoculation. We found that five of five amplicons tested contained C(8) tracts. Clearly, there was no in vivo selection for a frameshift in this tract of Cs, at least under the conditions we employed. However, a bvgAS clone from a chromosomal library prepared from strain 197N in 1996 was sequenced and had a C(7) tract and thus a predicted full-length protein product (95). (ii) A PCR amplicon of the bvgS gene from an older strain, WBA16 (102), constructed shortly after 197N was derived from 197 about 12 years ago, had a C(4) tract predicting the longer BvgS. (iii) Similarly, eight different B. avium isolates other than 197N derivatives, including ATCC type strain 35086, isolated at different times in different geographical regions all had C(4) tracts. It is important to note that we have not tested the virulence of those strains with C(4) or C(7) tracts under controlled in vivo conditions that we routinely employ.
TABLE 3.
Number of C residues in the bvgS poly(C) tract in several B. avium strains
| DNA source (year prepared) | Type of DNA sequenced | No. of C residues in poly(C) tract |
|---|---|---|
| 197N (2003) | Chromosomal library clone | 8 |
| 197N passaged through turkeys (2004) | Amplicons | 8 |
| 197N (1996) | Chromosomal library clone | 7 |
| DNT-negative mutant derived from 197N (1993) | Amplicon | 4 |
| 197N derivatives (five independent strains) (1995 to 2003) | Amplicons | 8 |
| B. avium strains (n = 8) other than 197N, minimally propagated in the lab | Amplicons | 4 |
These findings may warrant a testing of 197N constructed so as to have a bvgS C(4) or C(7) tract in vivo, since the shorter version of BvgS predicted in strain 197N would lack the third phosphorelay residue (H1172) that is essential for phosphorylation of BvgA (107) in the other bordetellae. It is possible that the C-terminal portion of BvgS could be provided in trans from an alternative translational start site; in trans complementation of phosphorylation of BvgA by the C-terminal peptide was shown in vitro by Uhl and Miller (106). A potential start codon occurs in the proper reading frame 18 amino acids before the histidine residue in question. At present, the simplest interpretation of the data suggests that B. avium, in contrast to the other members of the bordetellae, produces a BvgS product that functions in a unique manner, in which the terminal portion may be of less importance in the overall functioning of the molecule in the pathogenesis of bordetellosis.
In addition to the activation of a set of genes, the BvgAS two-component system of B. pertussis, B. parapertussis, and B. bronchiseptica represses the expression of another set of genes that are not expressed in the virulent state: virulence-repressed genes. This repression occurs via the activation of expression of a negative regulator, BvgR (68). In the previously sequenced bordetellae, the bvgR gene is located downstream of the bvgAS locus but is transcribed in the opposite orientation from it. In B. avium, no orthologue of bvgR is present. BvgR contains the EAL domain (PF00563) that is found in diverse bacterial signaling proteins. There are 10 CDSs in the B. avium genome that have this domain. One in particular, BAV1874, is immediately upstream of the bvgA orthologue. It is possible that this B. avium CDS encodes a regulatory protein that performs a function similar to BvgR of the other sequenced bordetellae.
Another group of genes expressed in the “intermediate” phase is best characterized for B. bronchiseptica and may represent a state essential for transmission between hosts (24). The best-studied member of this group is BipA, a cell surface protein with repetitive regions (99). The B. avium CDS, BAV0900, encodes a protein similar to BipA in B. bronchiseptica, but it differs from the latter in the number of internal repeats and does not appear to be a true bipA orthologue.
Secretion systems.
Secretion of proteins into the extracellular environment or target cells is required for various aspects of the lifestyle of pathogenic bacteria. In gram-negative bacteria, at least six major transport machineries have been identified (type I to VI; for a recent review, see reference 55). All of these systems except type II are present in the previously sequenced bordetellae (71). In contrast, B. avium possesses all of the systems except type III and IV, which are important to virulence of other bordetellae (type III for B. bronchiseptica [117] and type IV for B. pertussis [19, 85]). Unique in B. avium compared to the previously sequenced bordetellae is a gene cluster that potentially encodes a type II secretion system (BAV0331-BAV0345). However, the proteins that may be exported through the type II secretion system in B. avium are unknown. Also, the putative type I system genes are located next to large genes encoding proteins similar to virulence factors in other bacteria mentioned above.
Iron acquisition.
In animals, iron (Fe), an essential nutrient, is sequestered in high-affinity Fe-binding complexes, such as transferrin, lactoferrin, and hemoproteins (5), leaving the estimated free Fe concentration in biological fluids as low as 10−18 M (43, 115). To survive within the Fe-limited environment of the host, bacterial pathogens have evolved different mechanisms to acquire Fe (90), including contact-dependent mechanisms, which involve specific outer membrane (OM) receptors that directly bind the host Fe source (22, 23, 41), and contact-independent mechanisms, which involve specific OM receptors that bind to and transport Fe-bound siderophores. Energy required to transport host-derived Fe or heme across the OM is provided by the proton motive force of the cytoplasmic membrane via the TonB-ExbB-ExbD proteins (13). TonB has been shown to be essential for virulence in diverse organisms, including B. pertussis (77, 86, 101, 104).
