Abstract
In Pseudomonas aeruginosa, the GacS/GacA two-component system positively controls the quorum-sensing machinery and the expression of extracellular products via two small regulatory RNAs, RsmY and RsmZ. An rsmY rsmZ double mutant and a gacA mutant were similarly impaired in the synthesis of the quorum-sensing signal N-butanoyl-homoserine lactone, the disulfide bond-forming enzyme DsbA, and the exoproducts hydrogen cyanide, pyocyanin, elastase, chitinase (ChiC), and chitin-binding protein (CbpD). Both mutants showed increased swarming ability, azurin release, and early biofilm development.
In many gram-negative bacteria, the GacS/GacA two-component system regulates the expression of extracellular products, and these can be virulence determinants in pathogenic species. Although the signals that activate the GacS/GacA system have not been identified, it is evident from studies in Escherichia coli, Vibrio cholerae, Erwinia carotovora, Legionella pneumophila, Pseudomonas fluorescens, and other bacteria that activation takes place when cells grow to high densities, particularly during the transition from exponential to stationary phase (9, 13, 14, 16, 21, 45). Therefore, GacS/GacA-dependent gene regulation can be considered part of the quorum-sensing machinery (15, 16). The activated response regulator GacA strongly turns on the transcription of one or several genes encoding small RNAs, termed csrB/csrC (in E. coli and Salmonella enterica), csrB/csrC/csrD (in V. cholerae), rsmB (in E. carotovora), and rsmX/rsmY/rsmZ (in P. fluorescens) (7, 13, 14, 16, 45). These small regulatory RNAs have a high affinity for certain RNA-binding proteins that act as translational repressors, e.g., CsrA (in E. coli, S. enterica, and V. cholerae), RsmA (in E. carotovora), and RsmA/RsmE (in P. fluorescens) (7, 16, 17, 29, 30). Repeated ANGGA motifs (where N is any nucleotide) in target mRNAs are binding sites for CsrA, and when the most distal of these motifs coincides with the ribosome-binding site, translation of the mRNA can be repressed (4). The small Csr/Rsm RNAs have flower-like secondary structures with multiple GGA motifs in unpaired regions (10, 14, 16, 45). In the case of RsmY of P. fluorescens, it has been shown that the GGA repeats are essential for interaction with RsmA and RsmE (40).
In the opportunistic pathogen Pseudomonas aeruginosa, the Gac/Rsm system positively regulates the expression of the quorum-sensing signal N-butanoyl-homoserine lactone (C4-HSL) and of extracellular virulence factors, such as hydrogen cyanide (HCN), pyocyanin, and elastase (26, 28). P. aeruginosa gacA mutants are less virulent in animal and plant models in comparison with the wild type (27). In the Gac/Rsm signal transduction pathway, GacA is required for the transcription of the small RNA gene rsmZ. RsmZ RNA acts as an antagonist of the unique RNA-binding protein RsmA, which negatively controls the expression of quorum sensing and several extracellular products (2, 12, 26). During exoproduct expression, the Gac/Rsm system can have a dual mode of action. In the case of HCN biosynthesis, for example, about 30% of the Gac/Rsm input is due to enhanced expression of the rhlI gene encoding C4-HSL synthase. C4-HSL activates the transcription regulator RhlR which, together with the transcription factors LasR and ANR, turns on the transcription of the HCN biosynthetic genes hcnABC. About 70% of the Gac/Rsm input bypasses N-acyl-homoserine lactone-dependent transcriptional regulation and results directly in enhanced translation of hcnABC mRNA (24, 25).
Transcriptional regulation of rsmZ in P. aeruginosa is complex. In addition to GacS, the sensors RetS and LadS also determine the expression of the rsmZ gene. When RetS is deficient, P. aeruginosa cells clump and their type III secretion system is switched off. Deficiency of either GacS or LadS results in reduced biofilm maturation and increased type III secretion. GacS and LadS positively regulate rsmZ expression, whereas RetS has the opposite effect (8, 41). Furthermore, the rsmZ promoter is positively controlled by RsmA; this effect is probably indirect (12).
In V. cholerae and P. fluorescens, the GacS/GacA system activates the transcription of three functionally redundant small RNAs, whereas in E. coli and S. enterica two GacA-controlled small RNAs have been described (7, 14, 16, 45). By contrast, only one regulatory RNA that depends on GacS/GacA has been described in E. carotovora (17). Here we examine the situation in P. aeruginosa and we present evidence that the GacS/GacA system functions with two small RNAs, RsmZ and the newly characterized RsmY regulator (36).
Features of RsmY small RNA and construction of an rsmY mutant.
The rsmY gene of P. aeruginosa, which has 69% nucleotide sequence identity with that of P. fluorescens CHA0 (10, 39), is located between the dnr gene and open reading frame PA0528 on the chromosome of P. aeruginosa PAO1 (Fig. 1A). The rsmY promoter contains a conserved upstream activating sequence (UAS) which is characteristic for GacA-dependent small RNA genes (16, 39). The probable rsmY transcription start site can be deduced from that determined in P. fluorescens (10, 39). RsmY RNA (124 nucleotides) is predicted to contain four major stem-loop structures (10) and six GGA motifs (Fig. 1A) in single-stranded parts of the molecule. Unpaired GGA repeats are a hallmark of small RNAs that titrate RsmA/CsrA (10, 16, 30). A 96-bp deletion was introduced into the rsmY gene of P. aeruginosa PAO1 (Fig. 1A) by homologous recombination (26, 28), using E. coli S17-1/pME3087ΔrsmY (Table 1) as a donor and PAO1 as a recipient. In nutrient yeast broth (NYB), the resulting mutant PAO6420 had the same growth rate as the wild-type PAO1 (data not shown). According to genomic sequence data, strains PAO1 and PA14 (http://www.pseudomonas.com; http://ausubellab.mgh.harvard.edu/pa14sequencing) contain two genes encoding GacA-dependent small regulatory RNAs, rsmY and rsmZ (2, 12, 36), but no homolog of the rsmX gene, which codes for a third GacA-regulated small RNA in P. fluorescens (14).
FIG. 1.
Expression of the rsmY gene in P. aeruginosa. (A) Organization of the rsmY region in strain PAO1. The deduced −35 and −10 promoter sites are indicated with gray boxes. The palindromic sequence boxed from −75 to −58 denotes an upstream regulatory sequence (UAS), which is highly conserved in GacA-regulated genes (16, 39). Arrows indicate the putative rsmY transcription terminator. The 96-bp deletion in the rsmY gene (underlined) of the mutants PAO6420 and PAO6421 was verified by PCR and Southern blotting (data not shown). Conserved GGA motifs are shown in boldface. (B) Regulation of rsmY and rsmZ expression. The Northern blot shows the differential temporal accumulation of RsmY and RsmZ RNAs in PAO1 (wild type), PAO6354 (ΔrsmZ), PAO6420 (ΔrsmY), PAO6281 (gacA), and PAZH13 (rsmA). Total RNA was extracted from cells grown at 37°C in 200 ml of NYB, electrophoretically separated on a denaturing urea-polyacrylamide gel (8.3 M urea, 8% [wt/vol] acrylamide, 0.4% [wt/vol] bisacrylamide) in 50 mM Tris-borate, pH 8.3, 1 mM EDTA and transferred to a Hybond N membrane (39). Hybridizations were done as described elsewhere (39) with digoxigenin (DIG)-labeled DNA probes for RsmY and RsmZ, produced by PCR with Hotstar Taq polymerase (QIAGEN), DIG-labeled deoxynucleoside triphosphates (Roche), and primer pairs BH1/BH2 and PRSMZ1/PRSMZ2, respectively. Each lane was loaded with 3 μg of total RNA and checked by monitoring the intensities of 23S and 16S rRNAs (not shown).
TABLE 1.
Bacterial strains, plasmids, and primers
| Strain, plasmid, or primer | Genotype, phenotype, or sequencea | Reference or origin |
|---|---|---|
| E. coli strain | ||
| S17-1 | F−pro thi hsdR recA; chromosome::RP4-2 Tc::Mu Km::Tn7 Tpr Spr | 35 |
| P. aeruginosa strains | ||
| PAO1 | Wild type | ATCC 15692 |
| PAO6281 | gacA::Ω-Sm/Sp; Sm/Spr | 28 |
| PAO6354 | ΔrsmZ | 12 |
| PAO6420 | ΔrsmY | This study |
| PAO6421 | ΔrsmY ΔrsmZ | This study |
| PAO6554 | rsmZ-lacZ transcriptional fusion in mini-Tn7; Gmr | This study |
| PAO6555 | gacA::Ω-Sm/Sp, rsmZ-lacZ fusion in mini-Tn7; Sm/Spr Gmr | This study |
| PAO6556 | ΔrsmY ΔrsmZ, rsmZ-lacZ fusion in mini-Tn7; Gmr | This study |
| PAO6557 | ΔrsmA, rsmZ-lacZ fusion in mini-Tn7; Gmr | This study |
| PAO6558 | rsmY-lacZ transcriptional fusion in mini-Tn7; Gmr | This study |
| PAO6559 | gacA::Ω-Sm/Sp, rsmY-lacZ fusion in mini-Tn7; Sm/Spr Gmr | This study |
| PAO6567 | rsmY ΔrsmZ, rsmY-lacZ fusion in mini-Tn7; Gmr | This study |
| PAO6568 | ΔrsmA, rsmY-lacZ fusion in mini-Tn7; Gmr | This study |
| PAZH13 | ΔrsmA | 26 |
| Plasmids | ||
| pBluescript II SK, KS | Cloning vectors, ColE1 replicons; Apr | Stratagene |
| pME3087 | Suicide vector, ColE1 replicon, IncP-1, Mob; Tcr | 44 |
| pME3087ΔrsmY | pME3087 containing insert of pME3830ΔrsmY | This study |
| pME3280a | Mini-Tn7 gene delivery vector; Gmr | 48 |
| pME3328 | pBluescript II KS containing a 1.43-kb BamHI-XhoI fragment with ′rpoS rsmZ fdxA′; Apr | 12 |
| pME3331 | pME6016 derivative containing a transcriptional rsmZ-lacZ fusion; Tcr | 12 |
| pME3830 | pBluescript II SK containing a 1.3-kb PstI-StuI PCR fragment with ′dnr rsmY and PA0528′, obtained with primers PAOTRR1 and PAOTRR2; Apr | This study |
| pME3830ΔrsmY | pME3830 with a 96-bp deletion in rsmY, obtained by inverse PCR with primers Δ1 and Δ2 | This study |
| pME3843 | pME6010 containing a translational hcnA′-′lacZ fusion under the tac promoter | 25 |
| pME3859 | pME6010 containing a translational rsmA′-′lacZ fusion | 26 |
| pME3897 | pME6182 containing a 3.3-kb BamHI-XhoI fragment of pME3331 with a transcriptional rsmZ-lacZ fusion in the SmaI site of mini-Tn7; Gmr | This study |
| pME3898 | pME6182 containing a 3.3-kb EcoRI-XhoI fragment of pME7311 with a transcriptional rsmY-lacZ fusion in the SmaI site of mini-Tn7; Gmr | This study |
| pME6015 | Cloning vector for translational ′lacZ fusions; pVS1- p15A replicon; Tcr | 33 |
| pME6016 | Cloning vector for transcriptional lacZ fusions, pVS1-p15A replicon; Tcr | 33 |