The genome of B. bronchiseptica encodes 16 putative (71) or experimentally described (9-11, 14, 78, 109) TonB-dependent ferric complex receptors (42). In comparison, analysis of the B. avium genome revealed only eight genes encoding putative or described (69) TonB-dependent ferric complex receptors, which are listed in Table 4.
TABLE 4.
Genes predicted or shown to be involved in iron uptake by B. avium
| Locus | Homologue | Known or predicted function (reference) |
|---|---|---|
| BAV0191 | Unique | Transferrin binding protein (34) |
| BAV0634 | Unique; pbuA like | Siderophore receptor |
| BAV1197 | Unique; ptuA like | Siderophore receptor |
| BAV3134 | bhuR | OM heme receptor (69) |
| BAV3376 | hemC | OM heme receptor (103) |
| BAV1209 | bfrH | Siderophore receptor |
| BAV1854 | bfeA | Siderophore receptor (12) |
| BAV2512 | bfrB | Siderophore receptor (9) |
Since expression of bacterial Fe uptake systems is often regulated in response to the level of Fe in the microenvironment (61), we predicted that the TonB-dependent ferric complex receptors would be upregulated under conditions of Fe limitation. To address this hypothesis, outer membrane fractions of B. avium cultured under Fe-stressed conditions were analyzed, showing that five OMPs of between 70 and 100 kDa are upregulated by Fe-stressed growth (Fig. 6) (20). Three of these outer membrane proteins, BfrB, BfrH, and BfeA, have been identified by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) spectroscopy as homologues of ferric siderophore receptors (data not shown). The heme receptor BhuR, expressed as 91- and 82-kDa forms, is also expressed under Fe-stressed culture conditions (Fig. 6) (69). To confirm that the outer membrane proteins were correctly identified by MALDI-TOF analysis, genes encoding BfrB, BfrH, and BfeA were disrupted by plasmid integration in a B. avium bhuR mutant. The mutants KO1bfrB, KO1bfrH, and KO1bfeA failed to express the outer membrane proteins encoded by bfrB, bfrH, or bfeA, respectively (Fig. 6).
FIG. 6.
Outer membrane protein profiles of B. avium mutants. Outer membranes isolated from iron-replete (+Fe) (36 μM FeSO4) or iron-stressed (-Fe) (50 μM EDDHA) cultures of B. avium 4169, KO1 (the bhuR mutant), and KO1 derivatives with gene interruptions in bfrB (KO1bfrB), bfrH (KO1bfrH), and bfeA (KO1bfeA) were resolved by SDS-PAGE and stained with Coomassie brilliant blue. The two forms of BhuR (69) are denoted by arrows in the 4169 lane; asterisks (*) denote the expected locations of the polypeptides encoded by bfrB, bfrH, and bfeA. Molecular mass standards are denoted in kDa.
Interestingly, the four TonB-dependent ferric complex receptors that were not identified in this experiment are encoded by the three B. avium-specific genes and hemC. It is possible that expression of these genes requires another induction condition(s) in addition to Fe limitation. In B. pertussis, expression of the TonB-dependent receptor BfrD is regulated by BvgAS as well as Fe (6, 72). Identifying the conditions needed for these genes to be expressed and the Fe-bound substrate of their protein products may shed light on mechanisms of host specificity in the bordetellae.
Prophages and integrated plasmid.
There are three prophages in the B. avium genome compared to four in the sequenced strain of B. bronchiseptica.
The B. avium prophage A (BAVΦ-A; BAV0391-BAV0443) has an internal region (BAV0416-BAV0430) that is highly similar to an internal region (Bbp12-Bbp27) of the B. bronchiseptica bacteriophage BPP1 that undergoes tropism switching (62). However, the B. avium prophage lacks the major tropism determinant and the reverse transcriptase responsible for the tropism switching.
Adjacent to BAVΦ-A there is a plasmid-like conjugative element (BAV0364-BAV0389). A similar element is inserted at the same location in the B. bronchiseptica genome. Both elements share highly similar conjugal transfer genes (vir/tra), but the B. avium version carries three genes encoding components of a type I restriction-modification system.