| pME6182 | Mini-Tn7 gene delivery vector based on pME3280a, HindIII-SmaI-KpnI-NcoI-SphI MCS, ColE1 replicon; Gmr Apr | C. Reimmann |
| pME7311 | pME6016 derivative containing a 216-bp EcoRI-BamHI fragment (amplified with primers PrsmY1 and PrsmY2) carrying the rsmY promoter fused at the putative +5 site to the +1 site of lacZ; Tcr | This study |
| pME7321 | pBluescript II KS containing a 0.85-kb EcoRI-BamHI fragment with the rpsA upstream region and the first eight rpsA codons (amplified with primers PRPSA1 and PRPSA2); Apr | This study |
| pME7322 | pME6015 derivative with a 0.85-kb EcoRI-BamHI rpsA upstream fragment and the first eight codons fused in frame with the ′lacZ gene | This study |
| pUX-BF13 | Helper plasmid containing Tn7 transposition functions, R6K replicon; Apr | 1 |
| Primers (5′ → 3′) | ||
| PAOTRR1 | CTGTTCACTCGAAGCACTCC located in dnr, upstream of the rsmY region | |
| PAOTRR2 | TTCGCCAACTCCGCTATTTC located in PA0528, downstream of the rsmY region | |
| PrsmY1 | TTCCTGGAGCTGGACGGG located in dnr, upstream of the rsmY region | |
| PrsmY2 | CGCAGGATCCTGACGGTTTGAAGATTACGC with a BamHI restriction site (underlined), located in the +1 transcription start region of rsmY | |
| Δ1 | TAGAGATATCCAAAACCCCGCCCAAAAGGC with an EcoRV restriction site (underlined), located upstream of the rsmY terminator | |
| Δ2 | GCGTCTCTACGCATTAGAAGATATCCAGT with an EcoRV restriction site (underlined), located downstream of the rsmY transcription start site | |
| BH1 | ATCATCTTGACGCACGGCAAGC located upstream of rsmY (−148 to −128) | |
| BH2 | TCTGAGCGACGCGGTTTTCC located downstream of the rsmY terminator (+136 to +155) | |
| PRSMZ1 | CTAACAGGGAACACGCAACC, corresponding to the +1 site and the next 20 bp of rsmZ | |
| PRSMZ2 | AAAAAAAGGGGCGGGGTATT located in the terminator of rsmZ | |
| PRPSA1 | AAAAaGGATCCGAGTTCTGCGAAGCTTTCGCTC, with a BamHI restriction site (underlined), located downstream of the first codon of rpsA | |
| PRPSA2 | AAAAAGAATTCCGCGGTCAACCATGGTGTCGAC with an EcoRI restriction site (underlined), located in the cmk gene (PA3163) upstream of rpsA |
MCS, multiple cloning site; Apr, ampicillin resistance; Gmr, gentamicin resistance; Smr, streptomycin resistance; Spr, spectinomycin resistance; Tcr, tetracycline resistance; Tpr, trimethoprim resistance.
Expression of RsmY and RsmZ.
When hybridized to a specific rsmY probe in a Northern blot experiment (Fig. 1B), two bands of RsmY RNA were revealed in a denaturing gel, possibly corresponding to molecules with and without the terminator stem-loop structure. Mutations in gacA or rsmA markedly reduced the amount of RsmY transcript; the effect was even stronger in the rsmA mutant than in the gacA mutant. By contrast, deletion of rsmZ had little effect (Fig. 1B). The abundance of RsmZ, which was monitored in parallel, showed an analogous pattern (Fig. 1B).
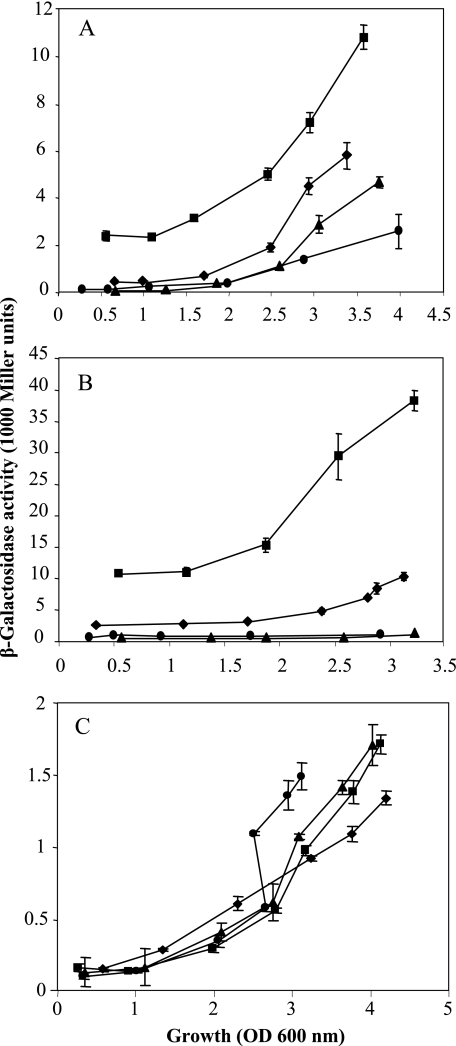
Transcriptional rsmY-lacZ and rsmZ-lacZ fusions were inserted into the P. aeruginosa chromosome of the wild type and of rsmA, rsmY rsmZ, and gacA mutants. This was achieved by transposition (1, 48) of mini-Tn7 constructs (Table 1) into the unique Tn7att site located downstream of the glmS gene. The rsmY-lacZ and rsmZ-lacZ fusions showed positive regulation by GacA and RsmA (Fig. 2A and B). In an rsmY rsmZ double mutant, the expression of both fusions was strongly elevated and, at the end of growth, about 40,000 Miller units were measured for the β-galactosidase activity of the rsmY-lacZ fusion (Fig. 2B). This value places the induced rsmY promoter in the category of the most highly expressed Pseudomonas promoters. The rsmA gene showed cell density-dependent expression which showed little variation in gacA, rsmA, or rsmY rsmZ mutants (Fig. 2C). In conclusion, transcription of both rsmY and rsmZ is positively regulated by RsmA and negatively by RsmY and RsmZ together. The mechanism by which the RNA-binding protein RsmA exerts this positive control is not understood. However, the data are consistent with a model in which the positive effects of RsmA on the rsmY and rsmZ promoters are antagonized by RsmY and RsmZ.