The B. avium prophage B (BAVΦ-B; BAV1280-BAV1342) is inserted at the 3′ end of the stable RNA gene ssrA. In the B. bronchiseptica genome, a different prophage occupies the same attachment site. There is evidence for a complete life cycle of the B. avium phage B due to the presence of phage-specific DNA in culture supernatants (data not shown). This analysis was done on DNA prepared after supernatants were treated with DNase, indicating that the DNA was protected by a capsid; however, electron microscopy on the supernatants has not been performed, and to date there has not been a sensitive strain identified for this phage. Phage Ba1 (91), which infects strain 197N, is >80% identical at the DNA level over two-thirds of its genome with prophage B (data not shown); Ba1 integrates at a different chromosomal site and confers a distinctly different immunity to Ba1 lysogens (91). Ba1 lysogens are still virulent, but 50% of spontaneous Ba1-resistant mutants are attenuated, and all resistant mutants have noticeable differences in LPS structure or amount (91).
The third B. avium prophage (BAVΦ-C; BAV1423-BAV1482) also shares an attachment site (tRNAleu) with another prophage in the B. bronchiseptica genome. These attachment sites therefore appear to represent preferred locations for the integration of mobile genetic elements in the bordetellae genomes.
Nutrition and metabolism.
B. avium, like the other sequenced bordetellae, cannot utilize glucose as a sole carbon or energy source (53). The genome reveals that the glycolytic pathway is incomplete due to the absence of genes encoding glucokinase and phosphofructokinase, similar to B. pertussis, B. parapertussis, and B. bronchiseptica. B. avium possesses genes that indicate a fully functional tricarboxylic acid cycle. Synthetic pathways for glutamate and aspartate appear to be complete, indicating that these amino acids are added to minimal medium for B. avium simply as sources of carbon and not to satisfy auxotrophic requirements (60). A nutritional requirement for cystine and methionine (53) is explained by the absence of a seven-gene cluster, present in other sequenced bordetellae, encoding components of the sulfate transport system (Sbp and CysUWA) and sulfate assimilation (CysHDN). The BAV3142-BAV3146 genes encode the components of a formate dehydrogenase, which requires selenocysteine. They are adjacent to CDSs encoding the components of the selenocysteine incorporation pathway; selenocysteinyl-tRNA(Sec) synthase (BAV3147; selA), selenocysteine-specific elongation factor (BAV3148; selB), selenide, water dikinase (BAV3149; selD), and the selenocysteil-tRNA (selC) itself. Both the formate dehydrogenase and the selenocysteine incorporation genes (except selD) are unique to B. avium. Other than these, there are few differences in the core metabolic genes, which probably reflects the similar nutritional environments of the niche inhabited by these organisms.
Conclusion.
The genome sequence of B. avium has confirmed the major differences previously observed between this organism and B. pertussis, B. parapertussis, and B. bronchiseptica. Most of the unique genes appear to encode surface or secreted elements, such as a variety of fimbriae, O antigen biosynthesis proteins, capsular or polysaccharide biosynthesis proteins, unique hemagglutinins, autotransporters, and two very large secreted proteins. These are most likely involved in attachment to and/or interactions with specific components of host cells and thus may provide the basis for avian host specificity. The surprising observation of a frameshift in a poly(C) tract in bvgS suggests that this region may provide a mechanism for phase variation, which is as yet poorly understood in this species and may operate differently from the BvgAS system previously characterized. Tantalizingly remaining to be elucidated are the components of B. avium that cause the clinical signs and pathology of bordetellosis, as the absence of genes encoding pertussis toxin and adenylate cyclase toxin was confirmed, but no other putative toxin genes were revealed by the sequence analysis. Nearly one-third of the predicted genes of B. avium do not have orthologues in B. bronchiseptica, confirming the previous phylogenetic analyses that place B. avium most distant from the previously sequenced Bordetella species and highlighting the diversity that can exist within named bacterial genera. As genome sequences of other bordetellae emerge, especially the environmental isolate B. petrii, the evolutionary relationships and history of the bacteria in this important genus will become clearer.
Acknowledgments
We acknowledge the support of the Wellcome Trust Sanger Institute core sequencing and informatics groups.
This work was supported by a grant to L. M. Temple and J. Parkhill from the USDA (539235), a grant to L. M. Temple and D. M. Miyamoto from NIH-AREA (526112), an NIH training grant (T32 DE007034) to N. D. King, and a grant to P. E. Orndorff and L. M. Temple from the USDA (2002-35204-12236).
REFERENCES
- 1.Akerley, B. J., P. A. Cotter, and J. F. Miller. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611-620. [DOI] [PubMed] [Google Scholar]
- 2.Akerley, B. J., D. M. Monack, S. Falkow, and J. F. Miller. 1992. The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J. Bacteriol. 174:980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, A., and D. Maskell. 1996. The identification, cloning and mutagenesis of a genetic locus required for lipopolysaccharide biosynthesis in Bordetella pertussis. Mol. Microbiol. 19:37-52. [DOI] [PubMed] [Google Scholar]
- 4.Allen, A. G., R. M. Thomas, J. T. Cadisch, and D. J. Maskell. 1998. Molecular and functional analysis of the lipopolysaccharide biosynthesis locus wlb from Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Mol. Microbiol. 29:27-38. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 6.Antoine, R., S. Alonso, D. Raze, L. Coutte, S. Lesjean, E. Willery, C. Locht, and F. Jacob-Dubuisson. 2000. New virulence-activated and virulence-repressed genes identified by systematic gene inactivation and generation of transcriptional fusions in Bordetella pertussis. J. Bacteriol. 182:5902-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arp, L. H., and N. F. Cheville. 1984. Tracheal lesions in young turkeys infected with Bordetella avium. Am. J. Vet. Res. 45:2196-2200. [PubMed] [Google Scholar]
- 8.Aussel, L., R. Chaby, K. Le Blay, J. Kelly, P. Thibault, M. B. Perry, and M. Caroff. 2000. Chemical and serological characterization of the Bordetella hinzii lipopolysaccharides. FEBS Lett. 485:40-46. [DOI] [PubMed] [Google Scholar]
- 9.Beall, B. 1998. Two iron-regulated putative ferric siderophore receptor genes in Bordetella bronchiseptica and Bordetella pertussis. Res. Microbiol. 149:189-201. [DOI] [PubMed] [Google Scholar]
- 10.Beall, B., P. K. Cassiday, and G. N. Sanden. 1995. Analysis of Bordetella pertussis isolates from an epidemic by pulsed-field gel electrophoresis. J. Clin. Microbiol. 33:3083-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beall, B., and T. Hoenes. 1997. An iron-regulated outer-membrane protein specific to Bordetella bronchiseptica and homologous to ferric siderophore receptors. Microbiology 143:135-145. [DOI] [PubMed] [Google Scholar]
- 12.Beall, B., and G. N. Sanden. 1995. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiology 141:3193-3205. [DOI] [PubMed] [Google Scholar]
- 13.Braun, V. 2003. Iron uptake by Escherichia coli. Front. Biosci. 8:s1409-s1421. [DOI] [PubMed] [Google Scholar]
- 14.Brickman, T. J., and S. K. Armstrong. 1999. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. J. Bacteriol. 181:5958-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carver, T. J., K. M. Rutherford, M. Berriman, M. A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422-3423. [DOI] [PubMed] [Google Scholar]
- 16.Cerdeno-Tarraga, A. M., S. Patrick, L. C. Crossman, G. Blakely, V. Abratt, N. Lennard, I. Poxton, B. Duerden, B. Harris, M. A. Quail, A. Barron, L. Clark, C. Corton, J. Doggett, M. T. Holden, N. Larke, A. Line, A. Lord, H. Norbertczak, D. Ormond, C. Price, E. Rabbinowitsch, J. Woodward, B. Barrell, and J. Parkhill. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307:1463-1465. [DOI] [PubMed] [Google Scholar]
- 17.Chahboune, A., M. Decaffmeyer, R. Brasseur, and B. Joris. 2005. Membrane topology of the Escherichia coli AmpG permease required for recycling of cell wall anhydromuropeptides and AmpC β-lactamase induction. Antimicrob. Agents Chemother. 49:1145-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung, A. M., K. M. Farizo, and D. L. Burns. 2004. Analysis of relative levels of production of pertussis toxin subunits and Ptl proteins in Bordetella pertussis. Infect. Immun. 72:2057-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connell, T. D., A. Dickenson, A. J. Martone, K. T. Militello, M. J. Filiatraut, M. L. Hayman, and J. Pitula. 1998. Iron starvation of Bordetella avium stimulates expression of five outer membrane proteins and regulates a gene involved in acquiring iron from serum. Infect. Immun. 66:3597-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cookson, B. T., P. Vandamme, L. C. Carlson, A. M. Larson, J. V. Sheffield, K. Kersters, and D. H. Spach. 1994. Bacteremia caused by a novel Bordetella species, “B. hinzii.” J. Clin. Microbiol. 32:2569-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornelissen, C. N., J. E. Anderson, I. C. Boulton, and P. F. Sparling. 2000. Antigenic and sequence diversity in gonococcal transferrin-binding protein A. Infect. Immun. 68:4725-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornelissen, C. N., and P. F. Sparling. 1994. Iron piracy: acquisition of transferrin-bound iron by bacterial pathogens. Mol. Microbiol. 14:843-850. [DOI] [PubMed] [Google Scholar]