FIG. 2.
Regulation of rsmY, rsmZ, and rsmA expression in P. aeruginosa PAO1 and mutants. (A) Expression of rsmZ was monitored by measuring β-galactosidase activities of a transcriptional rsmZ-lacZ fusion in the wild type (PAO6554; diamonds), a gacA mutant (PAO6555; triangles), an rsmY rsmZ double mutant (PAO6556; squares), and an rsmA mutant (PAO6557; circles). (B) Expression of rsmY was monitored by determining β-galactosidase activities of a transcriptional rsmY-lacZ fusion in the wild type (PAO6558; diamonds), a gacA mutant (PAO6559; triangles), an rsmY rsmZ double mutant (PAO6567; squares), and an rsmA mutant (PAO6568; circles). (C) Expression of rsmA was monitored by measuring β-galactosidase activities of a translational rsmA′-′lacZ fusion (on pME3859) in the wild type (PAO1; diamonds), a gacA mutant (PAO6281; triangles), an rsmY rsmZ double mutant (PAO6421; squares), and an rsmA mutant (PAZH13; circles). Cells were grown in 50-ml Erlenmeyer flasks containing 20 ml NYB supplemented with 0.05% (vol/vol) Triton X-100 with shaking at 37°C. β-Galactosidase specific activities were determined by the Miller method (20). The experiments were done in triplicate; average values ± standard deviation are shown.
Proteomic analysis of gacA and rsmY rsmZ mutants.
If the action of GacA is mediated by two small RNAs, RsmY and RsmZ, in P. aeruginosa, we expect that the global protein expression pattern should be similar in the gacA mutant PAO6281 and in the rsmY rsmZ double mutant PAO6421 (Table 1). This was found to be the case (see the supplemental material). Among the intracellular proteins detected, chitinase (the chiC product) and chitin-binding protein (the cbpD product) were the most strongly down-regulated proteins in both mutants, compared to the wild type (Table 2). The ChiC and CbpD proteins are expressed under quorum-sensing control and accumulate in the cytoplasm before they are slowly secreted (5, 6, 22, 46). Among the cell surface-associated proteins detected, DsbA, an enzyme forming disulfide bonds in periplasmic proteins, was clearly down-regulated in both the gacA and rsmY rsmZ mutants in comparison with the wild type (Table 2). DsbA function is important for type II secretion; a dsbA null mutant shows reduced secretion of extracellular enzymes such as elastase and lipase (18, 38).
TABLE 2.
Differentially expressed proteinsa
| Protein | PA no.b | Localizationc | Fold changed
|
|
|---|---|---|---|---|
| gacA/wt | rsmYZ/wt | |||
| Fructose-1,6-bisphosphate aldolase (Fda) | 0555 | IC | −3.1 | −2.5 |
| Chitin-binding protein (CbpD) | 0852 | IC | −2.1 | −3.6 |
| SF | −3.3 | −5.0 | ||
| EC | −5.9 | −7.4 | ||
| N-Succinylglutamate 5-semialdehyde dehydrogenase (AruC) | 0895 | IC | −2.5 | −2.7 |
| Flagellin (FliC) | 1092 | EC | +7.5 | +9.8 |
| Flagellar-capping protein (FliD) | 1094 | EC | +8.8 | +10.2 |
| Chitinase (ChiC) | 2300 | IC | −8.0 | −8.3 |
| Protein with hypothetical function | 3021 | SF | −4.6 | −5.0 |
| Protein with hypothetical function | 3022 | IC | −2.4 | −2.1 |
| Protein with hypothetical function | 3088 | EC | +4.6 | +6.7 |
| Probable peroxidase | 3529 | SF | +3.4 | +2.9 |
| Elastase (LasB) | 3724 | EC | −3.5 | −10.1 |
| Protein with hypothetical function | 3785 | EC | +4.1 | +4.7 |
| Protein with hypothetical function | 4453 | EC | +2.3 | +2.1 |
| Azurin | 4922 | EC | +2.3 | +3.4 |
| Thiol-disulfide interchange protein (DsbA) | 5489 | SF | −4.0 | −7.5 |
Only those changes in expression that were ≥2.0-fold are shown.