- 24.Cotter, P. A., and V. J. DiRita. 2000. Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54:519-565. [DOI] [PubMed] [Google Scholar]
- 25.Cotter, P. A., and J. F. Miller. 2001. Bordetella, p. 619-674. In E. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 26.Coutte, L., R. Antoine, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2001. Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. EMBO J. 20:5040-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craig, L., M. E. Pique, and J. A. Tainer. 2004. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2:363-378. [DOI] [PubMed] [Google Scholar]
- 28.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cummings, C. A., M. M. Brinig, P. W. Lepp, S. van de Pas, and D. A. Relman. 2004. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J. Bacteriol. 186:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diavatopoulos, D. A., C. A. Cummings, L. M. Schouls, M. M. Brinig, D. A. Relman, and F. R. Mooi. 2005. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 1:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Fabio, J. L., M. Caroff, D. Karibian, J. C. Richards, and M. B. Perry. 1992. Characterization of the common antigenic lipopolysaccharide O-chains produced by Bordetella bronchiseptica and Bordetella parapertussis. FEMS Microbiol. Lett. 76:275-281. [DOI] [PubMed] [Google Scholar]
- 32.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Economou, A., W. D. Hamilton, A. W. Johnston, and J. A. Downie. 1990. The Rhizobium nodulation gene nodO encodes a Ca2(+)-binding protein that is exported without N-terminal cleavage and is homologous to haemolysin and related proteins. EMBO J. 9:349-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekins, A., F. Bahrami, A. Sijercic, D. Maret, and D. F. Niven. 2004. Haemophilus somnus possesses two systems for acquisition of transferrin-bound iron. J. Bacteriol. 186:4407-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleckenstein, J. M., D. J. Kopecko, R. L. Warren, and E. A. Elsinghorst. 1996. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 64:2256-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gentry-Weeks, C. R., B. T. Cookson, W. E. Goldman, R. B. Rimler, S. B. Porter, and R. Curtiss III. 1988. Dermonecrotic toxin and tracheal cytotoxin, putative virulence factors of Bordetella avium. Infect. Immun. 56:1698-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gentry-Weeks, C. R., D. L. Provence, J. M. Keith, and R. Curtiss III. 1991. Isolation and characterization of Bordetella avium phase variants. Infect. Immun. 59:4026-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geuijen, C. A., R. J. Willems, M. Bongaerts, J. Top, H. Gielen, and F. R. Mooi. 1997. Role of the Bordetella pertussis minor fimbrial subunit, FimD, in colonization of the mouse respiratory tract. Infect. Immun. 65:4222-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldman, W. E., and L. A. Herwaldt. 1985. Bordetella pertussis tracheal cytotoxin. Dev. Biol. Stand. 61:103-111. [PubMed] [Google Scholar]
- 40.Goodnow, R. A. 1980. Biology of Bordetella bronchiseptica. Microbiol. Rev. 44:722-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gray-Owen, S. D., and A. B. Schryvers. 1996. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 4:185-191. [DOI] [PubMed] [Google Scholar]
- 42.Griffiths, E., and P. Williams. 1999. The iron-uptake systems of pathogenic bacteria, fungi and protozoa, p. 87-212. In J. J. Bullen and E. Griffiths (ed.), Iron and infection: molecular, physiological and clinical aspects, 2nd ed. John Wiley and Sons, Chichester, England.
- 43.Griffiths, E. 1999. Iron in biological systems, p. 1-26. In J. J. Bullen and E. Griffiths (ed.), Iron and infection: molecular, physiological and clinical aspects, 2nd ed. John Wiley and Sons, Chichester, England.
- 44.Handley, P. S., F. F. Correia, K. Russell, B. Rosan, and J. M. DiRienzo. 2005. Association of a novel high molecular weight, serine-rich protein (SrpA) with fibril-mediated adhesion of the oral biofilm bacterium Streptococcus cristatus. Oral Microbiol. Immunol. 20:131-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heiss, L. N., T. A. Flak, J. R. Lancaster, Jr., M. L. McDaniel, and W. E. Goldman. 1993. Nitric oxide mediates Bordetella pertussis tracheal cytotoxin damage to the respiratory epithelium. Infect. Agents Dis. 2:173-177. [PubMed] [Google Scholar]
- 46.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinz, K. H., and G. Glunder. 1985. Occurrence of Bordetella avium sp. nov. and Bordetella bronchiseptica in birds. Berl. Munch. Tierarztl. Wochenschr. 98:369-373. (In German.) [PubMed] [Google Scholar]