Mass spectrometry data (peptide mass fingerprints and tandem mass spectrometry results) were compared to the complete translated open reading frames of P. aeruginosa PAO1 (http://www.pseudomonas.com).
IC, intracellular; SF, surface associated; EC, extracellular.
Average values from six gels. When multiple spots were ascribed to the same protein, average values of all spots are given. Down-regulation in the mutants is indicated by a minus sign; up-regulation in the mutants is indicated by a plus sign.
In the extracellular medium (see the supplemental material), CbpD and elastase (LasB protein) were among those proteins that were present in markedly reduced amounts in gacA and rsmY rsmZ mutants in comparison with the wild type. In both mutants, several extracellular proteins were up-regulated in parallel, notably azurin, flagellin (FliC), and flagellum-capping protein (FliD) (Table 2). Previously, increased expression of flagellin and enhanced motility have been observed in gacA mutants of P. fluorescens F113 (32). Azurin is a periplasmic copper redox protein that has been implicated in anaerobic electron transport of P. aeruginosa. However, the physiological role of azurin is unknown (42). The azurin gene is induced by limited aeration and in stationary phase (42), and the azurin protein is released into the culture medium (47). Interestingly, in cell cultures azurin induces apoptosis of macrophages through binding to tumor suppressor protein p53 (47). The finding that azurin is overproduced in gacA and rsmY rsmZ mutants was unexpected. If azurin were produced during P. aeruginosa infection, one might speculate that these mutants could be more cytotoxic to macrophages than is the wild type. This would add a new facet to regulation of virulence by GacA.
Parallel effects of gacA and rsmY rsmZ mutations on quorum sensing.
The Gac/Rsm system activates the quorum-sensing machinery primarily by stimulating C4-HSL production (25, 28). As expected, the rsmY rsmZ double mutant was impaired in C4-HSL production, as was the gacA mutant (Table 3). Both mutants produced less HCN and pyocyanin in comparison with the wild type, whereas the single rsmY and rsmZ mutants showed intermediate levels of these quorum-sensing-regulated exoproducts. The repressing activity of RsmA on HCN and pyocyanin synthesis was lost in an rsmA mutant (Table 3), in agreement with the Gac/Rsm model. The negative effect of an rsmA mutation on swarming (12) was confirmed; lack of rsmA function causes cell clumping (12). Conversely, swarming motility was higher in gacA and rsmY rsmZ mutants than in the wild type (Table 3). The enhanced motility of the mutants might be facilitated by the absence of a putative clumping factor and by elevated synthesis of flagellar proteins.
TABLE 3.
Production of C4-HSL, HCN, and pyocyanin and swarming abilities of P. aeruginosa PAO1 and rsmA, gacA, rsmY, rsmZ, and rsmYZ mutants
| Strain | Genotype | C4-HSLa (μM) | HCNb (nmol/109 cells) | Pyocyaninc (μg/109 cells) | Swarming motilityd |
|---|---|---|---|---|---|
| PAO1 | Wild type | 2.1 ± 0.1 | 22.5 ± 2.0 | 10.4 ± 0.9 | + |
| PAZH13 | ΔrsmA | ND | 56.4 ± 10.0 | 28.2 ± 4.1 | − |
| PAO6354 | ΔrsmZ | ND | 10.1 ± 2.8 | 8.6 ± 1.9 | + |
| PAO6420 | ΔrsmY | ND | 8.2 ± 1.3 | 6.8 ± 0.9 | + |
| PAO6421 | ΔrsmYΔrsmZ | ≤0.5 | 5.1 ± 0.8 | 4.7 ± 1.0 | ++ |
| PAO6281 | gacA::ΩSp/Sm | ≤0.5 | 6.9 ± 0.8 | 4.8 ± 1.1 | ++ |
C4-HSL concentrations were estimated in culture supernatants of P. aeruginosa strains grown in NYB by thin-layer chromatography analysis (11) when cells reached an optical density at 600 nm of ≈2.0. The experiment was performed twice in triplicate. Values are means ± standard deviations. ND, not determined.
HCN was quantified in culture supernatants of strains grown semianaerobically for 24 h in sealed 120-ml flasks containing 60 ml of glycine minimal medium (43) supplemented with 0.05% (wt/vol) Triton X-100. Samples of 50 μl were mixed with 250 μl of 0.09 M NaOH and 1 ml of a reagent containing 0.05 M 1,2-dinitrobenzene and 0.1 M 4-nitrobenzaldehyde in 2-methoxyethanol (43). The reaction product was measured colorimetrically at 578 nm, and cyanide concentrations were estimated using a KCN standard curve. Average values of three measurements ± standard deviations are given.
Pyocyanin was quantified in supernatants of P. aeruginosa strains grown in glycerol-alanine medium (28). Supernatants (5 ml) were extracted with 3 ml of chloroform and centrifuged at 8,000 rpm. The chloroform layer was transferred to a new tube containing 1 ml 0.2 M HCl and mixed. After centrifugation, the absorption at 520 nm was measured in the top aqueous layer. Average values of three measurements ± standard deviations are given.
Swarming ability was tested on semisolid medium (12) after 8 h of incubation in high humidity at 37°C. ++, ability to swarm and invade ∼50% of the plate; +, ability to swarm and invade ∼40% of the plate; −, no swarming.
β-Galactosidase activities were determined in strains carrying a translational hcnA′-′lacZ fusion which was expressed from the tac promoter, i.e., uncoupled of the natural regulation by N-acyl-homoserine lactones and limited aeration. In gacA and rsmY rsmZ mutants, the activities were similarly low, whereas in an rsmA-negative background derepressed levels were found in comparison with the wild type (Fig. 3). Taken together, these findings indicate that in P. aeruginosa signal transduction from GacA to target genes for exoproducts is mediated by two small RNAs (RsmY and RsmZ) and one RNA-binding protein (RsmA).
FIG. 3.
Expression of HCN biosynthetic genes. β-Galactosidase activities of a translational hcnA′-′lacZ fusion under the control of the tac promoter (carried by plasmid pME3843) were determined in the wild type (PAO1; diamonds), a gacA mutant (PAO6281; triangles), an rsmY rsmZ double mutant (PAO6421; squares), and an rsmA mutant (PAZH13; circles). Cells were grown as indicated for Fig. 2. The experiment was done in triplicate; average values ± standard deviations are shown.
Role of RsmY and RsmZ in biofilm formation.
Recent evidence points to an important role of RsmZ in biofilm formation of P. aeruginosa strain PAK. The input from three sensors, GacS, RetS, and LadS, determines the expression of rsmZ and biofilm formation (8, 41). During early colonization of polystyrene microtiter plates by P. aeruginosa, mutation in gacS weakly enhances the biomass of attached bacteria, whereas mutation in retS strongly enhances this biofilm development. Mutation in ladS abrogates biofilm formation (8, 41). We tested biofilm formation of our PAO mutants. Cells were grown at 30°C in L broth (31) in polystyrene microtiter plates for 24 h. The medium was then removed, and 100 μl of a 1% (wt/vol) aqueous solution of crystal violet was added (23). Following staining at room temperature for 20 min, the dye was removed and the wells were washed thoroughly. For quantification of surface-attached cells, crystal violet was solubilized in a mixture of 200 μl dimethyl sulfoxide and 800 μl ethanol, and the absorbance was determined at 570 nm against an ethanol blank. In this assay, the relative biofilm masses were 0.5 ± 0.1 (mean ± standard deviation) A570 units for the wild-type PAO1, 1.1 ± 0.1 A570 units for the gacA mutant PAO6281, and 1.0 ± 0.1 A570 units for the rsmY rsmZ mutant PAO6421. Thus, the positive effect of the gacA mutation on biofilm formation is reproduced by the rsmY rsmZ double mutation, suggesting that RsmY and RsmZ together influence biofilm formation.
RsmA control of the rpsA gene product, ribosomal protein S1.
When RsmA acts as a translational repressor, it interferes with the access of ribosomes to mRNA by binding to the Shine-Dalgarno (SD) sequence (4). Ribosomal protein S1, an essential protein in E. coli and other gram-negative bacteria, binds to mRNAs upstream of the SD sequence, and this interaction is particularly important for translation initiation from weak or very strong SD sequences (19). The relative concentrations of free RsmA and S1 may therefore critically influence the rate of translation initiation of certain genes. In enteric bacteria, the 5′-untranslated region of rpsA mRNA contains two hairpins with GGA motifs in the loops. These hairpins and motifs are essential for translational repression of rpsA mRNA, a phenomenon which occurs when ribosomal protein S1 is overproduced (37). Conceivably, these mRNA structures might interact with CsrA/RsmA. Although this type of regulation of the rpsA gene is much weaker in P. aeruginosa and Pseudomonas putida than in enteric bacteria (37), we speculated that RsmA might nevertheless influence the expression of rpsA in P. aeruginosa. β-Galactosidase activities specified by a translational rpsA′-′lacZ fusion were significantly higher in an rsmA mutant than in the wild type (Fig. 4), indicating a negative effect of RsmA on rpsA expression. In a gacA mutant and in an rsmY rsmZ mutant, the rpsA′-′lacZ levels were slightly (but not significantly) below those of the wild type (Fig. 4).
FIG. 4.
Expression of the rpsA gene. β-Galactosidase activities of a translational rpsA′-′lacZ fusion carried by plasmid pME7322 were determined in the wild-type strain (PAO1; diamonds), a gacA mutant (PAO6281; triangles), an rsmY rsmZ double mutant (PAO6421; squares), and an rsmA mutant (PAZH13; circles). Cells were grown as indicated for Fig. 2. Average values ± standard deviations from four experiments are shown.
During growth of P. aeruginosa, the intracellular concentration of RsmA protein increases steadily and reaches a high level in stationary phase (26). By contrast, the S1 protein level declines in stationary phase (34). Overall, the RsmA/S1 ratio increases at least 10-fold during growth, and this trend can also be gleaned from the expression of rsmA′-′lacZ and rpsA′-′lacZ fusions in the wild-type PAO1. RsmA does not appear to regulate its own synthesis (Fig. 2C) but has a negative effect on S1 synthesis (Fig. 4). In this way, high RsmA/S1 ratios may be consolidated during late stages of growth and stationary phase.
Concluding remarks.
Our previous model of the GacS/GacA signal transduction pathway in P. aeruginosa (12) can now be refined by including RsmY. This small RNA has the typical attributes of a GacA-dependent regulatory RNA: it is transcribed from a promoter with a highly conserved palindromic UAS (TGTAAG… …CTTACA) (Fig. 1A), it has several stem-loop structures with unpaired GGA motifs (10), and it binds RsmA with high affinity (36). RsmY acts in parallel with RsmZ. When both rsmY and rsmZ are deleted, the phenotypic changes seen in various tests (Tables 2 and 3; Fig. 3 and 4) are very similar to those found in the gacA mutant. Both GacA and RsmA are needed for rsmY transcription (Fig. 1B and 2B). Whether GacA (in its phosphorylated form) physically interacts with the UAS remains to be demonstrated. The action of the RNA-binding protein RsmA, which is antagonized jointly by RsmY and RsmZ, is likely to be indirect and to involve one or several unknown transcription factors.