- 48.Hinz, K. H., G. Glunder, and H. Luders. 1978. Acute respiratory disease in turkey poults caused by Bordetella bronchiseptica-like bacteria. Vet. Rec. 103:262-263. [DOI] [PubMed] [Google Scholar]
- 49.Hopkins, B. A., J. K. Skeeles, G. E. Houghten, D. Slagle, and K. Gardner. 1990. A survey of infectious diseases in wild turkeys (Meleagridis gallopavo silvestris) from Arkansas. J. Wildl. Dis. 26:468-472. [DOI] [PubMed] [Google Scholar]
- 50.Jacob-Dubuisson, F., B. Kehoe, E. Willery, N. Reveneau, C. Locht, and D. A. Relman. 2000. Molecular characterization of Bordetella bronchiseptica filamentous haemagglutinin and its secretion machinery. Microbiology 146:1211-1221. [DOI] [PubMed] [Google Scholar]
- 51.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kattar, M. M., J. F. Chavez, A. P. Limaye, S. L. Rassoulian-Barrett, S. L. Yarfitz, L. C. Carlson, Y. Houze, S. Swanzy, B. L. Wood, and B. T. Cookson. 2000. Application of 16S rRNA gene sequencing to identify Bordetella hinzii as the causative agent of fatal septicemia. J. Clin. Microbiol. 38:789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kersters, K., K.-H. Hinz, A. Hertle, P. Segers, A. Lievens, O. Siegmann, and J. De Ley. 1984. Bordetella avium sp. nov., isolated from the respiratory tracts of turkeys and other birds. Int. J. Syst. Bacteriol. 34:56-70. [Google Scholar]
- 54.Kimura, A., K. T. Mountzouros, D. A. Relman, S. Falkow, and J. L. Cowell. 1990. Bordetella pertussis filamentous hemagglutinin: evaluation as a protective antigen and colonization factor in a mouse respiratory infection model. Infect. Immun. 58:7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kostakioti, M., C. L. Newman, D. G. Thanassi, and C. Stathopoulos. 2005. Mechanisms of protein export across the bacterial outer membrane. J. Bacteriol. 187:4306-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 57.Larocque, S., J. R. Brisson, H. Therisod, M. B. Perry, and M. Caroff. 2003. Structural characterization of the O-chain polysaccharide isolated from Bordetella avium ATCC 5086: variation on a theme(1). FEBS Lett. 535:11-16. [DOI] [PubMed] [Google Scholar]
- 58.Lesse, A. J., A. A. Campagnari, W. E. Bittner, and M. A. Apicella. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Immunol. Methods 126:109-117. [DOI] [PubMed] [Google Scholar]
- 59.Leyh, R., and R. W. Griffith. 1992. Characterization of the outer membrane proteins of Bordetella avium. Infect. Immun. 60:958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leyh, R. D., R. W. Griffith, and L. H. Arp. 1988. Transposon mutagenesis in Bordetella avium. Am. J. Vet. Res. 49:687-692. [PubMed] [Google Scholar]
- 61.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu, M., M. Gingery, S. R. Doulatov, Y. Liu, A. Hodes, S. Baker, P. Davis, M. Simmonds, C. Churcher, K. Mungall, M. A. Quail, A. Preston, E. T. Harvill, D. J. Maskell, F. A. Eiserling, J. Parkhill, and J. F. Miller. 2004. Genomic and genetic analysis of Bordetella bacteriophages encoding reverse transcriptase-mediated tropism-switching cassettes. J. Bacteriol. 186:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 64.Martin, D., M. S. Peppler, and B. R. Brodeur. 1992. Immunological characterization of the lipooligosaccharide B band of Bordetella pertussis. Infect. Immun. 60:2718-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mattoo, S., and J. D. Cherry. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin. Microbiol. Rev. 18:326-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mattoo, S., J. F. Miller, and P. A. Cotter. 2000. Role of Bordetella bronchiseptica fimbriae in tracheal colonization and development of a humoral immune response. Infect. Immun. 68:2024-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mazengia, E., E. A. Silva, J. A. Peppe, R. Timperi, and H. George. 2000. Recovery of Bordetella holmesii from patients with pertussis-like symptoms: use of pulsed-field gel electrophoresis to characterize circulating strains. J. Clin. Microbiol. 38:2330-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Merkel, T. J., and S. Stibitz. 1995. Identification of a locus required for the regulation of bvg-repressed genes in Bordetella pertussis. J. Bacteriol. 177:2727-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murphy, E. R., R. E. Sacco, A. Dickenson, D. J. Metzger, Y. Hu, P. E. Orndorff, and T. D. Connell. 2002. BhuR, a virulence-associated outer membrane protein of Bordetella avium, is required for the acquisition of iron from heme and hemoproteins. Infect. Immun. 70:5390-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neuhoff, V., N. Arold, D. Taube, and W. Ehrhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255-262. [DOI] [PubMed] [Google Scholar]