GacA is a major positive regulator of virulence in P. aeruginosa (27). According to the results of this study, most if not all major effects of GacA can be ascribed to the combined action of RsmY and RsmZ. The signal transduction pathway from GacA to individual virulence genes varies. For instance, the negative effect of gacA and rsmY rsmZ mutations on HCN production derives from reduced synthesis of the C4-HSL quorum-sensing signal (Table 3), which causes low hcn promoter activity, as well as from repressed hcn mRNA translation (Fig. 3), in agreement with earlier data (25). Impaired secretion of elastase (LasB protein) in gacA and rsmY rsmZ mutants can be explained by reduced C4-HSL synthesis (Table 3), which results in down-regulation of type II secretion (3), and by poor expression of DsbA (Table 2), a key enzyme involved in the folding of proteins released by type II secretion (18, 38). To some extent, RsmY and RsmZ appear to have redundant functions. However, they differ in at least one respect: the global regulator Hfq, a protein that facilitates base-pairing interactions between small RNAs and target mRNAs, stabilizes RsmY, but not RsmZ, and Hfq binds specifically to RsmY (36).
Supplementary Material
Acknowledgments
We thank Cornelia Reimmann for supplying plasmid pME6182 and Stephan Heeb for help during early phases of the project.
This work was supported by the Swiss National Foundation for Scientific Research (project 3100A0-100180) and the European Union (project QLK3-CT-2002-0286).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bao, Y., P. Lies, H. Fu, and G. P. Roberts. 1991. An improved Tn7-based system for single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167-168. [DOI] [PubMed] [Google Scholar]
- 2.Burrowes, E., A. Abbas, A. O'Neill, C. Adams, and F. O'Gara. 2005. Characterisation of the regulatory RNA RsmB from Pseudomonas aeruginosa PAO1. Res. Microbiol. 156:7-16. [DOI] [PubMed] [Google Scholar]
- 3.Chapon-Hervé, V., M. Akrim, A. Latifi, P. Williams, A. Lazdunski, and M. Bally. 1997. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol. Microbiol. 24:1169-1178. [DOI] [PubMed] [Google Scholar]
- 4.Dubey, A. K., C. S. Baker, T. Romeo, and P. Babitzke. 2005. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11:1579-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folders, J., J. Algra, M. S. Roelofs, L. C. van Loon, J. Tommassen, and W. Bitter. 2001. Characterization of Pseudomonas aeruginosa chitinase, a gradually secreted protein. J. Bacteriol. 183:7044-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Folders, J., J. Tommassen, L. C. van Loon, and W. Bitter. 2000. Identification of a chitin-binding protein secreted by Pseudomonas aeruginosa. J. Bacteriol. 182:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fortune, D. R., M. Suyemoto, and C. Altier. 2006. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect. Immun. 74:331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodman, A. L., B. Kulasekara, A. Rietsch, D. Boyd, R. S. Smith, and S. Lory. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745-754. [DOI] [PubMed] [Google Scholar]
- 9.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 14:1351-1363. [DOI] [PubMed] [Google Scholar]
- 10.Heeb, S., K. Heurlier, C. Valverde, M. Cámara, D. Haas, and P. Williams. 2004. Post-transcriptional regulation in Pseudomonas spp. via the Gac/Rsm regulatory network, p. 239-255. In J.-L. Ramos (ed.), Pseudomonas, vol. 2. Kluwer Academic/Plenum Publishers, New York, N.Y. [Google Scholar]
- 11.Heurlier, K., V. Dénervaud, G. Pessi, C. Reimmann, and D. Haas. 2003. Negative control of quorum sensing by RpoN (σ54) in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:2227-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heurlier, K., F. Williams, S. Heeb, C. Dormond, G. Pessi, D. Singer, M. Cámara, P. Williams, and D. Haas. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186:2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyytiäinen, H., M. Montesano, and E. T. Palva. 2001. Global regulators ExpA (GacA) and KdgR modulate extracellular enzyme gene expression through the RsmA-RsmB system in Erwinia carotovora subsp. carotovora. Mol. Plant-Microbe Interact. 14:931-938. [DOI] [PubMed] [Google Scholar]
- 14.Kay, E., C. Dubuis, and D. Haas. 2005. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc. Natl. Acad. Sci. USA 102:17136-17141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kay, E., C. Reimmann, and D. Haas. 2006. Small RNAs in bacterial cell-cell communication. Microbe 1:63-69. [Google Scholar]
- 16.Lenz, D. H., M. B. Miller, J. Zhu, R. V. Kulkarni, and B. L. Bassler. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 58:1186-1202. [DOI] [PubMed] [Google Scholar]
- 17.Liu, Y., Y. Cui, A. Mukherjee, and A. K. Chatterjee. 1998. Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol. Microbiol. 29:219-234. [DOI] [PubMed] [Google Scholar]
- 18.Malhotra, S., L. A. Silo-Suh, K. Mathee, and D. E. Ohman. 2000. Proteome analysis of the effect of mucoid conversion on global protein expression in Pseudomonas aeruginosa strain PAO1 shows induction of the disulfide bond isomerase DsbA. J. Bacteriol. 182:6999-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGinness, K. E., and R. T. Sauer. 2004. Ribosomal protein S1 binds mRNA and tmRNA similarly but plays distinct roles in translation of these molecules. Proc. Natl. Acad. Sci. USA 101:13454-13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Molofsky, A. B., and M. S. Swanson. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50:445-461. [DOI] [PubMed] [Google Scholar]