- 71.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 72.Passerini de Rossi, B. N., L. E. Friedman, C. B. Belzoni, S. Savino, B. Arico, R. Rappuoli, V. Masignani, and M. A. Franco. 2003. Vir90, a virulence-activated gene coding for a Bordetella pertussis iron-regulated outer membrane protein. Res. Microbiol. 154:443-450. [DOI] [PubMed] [Google Scholar]
- 73.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 74.Pilione, M. R., E. J. Pishko, A. Preston, D. J. Maskell, and E. T. Harvill. 2004. pagP is required for resistance to antibody-mediated complement lysis during Bordetella bronchiseptica respiratory infection. Infect. Immun. 72:2837-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Porter, J. F., R. Parton, and A. C. Wardlaw. 1991. Growth and survival of Bordetella bronchiseptica in natural waters and in buffered saline without added nutrients. Appl. Environ. Microbiol. 57:1202-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Porter, J. F., and A. C. Wardlaw. 1993. Long-term survival of Bordetella bronchiseptica in lakewater and in buffered saline without added nutrients. FEMS Microbiol. Lett. 110:33-36. [DOI] [PubMed] [Google Scholar]
- 77.Pradel, E., N. Guiso, F. D. Menozzi, and C. Locht. 2000. Bordetella pertussis TonB, a Bvg-independent virulence determinant. Infect. Immun. 68:1919-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pradel, E., and C. Locht. 2001. Expression of the putative siderophore receptor gene bfrZ is controlled by the extracytoplasmic-function sigma factor BupI in Bordetella bronchiseptica. J. Bacteriol. 183:2910-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Preston, A., A. G. Allen, J. Cadisch, R. Thomas, K. Stevens, C. M. Churcher, K. L. Badcock, J. Parkhill, B. Barrell, and D. J. Maskell. 1999. Genetic basis for lipopolysaccharide O-antigen biosynthesis in bordetellae. Infect. Immun. 67:3763-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Preston, A., and D. Maskell. 2001. The molecular genetics and role in infection of lipopolysaccharide biosynthesis in the bordetellae. J. Endotoxin Res. 7:251-261. [PubMed] [Google Scholar]
- 81.Preston, A., D. Maskell, A. Johnson, and E. R. Moxon. 1996. Altered lipopolysaccharide characteristic of the I69 phenotype in Haemophilus influenzae results from mutations in a novel gene, isn. J. Bacteriol. 178:396-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Preston, A., E. Maxim, E. Toland, E. J. Pishko, E. T. Harvill, M. Caroff, and D. J. Maskell. 2003. Bordetella bronchiseptica PagP is a Bvg-regulated lipid A palmitoyl transferase that is required for persistent colonization of the mouse respiratory tract. Mol. Microbiol. 48:725-736. [DOI] [PubMed] [Google Scholar]
- 83.Preston, A., R. Thomas, and D. J. Maskell. 2002. Mutational analysis of the Bordetella pertussis wlb LPS biosynthesis locus. Microb. Pathog. 33:91-95. [DOI] [PubMed] [Google Scholar]
- 84.Raffel, T. R., K. B. Register, S. A. Marks, and L. Temple. 2002. Prevalence of Bordetella avium infection in selected wild and domesticated birds in the eastern USA. J. Wildl. Dis. 38:40-46. [DOI] [PubMed] [Google Scholar]
- 85.Rambow-Larsen, A. A., and A. A. Weiss. 2004. Temporal expression of pertussis toxin and Ptl secretion proteins by Bordetella pertussis. J. Bacteriol. 186:43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reeves, S. A., A. G. Torres, and S. M. Payne. 2000. TonB is required for intracellular growth and virulence of Shigella dysenteriae. Infect. Immun. 68:6329-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Relman, D. A., M. Domenighini, E. Tuomanen, R. Rappuoli, and S. Falkow. 1989. Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc. Natl. Acad. Sci. USA 86:2637-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 89.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 90.Schaible, U. E., and S. H. Kaufmann. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2:946-953. [DOI] [PubMed] [Google Scholar]
- 91.Shelton, C. B., D. R. Crosslin, J. L. Casey, S. Ng, L. M. Temple, and P. E. Orndorff. 2000. Discovery, purification, and characterization of a temperate transducing bacteriophage for Bordetella avium. J. Bacteriol. 182:6130-6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shelton, C. B., L. M. Temple, and P. E. Orndorff. 2002. Use of bacteriophage Ba1 to identify properties associated with Bordetella avium virulence. Infect. Immun. 70:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sherlock, O., M. A. Schembri, A. Reisner, and P. Klemm. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 186:8058-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Skeeles, J. K., and L. H. Arp. 1997. Bordetellosis (turkey coryza), p. 275-288. In H. J. Barnes, B. W. Calnek, C. W. Beard, L. R. McDougald, and Y. M. Saif (ed.), Diseases of poultry. Iowa State University Press, Ames, Iowa.
- 95.Spears, P. A., L. M. Temple, D. M. Miyamoto, D. J. Maskell, and P. E. Orndorff. 2003. Unexpected similarities between Bordetella avium and other pathogenic bordetellae. Infect. Immun. 71:2591-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Spears, P. A., L. M. Temple, and P. E. Orndorff. 2000. A role for lipopolysaccharide in turkey tracheal colonization by Bordetella avium as demonstrated in vivo and in vitro. Mol. Microbiol. 36:1425-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Spiers, A. J., S. G. Kahn, J. Bohannon, M. Travisano, and P. B. Rainey. 2002. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 161:33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stibitz, S., W. Aaronson, D. Monack, and S. Falkow. 1989. Phase variation in Bordetella pertussis by frameshift mutation in a gene for a novel two-component system. Nature 338:266-269. [DOI] [PubMed] [Google Scholar]
- 99.Stockbauer, K. E., B. Fuchslocher, J. F. Miller, and P. A. Cotter. 2001. Identification and characterization of BipA, a Bordetella Bvg-intermediate phase protein. Mol. Microbiol. 39:65-78. [DOI] [PubMed] [Google Scholar]
- 100.Sweet, C. R., A. Preston, E. Toland, S. M. Ramirez, R. J. Cotter, D. J. Maskell, and C. R. Raetz. 2002. Relaxed acyl chain specificity of Bordetella UDP-N-acetylglucosamine acyltransferases. J. Biol. Chem. 277:18281-18290. [DOI] [PubMed] [Google Scholar]
- 101.Takase, H., H. Nitanai, K. Hoshino, and T. Otani. 2000. Requirement of the Pseudomonas aeruginosa tonB gene for high-affinity iron acquisition and infection. Infect. Immun. 68:4498-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Temple, L. M., A. A. Weiss, K. E. Walker, H. J. Barnes, V. L. Christensen, D. M. Miyamoto, C. B. Shelton, and P. E. Orndorff. 1998. Bordetella avium virulence measured in vivo and in vitro. Infect. Immun. 66:5244-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thomas, C. E., B. Olsen, and C. Elkins. 1998. Cloning and characterization of tdhA, a locus encoding a TonB-dependent heme receptor from Haemophilus ducreyi. Infect. Immun. 66:4254-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 106.Uhl, M. A., and J. F. Miller. 1996. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component phosphorelay. J. Biol. Chem. 271:33176-33180. [DOI] [PubMed] [Google Scholar]
- 107.Uhl, M. A., and J. F. Miller. 1996. Integration of multiple domains in a two-component sensor protein: the Bordetella pertussis BvgAS phosphorelay. EMBO J. 15:1028-1036. [PMC free article] [PubMed] [Google Scholar]
- 108.Vandamme, P., M. Heyndrickx, M. Vancanneyt, B. Hoste, P. De Vos, E. Falsen, K. Kersters, and K. H. Hinz. 1996. Bordetella trematum sp. nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans Rüger and Tan 1983. Int. J. Syst. Bacteriol. 46:849-858. [DOI] [PubMed] [Google Scholar]
- 109.Vanderpool, C. K., and S. K. Armstrong. 2001. The Bordetella bhu locus is required for heme iron utilization. J. Bacteriol. 183:4278-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.van der Zee, A., F. Mooi, J. Van Embden, and J. Musser. 1997. Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J. Bacteriol. 179:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vinogradov, E. 2002. Structure of the O-specific polysaccharide chain of the lipopolysaccharide of Bordetella hinzii. Carbohydr. Res. 337:961-963. [DOI] [PubMed] [Google Scholar]
- 112.von Wintzingerode, F., G. Gerlach, B. Schneider, and R. Gross. 2002. Phylogenetic relationships and virulence evolution in the genus Bordetella. Curr. Top. Microbiol. Immunol. 264:177-199. [PubMed] [Google Scholar]
- 113.von Wintzingerode, F., A. Schattke, R. A. Siddiqui, U. Rosick, U. B. Gobel, and R. Gross. 2001. Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int. J. Syst. Evol. Microbiol. 51:1257-1265. [DOI] [PubMed] [Google Scholar]
- 114.Weyant, R. S., D. G. Hollis, R. E. Weaver, M. F. Amin, A. G. Steigerwalt, S. P. O'Connor, A. M. Whitney, M. I. Daneshvar, C. W. Moss, and D. J. Brenner. 1995. Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J. Clin. Microbiol. 33:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wienk, K. J., J. J. Marx, and A. C. Beynen. 1999. The concept of iron bioavailability and its assessment. Eur. J. Nutr. 38:51-75. [DOI] [PubMed] [Google Scholar]
- 116.Yih, W. K., E. A. Silva, J. Ida, N. Harrington, S. M. Lett, and H. George. 1999. Bordetella holmesii-like organisms isolated from Massachusetts patients with pertussis-like symptoms. Emerg. Infect. Dis. 5:441-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yuk, M. H., E. T. Harvill, P. A. Cotter, and J. F. Miller. 2000. Modulation of host immune responses, induction of apoptosis and inhibition of NF-kappaB activation by the Bordetella type III secretion system. Mol. Microbiol. 35:991-1004. [DOI] [PubMed] [Google Scholar]