- 22.Nouwens, A. S., S. A. Beatson, C. B. Whitchurch, B. J. Walsh, H. P. Schweizer, J. S. Mattick, and S. J. Cordwell. 2003. Proteome analysis of extracellular proteins regulated by the las and rhl quorum sensing systems in Pseudomonas aeruginosa PAO1. Microbiology 149:1311-1322. [DOI] [PubMed] [Google Scholar]
- 23.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 24.Pessi, G., and D. Haas. 2000. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182:6940-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pessi, G., and D. Haas. 2001. Dual control of hydrogen cyanide biosynthesis by the global activator GacA in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 200:73-78. [DOI] [PubMed] [Google Scholar]
- 26.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. Holden, M. Cámara, D. Haas, and P. Williams. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahme, L. G., F. M. Ausubel, H. Cao, E. Drenkard, B. C. Goumnerov, G. W. Lau, S. Mahajan-Miklos, J. Plotnikova, M. W. Tan, J. Tsongalis, C. L. Walendziewicz, and R. G. Tompkins. 2000. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA 97:8815-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 29.Reimmann, C., C. Valverde, E. Kay, and D. Haas. 2005. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J. Bacteriol. 187:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321-1330. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sanchez-Contreras, M., M. Martin, M. Villacieros, F. O'Gara, I. Bonilla, and R. Rivilla. 2002. Phenotypic selection and phase variation occur during alfalfa root colonization by Pseudomonas fluorescens F113. J. Bacteriol. 184:1587-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schnider-Keel, U., A. Seematter, M. Maurhofer, C. Blumer, B. Duffy, C. Gigot-Bonnefoy, C. Reimmann, R. Notz, G. Défago, D. Haas, and C. Keel. 2000. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 182:1215-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sevo, M., E. Buratti, and V. Venturi. 2004. Ribosomal protein S1 specifically binds to the 5′ untranslated region of the Pseudomonas aeruginosa stationary-phase sigma factor rpoS mRNA in the logarithmic phase of growth. J. Bacteriol. 186:4903-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 36.Sonnleitner, E., M. Schuster, T. Sorger-Domenigg, E. P. Greenberg, and U. Bläsi. 2006. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 59:1542-1558. [DOI] [PubMed] [Google Scholar]
- 37.Tchufistova, L. S., A. V. Komarova, and I. V. Boni. 2003. A key role for the mRNA leader structure in translational control of ribosomal protein S1 synthesis in gamma-proteobacteria. Nucleic Acids Res. 31:6996-7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urban, A., M. Leipelt, T. Eggert, and K. E. Jaeger. 2001. DsbA and DsbC affect extracellular enzyme formation in Pseudomonas aeruginosa. J. Bacteriol. 183:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valverde, C., S. Heeb, C. Keel, and D. Haas. 2003. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 50:1361-1379. [DOI] [PubMed] [Google Scholar]
- 40.Valverde, C., M. Lindell, E. G. Wagner, and D. Haas. 2004. A repeated GGA motif is critical for the activity and stability of the riboregulator RsmY of Pseudomonas fluorescens. J. Biol. Chem. 279:25066-25074. [DOI] [PubMed] [Google Scholar]
- 41.Ventre, I., A. L. Goodman, I. Vallet-Gely, P. Vasseur, C. Soscia, S. Molin, S. Bleves, A. Lazdunski, S. Lory, and A. Filloux. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. USA 103:171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vijgenboom, E., J. E. Busch, and G. W. Canters. 1997. In vivo studies disprove an obligatory role of azurin in denitrification in Pseudomonas aeruginosa and show that azu expression is under control of RpoS and ANR. Microbiology 143:2853-2863. [DOI] [PubMed] [Google Scholar]
- 43.Voisard, C., C. Keel, D. Haas, and G. Défago. 1989. Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 8:351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voisard, C., C. T. Bull, C. Keel, J. Laville, M. Maurhofer, U. Schnider, G. Défago, and D. Haas. 1994. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches, p. 67-89. In F. O'Gara, D. N. Dowling, and B. Boesten (ed.), Molecular ecology of rhizosphere microorganisms. VCH, Weinheim, Germany.
- 45.Weilbacher, T., K. Suzuki, A. K. Dubey, X. Wang, S. Gudapaty, I. Morozov, C. S. Baker, D. Georgellis, P. Babitzke, and T. Romeo. 2003. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48:657-670. [DOI] [PubMed] [Google Scholar]
- 46.Winson, M. K., M. Cámara, A. Latifi, M. Foglino, S. R. Chhabra, M. Daykin, M. Bally, V. Chapon, G. P. Salmond, B. W. Bycroft, A. Lazdunski, G. S. A. B. Stewart, and P. Williams. 1995. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:9427-9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamada, T., M. Goto, V. Punj, O. Zaborina, K. Kimbara, T. K. Das Gupta, and A. M. Chakrabarty. 2002. The bacterial redox protein azurin induces apoptosis in J774 macrophages through complex formation and stabilization of the tumor suppressor protein p53. Infect. Immun. 70:7054-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuber, S., F. Carruthers, C. Keel, A. Mattart, C. Blumer, G. Pessi, C. Gigot-Bonnefoy, U. Schnider-Keel, S. Heeb, C. Reimmann, and D. Haas. 2003. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 16:634-644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.